Abstract
Liver toxicity (hepatotoxicity) is a critical issue in drug discovery and development. Standard preclinical evaluation of drug hepatotoxicity is generally performed using in vivo animal systems. However, only a small number of preselected compounds can be examined in vivo due to high experimental costs. A more efficient yet accurate screening technique which can identify potentially hepatotoxic compounds in the early stages of drug development would thus be valuable. Here, we develop and apply a novel genomic prediction technique for screening hepatotoxic compounds based on in vitro human liver cell tests. Using a training set of in vivo rodent experiments for drug hepatotoxicity evaluation, we discovered common biomarkers of drug-induced liver toxicity among six heterogeneous compounds. This gene set was further triaged to a subset of 32 genes that can be used as a multi-gene expression signature to predict hepatotoxicity. This multi-gene predictor was independently validated and showed consistently high prediction performance on five test sets of in vitro human liver cell and in vivo animal toxicity experiments. The predictor also demonstrated utility in evaluating different degrees of toxicity in response to drug concentrations which may be useful not only for discerning a compound’s general hepatotoxicity but also for determining its toxic concentration.
Keywords: Drug hepatocellular toxicity, Co-expression Extrapolation, Multi-gene Expression-based Predictor
Introduction
The liver is the primary organ of metabolism and detoxification in the body and a major target of drug toxicity (Bandara and Kennedy, 2002). Liver toxicity (hepatotoxicity) is one of the most critical issues in drug development and can lead to failure of drug candidates during preclinical or clinical studies (Jaeschke et al., 2002). Biochemically, drug-induced liver injury could be characterized into hepatocellular (predominantly initial Alanine transferase (ALT) elevation) and cholestatic (initial alkaline phosphatase (ALP) rise) types. Conventionally, hepatotoxic effects of novel compounds can be evaluated by in vivo experiments in rodent and other animal systems. An ALT level more than three times the upper limit of normal (ULN) is usually considered as significant liver injury even though histopathology is also a frequent tool to detect hepatotocixity without ALT elevations in animals. In vivo animal tests of hepatocellular toxicity can resemble physiological microenvironments in the human body. Nevertheless, these in vivo assays are not feasible for screening a large number of candidate compounds due to high costs and time.
Both cell culture and biochemical systems are also frequently used to evaluate the potential of drug-induced liver toxicity. These in vitro tests are cheaper, faster, and more convenient for screening many candidate drug compounds for their hepatotoxicity compared to in vivo analysis (Yang et al., 2004). However, even though such tests are widely used to examine the activity on important biomarkers such as P450 protein expression and activity, the in vitro systems generally cannot fully reflect hepatocellular toxic effects such as ALT induction and toxicity related to in vivo metabolites and mitochondria dysfunction.
In an attempt to circumvent the limitations of current in vitro systems, we sought to develop an in vitro cell-based prediction technique that can be effectively used for identifying hepatotoxic compounds. This technique is based on a multi-gene expression predictor which can discriminate a wide range of hepatotoxic compounds both in animals and in human liver cells by using expression-regulated biomarkers of liver toxicity that are shared between the two systems. Also, since the specific molecular mechanisms of hepatocellular toxicity among various compounds can often be different, we identify and use expression signatures which are commonly associated with the elevation of serum ALT levels among multiple heterogeneous compounds. We have used this predictor for testing a wide range of candidate compounds for their hepatocellular toxicity across in vivo rodent and in vitro human liver cell systems from five independent test sets with >160 structurally and mechanistically diverse chemical compounds and drugs.
Many studies have indicated that computational approaches, such as structural bioinformatics (Chou, 2004; Wang and Chou, 2011), molecular dynamics (Lian et al., 2011; Wang et al., 2009), molecular docking (Chou et al., 2003), predicting drug-target interaction (He et al., 2010), protein subcellular location prediction (Chou, 2001; Chou and Shen, 2008; Chou and Shen, 2010), antimicrobial peptide prediction (Wang et al., 2011), HIV protease cleavage site prediction (Chou, 1996), signal peptide prediction (Chou and Shen, 2007b), identifying GPCRs and their types (Xiao et al., 2011), estimating the upper-limit of enzyme-substrate reaction rate (Chou and Zhou, 1982), predicting the network of substrate-enzyme-product triads (Chen et al., 2010), as well as a series of user-friendly web-servers (Chou and Shen, 2009), can timely provide very useful information and insights for complex biological and biomedical investigations such as novel drug development. The present study is also attempted to develop a novel genomic prediction technique for screening hepatotoxic compounds in hopes that it may become a useful tool for early drug discovery and development.
Material and Methods
In order to develop a useful model or predictor for biological systems, the following steps are generally required: (i) benchmark dataset construction or selection, (ii) mathematical formulation for the statistical samples concerned, (iii) operating algorithm (or engine), (iv) anticipated accuracy, and (v) web-server establishment (Chou, 2011). We elaborate some of these procedures for our study as follows.
Hepatology and Microarray Data Sets
Six previously-published microarray sets from 4 in vivo rodent and 2 in vitro human hepatocellular toxicity experiments were used to construct and validate our prediction model (Table 1). The first in vivo data set, Rat1 (NCBI GEO GSE5509), consists of 39 rat liver samples after 48 hrs’ treatment with three hepatocellular toxic compounds (alpha-naphthylisothiocyanate, dimethylnitrosamine, or n-methylformamide), three low-toxic compounds (caerulein, dinitrophenol and rosiglitazone), and controls without treatment (Spicker et al., 2008). These compounds are quite heterogeneous in their structural and molecular mechanisms showing highly varying severities of cell death in the liver. Complete evaluation of liver histopathology indices such as serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were also available for these 39 rat samples. Two additional microarray data sets from in vivo animal liver cells after treatment with toxic compounds, Rat2 and Rat3, were obtained from the National Institute of Environmental Health Science (NIEHS, http://cebs.niehs.nih.gov) (Chou and Bushel, 2009). In these two studies, commonly-used drug compounds, Acetaminophen (Rat2) and 1,4-dichlorobenzene (Rat3) respectively, were administered for 6, 24, or 48 consecutive hrs at different dose concentrations to evaluate the levels of acute toxicity of in vivo animals. These two compounds have been shown to elevate ALT at a high dose (Bushel et al., 2007; Umemura et al., 1992). Based on the known toxicity of these two compounds, samples were classified into two groups---'low-toxic’ (dose<=150mg/kg, including untreated controls) and 'high-toxic' (dose=1500mg/kg) for each drug compound. The ALT data of the Rat3 set were also provided in the recent publication (Chou and Bushel, 2009).
Table 1. Rat liver (in vivo) and human liver cell (in vitro) microarray datasets were used for computational model derivation and evaluation.
Six previously-published microarray sets from four in vivo and two in vitro hepatocellular toxicity experiments were used to construct and validate our prediction model. Four microarray data sets of hepatology Rat1, Human1, and Human2 were from the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo). Two additional in vivo microarray data sets, Rat2 and Rat3, were obtained from the public web portal of the National Institute of Environmental Health Science (NIEHS, http://cebs.niehs.nih.gov). Rat1 set was chosen and used for our multi-gene model development. Human1 set was used to identify our COXEN genes which showed concordant gene expression networks between in vitro human liver cells and in vivo rat liver cells. The other four sets---Rat2, Rat3, Rat4, and Human2 were used only for our independent evaluation of the hepatocellular toxicity prediction model.
| Data Set | Sample Species | Study usage | Sourcea | ID | Array Platform | Samples | Drug |
|---|---|---|---|---|---|---|---|
| Rat1 | Rat liver (in vivo) | Training | GEO | GSE5509 | Affymetrix Rat 230 2.0 | 39 | 6 |
| Human1 | Human HepG2 (in vitro) | COXEN & Test | GEO | GSE6907 | Affymetrix Human HG-Focus | 30 | 9 |
| Rat2 | Rat Liver (in vivo) | Test | NIEHS | 839186700 | Affymetrix Rat 230 2.0 | 34 | 1 |
| Rat3 | Rat Liver (in vivo) | Test | NIEHS | 839173713 | Affymetrix Rat 230 2.0 | 48 | 1 |
| Rat4 | Rat Liver (in vivo) | Test | GEO | GSE8251 | GE | 990 | 147 |
| Healthcare/AmershamCodeLi | |||||||
| nkUniSet Rat I | |||||||
| Human2 | Human primary | Test | GEO | GSE10410 | Affymetrix HG U133 Plus 2.0 | 37 | 3 |
| hepatocyte (in vitro) | |||||||
GEO web address: http://www.ncbi.nlm.nih.gov/geo/
NIEHS web address: http://cebs.niehs.nih.gov/cebs-browser/cebsHome.do?enter=home
The final in vivo microarray data set, Rat4, was obtained from the public domain at the NCBI GEO site (GSE8251) (Fielden et al., 2007). This data set includes 703 rat liver samples treated with 147 structurally and mechanistically diverse chemical compounds including various FDA approved drugs, experimental compounds and environmental toxins, and 287 control samples from untreated animals. For our analysis, we first removed genes with a large proportion, e.g. >10% of missing expression values on the array data. We then selected 700 among the 990 samples after we excluded samples with any missing values. For the evaluation of our hepatocellular toxicity prediction, we classified the 147 compounds into two groups, “toxic” and “low-toxic”, based on reported toxicological information in the Micromedex database (http://www.micromedex.com/). Each compound was classified as a hepatotoxic agent if it was reported to elevate ALT level for both human and rat. Compounds without elevated ALT record or with rare hepatocellular toxicity cases reported in Micromedex were considered as “low-toxic” prior to our in silico toxicity prediction. In addition to the uncertainty of the Micromedex database information for some compounds, concentrations of the 147 compounds reported in the Micromedex database were not exactly the same as (or compared to) those used in the above animal experiments so their toxicity levels in the experiments were unclear. Nevertheless, since this set consisted of a large number of compounds, we tested whether there is a good statistical separation of our hepatotoxic prediction scores among these broadly-classified groups of low-toxic, toxic compound treated, and untreated controls.
