Abstract
Mammalian cancers depend on ‘multiple hits’, some of which promote growth, and some of which block apoptosis. We screened for mutations that require a synergistic block in apoptosis to promote tissue overgrowth, and identified myopic, the Drosophila homolog of the candidate tumor-suppressor and endosomal regulator His-domain-protein-tyrosine-phosphatase (HD-PTP). We find that Myopic regulates the Sav/Wts/Hpo (SWH) tumor suppressor pathway: Myopic PPxY motifs bind conserved residues in the WW domains of the transcriptional co-activator Yorkie and Myopic colocalizes with Yorkie at endosomes. Myopic controls Yorkie endosomal association and protein levels, ultimately influencing expression of some Yorkie target genes. However, the anti-apoptotic gene diap1 is not affected, which may explain the conditional nature of the myopic growth-phenotype. These data establish Myopic as a Yorkie-regulator and implicate Myopic-dependent association of Yorkie with endosomal compartments as a regulatory step in nuclear outputs of the SWH pathway.
Keywords: Myopic, Yorkie, endosome, Hippo, growth
Introduction
The link between unrestrained proliferation and the evasion of apoptosis in vertebrate tumors is well established (e.g Evan and Vousden, 2001). Many growth-promoting lesions such as amplification of c-Myc or the loss of Rb trigger compensatory apoptosis, which must then be overcome by anti-apoptotic lesions in order for tumorigenesis to proceed. Many of the molecular mechanisms that drive tumorigenesis are conserved in Drosophila melanogaster, and in recent years, Drosophila has proven itself amenable to the study of cooperating mutations that drive tumor progression and metastasis (Brumby and Richardson, 2005; Chi et al., 2010; Pagliarini and Xu, 2003; Wu et al., 2010). In some cases this cooperativity has been shown to arise from synergistic effects on cell proliferation and death pathways (Asano et al., 1996; Nicholson et al., 2009; Pellock et al., 2007; Staehling-Hampton et al., 1999), yet the extent to which compensatory apoptosis has hindered the identification of a conditional class of growth suppressor genes in Drosophila has not been comprehensively examined.
The conserved Salvador/Warts/Hippo (SWH) pathway controls a transcriptional program that includes both pro-growth and anti-apoptotic targets (Halder and Johnson, 2011; Pan, 2010). Pathway components include the protocadherin Fat, the apical membrane determinant Crumbs, and the FERM-domain proteins Expanded (Ex) and Merlin (Mer) (Bennett and Harvey, 2006; Chen et al., 2010; Cho et al., 2006; Grzeschik et al., 2010; Hamaratoglu et al., 2006; Ling et al., 2010; Robinson et al., 2010; Silva et al., 2006; Willecke et al., 2006). These factors regulate a core serine/threonine kinase cassette consisting of the Ste20-like kinase Hippo (Hpo) (Harvey et al., 2003; Jia et al., 2003; Pantalacci et al., 2003; Udan et al., 2003; Wu et al., 2003) which acts together with the scaffolding protein Salvador (Sav) (Kango-Singh et al., 2002; Tapon et al., 2002) to phosphorylate the NDR family kinase Warts (Wts) (Justice et al., 1995; Xu et al., 1995). Wts then (Lai et al., 2005) phosphorylates the co-transcriptional activator Yorkie (Yki) on sites including Ser168 (S168) (Dong et al., 2007; Huang et al., 2005). This modification anchors Yki in the cytoplasm by recruiting 14-3-3 proteins (Oh and Irvine, 2008). In the absence of SWH signaling, Yki shuttles into the nucleus and together with sequence-specific DNA binding factors (Goulev et al., 2008; Peng et al., 2009; Wu et al., 2008; Zhang et al., 2008; Zhao et al., 2008) activates a transcriptional program including the pro-growth microRNA bantam, the pro-division gene cyclin E (cycE), the anti-apoptotic gene diap1, and the upstream regulators ex and mer.
