Abstract
Yin Yang 1 (YY1) is a transcription factor with diverse and complex biological functions. YY1 either activates or represses gene transcription, depending on the stimuli received by the cells and its association with other cellular factors. Since its discovery, a biological role for YY1 in tumor development and progression has been suggested because of its regulatory activities toward multiple cancer-related proteins and signaling pathways and its overexpression in most cancers. In this review, we primarily focus on YY1 studies in cancer research, including the regulation of YY1 as a transcription factor, its activities independent of its DNA binding ability, the functions of its associated proteins, and mechanisms regulating YY1 expression and activities. We also discuss the correlation of YY1 expression with clinical outcomes of cancer patients and its target potential in cancer therapy. Although there is not a complete consensus about the role of YY1 in cancers based on its activities of regulating oncogene and tumor suppressor expression, most of the currently available evidence supports a proliferative or oncogenic role of YY1 in tumorigenesis.
Keywords: Yin Yang 1, transcription factor, protein modification, oncogene, tumorigenesis
I. INTRODUCTION
Yin Yang 1 (YY1) (also named nuclear factor [NF] E1, delta, and upstream control region binding protein) was first discovered as a DNA binding protein by several groups in 1991 and 1992.1–5 As a member of the GLI-Krüppel class proteins, the DNA binding domain of YY1 is located at its C-terminal zinc-finger region. YY1 is a ubiquitously expressed and evolutionarily conserved protein. Human YY1 consists of 414 amino acids and has multiple functional domains.6 As a transcription factor, YY1 consensus binding elements have been identified in more than 7% of vertebrate genes,7 suggesting its importance in modulating gene expression. Numerous studies consistently have indicated an essential role of YY1 in different biological processes, including cell growth, differentiation, and embryonic development.
The activities of YY1 in regulating gene expression were demonstrated in many early studies.1,3–5 Depending on the compositional differences of its recruited complexes, YY1 can either repress or activate the transcription of its target genes. Among these cofactors, histone deacetylases (HDACs) and methyltransferase enhancer of zeste homolog 2 (Ezh2) suppress gene expression by promoting histone deacetylation and histone H3 lysine 27 (H3-K27) methylation, respectively, whereas histone acetyltransferases p300, cyclic adenosine monophosphate response element binding (CREB) protein (CBP), and P300/CBP-associated factor (PCAF) stimulate gene expression by acetylating histones (see reviews6,8–10). However, several recent studies, including ours, demonstrated the DNA binding- or transcription-independent activities of YY1, which broadens the regulatory functions of YY1.11
Since its discovery, nearly a thousand YY1-related articles have been published. Several previous reviews have discussed the general functions of YY1.6,8–10 Although YY1 has a role in normal growth and development, most studies have focused on the regulatory roles of YY1 in various cancer-related signaling pathways, such as histone acetylation and methylation, and modulation of the activities or homeostasis of oncogenes and tumor suppressors. Most of these studies implicate a proliferative or promoting role of YY1 in cancer development, consistent with its overexpression status in cancers. In this review, we will focus on cancer-related studies of YY1.
The term Yin Yang is used to describe the interdependence of seemingly opposite forces in nature. The role of YY1 in tumorigenesis can also be viewed as “Yin Yang,” although its oncogenic role is clearly more prevalent than its tumor-suppressive potential, based on the available literature.
II. YIN YANG 1–REGULATED GENE EXPRESSION
When YY1 was first discovered more than 20 years ago, it was identified as a transcription factor that regulated the adeno-associated virus P5 promoter, and its overall effect depended on the viral oncogene E1A.1 Later, Shiet al8 and Thomas and Seto6 proposed the models of YY1-regulated gene expression. YY1 controls a plethora of genes, most of which are cancer-related. The zinc-finger domain at the C-terminal of YY1 recognizes the core sequences of ACAT and CCAT in its binding elements,12 which has been recently extended and further defined.13
The name Yin Yang represents its transcriptional activity in regulating gene expression. The seemingly contrary functions of YY1 depend on its recruited cofactors to target promoters, most of which are proteins that modulate the modifications of histones and DNA. The coactivators that can be recruited by YY1 include p300, CBP, PCAF, and protein arginine methyltransferase (PRMT) 4; the recruited corepressors include HDACs, Ezh2, Ezh1, and DNA methyltransferases (DNMTs). These will be discussed below in detail.
A. Yin Yang 1–Activated Gene Expression
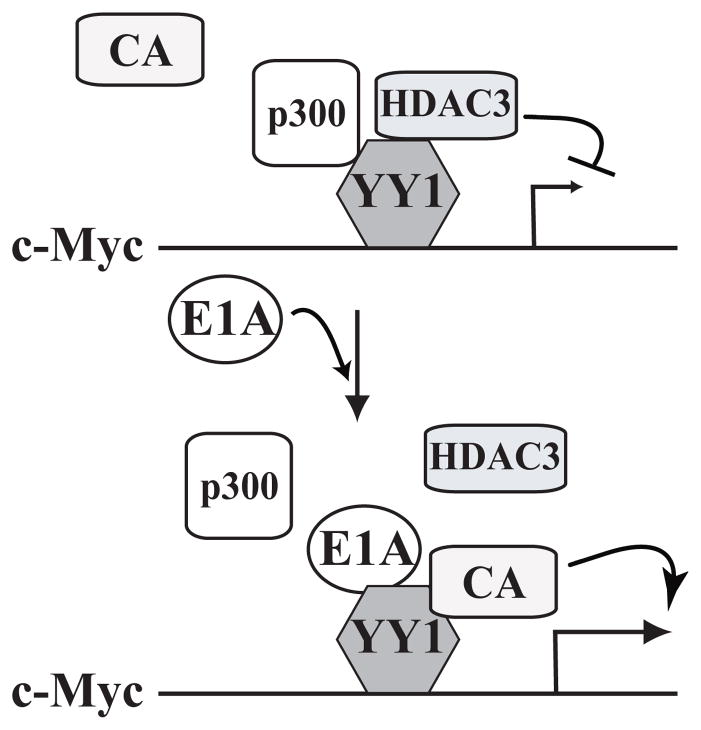
The YY1-activated genes known to be related to cancers are listed in Table 1. When Shi et al1 discovered the YY1 protein, its E1A-mediated “Yin Yang” effects, or gene repression and activation, on the adenovirus P5 promoter were reported. In the following studies, these seemingly contradictory effects of YY1’s transcriptional activities were observed repeatedly. In 1993, Riggs et al14 used YY1 overexpression and reporter assays to demonstrate that YY1 activated the expression of proto-oncogene c-Myc. In this study, transfected YY1 increased the levels of 2 major c-Myc messenger RNA (mRNA) transcript variants. Rezai-Zadehet al15 also showed that YY1 stimulated the c-Myc promoter and demonstrated that PRMT1-mediated methylation at the Arg3 of histone H4 (H4-R3) is important to this activation. However, 2 recent articles17,19 showed that YY1 could regulate c-Myc expression negatively in certain scenarios. Although p300 can both stabilize c-Myc and promote c-Myc-mediated transcription,16 it also can act as a co-repressor to form a p300-HDAC3-YY1 complex that inhibits c-Myc expression.17 Ablating any of these 3 proteins results in chromatin acetylation of the c-Myc promoter and induction of c-Myc transcription. This happens when oncogene E1A is present, which disrupts the YY1-p300-HDAC3 complex and leads to regional histone acetylation on the c-Myc promoter. Consequently, the c-Myc gene is activated.18 The stimulatory effect of E1A on YY1-mediated c-Myc gene activation is depicted in Fig. 1. Another study by Hu et al19 revealed a different mechanism of YY1-mediated c-Myc activation. In Burkitt lymphoma, an immunoglobulin heavy-chain gene HS3 enhancer positively regulates c-Myc expression. YY1 binds to this HS3 enhancer and recruits CBP to this region, which increases the histone acetylation of the c-Myc promoter and activates c-Myc gene expression.
TABLE 1.
Yin Yang 1–Activated Genes and Promoters
| Gene/Promoter | Function of the Gene Product | Mechanism/Observation | Reference |
|---|---|---|---|
| A. Oncogenic, proliferative and/or overexpressed genes in cancer | |||
| B23/nucleophosmin | Regulates nucleosome formation and inhibits tumor suppressors | Hepatitis C virus core, p300, and B23 itself are involved | 24,25 |
| c-Myc | Transcription factor and oncogene of various cancers | E1A converts YY1 from a repressor to an activator; p300 and HDAC3 also are involved | 14,186 |
| HER2/ERBB2/neu | Proto-oncogene in breast cancer | YY1 enhances AP-2 transcriptional activity on the human epidermal growth factor receptor 2 promoter | 30,31 |
| Cyclooxygenase-2 | Oncogene of various cancers | A model with YY1 recruiting p300 and HDAC1,2 was proposed | 38 |
| c-Fos | Proto-oncogene | E1A converts YY1 from a repressor to an activator in this regulation | 187 |
| Glucose regulating protein 78/binding immunoglobulin protein | Promotes tumor proliferation, survival, metastasis, and resistance to cancer therapies | p300 and protein arginine methyltransferase 1 are recruited | 34–36,188 |
| OTX2 | Oncogene of medulloblastoma | Acetylated YY1, but not the unmodified, can bind the enhancer | 39,189 |
| Snail | Enhances cell survival, movement and/ or EMT | YY1 binds a distal Snail 3′ enhancer | 40,42 |
| Msx2 | EMT and tumorigenesis | Three YY1 binding sites are involved | 43,190 |
| Mitochondrial genes: Cyto C, etc. | Cell respiration | Mammalian target of rapamycin regulates YY1-PGC-1α-mediated transcription | 60 |
| DR-α | Overexpressed in cancers | YY1 directly binds to the promoter | 191,192 |
| VASAP-60/PRKCSH/80K-H | Elevated in breast cancer | Two YY1-binding sites are involved. | 193 |
| TGF-β | Overexpressed in tumor; promotes invasiveness and metastasis | A polymorphism mutation in the TGF-β promoter creates a binding site of YY1 that activates TGF-β gene | 194,195 |
| B. Tumor suppression genes | |||
| Breast cancer 1 (BRCA1) | Tumor suppressor in breast cancer | YY1 directly binds to the promoter | 57 |
| ERGIC-53 | Transmembrane lectin facilitates the efficient export of a subset of secretory glycoproteins from the ER; induced by ER-stress | Activating transcription factor 6α-YY1 may associate to coactivate ERGIC-53 gene | 121 |
| HLJ1 | Tumor and invasion suppressor | AP-1 and p300 may be involved | 55,196 |
| p53 | Tumor suppressor | E1A and p300 can induce p53 expression further | 53 |
| p73 | A member of p53 family proteins | YY1 and E2F1 cooperate p73 transcription | 54 |
| Peg3 (via conserved sequence binding element 2) | Imprinted gene with tumor suppression function | CTCF is involved in this regulation | 197 |
| RIZ1 | A histone methyltransferase; altered expression in cancers; a potential tumor suppressor | Correlated with reduced H3-K9 dimethylation | 198 |
| C. Other regulatory proteins in tumorigenesis | |||
| α-SMA | Regulates pulmonary fibrosis that is associated with lung cancer | TGF-β modulates this regulation by inducing YY1 expression | 147 |
| EGFR | Cell signaling molecules involved in diverse cellular functions, including cell proliferation, differentiation, motility, and survival, and in tissue development | Sp1 and YY1 synergistically induce the epidermal growth factor receptor promoter; p53 suppresses this activation | 199 |
| GDAP1 | Broadly expressed in various cancer cell lines | YY1 directly binds to the promoter | 44 |
| Histone H2a and H3 | Aberrantly modified in cancers | Regulated by cell cycle | 58 |
| Histone H4 | Aberrantly modified in cancers | Multiple YY1 binding sites are involved | 59 |
| Heterochromatin protein 1α (HP1α) | Reduced in invasive breast cancer cells | Two YY1 binding sites are involved | 200 |
| Line-1 (promoter) | Altered methylation in cancers | YY1 determines the correct transcription of the full-length retrotransposed product | 136,201–203 |
| MCP-2 | Mediate inflammatory response | YY1 is an intermediate step of interleukin-15-regulated MCP-2 expression | 204 |
| M-opioid receptor | Cancer-related pain | Sp1 is involved | 148 |
| Myelin proteolipid protein | Primary constituent of myelin in the central nervous system | YY1 directly binds to the promoter | 205 |
| OTK18 | Induced by human immunodeficiency virus infection | YY1 directly binds to the promoter | 206 |
| OX40 | A therapeutic target in the treatment of autoimmunity and cancer | Sp1, Sp3, and nuclear factor κB are involved in this regulation | 207,208 |
| Poly(ADP-ribose) polymerase 1 (PARP) | Promotes poly(ADP-ribosyl)ation; related to DNA damage repair | YY1 directly binds the promoter | 209 |
| Proliferating cellular nuclear antigen (PCNA) | Involved in DNA synthesis and repair; cooperating with nucleophosmin/B23 | B23 is involved; accompanied by histone H4 deacetylation | 27,210 |
| RE-1 silencing transcription factor or neuron-restrictive silencer factor | Shows both tumor suppressor and oncogenic activities | YY1 directly binds to the promoter | 211,212 |
| gp91(phox) | Catalytic subunit of the nicotinamide adenine dinucleotide phosphate oxidase; potential target of cancer therapy | Multiple YY1-interacting sites are detected | 213,214 |
| B-type natriuretic peptide | Related to patients’ response to cancer therapy | HDAC2 is recruited | 215 |
| Transferrin receptor (CD71) | Related to poor prognosis and resistant to tamoxifen in breast cancer | YY1 directly binds to the promoter | 216 |
ADP, adenosine diphosphate; AP, activator protein; EMT, epithelial-mesenchymal transition; ER, endoplasmic reticulum; HDAC, histone deacetylase; MCP, monocyte chemotactic protein; TGF, transforming growth factor; YY1, Yin Yang 1.
FIGURE 1.
Schematic model for the stimulatory effect of E1A on Yin Yang 1 (YY1) -mediated c-Myc gene activation. YY1 forms a ternary complex with corepressors p300 and histone deacetylase (HDAC) 3, which represses c-Myc gene expression, probably in normal or resting cells. In transformed cells, the introduction of the viral oncogene E1A disrupts the YY1-p300-HDAC3 complex and leads to c-Myc gene activation, likely through recruiting a coactivator (CA) to promote histone acetylation.17 The stimulatory effects of E1A on YY1-mediated c-Fos gene activation21 and ORF5023 promoter activation, the hepatitis C virus core protein on YY1-mediated B23 gene activation24, and B23 on YY1-mediated proliferating cellular nuclear antigen (PCNA) gene activation26,27 employ the same model, although the YY1-recruited cofactors are different.
The regulation of another proto-oncogene, c-Fos, also follows the same pattern. Zhou et al20 reported that YY1 inhibits c-Fos gene expression through interaction with the transcription complex activating transcription factor (ATF)/CREB. However, 6 months later, they reported that E1A could reverse this inhibition by disrupting the ATF-CREB-YY1 complex, thereby converting YY1 from a repressor to an activator of the c-Fos gene.21 Another report also indicated that YY1 stimulated the association between serum response factor and the c-Fos serum response element,22 suggesting its role in activating c-Fos gene expression. A recent study provided another example of E1A-mediated conversion of YY1 from a repressor to an activator.23 ORF50 is a crucial gene responsible for the switch from latency to lytic replication of Kaposi’s sarcoma-associated herpesvirus. YY1 represses the ORF50 promoter, but the YY1-E1A fusion protein activates it.
In addition to E1A, YY1-regulated expression of B23/nucleophosmin can be modulated by a viral gene product, the hepatitis C virus (HCV) core, which plays an important role in liver oncogenesis.24 The interaction between the HCV core and YY1 leads to the recruitment of p300 and B23 to the YY1 consensus binding site on the B23 promoter and activates gene expression. However, in the absence of the HCV core, YY1 recruits HDAC1 to repress the B23 gene. Because B23 exhibits oncogenic effects by suppressing multiple tumor suppressors,25 this study strongly suggests a stimulatory role of YY1 in liver oncogenesis.
Interestingly, like E1A, B23 also can convert YY1 from a transcriptional repressor to an activator26 and promote the YY1-mediated activation of proliferating cellular nuclear antigen (PCNA).27 These studies provided additional evidence of oncogene-mediated YY1 activation. Overall, YY1 does not always activate, and may even repress, the expression of oncogenes; however, its activating or stimulatory effects on the expression of these oncogenes can be triggered by proliferative or oncogenic signals, such as E1A (Fig. 1).
The androgen receptor (AR) regulates cell differentiation in normal prostate development, but it also plays a crucial role in prostate cancer progression.28 We reported that YY1 interacted with AR and was essential for its optimal transcriptional activity. However, AR-mediated transcription was not enhanced when YY1 was expressed ectopically because YY1 already is overexpressed in prostate cancer cells. This result suggests that the overall levels of YY1 should be maintained within a certain range to achieve its regulatory function.29
YY1 also reportedly activates the expression of human epidermal growth factor receptor (HER) 2 (ERBB2, neu),30,31 which is overexpressed in approximately 30% of breast cancers and correlates with poor prognosis.32 Both gene amplification and transcriptional activation contribute to HER2 overexpression in cancers.33 However, YY1 overexpression is more prevalent in breast tumors without HER2 gene amplification, suggesting it has a role in promoting HER2 gene expression.31
Glucose regulating protein (GRP) 78/binding immunoglobulin protein is a prosurvival endoplasmic reticulum (ER) chaperone and exhibits oncogenic activities including promoting tumor growth, survival, metastasis, and resistance to therapeutic treatments.34 YY1 acts as a transcriptional coactivator of ATF6 and recruits PRMT1 to mediate histone H4 methylation, which leads to GRP78 gene activation.35 A later report also indicated that GATA-4 was essential to YY1-mediated GRP78 activation.36 Because YY1 binds only to the GRP78 promoter under ER stress conditions, this study indicates that YY1 promotes cell survival in response to cellular stresses.