The first in vitro human liver cell dataset (GSE6907), Human1, was obtained from the NCBI GEO (Gene Expression Omnibus) site (http://www.ncbi.nlm.nih.gov/geo/). This set includes 30 human hepatoma-derived cell line HepG2 samples under the exposure of six heavy metals (arsenic, cadmium, nickel, antimony, mercury, and chromium) and three other compounds, 2,3-dimethoxy-1,4-naphthoquinone (DMNQ), phenol, and N-nitrosodimethylamine (DMN) (Kawata et al., 2007). The HepG2 cell-line system is widely used as an experimental model for toxicological studies since it retains the activities of phase I and II enzymes and can induce activation and detoxification reactions (Kassie and Knasmuller, 2000). Gene expression profiling was conducted on the HepG2 cells following the treatments of these compounds. Based on the information in the Micromedex database, we classified phenol as low-toxic and the other eight compounds as hepatocellular toxic prior to our analysis. The second in vitro human liver cell dataset Human2 (GSE10410) performed by the US Environmental Protection Agency was reported with a toxicity evaluation of three triazole antifungals: myclobutanil (MYC), propiconazole (PPZ), and triadimefon (TDF) at the NCBI GEO site. Gene expression profiling was conducted on primary human hepatocytes (PHH), another widely-used human cell type for toxicity studies, following the treatments of these compounds at high (100µM), medium (30µM), and low doses (10µM). Based on the toxic information at the Pesticideinfo site (http://www.pesticideinfo.org) and Micromedex database, PPZ and TDF were classified as toxic compounds whereas MYC was considered a low-toxic compound for our analysis.
Of these six data sets, the Rat1 set was used for our multi-gene model development since the most accurate measurements of ALT levels were available for multiple structurally and molecular-mechanistically diverse compounds in this set. The microarray data of the Human1 set were used to further identify COXEN biomarkers which showed concordant gene expression networks between in vivo rat and in vitro human liver cells. The remaining sets (Rat2, Rat3, Rat4, and Human2) were not used for our biomarker discovery and modeling in any manner and used only for the independent evaluation of our multi-gene hepatotoxicity prediction model.
Model Development of Hepatotoxicity Prediction
The COXEN (CO-eXpression ExtrapolatioN) algorithm was originally developed to identify genes which were concordantly (expression) regulated between two independent cancer systems, e.g. the in vitro NCI 60 cell-line panel and an in vivo breast cancer patient set, and then to utilize the common gene set for cross-system (in vitro to in vivo) drug activity prediction. (Lee et al., 2007) In brief, COXEN is composed of six experimental and analysis steps—obtaining relevant drug activity data on the training set, molecular expression data both for the training and test sets, initial drug-activity biomarker discovery on the training set, sub-selection of COXEN biomarkers concordantly expressed between in vivo and in vitro systems, and multivariate prediction modeling with these COXEN biomarkers on the training set.
Adapting this algorithm to an in vitro hepatotoxicity prediction, our predictor development was made with five sequential analysis steps (Figure 1): the first was the identification of initial hepatotoxic biomarkers using the Rat1 set. Here, we identified the genes whose expression changes were consistently highly correlated with varying ALT levels upon treatments of multiple compounds using a rank-based (Spearman) correlation test with FDR (False Discovery Rate) q-value< 0.01 in order to reduce numerous false positives in high-throughput profiling-based discovery. The recent paper by Chou et al has also shown the potential of this correlation approach for identifying hepatotoxicity biomarker genes (Chou and Bushel, 2009). The second step was the homologous gene match between human and rat based on the HUGO gene nomenclature; the homologous genes on human microarrays, if represented, were identified for the hepatotoxicity biomarkers identified from the rat microarrays. Gene information of all microarray platforms of the six sets was also compared for this annotation match.
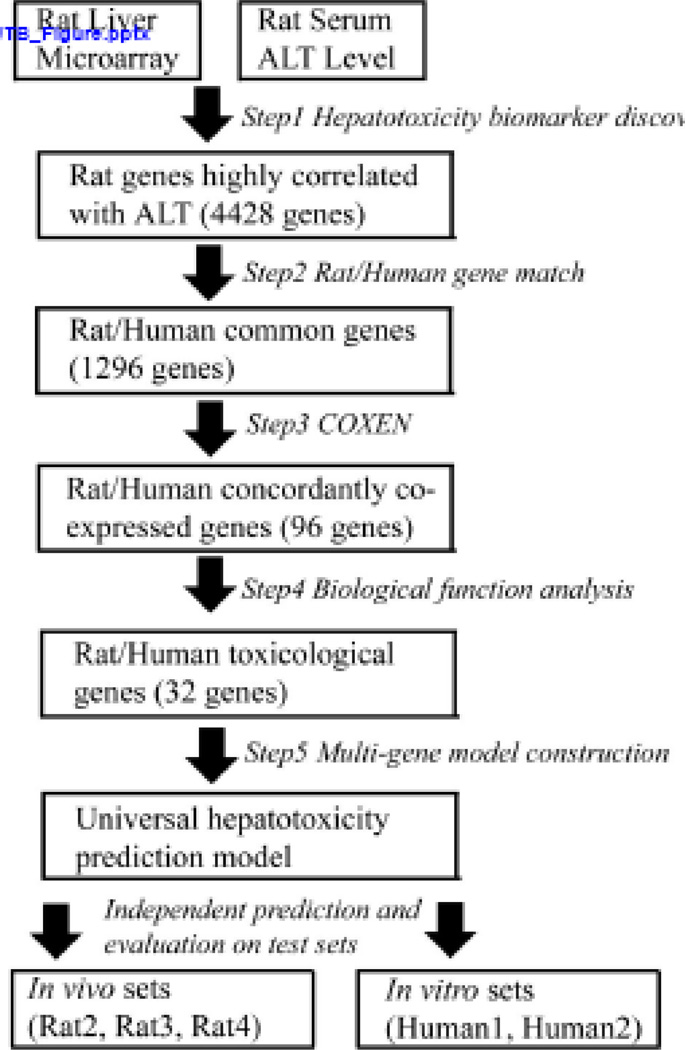
Figure 1. Schematic diagram of the computational model construction and validation processes.
Our hepatocellular toxicity prediction was developed by five distinct steps - the first was the identification of hepatocellular toxicity biomarker genes by correlation analysis with serum ALT. The second was the homologous gene match between human and rat while the third was the selection of the COXEN biomarkers among human-homologous hepatocellular toxic biomarkers obtained from the second step. In the fourth step, we further selected the biomarkers known to be directly related to liver toxicity and cell death performed by Ingenuity Pathway Analysis. This functional analysis step led to the final 32 biomarkers of hepatocellular toxicity. The last step was the multivariate prediction model construction based on the final 32 genes.
The third step was the selection of COXEN biomarkers among the human-homologous hepatotoxic biomarkers obtained from the second step. Among the initial hepatotoxic biomarkers discovered from the in vivo Rat1 set, we further triaged to the COXEN biomarkers which showed concordant expression network patterns between in vivo rat and in vitro human liver cells. In this step, we used a second-order correlation analysis to identify common gene networks which are similarly coexpressed due to similar coregulation between two independent systems. More technical details of this COXEN analysis can be found elsewhere (Lee et al., 2007). Note this step did not use any toxicity information of the second set Human1. In the fourth step, we identified the subset of the COXEN biomarkers whose molecular mechanisms have been known to be more directly related to liver toxicity. This functional gene selection was performed in order to develop a parsimonious prediction model, especially with biomarkers whose functions are directly relevant to hepatocellular toxicity mechanisms. This was possible because many different subsets of the COXEN biomarkers were found to be equally predictive in our preliminary analysis. Biological function analysis on these genes was performed by the Ingenuity Pathway Analysis software (IPA; www.ingenuity.com; Redwood City, CA), examining biological functions and toxicological mechanisms among the above COXEN biomarkers.
The final step (Step 5) was a multivariate prediction model construction. Microarray intensity values were first standardized within each gene for each set in order to reduce biased expression distributions of the six independent data sets obtained under different experimental settings. Based on the final biomarkers obtained in step 4, we then constructed a multivariate classification model to stratify liver injuries caused by chemical compounds, contrasting the toxic-compound treated animals and untreated control animals in the Rat1 set. We next applied principal component analysis to reduce the high dimension of the final biomarkers into a small number of multi-gene scores for subsequent classification modeling. Using these, we trained our prediction model based on the standard linear discriminant analysis (LDA) technique. The output of the LDA prediction model was then the posterior probabilities of hepatotoxicity between 0 (non-toxic) and 1 (toxic);a high predicted score, or posterior probability close to one, of a subject implies severely-injured liver cells or high in vivo toxicity whereas a low predicted score (close to zero) suggests uninjured liver cells or low in vivo toxicity. Logit scores of these LDA probabilities, log (p/1-p), were then plotted and compared between toxic and non-toxic compound groups for improved statistical comparison of predicted probability scores close to zero.