The SWH pathway has potent anti-growth activity (rev. in Pan, 2010; Zhao et al., 2010), and multiple mechanisms exist to limit Yki activity in developing tissues. While S168 is critical for Yki:14-3-3 binding, Wts phosphorylation of S111, S250 also contribute to Yki inhibition (Oh and Irvine, 2009; Ren et al., 2010). Yki is directly inhibited in a phosphorylation-independent manner via interactions between two WW domains within Yki that are bound by PPxY sequence motifs in Hpo, Wts and Ex (Badouel et al., 2009; Oh et al., 2009). Mammalian cells have additional regulatory mechanisms to control activity of the Yki homolog Yes-associated protein (YAP). YAP is tyrosine phosphorylated by c-Src/Yes kinases, which modulates the ability of YAP to recruit the Runx2 protein and control osteoblast differentiation (Zaidi et al., 2004). DNA damage triggers the c-Abl kinase to phosphorylate YAP on Y357, and this may bias YAP towards the promoters of apoptotic genes over growth-arrest genes (Levy et al., 2008; Strano et al., 2005). This complexity indicates that mutations in peripheral SWH pathway components may selectively affect the transcriptional specificity of nuclear Yki.
Here we apply a genetic approach to identify conditional growth suppressor mutants and identify Myopic (Mop) as a protein that physically interacts with Yki and restricts its activity. mop is a homolog of human His-domain protein tyrosine phosphatase gene (HD-PTP or PTPN23) (Toyooka et al., 2000), which is located in a region of the genome frequently deleted in cancers (Braga et al., 2002; Kok et al., 1997; Szeles et al., 1997). Mop and HD-PTP proteins contain an amino-terminal Bro1 domain and a carboxy-terminal domain with sequence homology to protein tyrosine phosphatase (PTPase) domains. HD-PTP interacts with endosomal proteins such as CHMP4b via the Bro1 domain, and interacts with the ESCRT-1 component Tsg101 via sites outside the Bro1 domain (Doyotte et al., 2008; Ichioka et al., 2007; Kim et al., 2005; Miura et al., 2008; Odorizzi et al., 2003)
We find that cells lacking Mop display overgrowth phenotypes only in the context of a block in cell death. This growth is accompanied by up-regulation of a subset of Yki transcriptional targets but not the anti-apoptotic gene diap1. mop interacts genetically with yki and acts downstream of wts, but at the level of ex and yki. Mop protein co-localizes with Yki in endosomes and physically interacts with Yki via PPxY motifs in Mop and WW domains in Yki. Moreover, loss of mop shifts the endosomal localization of endogenous Yki and elevates Yki protein levels. These data identify Mop as an inhibitor of Yki in developing tissues and implicate Mop-dependent control of Yki-containing endosomal complexes as a SWH regulatory mechanism in developing tissues.