Vascular endothelial growth factor (VEGF) is the key mediator of angiogenesis in cancer and is an essential target of cancer therapy. YY1 forms an active complex with hypoxia-inducible factor (HIF) 1α to activate VEGF gene expression, whereas YY1 depletion reduces systemic neoangiogenesis in vivo during metastasis.37 YY1 induces expression of cyclooxygenase-2 (COX-2), which is overexpressed in 40% of human invasive breast cancers and mediates bone metastasis.38 As a homeobox gene, OTX2 is essential in head development and acts as an important oncogenic driver in medulloblastoma. Acetylated YY1 binds to the enhancer sequence of the OTX2 gene and activates its expression.39
Several studies have suggested a regulatory role of YY1 in epithelial-mesenchymal transition (EMT). EMT can be induced by multiple oncogenic pathways and is inhibited by the tumor invasion suppressor E-cadherin. As a transcription factor, Snail inhibits E-cadherin expression during development and tumorigenesis. YY1 binds the 3′ enhancer of Snail and is essential to Snail expression in melanoma cells.40 However, YY1 does not directly regulate Snail-mediated E-cadherin transcription, although a YY1 binding region is present in the E-cadherin promoter.41,42 Another report demonstrated the activity of YY1 in mediating the transcriptional activation of a homeobox gene, Msx2, which plays an important role in promoting EMT in different cell types.43
Other recent studies also suggested the involvement of YY1 in cancer development. YY1 activates the GDAP1 gene, which is highly expressed in cancer cells originating from different tissues.44 In bladder cancer, YY1 seemed to regulate commonly the expression of down-regulated genes in T1-grade II disease and simultaneously in T1-grade III disease.45 YY1 and cyclin-dependent kinase 6 both were correlated positively with RNA levels of E6/E7, a classic oncogene complex.46
The studies summarized above describe oncogenes activated by YY1. There are also some reports indicating that YY1 mediates the activation of genes with tumor-suppressive functions. Although the negative regulation of p53 at the posttranslational level has been demonstrated by multiple groups,47–52 an early report suggested that overexpressed YY1 could stimulate the expression of a p53 promoter reporter and that this activation could be enhanced by co-transfected E1A.53 These results contradict the well-established oncogenic function of the E1A protein. Another report also indicated that YY1 and E2F1 cooperatively mediate the transcriptional activation of the p73 gene.54 Overexpressed YY1, together with activator protein 1, activated transcription of HLJ1, a suppressor of tumor invasion.55,56 A recent study from Lee et al57 indicated that YY1 transactivates the promoter of breast cancer 1 (BRCA1), a tumor suppressor gene. In breast cancer samples, YY1 positively correlates with BRCA1 expression.
The important role of YY1 in maintaining cellular activities including epigenetic changes and malignant transformation can be shown by its regulation in histone synthesis and cell respiration. YY1 promotes the expression of several histone proteins. Eliassen et al58 reported that YY1 binds a histone DNA element, H3.2α, and regulates the expression of histone H2a and H3 proteins. Last et al59 also demonstrated that YY1 contributes to histone H4 gene transcription. Cunningham et al60 investigated the role of YY1 in mediating cell respiration through mitochondria. They observed that YY1-binding elements are enriched greatly in mitochondrial genes, and YY1 silencing markedly decreases the expression of many mitochondrial genes and in turn compromises oxygen consumption. Consistently, YY1 protein is necessary for the rapamycin-mediated inhibition of mitochondrial genes. Mechanistic studies have indicated that the YY1-PGC-1α transcriptional complex is essential to the mitochondrial oxidative function, while mTOR interferes with this regulation through altering the YY1-PGC-1α complex. Overall, this study suggests that YY1 is involved in maintaining the basal respiration of cells. More YY1-activated genes related to tumorigenesis are shown in Table 1.
B. Yin Yang 1–Repressed Gene Expression
The YY1-repressed genes that are related to cancers are listed in Table 2. Most previous studies have indicated that YY1 represses gene expression through regulating histone deacetylation and H3-K27 methylation. Recently, Yu et al61 demonstrated that YY1 recruits BRCA1 associated protein 1 (BAP1), a de-ubiquitinating enzyme, to mediate gene silencing. In addition to histone modifications, YY1 also may regulate DNA methylation. As a tumor suppressor, CCAAT/enhancer-binding protein δ (CEBPD) inhibits cell differentiation and promotes apoptosis. In cancer cells, YY1 represses CEBPD gene expression through regulating the methylation of its promoter. Epigenetic silencing of CEBPD is achieved by YY1-mediated recruitment of a repressive complex (including SUZ12, Ezh2, DNMT1, DNMT3A, and DNMT3B) to the 2 YY1-binding motifs on the CEBPD promoter. Mutations of either of these motifs will abolish the regulation of these histone and DNA modifiers.62
TABLE 2.
Yin Yang 1–Repressed Genes/Promoters
| Gene/Promoter | Function of the Gene Product | Mechanism/Observation | Reference |
|---|---|---|---|
| A. Oncogenic and/or overexpressed genes | |||
| Lymphoid enhancer-binding factor-1 | Mediates tumor cell invasion | YY1 depletion activates the gene expression and reduces H3-K9 tri-methylation | 217 |
| interferon β | Potential target in cancer therapy | YY2 antagonizes the repression by YY1, Sin3A/NCoR/HDACs complex is recruited by YY1 | 108,218 |
| Has2 | Oncogenic role | Together with SP1; O-GlcNAc modification alteres this regulation | 139 |
| Hypoxia-inducible factor-2α (HIF-2α) | Oncogenic role | Phosphatase and tensin homolog released this repression | 97 |
| HOXB13 | Promotes cancer progression | HDAC4 is recruited | 83,219 |
| Hepatitis B virus pre-S mutants | Viral oncoproteins | A feedback inhibition of the mammalian target of rapamycin activation; HDAC1 is recruited | 220 |
| MAT2A | Upregulated in cancers; potential oncogenic role | Insulin-like growth factor-1 stimulates YY1-binding to the promoter | 145 |
| Matrix metalloproteinase-9 | Increasingly expressed in various cancers | Mono-ubiquitinated YY1 binds C-terminal binding protein; HDAC3 is recruited | 102 |
| ORF50 promoter | Regulates lytic replication of Kaposi sarcoma-associated herpesvirus | Fusion of E1A with YY1 activates the ORF50 promoter | 23 |
| PVT1 | Oncogenic role | A mutation leading to reduced YY1 causes PVT1 overexpression | 221 |
| Steroidogenic acute regulatory | Related to some cancers, e.g., glial tumors | HDAC1 is recruited | 222,223 |
| B. Tumor suppression genes | |||
| CCAAT/enhancer-binding protein δ (CEBPD) | Tumor suppressor | Recruit enhancer of zeste homolog 2, DNMT1, DNMT3A, DNMT3B | 62 |
| Chondromodulin-I | An angiogenesis inhibitor | HDAC2 is recruited by YY1 | 67 |
| Death receptor 5 (DR5) | Receptor in extrinsic apoptosis pathway | Rituximab inhibits DNA binding of YY1 and releases this repression | 68,154 |
| KISS1 | Metastasis suppressor | Sp1 is not involved. | 66 |
| microRNA-29 | Tumor suppressor of rhabdomyosarcoma | Through binding to a conserved regulatory region | 70 |
| microRNA-206 | Promotes cell apoptosis | c-Jun and c-Fos antagonize this regulation | 71 |
| p21 | Leads to cell cycle arrest | YY1 antagonizes p53-mediated transcription | 49,65 |
| p16(INK4a) | Tumor suppressor | HDAC3 and HDAC4 are recruited | 64 |
| Peg3 and Usp29 (via conserved sequence binding element 1) | Peg3: Tumor suppression Usp29: ubiquitin-specific protease 29 | Multiple YY1-interacting sites are detected | 197 |
| Retinoblastoma | Tumor suppressor | GA binding protein and host cell factor 1 are involved in this regulation | 63 |
| C. Other regulatory proteins related to cancers | |||
| α3β1-integrin | Contradictory role in cancer invasion | c-Myc, cyclic adenosine monophosphate response element binding, and HDACs are involved | 170 |
| CD30 | A member of the TNF receptor family; related to lymphoma | Directly binds the promoter | 224 |
| CXCR4, CXCR7 | Chemokine receptor; related to breast cancer cell migration | The regulation is regulated divergently by hypoxia | 225,226 |
| PPAR-δ | Nuclear receptor proteins regulate gene expression | Directly binds the promoter | 227 |
| ERCC5/XPG | DNA repair gene | E2F1 is involved in this regulation | 228 |
| Cyclin D1 | Regulates Cdk4 function | HDAC1 is recruited | 65,229 |
| Involucrin | A marker of differentiation | c-Jun is involved in this regulation | 230–232 |
| Hoxd4 | Regulates morphogenesis | MEL18 also recruited | 233 |
DNMT, DNA methyltransferase; HDAC, histone deacetylase; YY1, Yin Yang 1.
Many other YY1-inhibited genes exhibit tumor suppressive potential. YY1 binds the promoter of retinoblastoma (Rb) protein and cooperates with GA binding protein to repress Rb gene expression (Fig. 2A). During myogenesis, YY1 translocates from the nucleus to the cytoplasm, leading to Rb gene activation.63 YY1 negatively regulates tumor suppressors p16 and p21.49,64,65 In addition, YY1 interferes with p53-mediated transcriptional activation by attenuating the expression of its target genes.49 Recently, YY1 was shown to repress expression of the KiSS1 and chondromodulin-I genes, which inhibit tumor metastasis and angiogenesis, respectively.66,67 YY1 also down-regulates death receptor (DR) 5, a key component in the extrinsic pathway of apoptosis, and consequently confers therapeutic resistance to tumor cells.68 In hepatocellular carcinoma, overexpressed YY1 negatively correlates with Raf kinase inhibitor protein (RKIP), a metastasis-suppressor. The YY1/RKIP mRNA ratio was inverted profoundly in tumors compared with adjacent normal tissues.69
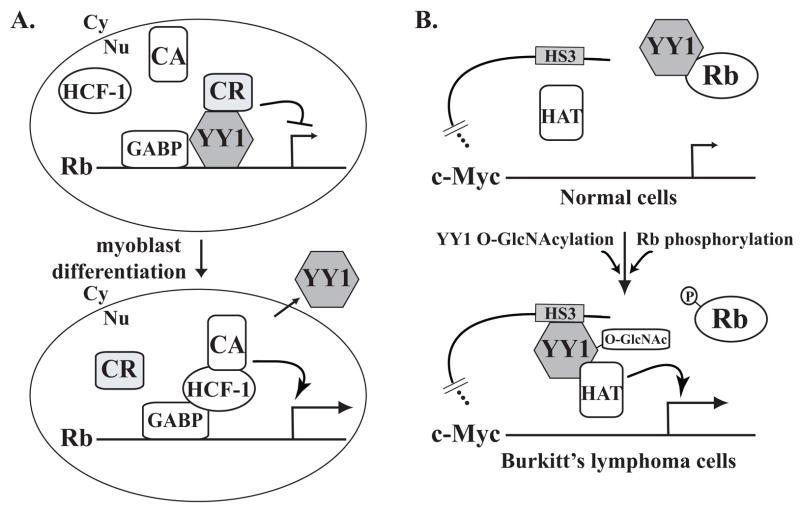
FIGURE 2.
Schematic model of the interplay between Yin Yang 1 (YY1) and retinoblastoma (Rb). A, YY1 negatively regulates Rb gene transcription. YY1 and GA binding protein (GABP) cooperatively repress Rb gene expression, likely through recruiting a co-repressor (CR). During myoblast differentiation, YY1 is translocated from nucleus (Nu) to cytoplasm (Cy). This leads to the recruitment of host cell factor (HCF)–1 and probably a co-activator (CA), and Rb gene activation.63 B, Rb negatively regulates YY1-activated c-Myc gene expression. In human Burkitt lymphoma cells, a translocated c-Myc allele is regulated by the immunoglobulin heavy-chain gene HS3 enhancer. In resting or normal cells, hypophosphorylated Rb binds YY1 and prevents its association with HS3. In Burkitt lymphoma cells, YY1 disassociates with Rb, binds to the HS3 enhancer, and recruits a histone acetyltransferase (HAT) to mediate the acetylation of the c-Myc promoter, which leads to c-Myc gene activation.19 Phosphorylation of Rb causes its disassociation with YY1.112 The O-GlcNAcylation of YY1 both disrupts the Rb-YY1 association and increases the DNA binding affinity of YY1.111,139,140
Several groups reported the role of YY1 in mediating microRNA (miR) expression. YY1 inhibits the expression of both miR-29 and miR-206.70,71 These 2 miRs possess tumor-suppressive activities through activating p53 and promoting cell apoptosis, respectively.72,73 On the other hand, YY1 promotes the expression of miR-190 that is upregulated in liver and pancreatic cancers and may play a role in AKT activation.74,75
YY1 binding elements are present in more than a thousand promoters of vertebrate genes.7 The overall effect of YY1 on the expression of a particular target gene depends on the extracellular stimuli and the availability of coactivators or corepressors, which may vary greatly in the cell lines employed in different studies. To interpret the data correctly, we should consider the following issues:
When YY1 is overexpressed robustly by transient transfection, the cofactors involved in the YY1-coordinated transcription can become saturated or even insufficient; some other proteins that are not involved in normal conditions or at regular YY1 levels may play a role to impact gene expression. Therefore, studies that examine only YY1 overexpression, typically through transfection, may generate artificial results. Such results should be confirmed by other approaches, such as different dosages of ectopic gene expression or gene silencing.
In the endogenous scenario, the transcriptional activities of YY1 largely are accomplished through mediating chromatin remodeling, including various histone modifications and promoter methylation. However, these chromatin modulations are unlikely to occur on transfected plasmid DNA in reporter assays, which have been used in many studies of YY1 transcriptional regulation. Studies of the endogenous genes or at least genome-integrated reporter cassettes are more reliable.
In some reports, YY1 was overexpressed ectopically in cancer cells, most of which already had increased levels of YY1. The antiproliferative effects observed in these conditions would not represent the physiologic consequences of YY1 alterations. Instead, ectopic YY1 expression in normal or non-tumorigenic cells using promoters with medium strengths, or its silencing in tumor cells, would be more accurate and logical in these studies.
Although it is difficult to judge YY1 solely as an oncogene or tumor suppressor based on the genes that YY1 transcriptionally regulates, the tumor-promoting effects of YY1-mediated transcriptional regulation clearly override its role in tumor suppression. It is likely that the overall regulation of YY1 in tumorigenesis may rely on the oncogenic stimuli, cell types, and the interplay with its recruited cofactors, the availability of which may alter at different physiologic conditions.
III. YIN YANG 1 ACTIVITIES INDEPENDENT OF ITS DNA BINDING ACTIVITY
YY1 initially was identified as a transcription factor to bind DNA directly and mediate gene expression.1 However, the regulatory activities of YY1 independent of its DNA binding increasingly have been appreciated.11 Certainly, these activities of YY1 also may play a role in regulating gene expression.
A. Yin Yang 1–Mediated Protein Posttranslational Modifications
YY1 interacts with many proteins that mediate posttranslational modifications, which largely has expanded the complexity of YY1-regulated biological processes. These YY1-interacting protein modifiers include HDACs, p300/CBP, Ezh2, Ezh1, protein inhibitor of activated STAT Y (PIASy), Ubc9, murine double minute 2 (Mdm2), and BAP1. Many of these YY1-interacting partners mediate YY1-regulated gene expression; however, they are involved in non-transcriptional activities. Meanwhile, the observations of YY1 translocation between nucleus and cytoplasm at different cell cycle stages strongly suggest that YY1 is involved in biological processes besides the regulation of gene expression.76,77
YY1 has been reported to interact with numerous proteins, including many protein modifiers that promote acetylation, deacetylation, methylation, ubiquitination, de-ubiquitination, and sumoylation of histone or nonhistone proteins. A well-established mechanism of YY1-regulated gene expression is through recruiting histone modifiers to the target promoters and modulating histone modifications.
1. Acetylation
YY1 was first demonstrated to interact with p300 by Lee et al78 in 1995. Importantly, YY1 and E1A bind to different domains of p300, whereas the binding sites of p300 and E1A on YY1 are also different, suggesting the presence of a YY1-E1A-p300 ternary complex. This explains the ability of E1A to convert YY1 from a transcriptional repressor to an activator by bringing in p300 to YY1 and acetylating histones on the YY1 target promoters, including adenovirus P5, c-Myc, and c-Fos.1,18,21 YY1 inhibits p300-mediated p53 acetylation, which is independent of the transcriptional activity of YY1.48 The acetylation of p53 prevents its ubiquitination79 and promotes p53 transcriptional activity through enhancing p53-DNA interaction.80 Therefore, the negative regulation of p53 acetylation by YY1 implicates its role in antagonizing p53 function.