Prediction Evaluation on Independent Sets
In statistical prediction modeling, different validation strategies are used to examine a predictor for its effectiveness and robustness in practical application: n-fold cross-validation, jackknife, or independent set-based tests (Chou, 2011; Chou and Zhang, 1995; Chou and Shen, 2007a; Gu et al., 2010; Kandaswamy et al., 2010; Masso and Vaisman, 2010; Mohabatkar, 2010; Zakeri et al., 2010; Zeng et al., 2009; Zhou et al., 2007b). When there is only one set both for model training and test, cross-validation or jackknife strategies can be useful; It would still be highly desirable to validate the final prediction model on a completely independent test set to avoid the multiple comparisons pitfall among an astronomically large number of competing models in high throughput-based prediction modeling. Since we here have multiple independent sets for our model validation, we tested the performance of our final prediction model simultaneously on six diverse test sets of in vivo animal and in vitro human liver cells across >160 diverse compounds.
The unaltered classification model established with the Rat1 set (and partially with the Human1 set for the COXEN step without its toxicity outcome) was then simultaneously applied to other sets---Rat2, Rat3, Rat4, Human1, and Human2. In this evaluation, we wanted to see if the predicted (posterior probability) scores of hepatotoxicity could statistically discriminate the animal and human liver cells treated with toxic compounds from the untreated controls or those treated with low-toxic compounds. The statistical significance of the difference in scores was first evaluated using the two-sample t-test between the toxic and low-toxic (or control) groups. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were then calculated to summarize its prediction performance. Sensitivity, specificity, PPV, NPV were defined as
A predefined cutoff value of prediction scores was required in order to evaluate these indices. To define the optimal cutoff value, a Receiver Operating Characteristic (ROC) curve analysis was performed for each set based on predicted scores of hepatotoxicity and reported toxicity information on each set. The optimal cutoff on each ROC curve was then determined by maximizing the so-called Youden index (= sensitivity+specificity-1) (Youden, 1950).
Results
Universal Biomarkers and a Prediction Model for Hepatotoxicity
The initial biomarker discovery (Step 1) resulted in 4428 hepatocellular toxicity biomarkers highly associated with ALT levels in the Rat1 set by Spearman correlation test FDR q-value<0.01 (Figure 1). The next annotation match step resulted in 1296 common genes between rat and human microarray data sets from the six studies (Step 2); a high proportion (>29%) of the initial toxicity biomarkers could be matched among the four different (cross-species genome-wide) array platforms from the six studies. Applying COXEN (Step 3), 96 of these common genes were identified with concordant gene expression network patterns between Rat1 and Human1 datasets with a Pearson correlation association p-value<0.05. Next, selecting genes known to play roles in liver toxicity and cell death based on the Ingenuity Pathway Analysis (IPA) software (Step 4), we obtained biomarkers whose known biological functions were closely related to hepatocellular toxicity. In particular, groups of cellular death biomarkers (29 genes; Hypergeometric test p-value=1.2E-06) and liver toxicity biomarkers (8 genes; Hypergeometric test p-value=1.3E-03) were found among the most significantly over-represented molecular functions and toxicity mechanisms of the 29 genes (Table 3). The selection of these hepatotoxicity functional genes led to the final 32 biomarkers (3 genes with 2 biomarkers)for our hepatotoxicity prediction modeling. In particular, we found eight biomarkers were well-known liver toxicity genes: CYP26B1, NR1H4, KDR, MGMT, CCL5, RAN, VTN, and MASP1 (highlighted in Table 3). Not surprisingly, CYP26B1, a member of the cytochrome P450 (CYP) gene family encoding a monooxygenase that catalyzes various reactions involved in drug metabolism (Estabrook, 2003), was included in the list. A majority of the 32 genes was also relevant to cellular death processes (29 genes) which appears to reflect the biological response to toxicants in the liver cells injured or which have undergone cell death. MMP2 and RAN are also known to be related to mitochondrial dysfunction (Zhou et al., 2007a).
Table 3. 32 selected toxicity related genes and their correlations with ALT level in data set Rat1.
These 32 genes were identified by correlation analysis (with ALT level), COXEN algorithm, and biological function analysis. These genes were sorted by q-value derived by FDR calculation. Eight genes directly related to liver toxicity were highlighted.
| Symbol | Correlation Coefficient |
P-value | q-value | Entrez Gene Name |
Description |
|---|---|---|---|---|---|
| 1373803_a_at | −0.78 | 5.87e-09 | 3.36e-06 | GHR | growth hormone receptor |
| 1370287_a_at | 0.77 | 1.06e-08 | 4.19e-06 | TPM1 | tropomyosin 1 (alpha) |
| O-6-methylguanine-DNA | |||||
| 1368311_at | 0.70 | 7.72e-07 | 4.23e-05 | MGMT | methyltransferase |
| 1369983_at | −0.69 | 1.04e-06 | 5.23e-05 | CCL5 | chemokine (C-C motif) ligand 5 |
| 1367590_at | 0.68 | 1.67e-06 | 6.97e-05 | RAN | RAN, member RAS oncogene family |
| cytochrome P450, family 26, | |||||
| 1376667_at | −0.64 | 1.29e-05 | 2.42e-04 | CYP26B1 | subfamily B, polypeptide 1 |
| 1369476_at | −0.62 | 2.48e-05 | 3.69e-04 | EFNB1 | ephrin-B1 |
| kinase insert domain receptor (a type | |||||
| 1367948_a_at | −0.61 | 3.25e-05 | 4.44e-04 | KDR | III receptor tyrosine kinase) |
| protein C (inactivator of coagulation | |||||
| 1369286_at | −0.61 | 4.03e-05 | 5.14e-04 | PROC | factors Va and VIIIa) |
| 1375789_at | −0.60 | 5.86e-05 | 6.61 e-04 | PTH1R | parathyroid hormone 1 receptor |
| 1387387_at | −0.56 | 2.08e-04 | 1.52e-03 | HPCA | Hippocalcin |
| 1393267_at | 0.56 | 2.08e-04 | 1.52e-03 | PSIP1 | PC4 and SFRS1 interacting protein 1 |