Results
slaughterhouse-five (sfv) alleles produce conditional tissue overgrowth
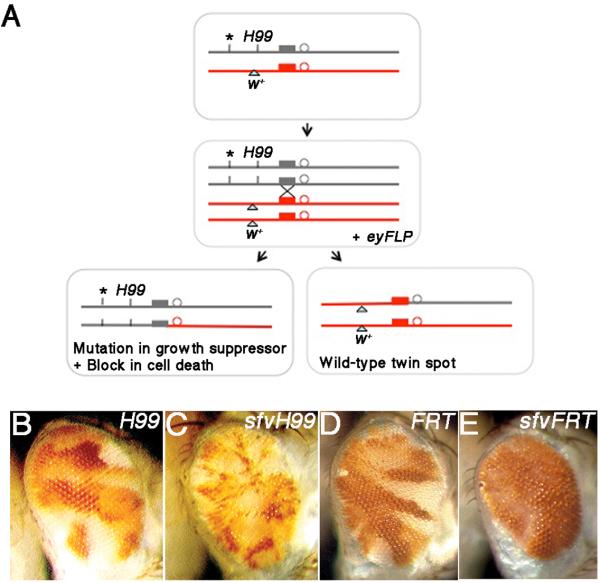
We designed and implemented a screen for survival-dependent growth suppressor genes on Drosophila chromosome 3L. The screen used the FLP recombinase driven by the promoter of the eyeless (ey) gene (Newsome et al., 2000) to produce a mixture of mutant clones (unpigmented) and wild-type twin spots (pigmented red) in the adult eye (Fig. 1A). To block cell death in mutant clones, mutagenesis was carried out using a parental FRT chromosome carrying the genomic deletion H99, which removes genes required for virtually all developmentally programmed cell death (White et al., 1994). H99 mutant clones display a block in developmental apoptosis in the pupal retina (30hrs after puparium formation (APF)) (Fig. S1A-A’) and exhibit minimal phenotypes in the adult eye (Fig. 1B vs. D). We used this background to screen for recessive mutations that could synergize with H99 to confer a growth advantage and identified a recessive-lethal complementation group that we named slaughterhouse-five (sfv) consisting of two alleles (F2.6.3 and F2.6.11) that exhibited a clonal growth advantage in the adult eye (Fig. 1C) relative to an H99 control. Experiments were carried out with the sfv3 allele (F2.6.3) unless otherwise indicated. To test the survival-dependent nature of the sfv phenotype, the H99 deletion was removed. Adult sfv mosaic eyes are small and rough, and contain little to no sfv mutant tissue (Fig. 1E). Clones of sfv mutant cells in the larval eye disc contain elevated levels of cleaved Caspase-3 (C3) (Fig. S1B-B’), indicating they are normally eliminated by apoptosis. Thus the overgrowth of sfv mutant cells is conditional on a synergistic block in cell death provided by the H99 deletion.
Figure 1. An eyFLP Mosaic Screen for Conditional Growth Suppressors.
(A) Clones of cells carrying EMS induced mutations (*) in the background of the H99,FRT80B chromosome were generated using the eyFLP transgene. Homozygous *H99,FRT mutant cells are marked by the absence of the P[m−w+] transgene and wild-type twin spots are marked by the presence of P[m−w+].
(B-E) Representative images of H99 alone (B), sfvH99 (C), control FRT80B (D) and sfvFRT80B (E) mosaic adult eyes.
sfv is allelic to myopic, the Drosophila homolog of HD-PTP
The sfv lesions were mapped by deficiency mapping and candidate gene sequencing. This identified a nonsense mutation in the seventh exon of the gene CG9311 on the sfv3 chromosome. The CG9311 gene corresponds to myopic (mop), which encodes the Drosophila homolog of vertebrate HD-PTP. The sfv3 mutation truncates Mop within its C-terminal region, just prior to the PTPase catalytic region (Fig. S1H). The lesion in the sfv11 allele was not identified, but staining with an anti-Mop antibody shows reduced Mop protein levels in both sfv3H99 and sfv11H99 eye clones (Fig. S1C-C’ and data not shown), indicating that both sfv alleles reduce Mop expression. The sfv3 and sfv11 alleles fail to complement existing mop alleles (data not shown) and will be referred to as mopsfv3 and mopsfv11.
Mop/HD-PTP is a conserved endosomal regulatory protein that contains an N-terminal Bro1 domain and a C-terminal predicted PTPase domain. The PTPase domain lacks activity in vitro due to an amino acid change in the phosphate-binding loop that diverges from all other active PTPases (Gingras et al., 2009), and this is change is conserved in Mop. The Mop Bro1 domain is required to promote the endocytic trafficking and activity of the EGF and Toll receptors (Huang et al., 2010; Miura et al., 2008) and defects in differentiation of Elav-positive neurons within mop eye clones have been attributed to a defect in EGF signaling (Miura et al., 2008). However, some Elav-positive cells remain in mopsfvH99 clones (Fig. S1D-E), and the R2/R5 photoreceptor marker Rough is increased in mopsfvH99 clones relative to mopsfv clones (Fig. S1F-G). Thus, the lack of photoreceptors in mop mutant clones may be due to excess apoptosis and impaired EGF-dependent photoreceptor recruitment.