Yang et al81 made a landmark discovery by demonstrating YY1-mediated histone deacetylation. They discovered that YY1 interacts with human RPD3, a homolog of yeast RPD3 that is the definition base for the class I HDACs. This human RPD3, determined to be HDAC2,6 contributes to YY1-mediated transcriptional repression. Many other studies indicated the involvement of HDACs in YY1-mediated gene suppression. For example, YY1 recruits HDAC2 to the HOXA11 target gene and the chondromodulin-I promoter to repress gene expression.67,82 YY1 also recruits HDAC4 and represses the expression of HOXB13 that mediates growth arrest in androgen receptor-negative prostate cancer cells.83
2. Methylation
Rezai-Zadeh et al15 first reported that YY1 recruited PRMT1 to the c-Myc promoter to mediate histone methylation of H4-R3. Later, Baumeister et al35 also demonstrated that YY1-recruited PRMT1 on the promoter of GRP78, a prosurvival ER chaperone protein, leads to histone H4-R3 methylation and GRP78 gene activation. YY1 interacts with 2 lysine-specific histone methyltransferases, Ezh1 and Ezh2, which mediate histone H3-K27 methylation, a hallmark of gene silencing in cancers.84 Caretti et al85 first demonstrated that YY1 recruited Ezh2 to mediate histone H3-K27 methylation in mouse skeletal muscle cells. Wilkinson et al86 conducted elegant studies to map the YY1-Ezh2 interaction and determine its biological roles. They carried out transgenic studies in Drosophila to demonstrate that the recruitment of Polycomb proteins motif, consisting of residues 201 to 226 of YY1, is necessary and sufficient to recruit Ezh2 and other Polycomb group (PcG) proteins to establish transcriptional repression.86 They also indicated that YY1 associated factor 2, a Drosophila ortholog of Ring1 and YY1 binding protein (RYBP), acts as a molecular bridge between YY1 and other PcG complex proteins.87 A more recent study indicated that the p38α kinase-mediated phosphorylation of Ezh2 at r372 promotes the recruitment of Polycomb repressive complex 2 by YY1.88
Many studies have suggested the concurrence of histone modifications and DNA methylation.89 YY1-mediated gene silencing of the tumor suppressor CEBPD is a good example of this multiple-level regulation. To inhibit CEBPD expression, YY1 recruits PcG proteins SUZ12 and Ezh2, as well as 3 DNMTs (DNMT1, DNMT3A, and DNMT3B), to mediate the methylation of both histone H3-K27 and the CEBPD promoter.62
3. Ubiquitination and Sumoylation
Several groups have demonstrated the negative regulation of p53 by YY1. YY1 directly interacts with both p53 and Mdm2, a ubiquitin E3 ligase, and enhances Mdm2-mediated p53 ubiquitination and degradation.47,48 YY1 depletion leads to either apoptosis or cell cycle arrest, depending on the cell types.47,50,90 Importantly, this regulation is independent of the transcriptional activity of YY1 because a YY1 mutant deficient in DNA binding retains the ability to stimulate p53 ubiquitination, and purified YY1 protein can promote p53 ubiquitination in vitro.47 A previous study indicated that YY1 promotes the expression of cytochrome c oxidase subunit 7C, regulating mitochondrial respiration.91 Recently, Yu et al61 demonstrated that this activation is mediated by the YY1-recruited deubiquitination enzyme BAP1 to the cytochrome c oxidase subunit 7C promoter.
Protein sumoylation may have consequences quite different from ubiquitination.92 The conjugation of small ubiquitin-related modifier (SUMO) may alter protein interaction, stability, and activity.93 We reported that the Lys288 residue of YY1 (Fig. 3) is a target for the conjugation of 3 different SUMO proteins (SUMO-1, -2 and -3).94 Sumoylation of YY1 can be catalyzed by a SUMO E3 ligase PIASy and can inhibit YY1 transcriptional activity. The SUMO conjugating enzyme Ubc9 also directly interacts with YY1. This study revealed the potential of YY1 in mediating sumoylation of its recruited proteins and thereby modulating expression of its target genes.
FIGURE 3.
Domain structure of human Yin Yang 1 (YY1) protein with indicated posttranslational modification sites or regions. Polo-like kinase 1 (Plk1) phosphorylates T39,137 while protein inhibitor of activated STAT Y (PIASy) stimulates the sumoylation at K288.94 p300 and PCAF mediate the acetylation of residues 171–200, while P300/CBP associated factor (PCAF) also acetylates the C-terminal.138 Phosphorylation of Thr348 and Thr378, but not Ser247, reduces the DNA binding affinity of YY1.77 The regions of the 32 lysine residues are indicated. The sequence of YY1 protein is based on the NCBI access number NM_003403.3. The figure is adapted from Sui10 with permission.
B. Yin Yang 1 as a Transcription Cofactor
Most YY1-related studies present YY1’s transcriptional activities on the genes or promoters containing YY1 binding elements. However, several recent reports, including ours, demonstrated the role of YY1 as a transcription cofactor independent of its DNA binding. We observed a YY1-dependent expression of prostate-specific antigen (PSA) in prostate cancer cells and a mutation of the YY1 binding element in the PSA promoter did not alter this regulation.29 Moreover, YY1 directly interacts with androgen response (AR) and promotes the association of AR with an AR element, but a YY1 mutant deficient in AR binding loses this effect. Importantly, AR binds to the YY1 C-terminal,29 its DNA binding domain. Therefore, it is unlikely that YY1 concurrently interacts with both the PSA promoter and AR protein. Overall, YY1 acts as a coactivator to promote AR-mediated PSA expression.
Several other studies also demonstrated the activities of YY1 independent of its DNA binding. YY1 represses the expression of the RNA methyltransferase like 1 gene without its own consensus binding site; this regulation depends on ATF/CREB.95 YY1 enhances the HOXA11-DNA association and is required for the recruitment of HDAC2 by HOXA11.82 YY1 acts as a co-repressor for the transcription factor GON4L.96 Also as a co-repressor, YY1 represses HIF-2α expression, which can be relieved by phosphatase and tensin homolog (PTEN)–mediated suppression of YY1.97
These studies indicate that YY1-mediated gene expression does not always require the direct binding of YY1 to target promoters, and they have extended our understanding of YY1’s role in gene transcription. Compared with a transcription factor, the cofactor YY1 will have different opportunities to expose its functional motifs and recruit other proteins to mediate gene expression. In this scenario, YY1 can provide a platform for the assembly of a scaffold with different transcriptional machineries where many other cofactors are recruited and assembled.90 Importantly, the involvement of multiple YY1-interacting protein modifiers may alter the posttranslational modification status of other factors and in turn determine the expression of a target gene.
C. Other Yin Yang 1–Regulated Processes With Unclear Mechanisms
A number of reports have described other YY1-involved biological processes, but the detailed mechanisms have not been studied. In a human keratinocyte cell line, YY1 inhibited cell differentiation in a 3-dimensional cell culture system.98 Wu et al99 also reported that YY1-depleted spermatocytes exhibited univalent formation, increased aneuploidy, and pachytene cell death, likely due to defects in DNA repair. Overall, YY1 is likely to be involved in many more signaling pathways not yet identified.
IV. YIN YANG 1-ASSOCIATED PROTEINS
Numerous proteins have been reported to associate with YY1; most are involved in different signaling pathways related to cancer development and progression. We categorize these YY1-associated proteins into 4 groups, as shown in Table 3.
TABLE 3.
Yin Yang 1 Interacting Proteins
| Protein | Function | Reference |
|---|---|---|
| A. Modifiers of histones and nonhistone proteins | ||
| p300, CREB binding protein | Acetylation of histone and nonhistone proteins | 78 |
| Histone deacetylases (HDACs) 1, 2, 3, 4, 5 | Deacetylation of histone and nonhistone proteins | 81,83,138,234,235 |
| Ezh1 | Histone methyltransferease | 84 |
| Ezh2 | Histone methyltransferease on H3-K27 and H1-K26 | 85,236 |
| Protein arginine methyltransferase 1 (PRMT1) | Histone methyltransferease on H4-R3 | 15 |
| Mdm2 | Ubiquitination of p53 and histones | 47,48 |
| Ubc9 | E2 conjugating enzyme of sumoylation | 94 |
| Protein inhibitor of activated STAT Y (PIASy) | E3 ligase of sumoylation | 94 |
| B. Other chromatin remodeling proteins | ||
| B23/Nucleophosmin | A histone chaperon involved in nucleosome formation | 26 |
| C-terminal binding protein1 (CtBP1) | A corepressor involved in chromatin remodeling | 100,101 |
| Ring1 and YY1 binding protein (RYBP) | A repressor present in PcG complex | 104 |
| CTCF | Chromatin remodeling; its deregulation causes epigenetic imbalance in cancer | 237,238 |
| INO80 | Chromatin remodeling and DNA repair | 109,110 |
| Sin3-associated protein 30 kDa (SAP30) | Involved in loss of heterozygosity | 107,108 |
| C. Tumorigenesis and apoptosis | ||
| p53 | Tumor suppression and genome stability | 47 |
| Retinoblastoma (Rb) | Tumor suppression and genome stability | 113 |
| p14ARF | Tumor suppression | 47 |
| E1A | Oncogene leading to tumorigenesis | 1 |
| Mdm2 | Oncogene enhancing p53 degradation | 47,239 |
| c-Myc | Oncogene transforming cells | 240 |
| c-Jun | Proto-oncogene | 116 |
| Caspases* 1, 3, 5, 6, 7 | Proteases activated during apoptosis | 117 |
| Poly (adenosine diphosphate-ribose) polymerase 1 (PARP-1) | Posttranslational modification, DNA repair | 142 |
| E2F2, E2F3 | Regulation of retinoblastoma pathway | 105 |
| MBP1 | c-Myc promoter binding protein 1 | 186 |
| α-enolase | Tumor antigen in lung cancer | 186 |
| Integrases of human immunodeficiency virus and leukemia-related virus | YY1 enhances integrase activity | 241 |
| D. General and other regulation | ||
| RNA Pol. II, TFIIB, Sp1, activating transcription factor/cyclic adenosine monophosphate response element binding, TAFII55 | General gene transcription | 20,118–121 |
| Notch1 receptor | Cell fate determination during embryonic development | 186,242 |
| SFMBT2 | A member of Polycomb group proteins | 243 |
| YY1AP (HCCA2) | Hepatocellular carcinoma-specific protein | 244–246 |
| YY1 associated factor 2 (YAF2) | Interacts with MycN in neuroblastoma | 247,248 |
| mTOR (FRAP1) | Key factor in transducing various stimuli to regulate a wide range of cellular functions | 60 |
| Raptor | Associated with mTOR; regulates its expression and kinase activity | 60 |
| PGC-1α | A coactivator of YY1 in regulating mitochondrial genes | 60 |
| Small and large hepatitis delta virus antigens (σ-virus antigen) | Involved in TGF-β and c-Jun-induced signaling cascade | 249 |
| HOXA11 | Regulates uterine development | 82 |
| Activator protein 2 (AP-2) | Acts as a cofactor to stimulate ERBB2 promoter | 30 |
| CP2 | Transcription factor; interacts with the HXPR motif of YY1 | 250 |
| SMAD1/4 | TGF-β signal pathway | 251 |
CREB, cyclic adenosine monophosphate response element binding; Ezh, enhancer of zeste homolog; Mdm2, murine double minute 2; mTOR, mammalian target of rapamycin; TGF, transforming growth factor; YY1, Yin Yang 1.
A. Proteins Regulating Modifications
As we have discussed, YY1 interacts with multiple modifiers that regulate protein acetylation/deacetylation (p300, CBP, PCAF); methylation (Ezh2, Ezh1, PRMT1); sumoylation (Ubc9, PIASy); and ubiquitination/deubiquitination (Mdm2, BAP1). The functional roles of these interactions are described in other sections in which these proteins and their regulation are discussed.
B. Other Chromatin Remodeling Regulators
Functional interplay between YY1 and many other proteins involved in chromatin remodeling has been demonstrated. As a nucleolar protein and histone chaperone, B23 regulates nucleosome formation and inhibits numerous tumor suppressors.25 B23 interacts with YY1 and relieves YY1-mediated transcriptional repression.26,27 YY1 is one of a few members in the PcG that directly bind DNA and recruit other PcG proteins to establish gene silencing. Using Drosophila as a model, Srinivasan and Atchison100 and Atchison et al101 demonstrated that the gene repression mediated by YY1 and other PcG proteins requires the transcriptional co-repressor C-terminal binding protein (CtBP). In rat hippocampal cells, CtBP specifically interacts with mono-ubiquitinated YY1 to repress matrix metalloproteinase-9 (MMP-9) gene transcription.102 Therefore, YY1 de-ubiquitination resulting from neuronal depolarization disrupts the repressive complex of YY1-CtBP-HDAC3 and in turn activates MMP-9 expression. A recent study from Basu and Atchison103 also indicated that CtBP levels regulate DNA binding affinity of YY1 and its PcG recruitment to DNA.
RYBP initially was identified as a co-repressor in the PcG complex.104 A later study indicated that RYBP mediates the interaction of YY1 with E2F proteins, which leads to activation of the Cdc6 gene.105 At the protein level, RYBP mediates the recruitment of PcG complex by YY1.87 Interestingly, RYBP antagonizes Mdm2-mediated p53 ubiquitination,106 in opposition to the effects of YY1.
As discussed above, YY1 can assemble a multifunctional complex to coordinate histone modification and DNA methylation. To repress tumor suppressor CEBPD, YY1-recruited SUZ12, Ezh2, DNMT1, DNMT3A, and DNMT3B form a repressive complex on the CEBPD promoter.62
Sin3-associated protein 30 kDa (SAP30) is a component of the human histone deacetylase complex. The interaction between SAP30 and YY1 can regulate YY1-mediated gene repression.107 SAP30 promotes the recruitment of HDAC1 by YY1 to repress gene expression. In addition, the recruitment of the Sin3A/nuclear receptor co-repressor/HDACs repressor complex by YY1 inhibits the expression of the interferon β gene.108
INO80 is a subfamily of switch 2/sucrose non-fermentable 2 chromatin remodeling proteins and plays regulatory roles in gene transcription, DNA repair, and DNA replication. Two recent studies demonstrated the functional interplay between YY1 and INO80. When YY1 activates transcription of its target genes, INO80 acts as an essential coactivator and helps YY1 to gain access to the target promoters.109 In addition, YY1 and INO80 are essential to homologous recombination-based DNA repair and therefore may regulate the cellular response to genotoxic stress.110
C. Proteins Involved in Tumor Suppression, Oncogenesis, Apoptosis, and DNA Damage
YY1 forms a ternary complex with Mdm2 and p53 to promote Mdm2-mediated p53 ubiquitination and degradation.47 In addition, YY1 interacts with many other proteins directly regulating tumorigenesis. YY1 binds to tumor suppressor Rb in vitro; both YY1 glycosylation and Rb phosphorylation can disrupt this interaction.111,112 In cell-based studies, only hypophosphorylated Rb interacted with YY1, and this interaction reduced DNA binding of YY1,112 implicating a regulatory role of Rb in YY1-mediated transcription. Interestingly, the YY1-Rb complex was observed only in resting cells, not in serum- or lipopolysaccharide-stimulated cells.112,113 The functional interplay between YY1 and Rb is schematically described in Fig. 2. Whether YY1 inhibits the tumor-suppressive activities of Rb and that YY1-Rb disassociation plays a role in cell proliferation or even malignant transformation is a logical prediction and worthy of further investigation.
YY1 also interacts with the tumor suppressor p14 alternate reading frame (ARF). In functional studies, YY1 competes with p53 to bind to p14ARF and therefore attenuates p14ARF-mediated p53 activation,47 which is a separate pathway to antagonize p53 function.
YY1 associates with many oncogene products, including E1A, c-Myc, c-Jun, Mdm2, and B23. As described above, viral oncogene E1A and nucleolar phosphoprotein B23 convert YY1 from a transcriptional repressor to an activator. As a YY1 interacting protein, c-Myc prevents YY1 from associating with its cofactors but does not block its binding to DNA.114 YY1–c-Myc interaction mediates the stimulation of Surf-1 in the mitogen-activated protein kinase cascade.115 Similarly, c-Jun also interacts with YY1 and decreases its binding affinity to its consensus DNA sequence.116
Krippner-Heidenreich et al117 provided evidence of YY1’s involvement in apoptosis. Several apoptotic stimuli could promote rapid translocation of YY1 into the cell nucleus and lead to cleavage of YY1 at Asp12-Gly and Asp119-Gly. Interestingly, one of these N-terminal truncated forms of YY1 could enhance Fas-induced apoptosis, suggesting that YY1 plays a role in positive feedback during apoptosis. An in vitro study showed that YY1 was cleaved by caspases 1, 3, 5, 6, and 7.
D. Other Regulatory Proteins
As a regulatory protein in gene expression, YY1 interacts with many general transcriptional factors and cofactors, such as RNA polymerase II, ATF/CREB, and Sp1.20,118–121 Consistent with our earlier discussion, this suggests that YY1 both can recruit cofactors to its target promoters and can act as a cofactor recruited by others. Importantly, the presence of YY1 in a transcriptional complex creates an interface for these YY1-interacting protein modifiers that may alter the function of transcriptional machinery by modulating their posttranslational modifications. Therefore, overexpressed YY1 in cancers may contribute to their aberrant epigenetic status and promote cancer development.
V. MECHANISMS REGULATING YIN YANG 1 EXPRESSION AND ACTIVITIES
Although most YY1-related reports have described its role in regulating gene expression and protein modifications, an increasing number of studies have revealed different mechanisms regulating YY1 expression (Table 4). YY1 expression is modulated at multiple levels. Sakhinia et al122 and Naidoo et al123 provided striking evidence of the inverse correlation between YY1 mRNA levels and protein expression in follicular lymphoma, suggesting these regulatory mechanisms may have contradictory effects.
TABLE 4.