| 1369693_a_at | −0.56 | 2.27e-04 | 1.61e-03 | SLC1A2 | solute carrier family 1, member 2 |
| Fas ligand (TNF superfamily, member | |||||
| 1387587_at | −0.55 | 3.01e-04 | 1.92e-03 | FASLG | 6) |
| eukaryotic translation initiation factor | |||||
| 1388390_at | 0.54 | 3.36e-04 | 2.06e-03 | EIF3H | 3, subunit H |
| 1368380_at | −0.54 | 3.47e-04 | 2.10e-03 | VTN | Vitronectin |
| glutamate receptor, ionotropic, AMPA | |||||
| 1368401_at | −0.54 | 3.99e-04 | 2.32e-03 | GRIA2 | 2 |
| 1369825_at | −0.53 | 5.28e-04 | 2.79e-03 | MMP2 | matrix metallopeptidase 2 |
| 1370875_at | 0.53 | 5.54e-04 | 2.89e-03 | EZR | Ezrin |
| mannan-binding lectin serine | |||||
| 1390585_at | −0.52 | 6.08e-04 | 3.09e-03 | MASP1 | peptidase 1 |
| angiotensinogen (serpin peptidase | |||||
| 1387811_at | −0.52 | 6.38e-04 | 3.17e-03 | AGT | inhibitor, clade A, member 8) |
| nuclear receptor subfamily 1, group H, | |||||
| 1369073_at | −0.50 | 1.32e-03 | 5.15e-03 | NR1H4 | member 4 |
| eukaryotic translation initiation factor | |||||
| 1398799_at | 0.49 | 1.55e-03 | 5.73e-03 | EIF4E | 4E |
| 1373466_at | 0.49 | 1.72e-03 | 6.14e-03 | CAST | Calpastatin |
| 1367994_at | −0.48 | 1.95e-03 | 6.64e-03 | DPYD | dihydropyrimidine dehydrogenase |
| potassium inwardly-rectifying channel, | |||||
| 1370598_a_at | −0.48 | 2.00e-03 | 6.73e-03 | KCNJ6 | subfamily J, member 6 |
| 1370369_at | −0.48 | 2.02e-03 | 2.02e-03 | GZMM | granzyme M (lymphocyte met-ase 1) |
| membrane-spanning 4-domains, | |||||
| 1370783_a_at | −0.47 | 2.25e-03 | 7.26e-03 | MS4A2 | subfamily A, member 2 |
| 1368923_at | −0.47 | 2.48e-03 | 7.79e-03 | ECEL1 | endothelin converting enzyme-like 1 |
| 1387660_at | −0.47 | 2.70e-03 | 8.21e-03 | IAPP | islet amyloid polypeptide |
| 1368919_at | −0.46 | 2.86e-03 | 8.53e-03 | PGF | placental growth factor |
| 1369877 at | −0.46 | 3.12e-03 | 9.02e-03 | CD8A | CD8a molecule |
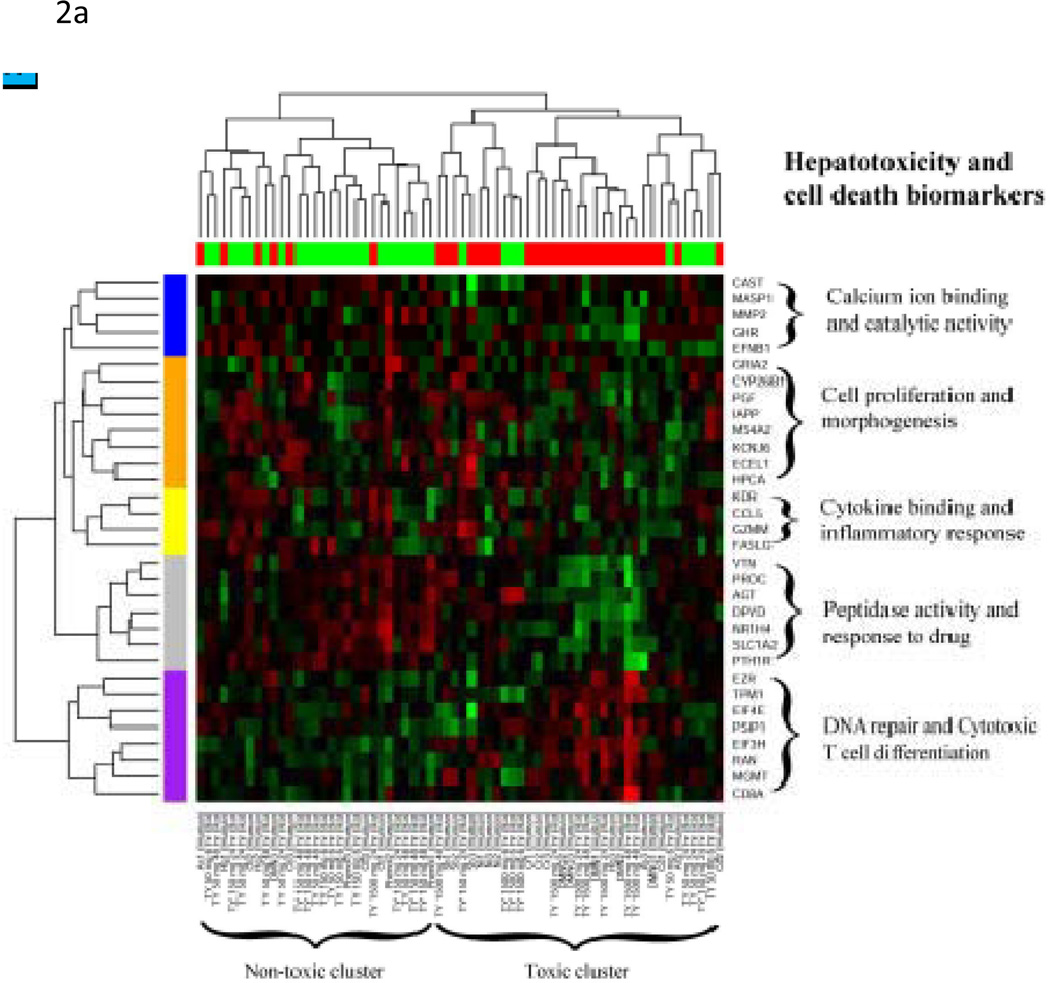
The 32 hepatotoxicity biomarkers showed consistent expression patterns within toxic and non-toxic compound groups despite their diverse toxicity characteristics and different biological systems in a clustering analysis (Figure 2A). That is, we performed a hierarchical clustering analysis of the 32 biomarkers simultaneously on the two test datasets (Rat2 and Human1) by standardizing each gene by subtracting the mean and dividing by the standard deviation within each set and then co-clustering the two sets. This within-species, within-gene normalization effectively enabled us to combine the two heterogeneous array datasets. The resulting co-clustering heatmap showed that the gene expression patterns of the clustered based on the reported compound toxicity regardless of the species and the potential different mechanisms of toxicity. The 32 biomarkers were also found to be further clustered into five sub-functional categories: calcium binding and catalytic activity, cell proliferation and morphogenesis, cytokine and inflammatory response, peptidase activity and response to drug, and DNA repair and cytotoxic T-cell differentiation.
Figure 2. Co-Clustering and pathway analysis of 32 universal hepatocellular toxicity biomarkers.
(A) Two test sets---Rat2 and Human1 of the 32 universal hepatocellular toxicity biomarkers---were standardized (subtracted by its mean and divided by its standard deviation) for each gene within each set and combined for a hierarchical clustering analysis on this combined set. Most of the samples from both in vivo rat or in vitro human liver cells were found to be well stratified between the toxic and non-toxic (control) compound-treated ones. Five functional subclusters of the 32 biomarkers were identified by examining common biological functions of these gene subclusters by the Affymetrix functional analysis tool (NetAffx). (B) The pathway analysis was also performed for these 32 genes in Ingenuity Pathway Analysis. The IPA network of the first pathway (Hepatic Fibrosis/Hepatic Stellate Cell Activation, p-value=2.90×10−7) is shown. Among our 32 biomarkers, GHR, SLC1A2, MMP2, VTN, DYPD, EIF4E, PGF, KDR, FASLG, EZR, CCL5, and NR1H4 were included in this network.
The pathway analysis was also performed for these 32 genes in Ingenuity Pathway Analysis. The top 5 pathways are Hepatic Fibrosis/Hepatic Stellate Cell Activation (p-value=2.90×10−7), Amyotrophic Lateral Sclerosis Signaling (p-value=9.77×10−4), IL-8 Signaling (p-value=4.57×10−3), Glutamate Receptor Signaling (p-value=6.16×10−3) and CCR5 Signaling in Macrophage (p-value=8.45×10−3); The IPA network of the first pathway is shown in Figure 2B.
Evaluation of Drug Hepatotoxicity Prediction in vivo
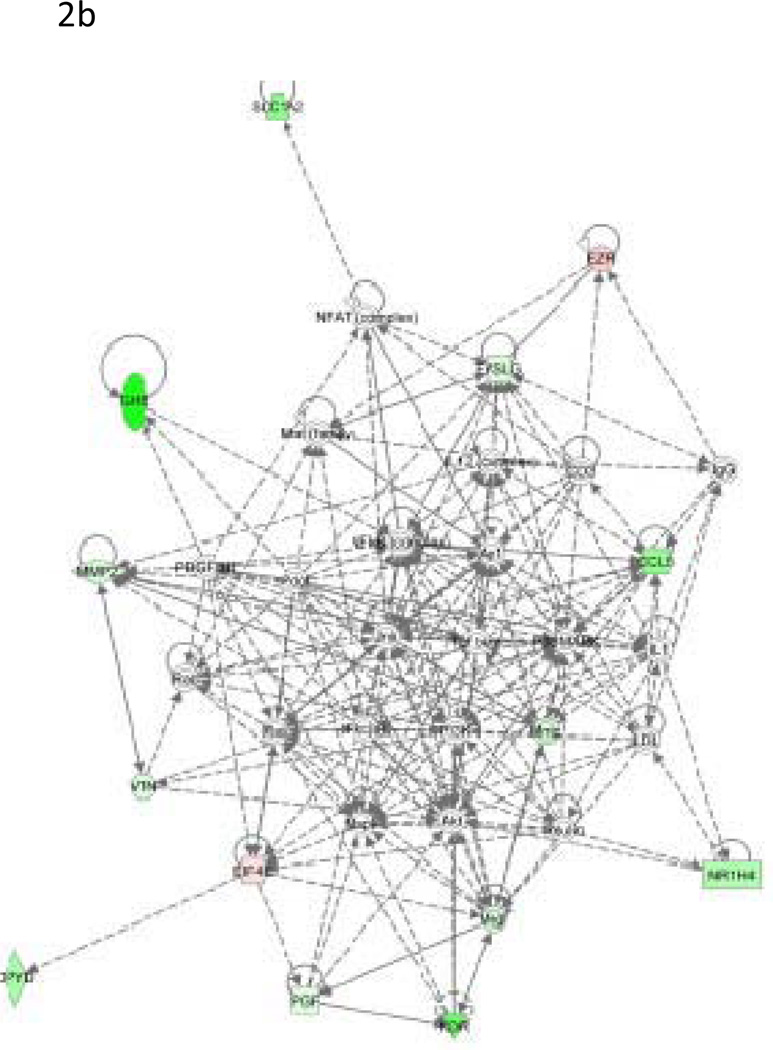
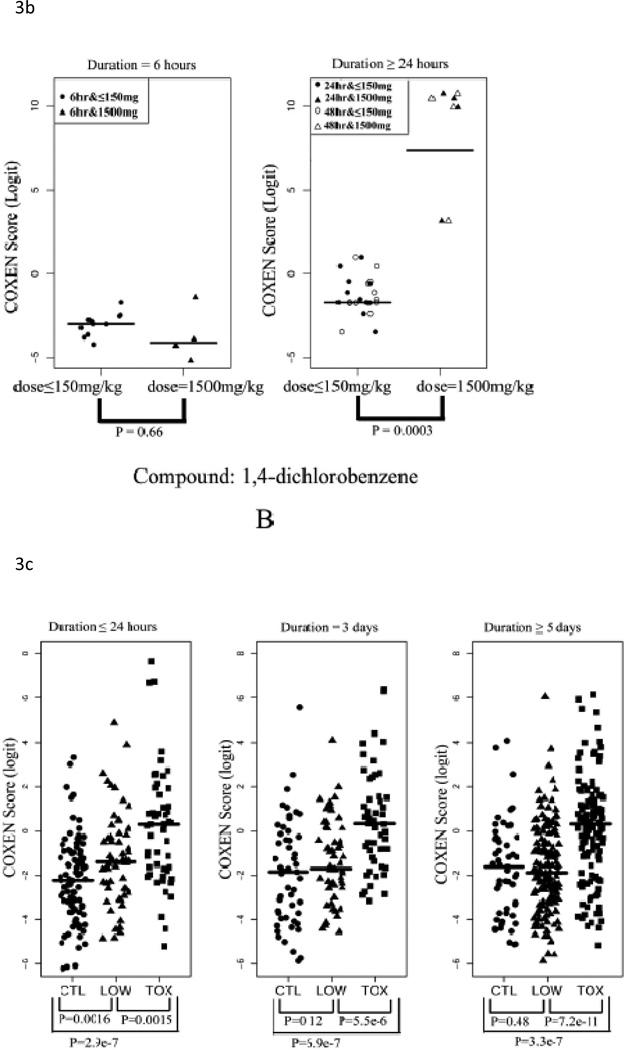
We evaluated the performance of our 32-gene model on the three in vivo rat datasets (Rat2, Rat3, and Rat4; Table 2). Toxicity of a compound strongly depends on its given dose. For example, acetaminophen (APAP) is quite safe at low doses but liver toxicity is observed at high doses. In fact, high doses of acetaminophen will cause hepatocellular toxicity and acetaminophen intoxication has been reported as one of the leading causes of liver failure in the US (Bushel et al., 2007). To examine our multi-gene model’s prediction ability to distinguish between non-toxic and toxic dose levels, we first applied our multi-gene prediction model to rat liver data sets Rat2 and Rat3. In these two studies, two compounds---acetaminophen (Rat2) and 1,4-dichlorobenzene (Rat3) were administered for 6, 24, or 48 consecutive hrs to rats at different doses to evaluate their acute toxicity. For each set, we compared our predicted scores between the high-dose (1500mg/kg) and low-dose groups (<=150mg/kg including control) with the Student’s two-sample t-test. In Figures 3A (Rat2) and 3B (Rat3), the predicted scores of these two compounds were plotted in each two sub-panels: 6 hrs and >24hrs after drug treatments. In both cases, highly significant differences were found between the high and low dose groups after 24 hrs of drug treatment in both sets (p<0.001). In particular, predicted scores of the low dose group were significantly lower than those of the high dose group and the two groups could be completely distinguished at optimal cutoff values (with sensitivity=1 and specificity=1) maximizing the Yuden index (= sensitivity+ specificity-1). On the contrary, no significant difference was found between the high and low dose group at 6 hrs after the drug administration (p = 0.91 for Rat2 and p = 0.66 for Rat3). These results appear to be consistent with pharmacokinetic observations indicating that it generally takes about 10 hrs to observe toxicity induced changes in gene transcription in liver cells and that significant elevation of ALT is generally observed at 12~24 hrs after administration of these compounds (http://emedicine.medscape.com/article/1008683-overview) (Stine et al., 1991).