mop mutations collaborate with a block in cell death to elicit organ overgrowth
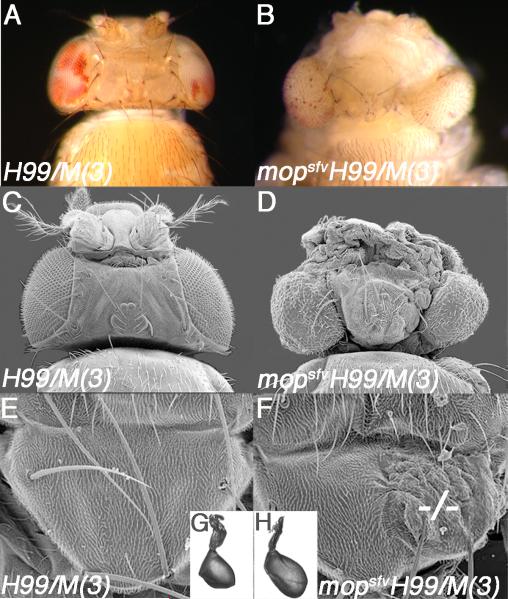
Genetic manipulations that simultaneously increase proliferation and reduce apoptosis often increase organ size. To test effect of the mopsfvH99 genotype on organ size, the recessive cell-lethal Minute technique was used to generate heads composed entirely of mopsfvH99 cells (mopsfvH99/M(3)). These animals die at the pharate adult stage with overgrown heads with extra folds of head cuticle relative to control heads, and eyes that are constricted at their margins, protrude from the head (Fig. 2A-D, 4K) and contain enlarged facets (data not shown). Generation of mopsfvH99 clones throughout the body produced outgrowths in the adult thorax (Fig. 2E-F) and increased haltere size (Fig. 2G-H) relative to H99 controls. Thus combined loss of Mop and the H99 pro-apoptotic genes increases the growth of multiple types of epithelia.
Figure 2. mop Loss Cooperates with a Block in Death to Increase Organ Size.
(A-B) Brightfield images of control H99/M(3) (A) and mopsfvH99/M(3) (B) eyes/heads.
(C-D) Scanning electron micrographs (SEM) of control H99/M(3) (C) and mopsfvH99/M(3) (D) eyes/heads.
(E-F) SEM of H99 (E) and mopsfvH99 (F) clones in adult thoraces. mopsfvH99 mutant tissue is marked by dashes (−/−).
(G-H) Images of mosaic H99 (G) and mopsfvH99 (H) halteres.
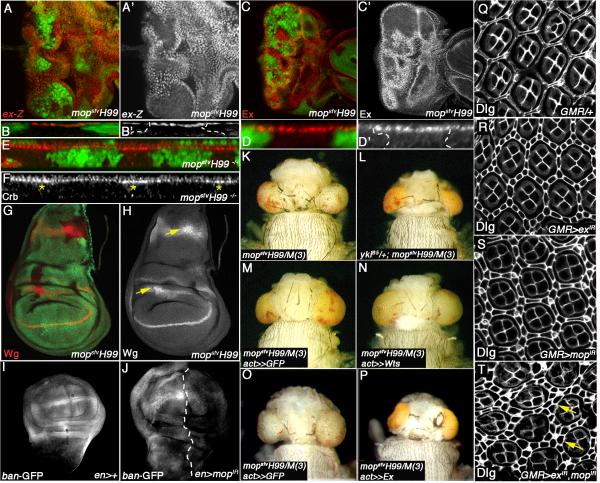
Figure 4. mop Promotes SWH Signaling and is Epistatic to warts.