Factors and Mechanisms Regulating the Function and Expression of Yin Yang 1
| Factor | Effect on YY1 | Reference |
|---|---|---|
| YY1 | Negative feedback; binding intron 1 | 124 |
| Insulin-like growth factor-1 | Activation | 143 |
| Fibroblast growth factor-2 | Activation | 146 |
| Hepatitis B virus protein | Activation | 56 |
| Tumor necrosis factor-α/NF-κB | Activation | 125,126 |
| Morphine | Stimulates YY1 expression | 148 |
| Lysophosphatidylcholine (it has proliferative and antiapoptotic effects) | Enhances YY1 expression | 149 |
| C/EBP-β | Induces YY1 activity | 252 |
| Estrogen-related receptor γ | Induces YY1 expression | 71 |
| Interleukin-15 | Represses YY1 expression | 204 |
| Prohibitin | Represses YY1 (through E2F1 binding site) | 129 |
| MicroRNA-29 | Inhibits YY1 translation | 46,70 |
| MicroRNA-34a | Inhibits YY1 translation | 156,157 |
| DETANONOate (nitric oxide donor) | Inhibits YY1 messenger RNA synthesis and YY1-DNA association by causing S-nitrosation of YY1 | 151,152 |
| Saquinavir | Down-regulates YY1 | 68,185 |
| Staphylococcal enterotoxin A | Reduces YY1 expression 15-fold in peripheral blood mononuclear cells | 153 |
| Naloxone | Down-regulates YY1 expression | 148 |
| Raf kinase inhibitor protein (RKIP, a signal transduction modulator and metastasis suppressor) | Down-regulates YY1 via NF-κB inhibition | 128,253 |
| Rituximab | Inhibits YY1 expression | 127,155 |
| Retinoblastoma Rb | Inhibits DNA binding of YY1 | 111,140 |
| p53 | Inhibits YY1-Sp1-mediated epidermal growth factor receptor activation | 103 |
| CtBP1 | Necessary for YY1-DNA association | 117 |
| Ring1 and YY1 binding protein (RYBP) | A molecular bridge between YY1 and other PcG complex proteins | 87 |
| Transforming growth factor-β (TGF-β) | Induces YY1 expression | 147 |
| Fentanyl | induces YY1 phosphorylation | 74 |
| Probucol | Induces YY1 expression | 254 |
| Lipopolysaccharide | Induces YY1 binding to cyclooxygenase-2 promoter | 38 |
| Sumoylation/PIASy | Enhances YY1 sumoylation | 94 |
| Ubiquitination | Enhances DNA binding and interaction with CtBP, histone deacetylase 3 | 102 |
| Different apoptotic stimuli and DNA synthesis inhibitor | Causes YY1 translocation and cleavage | 76,117 |
| Myeloid nuclear differentiation antigen (MNDA) | Enhances YY1 affinity to DNA | 150 |
CtBP, C-terminal binding protein; NF, nuclear factor; YY1, Yin Yang 1.
A. By Transcription Factors
As a transcription factor, YY1 regulates multiple genes that include itself. Kim et al124 recently indicated that YY1 is autoregulated through its own DNA-binding sites in its first intron. They determined that these YY1 binding sites are necessary for YY1 gene transcription, and exogenous YY1 inhibits the expression of the endogenous YY1 gene, suggesting a negative feedback loop.
YY1 expression is enhanced by transcription factor NF-κB that directly binds the YY1 promoter using its subunit p50/p65 heterodimer.125 As a result, tumor necrosis factor α (TNF-α) treatment in PC-3 cells stimulated YY1 expression and increased its DNA binding affinity. Meanwhile, genetic deletion of the p65 subunit of NF-κB correlated with reduced YY1 transcripts and protein.126 Consistently, rituximab, a chimeric antibody against CD20 that inhibits constitutive NF-κB activity, represses YY1 expression.127
RKIP is a metastasis suppressor gene and is expressed poorly in cancers. Baritaki et al128 demonstrated that RKIP inhibited YY1 gene transcription. Consistent with this observation, a recent study indicated a negative correlation between YY1 and RKIP expression in hepatocellular carcinoma.69 In addition, YY1 expression is inhibited by prohibitin through E2F1 binding sites.129
An in vitro study using colon cancer cells indicated that YY1 showed no evidence of gene amplification or chromosomal translocation.130 However, 2 YY1 mRNA isoforms (7.5 and 2.9 kb) were overexpressed substantially and aneuploidy was observed.
The promoter of mouse YY1 is guanosine and cytidine (G/C) rich.131 Our analyses also demonstrated a high G/C content in the human YY1 promoter and a high guanosine content in its 5′-untranslated region (UTR; of mRNA).132 Interestingly, the G/C contents of the human YY1 promoter increase monotonically as the analyzed region gets closer to the transcription start site. These features confer the YY1 promoter and 5′-UTR with the potential of forming G-quadruplex (G4) structures. G4 is a 4-stranded secondary structure of DNA or RNA stabilized by Hoogsteen hydrogen bonding of guanine quartets and the stacking of these planar quartets.133 G4 structures typically inhibit gene expression, which can be relieved by resolving enzymes.134 Recently, we demonstrated the presence of G4 structures in the YY1 promoter and 5′-UTR that inhibit the transcription mediated by them. Importantly, G4 Resolvase 1 resolves a G4 structure in the YY1 promoter and enhances YY1 expression. Consistently, YY1 levels positively correlated with G4 Resolvase 1 expression in a large cohort of breast cancer patients.132
The promoters of most proto-oncogenes, such as c-Myc and Bcl-2, are guanosine-rich and are regulated negatively by G4 structures in their promoters, whereas the promoters of tumor suppressors are diminished of closely linked guanosine-runs, relative to the genomic average.135 The G/C-rich feature and the presence of G4 structures in the YY1 promoter strongly suggest that YY1 is an oncogene.
B. By Posttranslational Modifications
YY1 regulates posttranslational modifications of multiple proteins. Meanwhile, YY1 itself is also a target of many modifications, including phosphorylation, acetylation, sumoylation, and ubiquitination. Among the amino acids that are modifiable, lysines are the targets of acetylation, methylation, ubiquitination, sumoylation, and neddylation. The high lysine content (8%) of YY1 makes it vulnerable to multiple modifications (Fig. 3). Interestingly, among the 414 residues of human YY1, all lysines are located among the 257 residues of its middle and C-terminal regions, but not in the first 157 residues.10 These modifications regulate the activities, stability, and subcellular localization of YY1.
Rizkallah and Hurt77 studied the effects of YY1 phosphorylation on its subcellular localization and transcriptional activities. They identified 3 phosphorylation sites in the YY1 protein: Ser247 in the spacer region and threonines 348 and 378 in the DNA binding domain. Phosphorylation of the 2 threonines, but not Ser247 (Fig. 3), abolished the DNA binding activity of YY1. Similar results were reported by Zheng et al.74 They indicated that YY1 phosphorylation induced by fentanyl, an agonist of the μ-opioid receptors, impaired the association of YY1 with the Talin2 promoter. However, a much earlier report indicated that the phosphatase treatment of YY1 could abolish its DNA binding affinity in vitro.136 The inconsistency between this study and the first 2 could be due to the different approaches or the presence of unidentified phosphorylation site(s) required for DNA binding of YY1. Very recently, Rizkallah et al137 demonstrated that Polo-like kinase 1 phosphorylates YY1 at threonine 39 (Fig. 3) and studied its fluctuation at different cell cycle stages; however, its role in YY1-mediated transcription was not discussed. Another recent study by de Nigris et al37 revealed the potential of AKT-mediated YY1 phosphorylation. YY1 and AKT were detected in the same immunocomplex. Importantly, when sarcoma osteogenic (SaOS) cells were treated by Ly29004, an inhibitor of phosphoinositide 3 kinase and AKT, YY1 exhibited decreased phosphorylation and cytoplasmic accumulation.37
Acetylation regulates the transcriptional activity of YY1.138 Both p300 and PCAF enhance the acetylation of the central region (residues 171–200) of YY1 (Fig. 3), and this modification augments YY1-mediated gene repression. PCAF also can cause acetylation of the YY1 C-terminal, which compromises the binding of YY1 to its consensus DNA sequence. Interestingly, HDACs can deacetylate YY1 residues in its central region but not at the C-terminal.
YY1 is a substrate of ubiquitination and sumoylation. YY1 degradation likely is regulated by ubiquitination and proteasomal degradation because the treatment of a proteasome inhibitor led to YY1 protein accumulation.47 In neuronal cells, mono-ubiquitinated YY1 interacts with CtBP and HDAC3 to establish a repressive complex that inhibits MMP-9 gene expression.102 We reported that YY1 can be conjugated by SUMO proteins, which is stimulated by PIASy, a SUMO-E3 ligase. Sumoylation of YY1 exhibited inhibitory effects on its target gene expression.94
YY1 is also a subject of other modifications. Some nuclear YY1 can be modified by O-linked N-acetylglucosamine regardless of the differentiation status of the cells, and this modification affects its transcriptional regulation of Has2 gene expression.139 Rb inhibits the DNA binding activity of YY1. However, the O-linked N-acetylglucosamine modification of YY1 caused by high glucose exposure disrupts the YY1-Rb association and consequently increases the ability of YY1 to bind to DNA (Fig. 2B).111,140 YY1 interacts with poly (adenosine diphosphate [ADP]–ribose) polymerase 1 and stimulates its function in catalyzing synthesis of ADP-ribose polymers.141 In addition, YY1 is poly(ADP-ribosyl)ated transiently after genotoxic treatment, which coincides with the activation of poly (ADP–ribose) polymerase 1.142
C. By Growth Factors
Multiple growth factors can increase YY1 levels. YY1 expression is stimulated by insulin-like growth factor-1 in fresh media.143–145 Depletion of insulin-like growth factor -1 by its antibody markedly decreases YY1 levels.143 YY1 expression also can be upregulated by fibroblast growth factor-2 during vascular cell injury.146 Recently, Caggia et al51 made an interesting observation about differential effects of transforming growth factor (TGF)–β3 treatment on YY1 and p53 expression in 2 prostate cell lines. In benign prostatic hyperplasia cells, TGF-β3 led to increased YY1 and decreased p53 expression; however, in DU-145 prostate cancer cells, TGF-β3 reduced YY1 levels and elevated p53 expression. A recent report also demonstrated the activation of YY1 by TGF-β and proposed a regulatory role of YY1 in pulmonary fibrosis.147 Proliferative drugs, such as morphine and lysophosphatidylcholine, also could enhance YY1 expression.148,149 Overall, YY1 levels generally are stimulated by proliferative stimuli, whereas antiproliferative signals tend to antagonize YY1, implicating YY1 as an oncogenic signal-responsive mediator that triggers multiple downstream pathways for malignant transformation.
D. By Other Biomolecules or Mechanisms
Many other molecules or mechanisms can modulate YY1 levels and activities. YY1 expression can be enhanced by lipopolysaccharide,38 and its transcriptional activities can be stimulated by myeloid nuclear differentiation antigen through enhanced YY1-DNA interaction.150
Several other biomolecules exert negative effects on YY1 expression or function. Different apoptotic stimuli, including the DNA synthesis inhibitor aphidicolin, can translocate and cleave YY1 protein.76,117 DETANONOate (a nitric oxide donor), naloxone (a drug used to counter the effects of opioid overdose), staphylococcal enterotoxin, probucol (an antihyperlipidemic drug), and rituximab (a chimeric monoclonal antibody against CD20) all inhibit YY1 expression.148,151–155
YY1 expression also can be suppressed by miRs. Although YY1 negatively regulates miR-29 expression, miR-29 also targets the 3′-UTR of the YY1 mRNA and blocks its translation.46,70 The interplay between YY1 and miR-29 implicates their function in skeletal myogenesis and rhabdomyosarcoma development. Recently, YY1 expression was shown to be down-regulated by miR-34a,156,157 which has a tumor-suppressive function.158
In human Burkitt lymphoma cells, YY1 promotes the expression of the translocated c-Myc allele by binding to the immunoglobulin heavy-chain gene HS3 enhancer, which in turn increases c-Myc promoter acetylation.19 Rb interacts with YY1 to prevent its binding to this enhancer region in normal B cells, but not in Burkitt lymphoma cells, which leads to elevated c-Myc expression in these tumor cells (Fig. 2B).
AKT and PTEN also regulate YY1 expression.97 Phosphorylated AKT positively correlates with YY1 levels. The tumor suppressor PTEN antagonizes phosphoinositide 3 kinase/AKT signaling, leading to YY1 down-regulation. Meanwhile, PTEN also may reduce YY1 levels via AKT independent mechanisms.97 In Drosophila, CtBP levels control the DNA binding of the YY1 ortholog pho and the recruitment of PcG proteins.103
Nguyen et al159 discovered human YY2 that has 65% similarity in complementary DNA (cDNA) and 56% similarity in protein to human YY1. Because the zinc-finger regions of these 2 proteins are similar, YY2 binds to the same consensus sequence as YY1 but with much lower affinity.160 Although it has been suggested that YY2 is a retroposed copy of YY1 inserted into another gene locus,161 a more recent study indicated that human YY2 is not wholly redundant to YY1 and could have opposite effects on their shared target genes.162 Interestingly, silencing of YY2 could reverse the antiproliferative effects of YY1 knockdown, and the depletion of these 2 proteins could cause inverse changes in ultraviolet sensitivity of cells.162
VI. YIN YANG 1 EXPRESSION IN CANCERS AND ITS CORRELATION WITH CLINICAL OUTCOMES
As discussed above, YY1 functional studies strongly implicate its regulatory role in tumorigenesis. Meanwhile, ample literature indicates its deregulated expression in different cancers.163,164 YY1 overexpression has been observed in human breast cancer,30 prostate carcinoma,165 ovarian cancer,166 brain cancer,167 acute myeloid leukemia,168 osteosarcoma,169,170 colon cancer,130 cervical cancer,52,171 large B-cell and follicular lymphoma,122 and hepatoblastoma.172 Some mechanisms regulating YY1 overexpression were discussed in the last section.
Seligson et al165 carried out a comprehensive study using a prostate cancer tissue microarray generated from 1364 representative tissues of 246 hormone-naive prostate cancer patients.165 They observed YY1 staining in both nucleus and cytoplasm in neoplastic tissues and prostatic intraepithelial neoplasia samples. YY1 exhibited increased expression in early malignancy and tumors of intermediate to high morphologic grade.
In 2008, Thomassen et al173 reported that YY1 is upregulated in metastatic breast cancer. In 2009, Powe et al174 studied a large breast cancer tissue microarray generated from 1176 patients to assess the correlation between YY1 and many biomarker genes. They discovered a positive correlation between YY1 staining and the expression of ER, progesterone receptor, and B-cell lymphoma 2. In agreement with its negative regulation toward p53, YY1 staining also showed inverse correlation to p53 levels. However, inconsistent with previous reports of the positive regulation of HER2 gene expression by YY1,30,31 Powe et al174 observed a significant inverse correlation between YY1 and HER2 levels. These conflicting results suggest that other regulatory pathways, in which YY1 may be involved, can counteract or override the stimulatory effects of YY1 on HER2 gene transcription.
Correlation between YY1 overexpression and tumor grades also has been reported in other cancers. In osteosarcoma, YY1 overexpression strongly and positively correlated with the degrees of malignancy.169,175 In colon cancer, YY1 staining intensity was more pronounced in poorly differentiated tumors than in moderately or well-differentiated colon cancers.130
Although YY1 frequently is overexpressed in most cancers, the correlation between YY1 levels and clinical outcomes varies among the diseases in different tissues. In prostate cancer, low nuclear YY1 staining correlated with a shorter time of recurrence, suggesting that YY1 increase was associated with better prognoses.165 In breast cancer tissues, YY1 staining also showed a significant association with improved breast cancer-specific survival and disease-free interval, but not for distant metastasis or tumor recurrence.174 In colon and ovarian cancers, YY1 expression positively correlated with the long-term survival periods of patients.130,166,176
On the contrary, hepatoblastoma patients with high YY1 levels showed a poor prognosis.172 Sakhinia et al122 carried out YY1 studies in follicular lymphoma. They first reported that increased YY1 mRNA levels were associated with a shorter survival interval. However, the later study by Naidoo et al123 indicated that elevated YY1 protein levels in follicular lymphoma patients could be used as a clinical prognostic marker for longer survival. Another group observed the association of YY1 overexpression with B-cell transformation and tumor progression in diffuse large B-cell lymphoma, but the clinical outcomes were not determined.177
Currently, the reasons for the apparent inconsistencies in YY1’s correlation with clinical outcomes among different cancers are unknown. We predict that the genes differentially regulated by YY1 in these cancers are important determinants of the outcomes or responses in cancer therapies. For example, Matsumura et al176 indicated that YY1 and E2F overexpression sensitized ovarian cancer cells to the treatment of taxanes, a group of anticancer drugs. Therefore, the enhanced therapeutic response caused by increased YY1 conferred longer survival to these ovarian cancer patients. In breast cancer, YY1 levels positively correlated with ER expression,174 which is a marker for better prognosis.
VII. YIN YANG 1 AND CANCER THERAPY
Because of its regulatory function in many cancer-related pathways and its overexpression in cancers, the potential of YY1 as a prognostic marker and therapeutic target has been suggested in multiple studies.
YY1 has functional interplay with several tumor suppressors. Importantly, YY1 antagonizes p53 through several distinct mechanisms,10 including promoting Mdm2-mediated p53 ubiquitination and degradation,47 blocking p53 acetylation,48 attenuating p14ARF-mediated p53 stabilization,47 and inhibiting p53-mediated transcription.49 These multiple and consistently negative regulatory mechanisms mediated by YY1 implicate p53 as a primary target of overexpressed YY1 in cancer cells. Although p53 mutations or deletions can be detected in more than 50% of cancers, more than 40% of tumors retain functional p53, and some cancers only have p53 inactivation at late stages. Therefore, most tumors still need to defeat p53-mediated tumor surveillance in their early stages. We predict that YY1 may play a role in this process. YY1 also negatively regulates p14ARF-mediated p53 activation and represses the expression of p16 and p2149,64,65. On the other hand, dephosphorylated Rb binds YY1 and mediates its DNA binding112, although it is still unclear whether YY1 has any impact on the tumor-suppressive function of Rb. PTEN also down-regulates YY1 expression and its transcriptional regulation toward HIF-2α.97
YY1 promotes the functions and expression of multiple oncogenes. Ezh2 has been identified as a bona fide oncogene178 and has been used as a marker of cancers with aggressive or metastatic potential. In functional studies, Ezh2 was required for cancer progression and invasion,179,180 and its overexpression increases the likelihood of therapeutic failure.181 YY1 recruits Ezh2 to target promoters; its overexpression in cancers may promote the methyltransferase activity of Ezh2 and establish aberrant epigenetics, which augments cancer progression. Meanwhile, YY1 activates the expression of c-Myc and c-Fos, especially in the presence of E1A. YY1 expression is stimulated by various growth factors. All these studies implicate a proliferative or oncogenic role of YY1 in cancers.