Table 2. Prediction results for 5 independent test sets.
Rat2, Rat3, Rat4, Human1, and Human2 were used independently to evaluate the classification performance of our hepatocellular toxicity prediction model. We wanted to see if the predicted scores of hepatocellular toxicity could statistically discriminate cells and animals treated with toxic compounds from the ones treated with non-toxic compounds (or untreated controls) in these sets. The statistical significance of the difference in scores was first evaluated using the twosample t-test between the toxic and non-toxic groups. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were then calculated.
| Subset | T-test P-value |
Sensitivity (%) |
Specificity (%) |
PPV (%) |
NPV (%) |
Youden index |
|
|---|---|---|---|---|---|---|---|
| In vivo rat liver cell data | |||||||
| Rat2 | Duration = 6 hrs | 0.91 | 0.00 | 100 | 0.00 | 62.5 | 0.00 |
| Duration ≥ 24 hrs | 6.0e-4 | 100 | 100 | 100 | 100 | 1.00 | |
| Rat3 | Duration = 6 hrs | 0.66 | 25.0 | 100 | 80.0 | 100 | 0.25 |
| Duration ≥ 24 hrs | 3.2e-4 | 100 | 100 | 100 | 100 | 1.00 | |
| Rat4 | Duration ≤ 1 day | 2.9e | 54.9 | 91.3 | 77.8 | 78.5 | 0.46 |
| Duration = 3 days | 7.0e | 75.0 | 64.7 | 67.2 | 71.1 | 0.40 | |
| Duration ≥ 5 days | 3.3e | 60.8 | 78.2 | 86.3 | 46.7 | 0.39 | |
| In vitro human liver cell data | |||||||
| Human1 | All samples | 8.4e-4 | 100 | 100 | 100 | 100 | 100 |
| Human2 | Propiconazole | 0.045 | 71.4 | 62.5 | 70.0 | 83.3 | 0.50 |
| Triadimefon | 6.0e-3 | 71.4 | 100 | 100 | 80.0 | 0.71 | |
| Myclobutanil | 0.13 | 71.4 | 71.4 | 71.4 | 71.4 | 0.43 |
Figure 3. Prediction of in vivo data sets Rat2, Rat3 and Rat4.
(A) Comparison of COXEN scores of rats in set Rat2 treated with low dose and high sublethal-dose of acetaminophen. (B) Comparison of COXEN scores of rats in set Rat3 treated with low dose and a high sublethal-dose of 1,4-dichlorobenzene. Predicted scores of these two compounds were plotted in each of two sub-panels: 6 hrs and ≥24hrs after drug treatments. In these applications, animals treated with high sublethal-doses for 24 hrs could be perfectly discriminated from those with low doses by our prediction model. (C) Comparison of COXEN predictive scores of control rats in set Rat4 (without any treatment), rats treated with low-toxic and toxic compounds. Toxic and low toxic compounds were defined by the Micromedex database (www.micromedex.com). Predicted scores of these compounds were plotted in three sub-panels: ≤24 hrs, 3 days, and 5 days after drug treatments. Statistical significance (p value) of the set of predictions was assessed by a two sample t-test.
In the next application, we used an independent large rat liver dataset (Rat4) to test the our multi-gene model. This set included 700 rat (liver) samples after treatment with 147 diverse chemical compounds which we pre-classified into 62 toxic and 85 low toxic compounds using the Micromedex database as described in Methods and Materials. We used our 32-gene model to predict hepatotoxicity of individual animals in three groups, those treated with toxic compounds, low-toxic compounds, or placebos (or untreated controls) at three time points after drug treatment: <24 hrs, 3 days, and >5 days. The prediction on this large set was complicated due to several uncertain factors such as compounds with ambiguous toxicity information in the Micromedex database and the absence of information describing the experimental doses of these compounds. In this validation study, we thus tested whether our predictor showed statistically significant differences of the predicted toxic scores among the broadly-classified groups of compounds. As expected, our toxicity prediction results on these groups were noisier, yet were able to significantly discriminate the three groups (Figure 3C). That is, predicted scores of the animals treated with toxic compounds were significantly higher than control samples and non-toxic compound treated samples (two sample t-test p-values < 0.005). Furthermore, gradually higher predicted scores were also observed among the control, low-toxic, and toxic animal groups for the samples within 24 hrs and 3 days after drug administration (Figure 3C). However, differences between the control and low-toxic groups were found to be gradually less distinguishable after 5 days’ drug administration. The time-dependent loss of predicted toxicity in the low toxic group may reflect the metabolism or clearance of these compounds over time. This was consistent with the literature that rats have been reported to adapt to these low toxins and their livers rapidly recover from injuries (Williams and Iatropoulos, 2002).
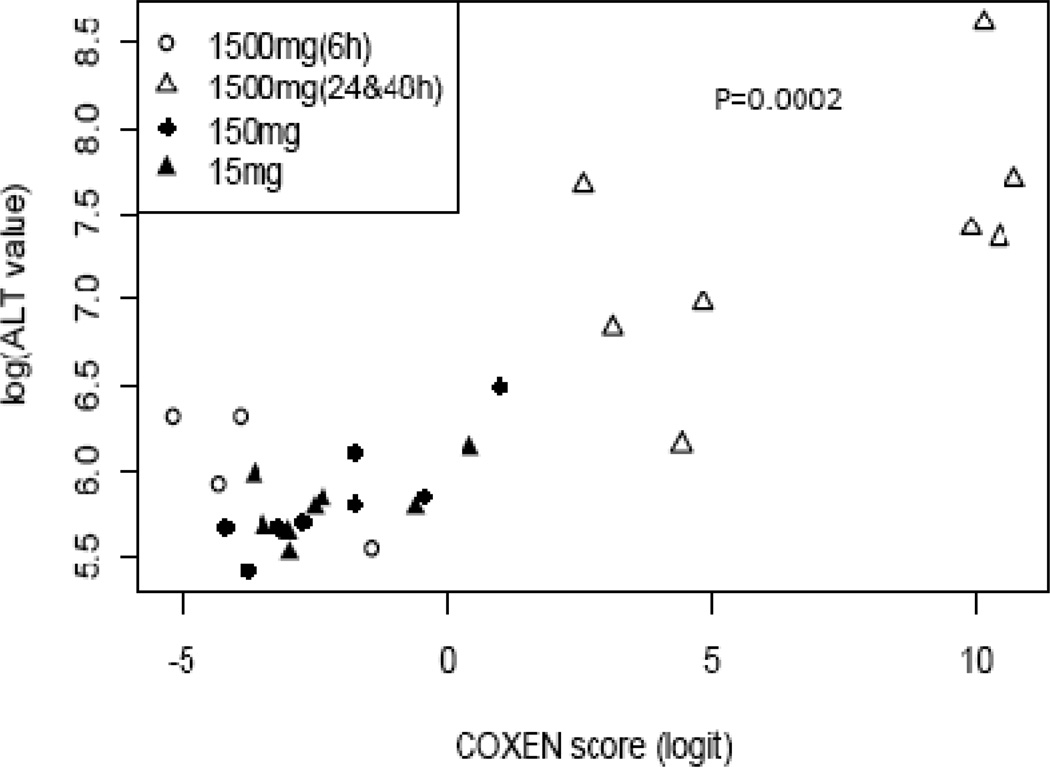
Since our hepatotoxicity predictor was built based on the common expression signatures highly associated with the elevation of serum ALT levels, we further quantitatively evaluated the reliability of the model against the actual reported rat ALT levels. As shown in Figure 4, there is a very high correlation between the predicted COXEN scores and experimental ALT levels (P value = 0.0002 and Spearman correlation coefficient is 0.66) on the independent Rat3 set. This result not only validates the prediction performance but also implies this predictor can be applied as a quantitative prediction of hepatotoxicity.