(A-H) Confocal images of mopsfvH99 clones (marked by the absence of GFP) in the larval eye (A-F) or wing (G-H) analyzed for expression of ex-lacZ (ex-Z) (optical section in A-A’ and transverse section in B-B’), Ex (optical section in C-C’ and transverse section in D-D’), Crb (transverse section in E-F), or Wg (G-H). Stars in (F) denote increased apical Crb in mopsfvH99 clones. Arrows in (H) denote increased Wg protein in mopsfvH99 clones in the proximal wing and the notum.
(I-J) Confocal images of bantam activity (visualized by ban-GFP) in control en>Gal4 (I) and en>Gal4,mopIR (J) discs. Dashed line denotes A:P boundary.
(K-P) Brightfield images of mopsfvH99/M(3) control heads (K,M,O) and mopsfvH99/M(3) heads that are either heterozygous for the ykiB5 allele (L), expressing UAS-Wts (N), or UAS-Expanded (full-length) (P). The ykiB5/+ experiment was carried out with the mopsfv11 allele.
(Q-T) Confocal images of control GMR/+(Q), GMR>exIR (R), GMR>mopIR (S), and GMR>exIR,mopIR (T) 48hr APF pupal eye discs stained with Dlg (white). Yellow arrows in (T) denote multilayered interommatidial cells in the exIR, mopIR double mutant.
mop loss increases cell proliferation rates and inhibits cell-cycle exit
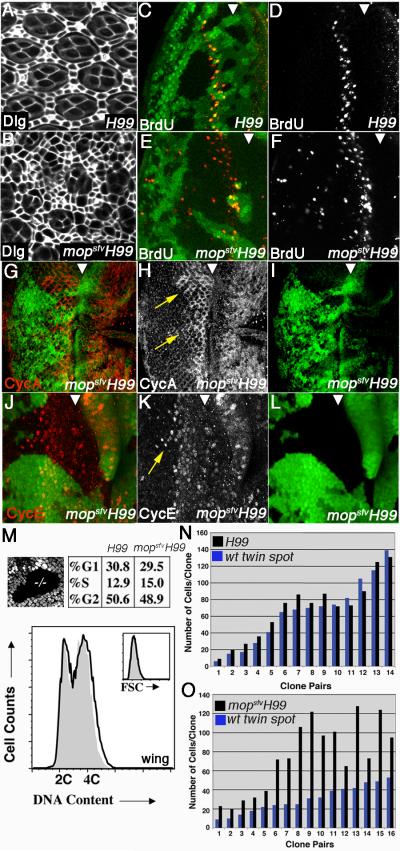
The synergy between mopsfv and H99 alleles indicates that mop loss may increase cell number conditional on a block in apoptosis. Staining with anti-Discs large (Dlg) to mark apical cell profiles reveals excess interommatidial cells (IOCs) in mopsfvH99 pupal eye clones relative to H99 mutant clones (Fig. 3A-B). Patterning defects are also evident within mopsfvH99 mutant pupal clones, which may be a consequence of additional roles for Mop in cell fate pathways (e.g. Miura et al., 2008). Scattered ectopic S-phase cells appear among mopsfvH99 mutant cells posterior to the second mitotic wave (SMW) (Fig. 3C-F), which normally marks the point at which become post-mitotic (Wolff and Ready, 1993); this phenotype is not evident in H99 control clones (Fig. 3D). mopsfvH99 mutant eye clones also show a perdurance of Cyclin A, and to a lesser extent Cyclin E, posterior to the morphogenetic furrow (MF) (Fig. 3G-L). To test whether mop limits the cell division, the rate of clonal expansion of H99 and mopsfvH99 clones were analyzed in the wing disc epithelium at fixed time points after clonal induction. mopsfvH99 clones are consistently larger and contain more cells than wild type ‘twin spots’ or control clones homozygous for the H99 deletion (Fig. 3N-O); the boundaries of mopsfvH99 clones tend to be regular (Fig. 3M, top left panel), suggesting that mop loss may affect cell adhesion. Fluorescence activated cell sorting (FACS) analysis of mopsfvH99 and H99 wing disc cells relative to a control ubi>GFP chromosome indicates that mopsfvH99 cells have a similar DNA-content profile and size as H99 control cells (Fig. 3M). Thus, although mopsfvH99 mutant cells appear to proliferate more rapidly than control cells, this is not accompanied by an overall shift in cell cycle phasing.