Multiple studies have suggested the therapeutic potential of YY1 in cancer treatment. The Bonavida group182 extensively studied the role of YY1 in chemo- and immunoresistance in cancer therapy and indicated that YY1 levels were related to therapeutic responsiveness. YY1 negatively regulates the expression of Fas, a transmembrane surface receptor of the tumor necrosis factor receptor family. Therefore, nitric oxide or rituximab treatment inhibited YY1 expression, which, consequently, could upregulate Fas and sensitize tumor cells to Fas-induced apoptosis.152,183 YY1 also represses the expression of DR5. In prostate cancer and B-cell non-Hodgkin lymphoma, DR5 down-regulation caused by YY1 overexpression makes the tumor cells resistant to the treatment with tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). Rituximab inhibits both the DNA binding and expression of YY1 and leads to DR5 upregulation, which also sensitizes TRAIL–induced apoptosis.154,155 Consistently, tumor cells with resistance to rituximab still maintained high levels of YY1.184 Recent studies from Mijatovic et al68 and Donia et al185 demonstrated that Saquinavir-nitric oxide, a drug with enhanced anticancer properties and less toxicity than nitric oxide, dominantly could reduce YY1 expression and consequently upregulate DR5. RKIP-mediated YY1 down-regulation also increased the sensitivity of tumor cells to rituximab.128
A study by de Nigris et al175 focused on the role of YY1 in cell invasion, angiogenesis, and metastasis. Their data indicated that YY1 depletion significantly decreased cell invasion and metastasis growth, which was associated with reduced VEGF and angiogenesis. This finding clearly suggests that YY1 is a promising and effective target in the therapy of bone cancer. A recent study by He et al52 demonstrated significantly elevated YY1 expression in cervical cancer tissues. They also observed that arsenic trioxide, an anti-cervical cancer agent, could reduce the levels of both YY1 mRNA and protein. Meanwhile, YY1 depletion significantly enhanced the apoptosis indicated by arsenic trioxide.52
Overall, YY1 has great potential as a target in cancer therapies. Theoretically, simultaneously targeting several pathways related to cancer development should result in a more efficient and prompt outcome than targeting each of them individually. If a regulatory protein contributing to the abnormality of multiple processes toward malignancy can be identified, targeting this key regulator may exhibit a substantial impact by concurrently reversing or adjusting multiple pathways. YY1 is an essential regulatory factor of numerous epigenetic events and its expression may affect many different biological processes leading to tumorigenesis. Therefore, YY1 is likely one of these key regulatory proteins in cancer development and can serve as an effective target in cancer therapies. Targeting or adjusting YY1 potentially can reverse the aberrant epigenetics of cancer cells and restore their normality. This will be especially important to the cancers in critical organs where radical surgery is not applicable.
VIII. CONCLUSION
As reviewed above, YY1 is a key regulator of multiple signaling pathways involved in cancer epigenetics. Although some discrepancies still exist, we conclude that, overall, YY1 is an oncogene based on the following properties: (1) Upon oncogenic stimulation, YY1 activates multiple well-characterized oncogenes, such as c-Myc, c-Fos, B23, and ERBB2; (2) YY1 mediates the functions of many proteins that contribute to aberrant epigenetic alterations in cancers, such as Ezh2, PRMT1, p300, HDACs, and DNMTs; (3) YY1 antagonizes the functions of several key tumor suppressors, such as p53 and p14ARF, through multiple mechanisms; (4) YY1 expression and function are stimulated by proliferative or oncogenic signals, such as various growth factors and AKT; (5) YY1 is antagonized by multiple tumor suppressors (such as PTEN, Rb, and several tumor suppressive miRs) and apoptotic stimuli; and (6) YY1 is overexpressed in most cancer types and its promoter contains G4-forming structures that typically are present in the promoters of oncogenes. However, because of the complex regulatory role of YY1 in multiple signaling pathways, YY1 levels may correlate with either good or poor clinical outcomes of patients, depending on cancer types.
Acknowledgments
We thank Ms. Karen Klein for reading the manuscript. This work was supported in part by the Golfers Against Cancer Fund and the Research Scholar Grants (116403-RSG-09-082-01-MGO) from the American Cancer Society (Dr Sui); by National Cancer Institute training grant 5T32CA079448 (Mr Stovall); in part by the National Institutes of Health (R01 CA106314) and the Research Scholar Grants (113897-RSG-07-207-01-MGO) from the American Cancer Society (Dr Inoue).
ABBREVIATIONS
- AP-1
activator protein 1
- AR
androgen receptor
- ARE
androgen response element
- BAP1
BRCA1 associated protein-1
- BRCA1
breast cancer 1 (a tumor suppressor gene)
- CEBPD
CCAAT/enhancer-binding protein δ
- ChM-I
chondromodulin-I
- COX-2
cyclooxygenase-2
- COX7c
cytochrome c oxidase subunit 7C
- CtBP
C-terminal binding protein (a transcriptional co-repressor)
- DNMT
DNA methyltransferases
- DR5
death receptor 5
- EMT
epithelial-mesenchymal transition
- ER
estrogen receptor
- Ezh2
enhancer of zeste homolog 2
- G4
G-quadruplex
- H3-K27
histone H3 lysine 27
- HCV
hepatitis C virus
- HDAC
histone deacetylase
- HER2 (neu,ERBB2)
human epidermal growth factor receptor 2
- HIF-1α
hypoxia-inducible factor 1-alpha
- Mdm2
murine double minute 2
- mTOR
mammalian target of rapamycin
- NF-κB
nuclear factor-kappaB
- PARP
poly (ADP-ribose) polymerase 1
- PCAF
P300/CBP-associated factor
- PcG
polycomb group
- PCNA
proliferating cellular nuclear antigen
- PgR
progesterone receptor
- Plk1
Polo-like kinase 1
- PRC
polycomb repressive complex
- PRMT
protein arginine methyltransferases
- PSA
prostate-specific antigen
- PTEN
phosphatase and tensin homolog (a tumor suppressor gene)
- Rb
Retinoblastoma
- REPO
recruitment of polycomb proteins
- RKIP
Raf kinase inhibitor protein
- RNMTL1
RNA methyl-transferase like 1
- RYBP
Ring1 and YY1 binding protein
- SRF
serum response factor
- SUMO
Small Ubiquitin-related Modifier
- TGF
transforming growth factor
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
- UTR
untranslated region (of mRNA)
- VEGF
vascular endothelial growth factor
- YY1
Yin Yang 1
References
- 1.Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67(2):377–88. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 2.Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354(6350):241–5. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 3.Park K, Atchison ML. Isolation of a candidate repressor/activator, NF-E1 (YY-1, delta), that binds to the immunoglobulin kappa 3′ enhancer and the immunoglobulin heavy-chain mu E1 site. Proc Natl Acad Sci U S A. 1991;88(21):9804–8. doi: 10.1073/pnas.88.21.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hariharan N, Kelley DE, Perry RP. Delta, a transcription factor that binds to downstream elements in several polymerase II promoters, is a functionally versatile zinc finger protein. Proc Natl Acad Sci U S A. 1991;88(21):9799–803. doi: 10.1073/pnas.88.21.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flanagan JR, Becker KG, Ennist DL, Gleason SL, Driggers PH, Levi BZ, et al. Cloning of a negative transcription factor that binds to the upstream conserved region of Moloney murine leukemia virus. Mol Cell Biol. 1992;12(1):38–44. doi: 10.1128/mcb.12.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999;236(2):197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 7.Hyde-DeRuyscher RP, Jennings E, Shenk T. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 1995;23(21):4457–65. doi: 10.1093/nar/23.21.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997;1332(2):F49–66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 9.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25(8):1125–42. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 10.Sui G. The regulation of YY1 in tumorigenesis and its targeting potential in cancer therapy. Mol Cell Pharmacol. 2009;1(3):157–76. [Google Scholar]
- 11.Deng Z, Cao P, Wan M, Sui G. Yin Yang 1: a multifaceted protein beyond a transcription factor. Transcription. 2010;1(2):81–4. doi: 10.4161/trns.1.2.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yant SR, Zhu W, Millinoff D, Slightom JL, Goodman M, Gumucio DL. High affinity YY1 binding motifs: identification of two core types (ACAT and CCAT) and distribution of potential binding sites within the human beta globin cluster. Nucleic Acids Res. 1995;23(21):4353–62. doi: 10.1093/nar/23.21.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J, Kim J. YY1’s longer DNA-binding motifs. Genomics. 2009;93(2):152–8. doi: 10.1016/j.ygeno.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riggs KJ, Saleque S, Wong KK, Merrell KT, Lee JS, Shi Y, et al. Yin-yang 1 activates the c-myc promoter. Mol Cell Biol. 1993;13(12):7487–95. doi: 10.1128/mcb.13.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rezai-Zadeh N, Zhang X, Namour F, Fejer G, Wen YD, Yao YL, et al. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 2003;17(8):1019–29. doi: 10.1101/gad.1068003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faiola F, Liu X, Lo S, Pan S, Zhang K, Lymar E, et al. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol Cell Biol. 2005;25(23):10220–34. doi: 10.1128/MCB.25.23.10220-10234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sankar N, Baluchamy S, Kadeppagari RK, Singhal G, Weitzman S, Thimmapaya B. p300 provides a corepressor function by cooperating with YY1 and HDAC3 to repress c-Myc. Oncogene. 2008;27(43):5717–28. doi: 10.1038/onc.2008.181. [DOI] [PubMed] [Google Scholar]
- 18.Kadeppagari RK, Sankar N, Thimmapaya B. Adenovirus transforming protein E1A induces c-Myc in quiescent cells by a novel mechanism. J Virol. 2009;83(10):4810–22. doi: 10.1128/JVI.02145-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu HM, Kanda K, Zhang L, Boxer LM. Activation of the c-myc p1 promoter in Burkitt’s lymphoma by the hs3 immunoglobulin heavy-chain gene enhancer. Leukemia. 2007;21(4):747–53. doi: 10.1038/sj.leu.2404583. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Q, Gedrich RW, Engel DA. Transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB. J Virol. 1995;69(7):4323–30. doi: 10.1128/jvi.69.7.4323-4330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Q, Engel DA. Adenovirus E1A243 disrupts the ATF/CREB-YY1 complex at the mouse c-fos promoter. J Virol. 1995;69(12):7402–9. doi: 10.1128/jvi.69.12.7402-7409.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Natesan S, Gilman M. YY1 facilitates the association of serum response factor with the c-fos serum response element. Mol Cell Biol. 1995;15(11):5975–82. doi: 10.1128/mcb.15.11.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang PJ, Chen LW, Shih YC, Tsai PH, Liu AC, Hung CH, et al. Role of the cellular transcription factor YY1 in the latent-lytic switch of Kaposi’s sarcoma-associated herpesvirus. Virology. 2011;413(2):194–204. doi: 10.1016/j.virol.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Mai RT, Yeh TS, Kao CF, Sun SK, Huang HH, Wu Lee YH. Hepatitis C virus core protein recruits nucleolar phosphoprotein B23 and coactivator p300 to relieve the repression effect of transcriptional factor YY1 on B23 gene expression. Oncogene. 2006;25(3):448–62. doi: 10.1038/sj.onc.1209052. [DOI] [PubMed] [Google Scholar]
- 25.Ye K. Nucleophosmin/B23, a multifunctional protein that can regulate apoptosis. Cancer Biol Ther. 2005;4(9):918–23. doi: 10.4161/cbt.4.9.2072. [DOI] [PubMed] [Google Scholar]
- 26.Inouye CJ, Seto E. Relief of YY1-induced transcriptional repression by protein-protein interaction with the nucleolar phosphoprotein B23. J Biol Chem. 1994;269(9):6506–10. [PubMed] [Google Scholar]
- 27.Weng JJ, Yung BY. Nucleophosmin/B23 regulates PCNA promoter through YY1. Biochem Biophys Res Commun. 2005;335(3):826–31. doi: 10.1016/j.bbrc.2005.07.150. [DOI] [PubMed] [Google Scholar]
- 28.Sharifi N, Farrar WL. Androgen receptor as a therapeutic target for androgen independent prostate cancer. Am J Ther. 2006;13(2):166–70. doi: 10.1097/00045391-200603000-00013. [DOI] [PubMed] [Google Scholar]
- 29.Deng Z, Wan M, Cao P, Rao A, Cramer SD, Sui G. Yin Yang 1 regulates the transcriptional activity of androgen receptor. Oncogene. 2009;28(42):3746–57. doi: 10.1038/onc.2009.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Begon DY, Delacroix L, Vernimmen D, Jackers P, Winkler R. Yin Yang 1 cooperates with activator protein 2 to stimulate ERBB2 gene expression in mammary cancer cells. J Biol Chem. 2005;280(26):24428–34. doi: 10.1074/jbc.M503790200. [DOI] [PubMed] [Google Scholar]
- 31.Allouche A, Nolens G, Tancredi A, Delacroix L, Mardaga J, Fridman V, et al. The combined immunodetection of AP-2alpha and YY1 transcription factors is associated with ERBB2 gene overexpression in primary breast tumors. Breast Cancer Res. 2008;10(1):R9. doi: 10.1186/bcr1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harari D, Yarden Y. Molecular mechanisms underlying ErbB2/HER2 action in breast cancer. Oncogene. 2000;19(53):6102–14. doi: 10.1038/sj.onc.1203973. [DOI] [PubMed] [Google Scholar]
- 33.Seshadri R, Matthews C, Dobrovic A, Horsfall DJ. The significance of oncogene amplification in primary breast cancer. Int J Cancer. 1989;43(2):270–2. doi: 10.1002/ijc.2910430218. [DOI] [PubMed] [Google Scholar]
- 34.Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67(8):3496–9. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- 35.Baumeister P, Luo S, Skarnes WC, Sui G, Seto E, Shi Y, et al. Endoplasmic reticulum stress induction of the Grp78/BiP promoter: activating mechanisms mediated by YY1 and its interactive chromatin modifiers. Mol Cell Biol. 2005;25(11):4529–40. doi: 10.1128/MCB.25.11.4529-4540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao C, Tai WC, Bai Y, Poizat C, Lee AS. In vivo regulation of Grp78/BiP transcription in the embryonic heart: role of the endoplasmic reticulum stress response element and GATA-4. J Biol Chem. 2006;281(13):8877–87. doi: 10.1074/jbc.M505784200. [DOI] [PubMed] [Google Scholar]
- 37.de Nigris F, Crudele V, Giovane A, Casamassimi A, Giordano A, Garban HJ, et al. CXCR4/YY1 inhibition impairs VEGF network and angiogenesis during malignancy. Proc Natl Acad Sci U S A. 2010;107(32):14484–9. doi: 10.1073/pnas.1008256107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joo M, Wright JG, Hu NN, Sadikot RT, Park GY, Blackwell TS, et al. Yin Yang 1 enhances cyclooxygenase-2 gene expression in macrophages. Am J Physiol Lung Cell Mol Physiol. 2007;292(5):L1219–26. doi: 10.1152/ajplung.00474.2006. [DOI] [PubMed] [Google Scholar]
- 39.Takasaki N, Kurokawa D, Nakayama R, Nakayama J, Aizawa S. Acetylated YY1 regulates Otx2 expression in anterior neuroectoderm at two cis-sites 90 kb apart. EMBO J. 2007;26(6):1649–59. doi: 10.1038/sj.emboj.7601619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer MB, Majumder P, Cooper JC, Yoon H, Wade PA, Boss JM. Yin Yang 1 regulates the expression of Snail through a distal enhancer. Mol Cancer Res. 2009;7(2):221–9. doi: 10.1158/1541-7786.MCR-08-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tong ZT, Cai MY, Wang XG, Kong LL, Mai SJ, Liu YH, et al. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene. 2011 doi: 10.1038/onc.2011.254. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 42.Bonavida B, Baritaki S. Dual role of NO donors in the reversal of tumor cell resistance and EMT: Downregulation of the NF-kappaB/Snail/YY1/RKIP circuitry. Nitric Oxide. 2011;24(1):1–7. doi: 10.1016/j.niox.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 43.Tan DP, Nonaka K, Nuckolls GH, Liu YH, Maxson RE, Slavkin HC, et al. YY1 activates Msx2 gene independent of bone morphogenetic protein signaling. Nucleic Acids Res. 2002;30(5):1213–23. doi: 10.1093/nar/30.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ratajewski M, Pulaski L. YY1-dependent transcriptional regulation of the human GDAP1 gene. Genomics. 2009;94(6):407–13. doi: 10.1016/j.ygeno.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 45.Zaravinos A, Lambrou GI, Boulalas I, Delakas D, Spandidos DA. Identification of common differentially expressed genes in urinary bladder cancer. PLoS ONE. 2011;6(4):e18135. doi: 10.1371/journal.pone.0018135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Wang F, Xu J, Ye F, Shen Y, Zhou J, et al. Progressive miRNA expression profiles in cervical carcinogenesis and identification of HPV-related target genes for miR-29. J Pathol. 2011;224(4):484–95. doi: 10.1002/path.2873. [DOI] [PubMed] [Google Scholar]