Figure 4. High correlation between predicted scores and experimental ALT levels in independent set Rat2.
Our predicted scores of hepatotoxicity were highly associated with the experimentally observed ALT levels of animals across the entire range of liver toxicity (Spearman correlation=0.66 with P-value = 0.0002).
Evaluation of in vitro Human Liver Cells
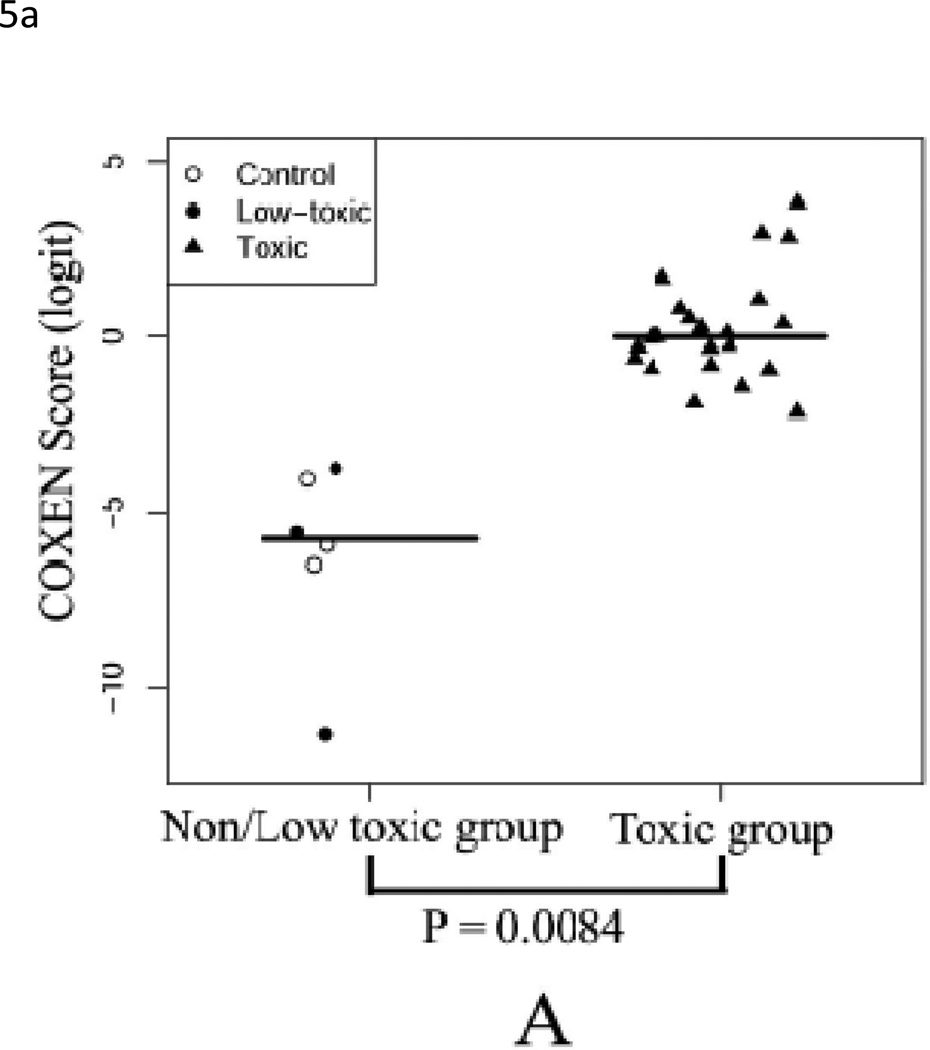
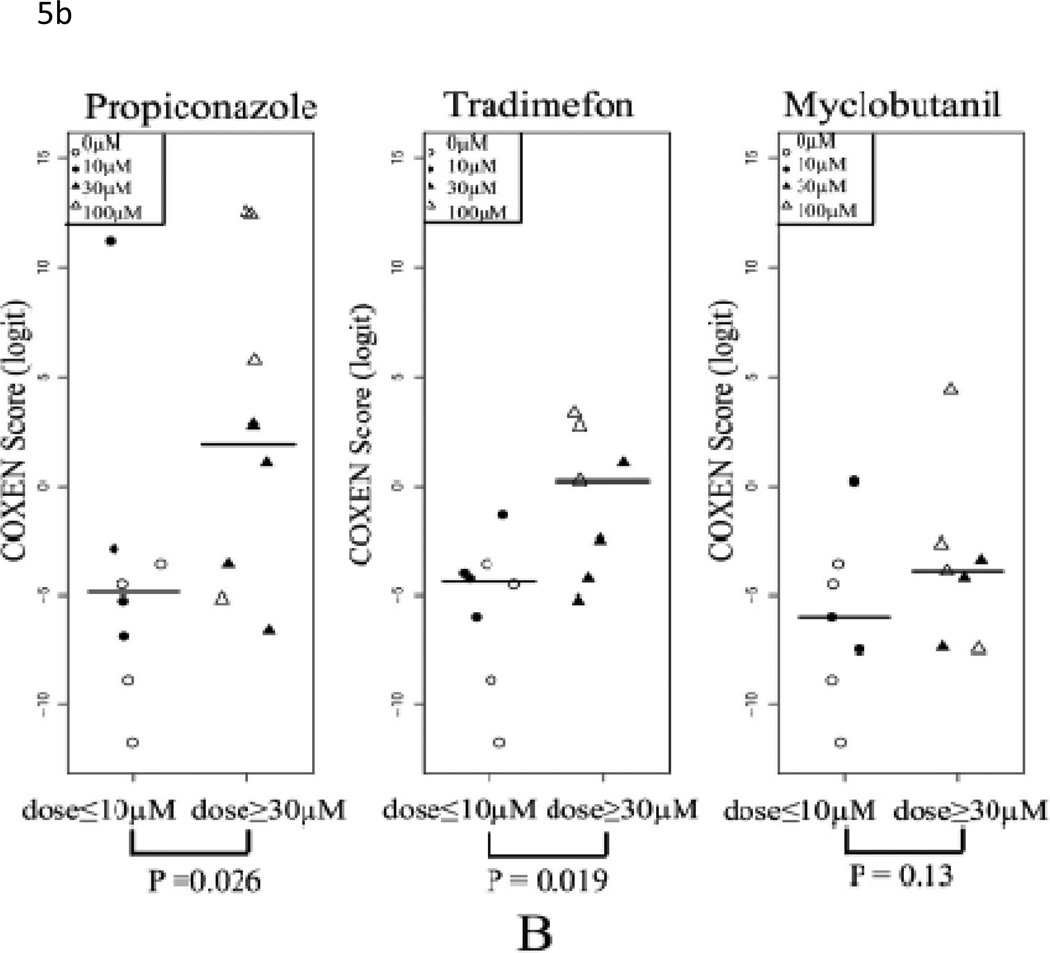
Since our goal is to build a prediction model which could be used in an in vitro system to simultaneously reflect in vivo hepatotoxicity, we applied the identical predictor to the two in vitro human liver cell datasets, Human1 and Human2. The Human1 set included 30 HepG2 cell samples treated with eight toxic compounds, one low-toxic compound and control group. For this set, our predicted hepatotoxicity scores of samples treated with toxic compounds were significantly higher than those treated by low-toxic compound and control samples (t-test p-value<0.001, Table 2) with perfect stratification of specificity=1 and sensitivity=1 (Figure 5A). In the Human2 set, primary hepatocytes were treated with two toxic (PPZ and TDF) and one low-toxic (MYC) anti-fungal compounds at 3 doses (10, 30, and 100 µM); untreated control cells were also included. In order to evaluate our prediction, we divided each drug set into high (30µM and 100µM) and low dose groups (control and 10µM). As shown in Figure 5B, predicted scores were significantly different for toxic compounds PPZ and TDF (t-test p-values=0.026 and 0.019, respectively). Furthermore, the predicted hepatotoxicity scores at 100µM were higher than those of 30µM for the 2 toxic compounds (PPZ and TDF). On the contrary, there was no dose-dependent effect seen with MYC, the non-toxic compound (p-value = 0.13). These results again indicate our multi-gene model could distinguish between different doses of the same compounds as well as differences between toxic and non-toxic compounds.