Figure 3. mop Limits Cell Division.
(A-B) Confocal images of H99 (A) and mopsfvH99 (B) pupal eye clones (40hr APF) stained with anti-Dlg.
(C-L) Images of H99 (C,D) or mopsfvH99 (E-L) eye clones marked by the absence of GFP (green) and stained for BrdU incorporation (C-F), Cyclin A (G-I), or Cyclin E (J-L). The morphogenetic furrow (MF) is marked by an arrowhead in (C-L). Yellow arrows in (H) denote perdurance of Cyclin A posterior to the MF. Yellow arrow in (K) denotes perdurance of Cyclin E in basal nuclei posterior to the MF.
(M) Confocal image of a mopsfvH99 mutant wing clone (black, −/−) and its wild-type twin spot (white) and corresponding flow cytometric analysis of DNA content and cell size (‘FCS’; see inset) in H99/M(3) (grey fill) and mopsfvH99/M(3) (black line) early third instar wing discs.
(N-O) Cell number in individual H99 (N) or mopsfvH99 (O) clone:twin spot pairs. Data is arranged in order of increasing clone size. Variation in mopsfvH99 cell counts is due to position-specific effects in the wing.
mop promotes SWH signaling and functions downstream of the core SWH kinase cassette
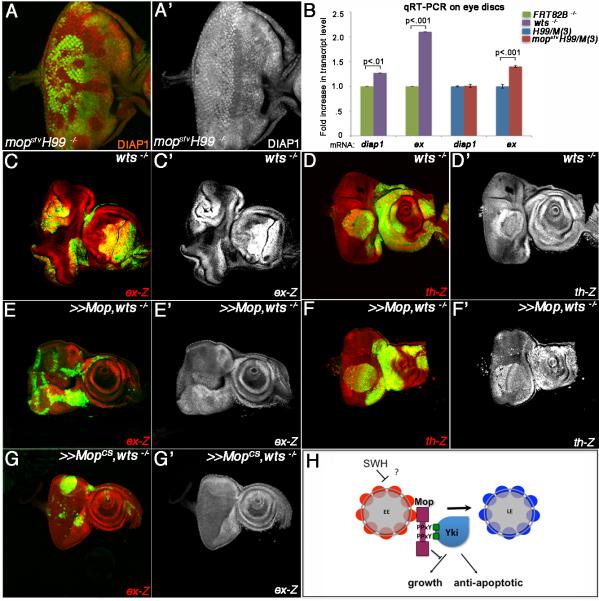
The mopsfvH99 mutant growth phenotypes resemble those associated with Salvador/Warts/Hippo (SWH) pathway mutants.(rev. in Pan, 2010). We therefore tested the effect of mop alleles on expression of SWH target genes and proteins. Expression of the ex-lacZ transcriptional reporter (ex-Z) is upregulated in apically located mopsfvH99 mutant cells posterior to the MF (Fig. 4A-A’); this effect is most apparent in transverse sections of peripodial cells (Fig. 4B-B’) but is also detectable by qRT-PCR analysis of ex mRNA in intact discs (Fig. 7B). Ex protein levels are increased in tangential (Fig. 4C-C’) and transverse (Fig 4. D-D’) sections of mopsfvH99 clones. Control H99 clones have no effect on ex-Z or Ex (Fig. 2SA-A’, B-B’, C-D). mopsfvH99 mutant disc cells accumulate the apical membrane protein Crumbs (Crb) (Fig. 4E-F), which occurs in wts mutant cells via a Yki-dependent mechanism (Genevet et al., 2009; Hamaratoglu et al., 2009). Wg protein accumulates in mopsfvH99 clones located in the hinge and notum regions of the larval wing (Fig. 4G-H), which also occurs in some SWH mutants (Cho et al., 2006; Hamaratoglu et al., 2006; Maitra et al., 2006; Pellock et al., 2007). Expression of a mop inverted repeat (UAS-mopIR) transgene in the posterior wing disc reduces expression of the bantam-GFP sensor (ban-GFP) (Fig. 4I-J), indicating that levels of the ban miRNA (Oh and Irvine, 2011) are elevated in Mop-depleted cells. Parallel analysis of the effect of mopIR on epitope tagged Yki (Yki:V5;(Oh and Irvine, 2009)) shows that knockdown of mop causes Yki:V5 to accumulate in the cell cytoplasm (Fig. S2I-J). Consistent with an inhibitory role for Mop in the SWH pathway, heterozygosity for a yki mutant allele efficiently suppressed the overgrowth of mopsfvH99/M(3) eyes and heads (Fig. 4K-L). Thus, although Mop loss does not stabilize Yki in the nucleus, its effect on the expression of SWH targets and its genetic dependence on a diploid dose of yki, argues that the mopsfvH99 growth phenotype is mediated at least in part by elevated Yki activity.