- 47.Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117(7):859–72. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 48.Gronroos E, Terentiev AA, Punga T, Ericsson J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc Natl Acad Sci U S A. 2004;101(33):12165–70. doi: 10.1073/pnas.0402283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yakovleva T, Kolesnikova L, Vukojevic V, Gileva I, Tan-No K, Austen M, et al. YY1 binding to a subset of p53 DNA-target sites regulates p53-dependent transcription. Biochem Biophys Res Commun. 2004;318(2):615–24. doi: 10.1016/j.bbrc.2004.04.065. [DOI] [PubMed] [Google Scholar]
- 50.Bain M, Sinclair J. Targeted inhibition of the transcription factor YY1 in an embryonal carcinoma cell line results in retarded cell growth, elevated levels of p53 but no increase in apoptotic cell death. Eur J Cell Biol. 2005;84(5):543–53. doi: 10.1016/j.ejcb.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 51.Caggia S, Libra M, Malaponte G, Cardile V. Modulation of YY1 and p53 expression by transforming growth factor-beta3 in prostate cell lines. Cytokine. 2011 doi: 10.1016/j.cyto.2011.06.024. Epub 2011 July 31. [DOI] [PubMed] [Google Scholar]
- 52.He G, Wang Q, Zhou Y, Wu X, Wang L, Duru N, et al. YY1 is a novel potential therapeutic target for the treatment of HPV infection-induced cervical cancer by arsenic trioxide. Int J Gynecol Cancer. 2011;21(6):1097–104. doi: 10.1097/IGC.0b013e31821d2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Furlong EE, Rein T, Martin F. YY1 and NF1 both activate the human p53 promoter by alternatively binding to a composite element, and YY1 and E1A cooperate to amplify p53 promoter activity. Mol Cell Biol. 1996;16(10):5933–45. doi: 10.1128/mcb.16.10.5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu S, Murai S, Kataoka K, Miyagishi M. Yin Yang 1 induces transcriptional activity of p73 through cooperation with E2F1. Biochem Biophys Res Commun. 2008;365(1):75–81. doi: 10.1016/j.bbrc.2007.10.145. [DOI] [PubMed] [Google Scholar]
- 55.Wang CC, Tsai MF, Dai TH, Hong TM, Chan WK, Chen JJ, et al. Synergistic activation of the tumor suppressor, HLJ1, by the transcription factors YY1 and activator protein 1. Cancer Res. 2007;67(10):4816–26. doi: 10.1158/0008-5472.CAN-07-0504. [DOI] [PubMed] [Google Scholar]
- 56.Zhang L, Cai X, Chen K, Wang Z, Wang L, Ren M, et al. Hepatitis B virus protein up-regulated HLJ1 expression via the transcription factor YY1 in human hepatocarcinoma cells. Virus Res. 2011;157(1):76–81. doi: 10.1016/j.virusres.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 57.Lee MH, Lahusen T, Wang RH, Xiao C, Xu X, Hwang YS, et al. Yin Yang 1 positively regulates BRCA1 and inhibits mammary cancer formation. Oncogene. 2011 doi: 10.1038/onc.2011.217. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eliassen KA, Baldwin A, Sikorski EM, Hurt MM. Role for a YY1-binding element in replication-dependent mouse histone gene expression. Mol Cell Biol. 1998;18(12):7106–18. doi: 10.1128/mcb.18.12.7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Last TJ, van Wijnen AJ, Birnbaum MJ, Stein GS, Stein JL. Multiple interactions of the transcription factor YY1 with human histone H4 gene regulatory elements. J Cell Biochem. 1999;72(4):507–16. [PubMed] [Google Scholar]
- 60.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450(7170):736–40. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 61.Yu H, Mashtalir N, Daou S, Hammond-Martel I, Ross J, Sui G, et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol Cell Biol. 2010;30(21):5071–85. doi: 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ko CY, Hsu HC, Shen MR, Chang WC, Wang JM. Epigenetic silencing of CCAAT/enhancer-binding protein delta activity by YY1/polycomb group/DNA methyltransferase complex. J Biol Chem. 2008;283(45):30919–32. doi: 10.1074/jbc.M804029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Delehouzee S, Yoshikawa T, Sawa C, Sawada J, Ito T, Omori M, et al. GABP, HCF-1 and YY1 are involved in Rb gene expression during myogenesis. Genes Cells. 2005;10(7):717–31. doi: 10.1111/j.1365-2443.2005.00873.x. [DOI] [PubMed] [Google Scholar]
- 64.Wang X, Feng Y, Xu L, Chen Y, Zhang Y, Su D, et al. YY1 restrained cell senescence through repressing the transcription of p16. Biochim Biophys Acta. 2008;1783(10):1876–83. doi: 10.1016/j.bbamcr.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 65.Santiago FS, Ishii H, Shafi S, Khurana R, Kanellakis P, Bhindi R, et al. Yin Yang-1 inhibits vascular smooth muscle cell growth and intimal thickening by repressing p21WAF1/Cip1 transcription and p21WAF1/Cip1-Cdk4-cyclin D1 assembly. Circ Res. 2007;101(2):146–55. doi: 10.1161/CIRCRESAHA.106.145235. [DOI] [PubMed] [Google Scholar]
- 66.Mueller JK, Dietzel A, Lomniczi A, Loche A, Tefs K, Kiess W, et al. Transcriptional regulation of the human KiSS1 gene. Mol Cell Endocrinol. 2011;342(1–2):8–19. doi: 10.1016/j.mce.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aoyama T, Okamoto T, Fukiage K, Otsuka S, Furu M, Ito K, et al. Histone modifiers, YY1 and p300, regulate the expression of cartilage-specific gene, chondromodulin-I, in mesenchymal stem cells. J Biol Chem. 2010;285(39):29842–50. doi: 10.1074/jbc.M110.116319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mijatovic S, Maksimovic-Ivanic D, Mojic M, Timotijevic G, Miljkovic D, Mangano K, et al. Cytotoxic and immune-sensitizing properties of nitric oxide-modified Saquinavir in iNOS-positive human melanoma cells. J Cell Physiol. 2011;226(7):1803–12. doi: 10.1002/jcp.22513. [DOI] [PubMed] [Google Scholar]
- 69.Notarbartolo M, Giannitrapani L, Vivona N, Poma P, Labbozzetta M, Florena AM, et al. Frequent alteration of the Yin Yang 1/Raf-1 kinase inhibitory protein ratio in hepatocellular carcinoma. OMICS. 2011;15(5):267–72. doi: 10.1089/omi.2010.0096. [DOI] [PubMed] [Google Scholar]
- 70.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14(5):369–81. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song G, Wang L. Nuclear receptor SHP activates miR-206 expression via a cascade dual inhibitory mechanism. PLoS ONE. 2009;4(9):e6880. doi: 10.1371/journal.pone.0006880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16(1):23–9. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 73.Wang X, Ling C, Bai Y, Zhao J. MicroRNA-206 is associated with invasion and metastasis of lung cancer. Anat Rec (Hoboken) 2011;294(1):88–92. doi: 10.1002/ar.21287. [DOI] [PubMed] [Google Scholar]
- 74.Zheng H, Chu J, Zeng Y, Loh HH, Law PY. Yin Yang 1 phosphorylation contributes to the differential effects of mu-opioid receptor agonists on microRNA-190 expression. J Biol Chem. 2010;285(29):21994–2002. doi: 10.1074/jbc.M110.112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beezhold K, Liu J, Kan H, Meighan T, Castranova V, Shi X, et al. miR-190-mediated downregulation of PHLPP contributes to arsenic-induced Akt activation and carcinogenesis. Toxicol Sci. 2011;123(2):411–20. doi: 10.1093/toxsci/kfr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Palko L, Bass HW, Beyrouthy MJ, Hurt MM. The Yin Yang-1 (YY1) protein undergoes a DNA-replication-associated switch in localization from the cytoplasm to the nucleus at the onset of S phase. J Cell Sci. 2004;117(Pt 3):465–76. doi: 10.1242/jcs.00870. [DOI] [PubMed] [Google Scholar]
- 77.Rizkallah R, Hurt MM. Regulation of the transcription factor YY1 in mitosis through phosphorylation of its DNA-binding domain. Mol Biol Cell. 2009;20(22):4766–76. doi: 10.1091/mbc.E09-04-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee JS, Galvin KM, See RH, Eckner R, Livingston D, Moran E, et al. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995;9(10):1188–98. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 79.Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem. 2002;277(52):50607–11. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- 80.Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci U S A. 2004;101(8):2259–64. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang WM, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci U S A. 1996;93(23):12845–50. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luke MP, Sui G, Liu H, Shi Y. Yin Yang 1 physically interacts with HOXA11 and represses HOXA11-dependent transcription. J Biol Chem. 2006;281(44):33226–32. doi: 10.1074/jbc.M606584200. [DOI] [PubMed] [Google Scholar]
- 83.Ren G, Zhang G, Dong Z, Liu Z, Li L, Feng Y, et al. Recruitment of HDAC4 by transcription factor YY1 represses HOXB13 to affect cell growth in AR-negative prostate cancers. Int J Biochem Cell Biol. 2009;41(5):1094–101. doi: 10.1016/j.biocel.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 84.Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14(5):637–46. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 85.Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18(21):2627–38. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wilkinson FH, Park K, Atchison ML. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci U S A. 2006;103(51):19296–301. doi: 10.1073/pnas.0603564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wilkinson F, Pratt H, Atchison ML. PcG recruitment by the YY1 REPO domain can be mediated by Yaf2. J Cell Biochem. 2010;109(3):478–86. doi: 10.1002/jcb.22424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, et al. TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell stem cell. 2010;7(4):455–69. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 90.Affar el B, Gay F, Shi Y, Liu H, Huarte M, Wu S, et al. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol. 2006;26(9):3565–81. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Seelan RS, Grossman LI. Structural organization and promoter analysis of the bovine cytochrome c oxidase subunit VIIc gene. A functional role for YY1. J Biol Chem. 1997;272(15):10175–81. doi: 10.1074/jbc.272.15.10175. [DOI] [PubMed] [Google Scholar]
- 92.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18(17):2046–59. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 93.Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15(5):536–41. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 94.Deng Z, Wan M, Sui G. PIASy-mediated sumoylation of Yin Yang 1 depends on their interaction but not the RING finger. Mol Cell Biol. 2007;27(10):3780–92. doi: 10.1128/MCB.01761-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu J, De Zhu J, Ni M, Wan F, Gu JR. The ATF/CREB site is the key element for transcription of the human RNA methyltransferase like 1(RNMTL1) gene, a newly discovered 17p13.3 gene. Cell Res. 2002;12(3–4):177–97. doi: 10.1038/sj.cr.7290124. [DOI] [PubMed] [Google Scholar]
- 96.Lu P, Hankel IL, Hostager BS, Swartzendruber JA, Friedman AD, Brenton JL, et al. The developmental regulator protein Gon4l associates with protein YY1, co-repressor Sin3a, and histone deacetylase 1 and mediates transcriptional repression. J Biol Chem. 2011;286(20):18311–9. doi: 10.1074/jbc.M110.133603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Petrella BL, Brinckerhoff CE. PTEN suppression of YY1 induces HIF-2 activity in von-Hippel-Lindau-null renal-cell carcinoma. Cancer Biol Ther. 2009;8(14):1389–401. doi: 10.4161/cbt.8.14.8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taguchi S, Kawachi Y, Ishitsuka Y, Fujisawa Y, Furuta J, Nakamura Y, et al. Overexpression of the transcription factor Yin-Yang-1 suppresses differentiation of HaCaT cells in three-dimensional cell culture. J Invest Dermatol. 2011;131(1):37–45. doi: 10.1038/jid.2010.229. [DOI] [PubMed] [Google Scholar]
- 99.Wu S, Hu YC, Liu H, Shi Y. Loss of YY1 impacts the heterochromatic state and meiotic double-strand breaks during mouse spermatogenesis. Mol Cell Biol. 2009;29(23):6245–56. doi: 10.1128/MCB.00679-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Srinivasan L, Atchison ML. YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 2004;18(21):2596–601. doi: 10.1101/gad.1228204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 2003;22(6):1347–58. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rylski M, Amborska R, Zybura K, Mioduszewska B, Michaluk P, Jaworski J, et al. Yin Yang 1 is a critical repressor of matrix metalloproteinase-9 expression in brain neurons. J Biol Chem. 2008;283(50):35140–53. doi: 10.1074/jbc.M804540200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Basu A, Atchison ML. CtBP levels control intergenic transcripts, PHO/YY1 DNA binding, and PcG recruitment to DNA. J Cell Biochem. 2010;110(1):62–9. doi: 10.1002/jcb.22487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Garcia E, Marcos-Gutierrez C, del Mar Lorente M, Moreno JC, Vidal M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 1999;18(12):3404–18. doi: 10.1093/emboj/18.12.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schlisio S, Halperin T, Vidal M, Nevins JR. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002;21(21):5775–86. doi: 10.1093/emboj/cdf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen D, Zhang J, Li M, Rayburn ER, Wang H, Zhang R. RYBP stabilizes p53 by modulating MDM2. EMBO Rep. 2009;10(2):166–72. doi: 10.1038/embor.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang NE, Lin CH, Lin YS, Yu WC. Modulation of YY1 activity by SAP30. Biochem Biophys Res Commun. 2003;306(1):267–75. doi: 10.1016/s0006-291x(03)00966-5. [DOI] [PubMed] [Google Scholar]
- 108.Le May N, Mansuroglu Z, Leger P, Josse T, Blot G, Billecocq A, et al. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS pathogens. 2008;4(1):e13. doi: 10.1371/journal.ppat.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, et al. YY1 functions with INO80 to activate transcription. Nat Struct Mol Biol. 2007;14(9):872–4. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- 110.Wu S, Shi Y, Mulligan P, Gay F, Landry J, Liu H, et al. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat Struct Mol Biol. 2007;14(12):1165–72. doi: 10.1038/nsmb1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hiromura M, Choi CH, Sabourin NA, Jones H, Bachvarov D, Usheva A. YY1 is regulated by O-linked N-acetylglucosaminylation (O-glcNAcylation) J Biol Chem. 2003;278(16):14046–52. doi: 10.1074/jbc.M300789200. [DOI] [PubMed] [Google Scholar]
- 112.Gordon SJ, Saleque S, Birshtein BK. Yin Yang 1 is a lipopolysaccharide-inducible activator of the murine 3′ Igh enhancer, hs3. J Immunol. 2003;170(11):5549–57. doi: 10.4049/jimmunol.170.11.5549. [DOI] [PubMed] [Google Scholar]
- 113.Petkova V, Romanowski MJ, Sulijoadikusumo I, Rohne D, Kang P, Shenk T, et al. Interaction between YY1 and the retinoblastoma protein. Regulation of cell cycle progression in differentiated cells. J Biol Chem. 2001;276(11):7932–6. doi: 10.1074/jbc.M007411200. [DOI] [PubMed] [Google Scholar]
- 114.Shrivastava A, Yu J, Artandi S, Calame K. YY1 and c-Myc associate in vivo in a manner that depends on c-Myc levels. Proc Natl Acad Sci U S A. 1996;93(20):10638–41. doi: 10.1073/pnas.93.20.10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vernon EG, Gaston K. Myc and YY1 mediate activation of the Surf-1 promoter in response to serum growth factors. Biochim Biophys Acta. 2000;1492(1):172–9. doi: 10.1016/s0167-4781(00)00116-0. [DOI] [PubMed] [Google Scholar]
- 116.Kang JH, Chang SY, Yeom DH, Kim SA, Um SJ, Hong KJ. Weakening of the repressive YY-1 site on the thrombospondin-1 promoter via c-Jun/YY-1 interaction. Exp Mol Med. 2004;36(4):300–10. doi: 10.1038/emm.2004.41. [DOI] [PubMed] [Google Scholar]
- 117.Krippner-Heidenreich A, Walsemann G, Beyrouthy MJ, Speckgens S, Kraft R, Thole H, et al. Caspase-Dependent Regulation and Subcellular Redistribution of the Transcriptional Modulator YY1 during Apoptosis. Mol Cell Biol. 2005;25(9):3704–14. doi: 10.1128/MCB.25.9.3704-3714.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Usheva A, Shenk T. YY1 transcriptional initiator: protein interactions and association with a DNA site containing unpaired strands. Proc Natl Acad Sci U S A. 1996;93(24):13571–6. doi: 10.1073/pnas.93.24.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Seto E, Lewis B, Shenk T. Interaction between transcription factors Sp1 and YY1. Nature. 1993;365(6445):462–4. doi: 10.1038/365462a0. [DOI] [PubMed] [Google Scholar]
- 120.Austen M, Luscher B, Luscher-Firzlaff JM. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J Biol Chem. 1997;272(3):1709–17. doi: 10.1074/jbc.272.3.1709. [DOI] [PubMed] [Google Scholar]
- 121.Renna M, Caporaso MG, Bonatti S, Kaufman RJ, Remondelli P. Regulation of ergic-53 gene transcription in response to endoplasmic reticulum stress. J Biol Chem. 2007;282(31):22499–512. doi: 10.1074/jbc.M703778200. [DOI] [PubMed] [Google Scholar]