Figure 5. Prediction of in vitro data sets Human1 and Human2.
(A) Comparison of COXEN scores of HepG2 cells treated with low toxic (include control) and toxic compounds. Predicted scores of samples treated with toxic compounds were significantly higher than those treated by low-toxic compound and control samples (p-value<0.001). (B) Comparison of COXEN scores of primary human hepatocytes treated with high doses (≥ 30 µM) and low doses (≤10 µM) of three compounds, myclobutanil (MYC), propiconazole (PPZ), and triadimefon (TDF). Based on toxic information at the Pesticideinfo site (http://www.pesticideinfo.org), PPZ and TDF were classified as toxic compounds whereas MYC was considered as a low-toxic compound. Predicted scores were significantly different for toxic compounds PPZ and TDF (p-values=0.026 and 0.019, respectively). Furthermore, predicted hepatocellular toxicity scores of 100µM were higher than those of 30µM among the toxic dose group. On the contrary, predicted scores of the low and high dose groups of the low-toxic compound MYC were not statistically different (p-value = 0.13). Statistical significance (p value) of the set of predictions was assessed by a two-sample t-test.
Discussion
In this study, we developed an in vitro cell-based molecular predictor of hepatotoxicity and demonstrated its performance on diverse independent toxicity test sets of in vivo animal and in vitro human liver models across >160heterogeneous compounds. Our in vitro-based predictor also showed the ability to distinguish between toxic and non-toxic doses of single drugs. Thus we believe this in vitro test could potentially be used in early phases of drug discovery to examine the putative hepatotoxicity of a relatively large number of small molecules facilitating the selection of candidates for in vivo toxicity studies. We, however, note that our predictor is not developed to replace in vivo animal systems but to provide an efficient in vitro test for predicting drug hepatotoxicity that complements standard in vivo animal-based liver injury tests. Also, our predictor is not constructed to predict drug-induced idiosyncratic liver injury which should be developed based on a study of a large patient population and would require extremely high sensitivity.
Our prediction model utilizes consistent expression signatures across two systems (in vivo rats and in vitro human liver cells) resulting in its high prediction performance on both systems. Using this cross-species and inter-system predictability, the model’s prediction performance was consistently shown with five in vitro and in vivo test sets treated with diverse chemical and drug compounds and different array platforms. This multi-gene predictor also showed its ability in stratifying different toxic doses and different degrees of toxicity at elapsed time points after treatment. Our work differs from earlier studies describing molecular signatures specific to one biological system (Fielden et al., 2007; Zidek et al., 2007). Additionally, in some previous toxicogenomic studies, the training set and test sets were from the same experiments. Such model training and internal validation, however, can often be subject to selection bias due to a bias of specific microarray or assay platforms used and a multiple comparisons pitfall in the high-throughput profiling-based prediction modeling. These models frequently fail to perform well on independent toxicity studies (Fielden et al., 2007). We stress our current study has avoided this pitfall by validating five independent data sets from different experimental settings and microarray platforms. These test sets include more than 160 structurally and molecular-mechanistically diverse chemical compounds including various FDA approved drugs, experimental compounds, and environmental toxins.
There still exist several critical limitations of this in vitro-based cell system. For instance, different compounds may induce different forms of liver toxicity (e.g. cell death, metabolic perturbations and/or mitochondria dysfunction) which could result in unique gene expression profiles. Even though we selected the genomic biomarkers commonly associated with multiple heterogeneous toxic and clinically-used compounds to circumvent this difficulty, our predictor would not be able to accurately predict many different mechanisms of liver toxicity in the same degree. Nevertheless, we believe our study provides a proof of concept for the use of an in vitro cell test for the evaluation of hepatotoxicity, hoping to stimulate additional work in this area. We also believe our gene signature can be further refined as more data on liver toxicity becomes available. A second limitation is the use of HepG2 data for the development of our predictive gene set. HepG2 cells lack metabolic mechanisms and differ in many other biological characteristics from in vivo models. Despite this limitation, our predictor could predict the toxicity of many compounds in human primary hepatocytes suggesting that this molecular expression signature derived from common genes induced by toxic compounds in both rat liver and human cells may provide an accurate reflection of human liver.
Several additional limitations may exist for our prediction model: First, we developed and applied our predictor to 48-hr in vivo systems to demonstrate our model's prediction performance on such in vivo injury models (due to the data sets available for our training and independent evaluation). Therefore, changes in gene expression that occur at early time points (e.g. 3 and 5 hours after dosing) were not captured and this model may only be useful to predict toxic compounds which elevate ALT at 48 hrs after treatment. Nevertheless, since our goal is to develop an efficient in vitro model which can be used to screen early stages of candidate drug compounds, we believe this48-hr toxicity predictor can still be highly useful and effective unless a compound’s toxicity is greatly diminished in 48 hrs.
Cytochrome P450 enzymes play important roles in the detoxification and clearance of toxic compounds and the expression levels and activity of these enzymes are often used to predict potential problems with compound metabolism or drug-drug interactions. Our study indicates that there are additional functional classes of genes that are highly predictive of hepatotoxicity such as inflammatory genes and genes encoding proteins involved in DNA repair. Consequently, our multi-gene predictor is not heavily biased toward genes encoding p450 enzymes activities. The lack of emphasis on p450 gene expression in the gene signature raises the possibility that in the future it may be possible to predict hepatoxicity using cultured cell lines. Interestingly, our gene signature also includes genes such as MMP2 and RAN that may be directly associated with mitochondrial dysfunction (Xia et al., 2008; Zhou et al., 2007a).
The selection of the final 32 hepatotoxic functional biomarkers (among the 96 COXEN biomarkers) was somewhat subjective within the limited knowledge of genes currently known to be involved in hepatocellular toxicity and there may exist many other equally or more highly predictive multi-gene models of hepatotoxicity. However, a search for the optimal model with the highest prediction power is not computationally feasible due to an astronomical number of candidate multi-gene models and the over-fitting pitfalls on a relatively small number of training samples. When we compared performance of our functionally-selected 32-gene model with that of 100 randomly selected 32-gene models, the predictability of our 32-gene model was found to be within the top 10% based on the AUC (area under the curve) comparison for overall predictability. Furthermore, we found the 32 biomarkers showed distinctive expression patterns among their different sub-functional categories (Figure 2). For example, DNA repair and cytotoxic T-cell differentiation biomarkers were over-expressed among toxic-treated animals and human liver cells while the biomarkers of peptidase activity and drug response were under-expressed in the same group of animal livers and human liver cells. Therefore, even though our 32-gene model may not be the most highly performing prediction model, we believe it performs consistently well for the two systems on various toxic compounds and can provide more direct biological insight on drug liver toxicity for further investigation and understanding on hepatocellular toxic gene mechanisms.
Since the number of biomarkers in our final model may still be relatively large, we will need to investigate whether more parsimonious models (with fewer biomarkers) can provide equally, or even more highly predictive for stratifying toxic compounds. Also, as often done in similar studies (Chou and Shen, 2009), a user-friendly and publicly accessible web-server for our bioinformatics algorithm can be quite useful for developing many more predictor models, so we will make our continuous efforts to develop such a web-server.
Highlights.
The in vitro-derived models are effective in predicting drug hepatotoxicity on in vivo animals and in vitro liver cells.
Our prediction model accurately stratified rat and human liver cells treated with various compounds of liver toxicity.
Introducing a novel methodology that can be readily generalized to develop assays for many other types of drug toxicity.
Accelerating the “personalized toxicogenomics” to tailor drug treatments based on patients’ toxicity profiles.