Figure 7. Mop Exhibits Specificity in the Regulation of Yki Activity.
(A-A’) Confocal image of a mopsfvH99 mosaic eye disc stained with an antibody to DIAP1.
(B) qRT-PCR analysis of diap1 and ex mRNA levels in wts mosaic, H99/M(3) and mopsfvH99/M(3) eye imaginal discs. p-values were calculated using an unpaired t-test and error bars represent standard deviation from the mean of two independent experiments performed in triplicate.
(C-G) Confocal images of wts mosaic eye discs (C-C’, D-D’), or wts discs with MARCM-mediated expression of UAS-Mop (E-E’, F-F’) or UAS-MopCS (G-G’) stained for expression of ex-Z (C-C’, E-E’, G-G’) or th-Z (diap1) (D-D’, F-F’). In all genotypes, wts clones are positively marked by GFP (green).
(H) Model of the Mop:Yki interaction. The PPxY motifs in Mop are shown, as are the Yki WW domains (green squares). EE=early endosome; LE=late endosome.
Genetic analysis was used to place mop into a functional hierarchy with other SWH pathway genes. Expression of Wts suppresses disc growth (Lai et al., 2005; Wei et al., 2007), yet overexpression of Wts in the mopsfvH99/M(3) background has no effect on the size or morphology of pharate adult heads (Fig. 4M-N). By contrast expression of full-length Ex (either UAS-Ex or UAS-Ex:GFP), which can act downstream of Wts via direct binding to Yki (Badouel et al., 2009), suppresses the size of mopsfvH99/M(3) heads (Fig. 4O-P). The C-terminus of Ex (CT:GFP), which contains PPxY motifs that bind Yki, is a more potent suppressor of the mopsfvH99/M(3) growth phenotype than full-length Ex:GFP, while the control linker domain of Ex (linker:GFP) has no effect on mopsfvH99/M(3) heads (Fig. S2K-P). CT:GFP had only a slight effect on the size of control heads, indicating that loss of mop sensitizes cells to elevated levels of Ex. To test synergy between Mop and Ex loss, these factors were knocked down individually or in combination in the developing eye using UAS-exIR and UAS-mopIR transgenes. While exIR or mopIR individually produce a mild increase in IOCs in the pupal eye, combined expression of both enhances the IOC phenotype (Fig. 4Q-T). This genetic interaction between Mop and Ex in control of IOC numbers mirrors other peripheral regulators of the SWH pathway, which show mild IOC phenotypes when lost individually, but synergistic effects when combined (e.g. Baumgartner et al., 2010; Genevet et al., 2010; Ling et al., 2010; Yu et al., 2010).