- 122.Sakhinia E, Glennie C, Hoyland JA, Menasce LP, Brady G, Miller C, et al. Clinical quantitation of diagnostic and predictive gene expression levels in follicular and diffuse large B-cell lymphoma by RT-PCR gene expression profiling. Blood. 2007;109(9):3922–8. doi: 10.1182/blood-2006-09-046391. [DOI] [PubMed] [Google Scholar]
- 123.Naidoo K, Clay V, Hoyland JA, Swindell R, Linton K, Illidge T, et al. YY1 expression predicts favourable outcome in follicular lymphoma. J Clin Pathol. 2011;64(2):125–9. doi: 10.1136/jcp.2010.078188. [DOI] [PubMed] [Google Scholar]
- 124.Kim JD, Yu S, Kim J. YY1 is autoregulated through its own DNA-binding sites. BMC Mol Biol. 2009;10:85. doi: 10.1186/1471-2199-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang H, Hertlein E, Bakkar N, Sun H, Acharyya S, Wang J, et al. NF-kappaB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol Cell Biol. 2007;27(12):4374–87. doi: 10.1128/MCB.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huerta-Yepez S, Vega M, Garban H, Bonavida B. Involvement of the TNF-alpha autocrine-paracrine loop, via NF-kappaB and YY1, in the regulation of tumor cell resistance to Fas-induced apoptosis. Clin Immunol. 2006;120(3):297–309. doi: 10.1016/j.clim.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 127.Vega MI, Jazirehi AR, Huerta-Yepez S, Bonavida B. Rituximab-induced inhibition of YY1 and Bcl-xL expression in Ramos non-Hodgkin’s lymphoma cell line via inhibition of NF-kappaB activity: role of YY1 and Bcl-xL in Fas resistance and chemoresistance, respectively. J Immunol. 2005;175(4):2174–83. doi: 10.4049/jimmunol.175.4.2174. [DOI] [PubMed] [Google Scholar]
- 128.Baritaki S, Katsman A, Chatterjee D, Yeung KC, Spandidos DA, Bonavida B. Regulation of tumor cell sensitivity to TRAIL-induced apoptosis by the metastatic suppressor Raf kinase inhibitor protein via Yin Yang 1 inhibition and death receptor 5 up-regulation. J Immunol. 2007;179(8):5441–53. doi: 10.4049/jimmunol.179.8.5441. [DOI] [PubMed] [Google Scholar]
- 129.Joshi B, Rastogi S, Morris M, Carastro LM, DeCook C, Seto E, et al. Differential regulation of human YY1 and caspase 7 promoters by prohibitin through E2F1 and p53 binding sites. Biochem J. 2007;401(1):155–66. doi: 10.1042/BJ20060364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chinnappan D, Xiao D, Ratnasari A, Andry C, King TC, Weber HC. Transcription factor YY1 expression in human gastrointestinal cancer cells. Int J Oncol. 2009;34(5):1417–23. [PubMed] [Google Scholar]
- 131.Safrany G, Perry RP. Characterization of the mouse gene that encodes the delta/YY1/NF-E1/UCRBP transcription factor. Proc Natl Acad Sci U S A. 1993;90(12):5559–63. doi: 10.1073/pnas.90.12.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang W, Smaldino PJ, Zhang Q, Miller LD, Cao P, Stadelman K, et al. Yin Yang 1 contains G-quadruplex structures in its promoter and 5′-UTR and its expression is modulated by G4 resolvase 1. Nucleic Acids Res. 2011 Oct 12; doi: 10.1093/nar/gkr849. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Brooks TA, Kendrick S, Hurley L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010;277(17):3459–69. doi: 10.1111/j.1742-4658.2010.07759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vaughn JP, Creacy SD, Routh ED, Joyner-Butt C, Jenkins GS, Pauli S, et al. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J Biol Chem. 2005;280(46):38117–20. doi: 10.1074/jbc.C500348200. [DOI] [PubMed] [Google Scholar]
- 135.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34(14):3887–96. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Becker KG, Jedlicka P, Templeton NS, Liotta L, Ozato K. Characterization of hUCRBP (YY1, NF-E1, delta): a transcription factor that binds the regulatory regions of many viral and cellular genes. Gene. 1994;150(2):259–66. doi: 10.1016/0378-1119(94)90435-9. [DOI] [PubMed] [Google Scholar]
- 137.Rizkallah R, Alexander KE, Kassardjian A, Luscher B, Hurt MM. The transcription factor YY1 is a substrate for Polo-like kinase 1 at the G2/M transition of the cell cycle. PLoS ONE. 2011;6(1):e15928. doi: 10.1371/journal.pone.0015928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21(17):5979–91. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jokela TA, Makkonen KM, Oikari S, Karna R, Koli E, Hart GW, et al. Cellular content of UDP-N-Acetylhexosamines controls hyaluronan synthase 2 expression and correlates with O-GlcNAc modification of transcription factors YY1 and SP1. J Biol Chem. 2011;286(38):33632–40. doi: 10.1074/jbc.M111.265637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ozcan S, Andrali SS, Cantrell JE. Modulation of transcription factor function by O-GlcNAc modification. Biochim Biophys Acta. 2010;1799(5–6):353–64. doi: 10.1016/j.bbagrm.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Griesenbeck J, Ziegler M, Tomilin N, Schweiger M, Oei SL. Stimulation of the catalytic activity of poly(ADP-ribosyl) transferase by transcription factor Yin Yang 1. FEBS Lett. 1999;443(1):20–4. doi: 10.1016/s0014-5793(98)01671-8. [DOI] [PubMed] [Google Scholar]
- 142.Oei SL, Shi Y. Poly(ADP-ribosyl)ation of transcription factor Yin Yang 1 under conditions of DNA damage. Biochem Biophys Res Commun. 2001;285(1):27–31. doi: 10.1006/bbrc.2001.5115. [DOI] [PubMed] [Google Scholar]
- 143.Flanagan JR. Autologous stimulation of YY1 transcription factor expression: role of an insulin-like growth factor. Cell Growth Differ. 1995;6(2):185–90. [PubMed] [Google Scholar]
- 144.Huang HY, Li X, Liu M, Song TJ, He Q, Ma CG, et al. Transcription factor YY1 promotes adipogenesis via inhibiting CHOP-10 expression. Biochem Biophys Res Commun. 2008;375(4):496–500. doi: 10.1016/j.bbrc.2008.07.151. [DOI] [PubMed] [Google Scholar]
- 145.Yang H, Li TW, Peng J, Mato JM, Lu SC. Insulin-like growth factor 1 activates methionine adenosyltransferase 2A transcription by multiple pathways in human colon cancer cells. Biochem J. 2011;436(2):507–16. doi: 10.1042/BJ20101754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Santiago FS, Lowe HC, Bobryshev YV, Khachigian LM. Induction of the transcriptional repressor Yin Yang-1 by vascular cell injury. Autocrine/paracrine role of endogenous fibroblast growth factor-2. J Biol Chem. 2001;276(44):41143–9. doi: 10.1074/jbc.M104913200. [DOI] [PubMed] [Google Scholar]
- 147.Lin X, Sime PJ, Xu H, Williams MA, LaRussa L, Georas SN, et al. Yin Yang 1 is a novel regulator of pulmonary fibrosis. Am J Respir Crit Care Med. 2011;183(12):1689–97. doi: 10.1164/rccm.201002-0232OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li H, Liu H, Wang Z, Liu X, Guo L, Huang L, et al. The role of transcription factors Sp1 and YY1 in proximal promoter region in initiation of transcription of the mu opioid receptor gene in human lymphocytes. J Cell Biochem. 2008;104(1):237–50. doi: 10.1002/jcb.21616. [DOI] [PubMed] [Google Scholar]
- 149.Hara Y, Kusumi Y, Mitsumata M, Li XK, Fujino M. Lysophosphatidylcholine upregulates LOX-1, chemokine receptors, and activation-related transcription factors in human T-cell line Jurkat. J romb rombolysis. 2008;26(2):113–8. doi: 10.1007/s11239-007-0158-x. [DOI] [PubMed] [Google Scholar]
- 150.Xie J, Briggs JA, Briggs RC. Human hematopoietic cell specific nuclear protein MNDA interacts with the multifunctional transcription factor YY1 and stimulates YY1 DNA binding. J Cell Biochem. 1998;70(4):489–506. [PubMed] [Google Scholar]
- 151.Huerta-Yepez S, Vega M, Escoto-Chavez SE, Murdock B, Sakai T, Baritaki S, et al. Nitric oxide sensitizes tumor cells to TRAIL-induced apoptosis via inhibition of the DR5 transcription repressor Yin Yang 1. Nitric Oxide. 2009;20(1):39–52. doi: 10.1016/j.niox.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 152.Hongo F, Garban H, Huerta-Yepez S, Vega M, Jazirehi AR, Mizutani Y, et al. Inhibition of the transcription factor Yin Yang 1 activity by S-nitrosation. Biochem Biophys Res Commun. 2005;336(2):692–701. doi: 10.1016/j.bbrc.2005.08.150. [DOI] [PubMed] [Google Scholar]
- 153.Porichis F, Morou A, Baritaki S, Spandidos DA, Krambovitis E. Activation-induced cell death signalling in CD4+ T cells by staphylococcal enterotoxin A. Toxicol Lett. 2008;176(1):77–84. doi: 10.1016/j.toxlet.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 154.Baritaki S, Huerta-Yepez S, Sakai T, Spandidos DA, Bonavida B. Chemotherapeutic drugs sensitize cancer cells to TRAIL-mediated apoptosis: up-regulation of DR5 and inhibition of Yin Yang 1. Mol Cancer Ther. 2007;6(4):1387–99. doi: 10.1158/1535-7163.MCT-06-0521. [DOI] [PubMed] [Google Scholar]
- 155.Vega MI, Baritaki S, Huerta-Yepez S, Martinez-Paniagua MA, Bonavida B. A potential mechanism of rituximab-induced inhibition of tumor growth through its sensitization to tumor necrosis factor-related apoptosis-inducing ligand-expressing host cytotoxic cells. Leuk Lymphoma. 2011;52(1):108–21. doi: 10.3109/10428194.2010.531408. [DOI] [PubMed] [Google Scholar]
- 156.Chen QR, Yu LR, Tsang P, Wei JS, Song YK, Cheuk A, et al. Systematic proteome analysis identifies transcription factor YY1 as a direct target of miR-34a. J Proteome Res. 2011;10(2):479–87. doi: 10.1021/pr1006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kaller M, Liffers ST, Oeljeklaus S, Kuhlmann K, Roh S, Hoffmann R, et al. Genome-wide characterization of miR-34a Induced changes in protein and mRNA expression by a Combined pulsed SILAC and microarray analysis. Mol Cell Proteomics. 2011;10(8):M111.010462. doi: 10.1074/mcp.M111.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17(2):193–9. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 159.Nguyen N, Zhang X, Olashaw N, Seto E. Molecular cloning and functional characterization of the transcription factor YY2. J Biol Chem. 2004;15:15. doi: 10.1074/jbc.M402525200. [DOI] [PubMed] [Google Scholar]
- 160.Kim JD, Faulk C, Kim J. Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX1. Nucleic Acids Res. 2007;35(10):3442–52. doi: 10.1093/nar/gkm235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Luo C, Lu X, Stubbs L, Kim J. Rapid evolution of a recently retroposed transcription factor YY2 in mammalian genomes. Genomics. 2006;87(3):348–55. doi: 10.1016/j.ygeno.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 162.Chen L, Shioda T, Coser KR, Lynch MC, Yang C, Schmidt EV. Genome-wide analysis of YY2 versus YY1 target genes. Nucleic Acids Res. 2010;38(12):4011–26. doi: 10.1093/nar/gkq112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Castellano G, Torrisi E, Ligresti G, Malaponte G, Militello L, Russo AE, et al. The involvement of the transcription factor Yin Yang 1 in cancer development and progression. Cell Cycle. 2009;8(9):1367–72. doi: 10.4161/cc.8.9.8314. [DOI] [PubMed] [Google Scholar]
- 164.Zaravinos A, Spandidos DA. Yin yang 1 expression in human tumors. Cell Cycle. 2010;9(3):512–22. doi: 10.4161/cc.9.3.10588. [DOI] [PubMed] [Google Scholar]
- 165.Seligson D, Horvath S, Huerta-Yepez S, Hanna S, Garban H, Roberts A, et al. Expression of transcription factor Yin Yang 1 in prostate cancer. Int J Oncol. 2005;27(1):131–41. [PubMed] [Google Scholar]
- 166.Berchuck A, Iversen ES, Lancaster JM, Pittman J, Luo J, Lee P, et al. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin Cancer Res. 2005;11(10):3686–96. doi: 10.1158/1078-0432.CCR-04-2398. [DOI] [PubMed] [Google Scholar]
- 167.Baritaki S, Chatzinikola AM, Vakis AF, Soulitzis N, Karabetsos DA, Neonakis I, et al. YY1 over-expression in human brain gliomas and meningiomas correlates with TGF-beta1, IGF-1 and FGF-2 mRNA levels. Cancer Invest. 2009;27(2):184–92. doi: 10.1080/07357900802210760. [DOI] [PubMed] [Google Scholar]
- 168.Erkeland SJ, Valkhof M, Heijmans-Antonissen C, Delwel R, Valk PJ, Hermans MH, et al. The gene encoding the transcriptional regulator Yin Yang 1 (YY1) is a myeloid transforming gene interfering with neutrophilic differentiation. Blood. 2003;101(3):1111–7. doi: 10.1182/blood-2002-04-1207. [DOI] [PubMed] [Google Scholar]
- 169.de Nigris F, Botti C, de Chiara A, Rossiello R, Apice G, Fazioli F, et al. Expression of transcription factor Yin Yang 1 in human osteosarcomas. Eur J Cancer. 2006;42(15):2420–4. doi: 10.1016/j.ejca.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 170.de Nigris F, Botti C, Rossiello R, Crimi E, Sica V, Napoli C. Cooperation between Myc and YY1 provides novel silencing transcriptional targets of alpha3beta1-integrin in tumour cells. Oncogene. 2007;26:382–94. doi: 10.1038/sj.onc.1209804. [DOI] [PubMed] [Google Scholar]
- 171.Baritaki S, Sifakis S, Huerta-Yepez S, Neonakis IK, Soufla G, Bonavida B, et al. Overexpression of VEGF and TGF-beta1 mRNA in Pap smears correlates with progression of cervical intraepithelial neoplasia to cancer: implication of YY1 in cervical tumorigenesis and HPV infection. Int J Oncol. 2007;31(1):69–79. [PubMed] [Google Scholar]
- 172.Shin E, Lee KB, Park SY, Kim SH, Ryu HS, Park YN, et al. Gene expression profiling of human hepatoblastoma using archived formalin-fixed and paraffin-embedded tissues. Virchows Arch. 2011;458(4):453–65. doi: 10.1007/s00428-011-1043-8. [DOI] [PubMed] [Google Scholar]
- 173.Thomassen M, Tan Q, Kruse TA. Gene expression meta-analysis identifies metastatic pathways and transcription factors in breast cancer. BMC Cancer. 2008;8:394. doi: 10.1186/1471-2407-8-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Powe DG, Akhtar G, Habashy HO, Abdel-Fatah T, Rakha EA, Green AR, et al. Investigating AP-2 and YY1 protein expression as a cause of high HER2 gene transcription in breast cancers with discordant HER2 gene amplification. Breast Cancer Res. 2009;11(6):R90. doi: 10.1186/bcr2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.de Nigris F, Rossiello R, Schiano C, Arra C, Williams-Ignarro S, Barbieri A, et al. Deletion of Yin Yang 1 protein in osteosarcoma cells on cell invasion and CXCR4/angiogenesis and metastasis. Cancer Res. 2008;68(6):1797–808. doi: 10.1158/0008-5472.CAN-07-5582. [DOI] [PubMed] [Google Scholar]
- 176.Matsumura N, Huang Z, Baba T, Lee PS, Barnett JC, Mori S, et al. Yin yang 1 modulates taxane response in epithelial ovarian cancer. Mol Cancer Res. 2009;7(2):210–20. doi: 10.1158/1541-7786.MCR-08-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Castellano G, Torrisi E, Ligresti G, Nicoletti F, Malaponte G, Traval S, et al. Yin Yang 1 overexpression in diffuse large B-cell lymphoma is associated with B-cell transformation and tumor progression. Cell Cycle. 2010;9(3):557–63. doi: 10.4161/cc.9.3.10554. [DOI] [PubMed] [Google Scholar]
- 178.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22(20):5323–35. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647(1–2):21–9. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 180.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 181.Berezovska OP, Glinskii AB, Yang Z, Li XM, Hoffman RM, Glinsky GV. Essential role for activation of the Polycomb group (PcG) protein chromatin silencing pathway in metastatic prostate cancer. Cell Cycle. 2006;5(16):1886–901. doi: 10.4161/cc.5.16.3222. [DOI] [PubMed] [Google Scholar]
- 182.Bonavida B, Huerta-Yepez S, Goodglick L, Mizutani Y, Miki T. Can we develop biomarkers that predict response of cancer patients to immunotherapy? Biomarkers. 2005;10 (Suppl 1):69–76. doi: 10.1080/13547500500216827. [DOI] [PubMed] [Google Scholar]
- 183.Vega MI, Huerta-Yepez S, Jazirehi AR, Garban H, Bonavida B. Rituximab (chimeric anti-CD20) sensitizes B-NHL cell lines to Fas-induced apoptosis. Oncogene. 2005;24(55):8114–27. doi: 10.1038/sj.onc.1208954. [DOI] [PubMed] [Google Scholar]
- 184.Dalle S, Dupire S, Brunet-Manquat S, Reslan L, Plesa A, Dumontet C. In vivo model of follicular lymphoma resistant to rituximab. Clin Cancer Res. 2009;15(3):851–7. doi: 10.1158/1078-0432.CCR-08-1685. [DOI] [PubMed] [Google Scholar]
- 185.Donia M, Maksimovic-Ivanic D, Mijatovic S, Mojic M, Miljkovic D, Timotijevic G, et al. In vitro and in vivo anticancer action of Saquinavir-NO, a novel nitric oxide-derivative of the protease inhibitor saquinavir, on hormone resistant prostate cancer cells. Cell Cycle. 2011;10(3):492–9. doi: 10.4161/cc.10.3.14727. [DOI] [PubMed] [Google Scholar]
- 186.Hsu KW, Hsieh RH, Lee YH, Chao CH, Wu KJ, Tseng MJ, et al. The activated Notch1 receptor cooperates with alpha-enolase and MBP-1 in modulating c-myc activity. Mol Cell Biol. 2008;28(15):4829–42. doi: 10.1128/MCB.00175-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Natesan S, Gilman MZ. DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes Dev. 1993;7(12B):2497–509. doi: 10.1101/gad.7.12b.2497. [DOI] [PubMed] [Google Scholar]