ACKNOWLEDGMENTS
This work was supported in part by National Institutes of Health grant R01HL081690 to JKL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests’ statement: None
References
- Bandara LR, Kennedy S. Toxicoproteomics – a new preclinical tool. Drug Discov Today. 2002;7:411–418. doi: 10.1016/s1359-6446(02)02211-0. [DOI] [PubMed] [Google Scholar]
- Bushel PR, Heinloth AN, Li J, Huang L, Chou JW, Boorman GA, Malarkey DE, Houle CD, Ward SM, Wilson RE, Fannin RD, Russo MW, Watkins PB, Tennant RW, Paules RS. Blood gene expression signatures predict exposure levels. Proc Natl Acad Sci U S A. 2007;104:18211–18216. doi: 10.1073/pnas.0706987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Feng KY, Cai YD, Chou KC, Li HP. Predicting the network of substrate-enzyme-product triads by combining compound similarity and functional domain composition. BMC Bioinformatics. 2010;11:293. doi: 10.1186/1471-2105-11-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JW, Bushel PR. Discernment of possible mechanisms of hepatotoxicity via biological processes over represented by co expressed genes. BMC Genomics. 2009;10:272. doi: 10.1186/1471-2164-10-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou KC. Review: Prediction of HIV protease cleavage sites in proteins. Anal Biochem. 1996;233:1–14. doi: 10.1006/abio.1996.0001. [DOI] [PubMed] [Google Scholar]
- Chou KC. Prediction of protein cellular attributes using pseudo amino acid composition. Proteins. 2001;43:246–255. doi: 10.1002/prot.1035. [DOI] [PubMed] [Google Scholar]
- Chou KC. Structural bioinformatics and its impact to biomedical science. Curr Med Chem. 2004;11:2105–2134. doi: 10.2174/0929867043364667. [DOI] [PubMed] [Google Scholar]
- Chou KC. Some remarks on protein attribute prediction and pseudo amino acid composition. J Theor Biol. 2011;273:236–247. doi: 10.1016/j.jtbi.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou KC, Zhou GP. Role of the protein outside active site on the diffusion controlled reaction of enzyme. Journal of American Chemical Society. 1982;104:1409–1413. [Google Scholar]
- Chou KC, Zhang CT. Prediction of protein structural classes. Crit Rev Biochem Mol Biol. 1995;30:275–349. doi: 10.3109/10409239509083488. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Review: Recent progresses in protein subcellular location prediction. Anal Biochem. 2007a:370. doi: 10.1016/j.ab.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Signal CF: a subsite coupled and window fusing approach for predicting signal peptides. Biochem Biophys Res Commun. 2007b;357:633–640. doi: 10.1016/j.bbrc.2007.03.162. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Cell PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat Protoc. 2008;3:153–162. doi: 10.1038/nprot.2007.494. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Review: recent advances in developing web servers for predicting protein attributes. Natural Science. 2009;2:63–92. [Google Scholar]
- Chou KC, Shen HB. Cell PLoc 2.0: An improved package of web servers for predicting subcellular localization of proteins in various organisms. Natural Science. 2010;2:1090–1103. doi: 10.1038/nprot.2007.494. [DOI] [PubMed] [Google Scholar]
- Chou KC, Wei DQ, Zhong WZ. Binding mechanism of coronavirus main proteinase with ligands and its implication to drug design against SARS. Biochem Biophys Res Commun. 2003;308:148–151. doi: 10.1016/S0006-291X(03)01342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estabrook RW. A passion for P450s (rememberances of the early history of research on cytochrome P450) Drug Metab Dispos. 2003;31:1461–1473. doi: 10.1124/dmd.31.12.1461. [DOI] [PubMed] [Google Scholar]
- Fielden MR, Brennan R, Gollub J. A gene expression biomarker provides early prediction and mechanistic assessment of hepatic tumor induction by nongenotoxic chemicals. Toxicol Sci. 2007;99:90–100. doi: 10.1093/toxsci/kfm156. [DOI] [PubMed] [Google Scholar]
- Gu Q, Ding YS, Zhang TL. Prediction of G protein coupled receptor classes in low homology using Chou's pseudo amino acid composition with approximate entropy and hydrophobicity patterns. Protein Pept Lett. 2010;17:559–567. doi: 10.2174/092986610791112693. [DOI] [PubMed] [Google Scholar]
- He Z, Zhang J, Shi XH, Hu LL, Kong X, Cai YD, Chou KC. Predicting drug target interaction networks based on functional groups and biological features. PLoS One. 2010;5:e9603. doi: 10.1371/journal.pone.0009603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeschke H, Gores GJ, Cederbaum AI, Hinson JA, Pessayre D, Lemasters JJ. Mechanisms of hepatotoxicity. Toxicol Sci. 2002;65:166–176. doi: 10.1093/toxsci/65.2.166. [DOI] [PubMed] [Google Scholar]
- Kandaswamy KK, Chou KC, Martinetz T, Moller S, Suganthan PN, Sridharan S, Pugalenthi G. AFP Pred: A random forest approach for predicting antifreeze proteins from sequence derived properties. J Theor Biol. 2010;270:56–62. doi: 10.1016/j.jtbi.2010.10.037. [DOI] [PubMed] [Google Scholar]
- Kassie F, Knasmuller S. Genotoxic effects of allyl isothiocyanate (AITC) and phenethyl isothiocyanate (PEITC) Chem Biol Interact. 2000;127:163–180. doi: 10.1016/s0009-2797(00)00178-2. [DOI] [PubMed] [Google Scholar]
- Kawata K, Yokoo H, Shimazaki R, Okabe S. Classification of heavy metal toxicity by human DNA microarray analysis. Environ Sci Technol. 2007;41:3769–3774. doi: 10.1021/es062717d. [DOI] [PubMed] [Google Scholar]
- Lee JK, Havaleshko DM, Cho H, Weinstein JN, Kaldjian EP, Karpovich J, Grimshaw A, Theodorescu D. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci U S A. 2007;104:13086–13091. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian P, Wei DQ, Wang JF, Chou KC. An allosteric mechanism inferred from molecular dynamics simulations on phospholamban pentamer in lipid membranes. PLoS One. 2011;6:e18587. doi: 10.1371/journal.pone.0018587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masso M, Vaisman II. Knowledge based computational mutagenesis for predicting the disease potential of human non synonymous single nucleotide polymorphisms. J Theor Biol. 2010;266:560–568. doi: 10.1016/j.jtbi.2010.07.026. [DOI] [PubMed] [Google Scholar]
- Mohabatkar H. Prediction of cyclin proteins using Chou's pseudo amino acid composition. Protein Pept Lett. 2010;17:1207–1214. doi: 10.2174/092986610792231564. [DOI] [PubMed] [Google Scholar]
- Spicker JS, Brunak S, Frederiksen KS, Toft H. Integration of clinical chemistry, expression, and metabolite data leads to better toxicological class separation. Toxicol Sci. 2008;102:444–454. doi: 10.1093/toxsci/kfn001. [DOI] [PubMed] [Google Scholar]
- Stine ER, Gunawardhana L, Sipes IG. The acute hepatotoxicity of the isomers of dichlorobenzene in Fischer 344 and Sprague Dawley rats: isomer specific and strain specific differential toxicity. Toxicol Appl Pharmacol. 1991;109:472–481. doi: 10.1016/0041-008x(91)90010-c. [DOI] [PubMed] [Google Scholar]
- Umemura T, Tokumo K, Williams GM. Cell proliferation induced in the kidneys and livers of rats and mice by short term exposure to the carcinogen p dichlorobenzene. Arch Toxicol. 1992;66:503–507. doi: 10.1007/BF01970676. [DOI] [PubMed] [Google Scholar]
- Wang JF, Chou KC. Insights from modeling the 3D structure of New Delhi metallo beta lactamse and its binding interactions with antibiotic drugs. PLoS One. 2011;6:e18414. doi: 10.1371/journal.pone.0018414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JF, Gong K, Wei DQ, Li YX, Chou KC. Molecular dynamics studies on the interactions of PTP1B with inhibitors: from the first phosphate binding site to the second one. Protein Eng Des Sel. 2009;22:349–355. doi: 10.1093/protein/gzp012. [DOI] [PubMed] [Google Scholar]
- Wang P, Hu L, Liu G, Jiang N, Chen X, Xu J, Zheng W, Li L, Tan M, Chen Z, Song H, Cai YD, Chou KC. Prediction of antimicrobial peptides based on sequence alignment and feature selection methods. PLoS One. 2011;6:e18476. doi: 10.1371/journal.pone.0018476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GM, Iatropoulos MJ. Alteration of liver cell function and proliferation: differentiation between adaptation and toxicity. Toxicol Pathol. 2002;30:41–53. doi: 10.1080/01926230252824699. [DOI] [PubMed] [Google Scholar]
- Xia F, Lee CW, Altieri DC. Tumor cell dependence on Ran GTP directed mitosis. Cancer Res. 2008;68:1826–1833. doi: 10.1158/0008-5472.CAN-07-5279. [DOI] [PubMed] [Google Scholar]
- Xiao X, Wang P, Chou KC. Quat 2L: a web server for predicting protein quaternary structural attributes. Mol Divers. 2011;15:149–155. doi: 10.1007/s11030-010-9227-8. [DOI] [PubMed] [Google Scholar]
- Yang Y, Blomme EA, Waring JF. Toxicogenomics in drug discovery: from preclinical studies to clinical trials. Chem Biol Interact. 2004;150:71–85. doi: 10.1016/j.cbi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Zakeri P, Moshiri B, Sadeghi M. Prediction of protein submitochondria locations based on data fusion of various features of sequences. J Theor Biol. 2010;269:208–216. doi: 10.1016/j.jtbi.2010.10.026. [DOI] [PubMed] [Google Scholar]
- Zeng YH, Guo YZ, Xiao RQ, Yang L, Yu LZ, Li ML. Using the augmented Chou's pseudo amino acid composition for predicting protein submitochondria locations based on auto covariance approach. J Theor Biol. 2009;259:366–372. doi: 10.1016/j.jtbi.2009.03.028. [DOI] [PubMed] [Google Scholar]
- Zhou HZ, Ma X, Gray MO, Zhu BQ, Nguyen AP, Baker AJ, Simonis U, Cecchini G, Lovett DH, Karliner JS. Transgenic MMP 2 expression induces latent cardiac mitochondrial dysfunction. Biochem Biophys Res Commun. 2007a;358:189–195. doi: 10.1016/j.bbrc.2007.04.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XB, Chen C, Li ZC, Zou XY. Using Chou's amphiphilic pseudo amino acid composition and support vector machine for prediction of enzyme subfamily classes. J Theor Biol. 2007b;248:546–551. doi: 10.1016/j.jtbi.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Zidek N, Hellmann J, Kramer PJ, Hewitt PG. Acute hepatotoxicity: a predictive model based on focused illumina microarrays. Toxicol Sci. 2007;99:289–302. doi: 10.1093/toxsci/kfm131. [DOI] [PubMed] [Google Scholar]