- 188.Li WW, Hsiung Y, Zhou Y, Roy B, Lee AS. Induction of the mammalian GRP78/BiP gene by Ca2+ depletion and formation of aberrant proteins: activation of the conserved stress-inducible grp core promoter element by the human nuclear factor YY1. Mol Cell Biol. 1997;17(1):54–60. doi: 10.1128/mcb.17.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Di C, Liao S, Adamson DC, Parrett TJ, Broderick DK, Shi Q, et al. Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans retinoic acid. Cancer Res. 2005;65(3):919–24. [PubMed] [Google Scholar]
- 190.Satoh K, Ginsburg E, Vonderhaar BK. Msx-1 and Msx-2 in mammary gland development. J Mammary Gland Biol Neoplasia. 2004;9(2):195–205. doi: 10.1023/B:JOMG.0000037162.84758.b5. [DOI] [PubMed] [Google Scholar]
- 191.Hehlgans T, Strominger JL. Activation of transcription by binding of NF-E1 (YY1) to a newly identified element in the first exon of the human DR alpha gene. J Immunol. 1995;154(10):5181–7. [PubMed] [Google Scholar]
- 192.Rangel LB, Agarwal R, Sherman-Baust CA, Mello-Coelho V, Pizer ES, Ji H, et al. Anomalous expression of the HLA-DR alpha and beta chains in ovarian and other cancers. Cancer Biol Ther. 2004;3(10):1021–7. doi: 10.4161/cbt.3.10.1142. [DOI] [PubMed] [Google Scholar]
- 193.Brule S, Sayasith K, Sirois J, Silversides DW, Lussier JG. Structure of the bovine VASAP-60/PRKCSH gene, functional analysis of the promoter, and gene expression analysis. Gene. 2007;391(1–2):63–75. doi: 10.1016/j.gene.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 194.Hobbs K, Negri J, Klinnert M, Rosenwasser LJ, Borish L. Interleukin-10 and transforming growth factor-beta promoter polymorphisms in allergies and asthma. Am J Respir Crit Care Med. 1998;158(6):1958–62. doi: 10.1164/ajrccm.158.6.9804011. [DOI] [PubMed] [Google Scholar]
- 195.de Souza AP, Trevilatto PC, Scarel-Caminaga RM, de Brito RB, Line SR. Analysis of the TGF-beta1 promoter polymorphism (C-509T) in patients with chronic periodontitis. J Clin Periodontol. 2003;30(6):519–23. doi: 10.1034/j.1600-051x.2003.00323.x. [DOI] [PubMed] [Google Scholar]
- 196.Wang CC, Tsai MF, Hong TM, Chang GC, Chen CY, Yang WM, et al. The transcriptional factor YY1 upregulates the novel invasion suppressor HLJ1 expression and inhibits cancer cell invasion. Oncogene. 2005;24(25):4081–93. doi: 10.1038/sj.onc.1208573. [DOI] [PubMed] [Google Scholar]
- 197.Kim JD, Yu S, Choo JH, Kim J. Two evolutionarily conserved sequence elements for Peg3/Usp29 transcription. BMC Mol Biol. 2008;9:108. doi: 10.1186/1471-2199-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Abbondanza C, de Nigris F, De Rosa C, Rossiello R, Puca GA, Napoli C. Silencing of YY1 down-regulates RIZ1 promoter in human osteosarcoma. Oncol Res. 2008;17(1):33–41. doi: 10.3727/096504008784046081. [DOI] [PubMed] [Google Scholar]
- 199.Bheda A, Creek KE, Pirisi L. Loss of p53 induces epidermal growth factor receptor promoter activity in normal human keratinocytes. Oncogene. 2008;27(31):4315–23. doi: 10.1038/onc.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lieberthal JG, Kaminsky M, Parkhurst CN, Tanese N. The role of YY1 in reduced HP1alpha gene expression in invasive human breast cancer cells. Breast Cancer Res. 2009;11(3):R42. doi: 10.1186/bcr2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Becker KG, Swergold GD, Ozato K, Thayer RE. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum Mol Genet. 1993;2(10):1697–702. doi: 10.1093/hmg/2.10.1697. [DOI] [PubMed] [Google Scholar]
- 202.Singer MF, Krek V, McMillan JP, Swergold GD, Thayer RE. LINE-1: a human transposable element. Gene. 1993;135(1–2):183–8. doi: 10.1016/0378-1119(93)90064-a. [DOI] [PubMed] [Google Scholar]
- 203.Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 2004;32(13):3846–55. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mirghomizadeh F, Bullwinkel J, Orinska Z, Janssen O, Petersen A, Singh PB, et al. Transcriptional regulation of mouse mast cell protease-2 by interleukin-15. J Biol Chem. 2009;284(47):32635–41. doi: 10.1074/jbc.M109.015446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Berndt JA, Kim JG, Tosic M, Kim C, Hudson LD. The transcriptional regulator Yin Yang 1 activates the myelin PLP gene. J Neurochem. 2001;77(3):935–42. doi: 10.1046/j.1471-4159.2001.00307.x. [DOI] [PubMed] [Google Scholar]
- 206.Buescher JL, Martinez LB, Sato S, Okuyama S, Ikezu T. YY1 and FoxD3 regulate antiretroviral zinc finger protein OTK18 promoter activation induced by HIV-1 infection. J Neuroimmune Pharmacol. 2009;4(1):103–15. doi: 10.1007/s11481-008-9139-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tone Y, Kojima Y, Furuuchi K, Brady M, Yashiro-Ohtani Y, Tykocinski ML, et al. OX40 Gene Expression Is Up-Regulated by Chromatin Remodeling in Its Promoter Region Containing Sp1/Sp3, YY1, and NF-{kappa}B Binding Sites. J Immunol. 2007;179(3):1760–7. doi: 10.4049/jimmunol.179.3.1760. [DOI] [PubMed] [Google Scholar]
- 208.Redmond WL, Weinberg AD. Targeting OX40 and OX40L for the treatment of autoimmunity and cancer. Critical reviews in immunology. 2007;27(5):415–36. doi: 10.1615/critrevimmunol.v27.i5.20. [DOI] [PubMed] [Google Scholar]
- 209.Oei SL, Shi Y. Transcription factor Yin Yang 1 stimulates poly(ADP-ribosyl)ation and DNA repair. Biochem Biophys Res Commun. 2001;284(2):450–4. doi: 10.1006/bbrc.2001.4985. [DOI] [PubMed] [Google Scholar]
- 210.Labrie C, Lee BH, Mathews MB. Transcription factors RFX1/EF-C and ATF-1 associate with the adenovirus E1A-responsive element of the human proliferating cell nuclear antigen promoter. Nucleic Acids Res. 1995;23(18):3732–41. doi: 10.1093/nar/23.18.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jiang L, Yao M, Shi J, Shen P, Niu G, Fei J. Yin yang 1 directly regulates the transcription of RE-1 silencing transcription factor. J Neurosci Res. 2008;86(6):1209–16. doi: 10.1002/jnr.21595. [DOI] [PubMed] [Google Scholar]
- 212.Majumder S. REST in good times and bad: roles in tumor suppressor and oncogenic activities. Cell Cycle. 2006;5(17):1929–35. doi: 10.4161/cc.5.17.2982. [DOI] [PubMed] [Google Scholar]
- 213.Jacobsen BM, Skalnik DG. YY1 binds five cis-elements and trans-activates the myeloid cell-restricted gp91(phox) promoter. J Biol Chem. 1999;274(42):29984–93. doi: 10.1074/jbc.274.42.29984. [DOI] [PubMed] [Google Scholar]
- 214.Ushio-Fukai M, Nakamura Y. Reactive oxygen species and angiogenesis: NADPH oxidase as target for cancer therapy. Cancer Lett. 2008;266(1):37–52. doi: 10.1016/j.canlet.2008.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Glenn DJ, Wang F, Chen S, Nishimoto M, Gardner DG. Endothelin-stimulated human B-type natriuretic peptide gene expression is mediated by Yin Yang 1 in association with histone deacetylase 2. Hypertension. 2009;53(3):549–55. doi: 10.1161/HYPERTENSIONAHA.108.125088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Holloway K, Sade H, Romero IA, Male D. Action of transcription factors in the control of transferrin receptor expression in human brain endothelium. J Mol Biol. 2007;365(5):1271–84. doi: 10.1016/j.jmb.2006.10.071. [DOI] [PubMed] [Google Scholar]
- 217.Yokoyama NN, Pate KT, Sprowl S, Waterman ML. A role for YY1 in repression of dominant negative LEF-1 expression in colon cancer. Nucleic Acids Res. 2010;38(19):6375–88. doi: 10.1093/nar/gkq492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Klar M, Bode J. Enhanceosome formation over the beta interferon promoter underlies a remote-control mechanism mediated by YY1 and YY2. Mol Cell Biol. 2005;25(22):10159–70. doi: 10.1128/MCB.25.22.10159-10170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Miao J, Wang Z, Provencher H, Muir B, Dahiya S, Carney E, et al. HOXB13 promotes ovarian cancer progression. Proc Natl Acad Sci U S A. 2007;104(43):17093–8. doi: 10.1073/pnas.0707938104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Teng CF, Wu HC, Tsai HW, Shiah HS, Huang W, Su IJ. Novel feedback inhibition of surface antigen synthesis by Mammalian Target of Rapamycin (mTOR) signal and its implication for hepatitis B virus tumorigenesis and therapy. Hepatology. 2011;54:1199–207. doi: 10.1002/hep.24529. [DOI] [PubMed] [Google Scholar]
- 221.Meyer KB, Maia AT, O’Reilly M, Ghoussaini M, Prathalingam R, Porter-Gill P, et al. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS Genet. 2011;7(7):e1002165. doi: 10.1371/journal.pgen.1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Liu Q, Merkler KA, Zhang X, McLean MP. Prostaglandin F2alpha suppresses rat steroidogenic acute regulatory protein expression via induction of Yin Yang 1 protein and recruitment of histone deacetylase 1 protein. Endocrinology. 2007;148(11):5209–19. doi: 10.1210/en.2007-0326. [DOI] [PubMed] [Google Scholar]
- 223.Kim HJ, Kim JE, Ha M, Kang SS, Kim JT, Park IS, et al. Steroidogenic acute regulatory protein expression in the normal human brain and intracranial tumors. Brain Res. 2003;978(1–2):245–9. doi: 10.1016/s0006-8993(03)02840-3. [DOI] [PubMed] [Google Scholar]
- 224.Franchina M, Woo AJ, Dods J, Karimi M, Ho D, Watanabe T, et al. The CD30 gene promoter micro-satellite binds transcription factor Yin Yang 1 (YY1) and shows genetic instability in anaplastic large cell lymphoma. J Pathol. 2008;214(1):65–74. doi: 10.1002/path.2258. [DOI] [PubMed] [Google Scholar]
- 225.Lee BC, Lee TH, Zagozdzon R, Avraham S, Usheva A, Avraham HK. Carboxyl-terminal Src kinase homologous kinase negatively regulates the chemokine receptor CXCR4 through YY1 and impairs CXCR4/CXCL12 (SDF-1alpha)-mediated breast cancer cell migration. Cancer Res. 2005;65(7):2840–5. doi: 10.1158/0008-5472.CAN-04-3309. [DOI] [PubMed] [Google Scholar]
- 226.Tarnowski M, Grymula K, Reca R, Jankowski K, Maksym R, Tarnowska J, et al. Regulation of expression of stromal-derived factor-1 receptors: CXCR4 and CXCR7 in human rhabdomyosarcomas. Mol Cancer Res. 2010;8(1):1–14. doi: 10.1158/1541-7786.MCR-09-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.He CQ, Ding NZ, Fan W. YY1 repressing peroxisome proliferator-activated receptor delta promoter. Mol Cell Biochem. 2008;308(1–2):247–52. doi: 10.1007/s11010-007-9632-1. [DOI] [PubMed] [Google Scholar]
- 228.Crawford EL, Blomquist T, Mullins DN, Yoon Y, Hernandez DR, Al-Bagdhadi M, et al. CEBPG regulates ERCC5/XPG expression in human bronchial epithelial cells and this regulation is modified by E2F1/YY1 interactions. Carcinogenesis. 2007;28(12):2552–9. doi: 10.1093/carcin/bgm214. [DOI] [PubMed] [Google Scholar]
- 229.Cicatiello L, Addeo R, Sasso A, Altucci L, Petrizzi VB, Borgo R, et al. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter. Mol Cell Biol. 2004;24(16):7260–74. doi: 10.1128/MCB.24.16.7260-7274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Lopez-Bayghen E, Vega A, Cadena A, Granados SE, Jave LF, Gariglio P, et al. Transcriptional analysis of the 5′-noncoding region of the human involucrin gene. J Biol Chem. 1996;271(1):512–20. doi: 10.1074/jbc.271.1.512. [DOI] [PubMed] [Google Scholar]
- 231.Xu X, Kawachi Y, Nakamura Y, Sakurai H, Hirota A, Banno T, et al. Yin-yang 1 negatively regulates the differentiation-specific transcription of mouse loricrin gene in undifferentiated keratinocytes. J Invest Dermatol. 2004;123(6):1120–6. doi: 10.1111/j.0022-202X.2004.23492.x. [DOI] [PubMed] [Google Scholar]
- 232.Alvarez-Salas LM, Benitez-Hess ML, Dipaolo JA. YY-1 and c-Jun transcription factors participate in the repression of the human involucrin promoter. Int J Oncol. 2005;26(1):259–66. doi: 10.3892/ijo.26.1.259. [DOI] [PubMed] [Google Scholar]
- 233.Kobrossy L, Rastegar M, Featherstone M. Interplay between chromatin and trans-acting factors regulating the Hoxd4 promoter during neural differentiation. J Biol Chem. 2006;281(36):25926–39. doi: 10.1074/jbc.M602555200. [DOI] [PubMed] [Google Scholar]
- 234.Yang WM, Yao YL, Sun JM, Davie JR, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272(44):28001–7. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 235.Sucharov CC, Langer S, Bristow M, Leinwand L. Shuttling of HDAC5 in H9C2 cells regulates YY1 function through CaMKIV/PKD and PP2A. Am J Physiol Cell Physiol. 2006;291(5):C1029–37. doi: 10.1152/ajpcell.00059.2006. [DOI] [PubMed] [Google Scholar]
- 236.Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14(2):183–93. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- 237.Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT. Identification of a ctcf cofactor, yy1, for the x chromosome binary switch. Mol Cell. 2007;25(1):43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 238.Filippova GN. Genetics and epigenetics of the multifunctional protein CTCF. Cur Top Dev Biol. 2008;80:337–60. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- 239.Liao WR, Hsieh RH, Hsu KW, Wu MZ, Tseng MJ, Mai RT, et al. The CBF1-independent Notch1 signal pathway activates human c-myc expression partially via transcription factor YY1. Carcinogenesis. 2007;28(9):1867–76. doi: 10.1093/carcin/bgm092. [DOI] [PubMed] [Google Scholar]
- 240.Shrivastava A, Saleque S, Kalpana GV, Artandi S, Goff SP, Calame K. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science. 1993;262(5141):1889–92. doi: 10.1126/science.8266081. [DOI] [PubMed] [Google Scholar]
- 241.Inayoshi Y, Okino Y, Miyake K, Mizutani A, Yamamoto-Kishikawa J, Kinoshita Y, et al. Transcription factor YY1 interacts with retroviral integrases and facilitates integration of moloney murine leukemia virus cDNA into the host chromosomes. J Virol. 2010;84(16):8250–61. doi: 10.1128/JVI.02681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Yeh TS, Lin YM, Hsieh RH, Tseng MJ. Association of transcription factor YY1 with the high molecular weight Notch complex suppresses the transactivation activity of Notch. J Biol Chem. 2003;278(43):41963–9. doi: 10.1074/jbc.M304353200. [DOI] [PubMed] [Google Scholar]
- 243.Kuzmin A, Han Z, Golding MC, Mann MR, Latham KE, Varmuza S. The PcG gene Sfmbt2 is paternally expressed in extraembryonic tissues. Gene Expr Patterns. 2008;8(2):107–16. doi: 10.1016/j.modgep.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ohtomo T, Horii T, Nomizu M, Suga T, Yamada J. Molecular cloning of a structural homolog of YY1AP, a coactivator of the multifunctional transcription factor YY1. Amino Acids. 2007;33(4):645–52. doi: 10.1007/s00726-006-0482-z. [DOI] [PubMed] [Google Scholar]
- 245.Wang CY, Liang YJ, Lin YS, Shih HM, Jou YS, Yu WC. YY1AP, a novel co-activator of YY1. J Biol Chem. 2004;279(17):17750–5. doi: 10.1074/jbc.M310532200. [DOI] [PubMed] [Google Scholar]
- 246.Li L, Shi Y, Wu H, Wan B, Li P, Zhou L, et al. Hepatocellular carcinoma-associated gene 2 interacts with MAD2L2. Mol Cell Biochem. 2007;304(1–2):297–304. doi: 10.1007/s11010-007-9512-8. [DOI] [PubMed] [Google Scholar]
- 247.Kalenik JL, Chen D, Bradley ME, Chen SJ, Lee TC. Yeast two-hybrid cloning of a novel zinc finger protein that interacts with the multifunctional transcription factor YY1. Nucleic Acids Res. 1997;25(4):843–9. doi: 10.1093/nar/25.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Bannasch D, Madge B, Schwab M. Functional interaction of Yaf2 with the central region of MycN. Oncogene. 2001;20(41):5913–9. doi: 10.1038/sj.onc.1204747. [DOI] [PubMed] [Google Scholar]
- 249.Huang WH, Mai RT, Lee YH. Transcription factor YY1 and its associated acetyltransferases CBP and p300 interact with hepatitis delta antigens and modulate hepatitis delta virus RNA replication. J Virol. 2008;82(15):7313–24. doi: 10.1128/JVI.02581-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Kang HC, Chung BM, Chae JH, Yang SI, Kim CG, Kim CG. Identification and characterization of four novel peptide motifs that recognize distinct regions of the transcription factor CP2. FEBS J. 2005;272(5):1265–77. doi: 10.1111/j.1742-4658.2005.04564.x. [DOI] [PubMed] [Google Scholar]
- 251.Lee KH, Evans S, Ruan TY, Lassar AB. SMAD-mediated modulation of YY1 activity regulates the BMP response and cardiac-specific expression of a GATA4/5/6-dependent chick Nkx2.5 enhancer. Development. 2004;131(19):4709–23. doi: 10.1242/dev.01344. [DOI] [PubMed] [Google Scholar]
- 252.Ralph WM, Jr, Liu K, Auborn KJ. CCAAT/enhancer-binding protein beta represses human papillomavirus 11 upstream regulatory region expression through a promoter-proximal YY1-binding site. J Gen Virol. 2006;87(Pt 1):51–9. doi: 10.1099/vir.0.81207-0. [DOI] [PubMed] [Google Scholar]
- 253.Granovsky AE, Rosner MR. Raf kinase inhibitory protein: a signal transduction modulator and metastasis suppressor. Cell Res. 2008;18(4):452–7. doi: 10.1038/cr.2008.43. [DOI] [PubMed] [Google Scholar]
- 254.Beck K, Wu BJ, Ni J, Santiago FS, Malabanan KP, Li C, et al. Interplay between heme oxygenase-1 and the multifunctional transcription factor yin yang 1 in the inhibition of intimal hyperplasia. Circ Res. 2010;107(12):1490–7. doi: 10.1161/CIRCRESAHA.110.231985. [DOI] [PubMed] [Google Scholar]