Mop interacts with Yki via residues in the Yki WW-domains
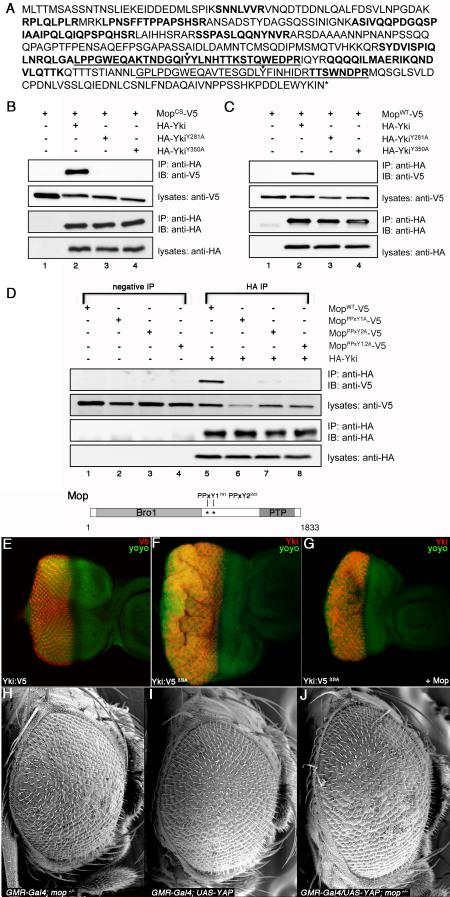
To identify the protein target of Mop within the SWH pathway, we undertook an affinity purification/mass spectrometry (AP/MS) analysis of Mop-containing complexes (Kyriakakis et al., 2008; Veraksa et al., 2005) purified from cultured S2 cells. A form of Mop with a point mutation in the putative PTPase catalytic domain (Cys1728 to Ala; MopCS) was used for this analysis in order to enhance interactions with endogenous proteins. This technique identified 14 partially overlapping peptides derived from the endogenous Yki protein (bolded in Fig. 5A). Co-immunoprecipitation (co-IP) analysis from S2 cells expressing HA-Yki and Mop-V5 confirmed the interaction in the reciprocal orientation: HA-Yki is able to efficiently co-IP both MopCS and wild-type Mop (MopWT) (Fig. 5B-C). To study the Mop:Yki interaction further, two conserved tyrosines (Y281 and Y350) within the Yki WW domains (see Fig. 5A) were individually altered to Alanine (A) (HA-YkiY281A and HA-YkiY350A). Mutation of an equivalent Y residue within the WW-domain of Polyglutamine tract-binding protein-1 (PQBP1) disrupts binding to PPxY-containing peptides (Tapia et al., 2010). Although expressed at similar levels to HA-Yki, both of the Y-to-A WW mutants are defective in binding to MopWT and MopCS (Fig 5B-C). Mop contains two candidate PPxY motifs located in its linker region between the Bro1 domain and the PTPase-like domain (Fig. 5D), indicating that Mop may bind Yki via a WW:PPxY interaction module similar to the interaction between the Yki WW domains and PPxY motifs in Ex, Hpo and Wts (Badouel et al., 2009; Oh et al., 2009). To test this, each of the Mop PPxY motifs were changed to PPxA (PPxY1A, PPxY2A, or PPxY1,2A) and tested for Yki binding. The Mop-PPxY1,2A mutant showed a loss of Yki binding, while each single mutant showed minor residual binding (Fig. 5D and data not shown). Thus, Mop can bind to Yki via a WW:PPxY interaction mechanism that is conserved in other SWH proteins that bind directly to Yki.
Figure 5. Mop Physically Interacts with Yki via a WW:PPxY mechanism.
(A) The Yki-PF isoform. Fourteen distinct, partially overlapping peptides (indicated in bold) were recovered from affinity purification/mass spectrometry analysis of MopCS-containing complexes. The WW domains of Yki are underlined and asterisks denote the conserved tyrosine residues required for the Mop:Yki interaction (see below).
(B) Physical association between MopCS and Yki. Lanes 1-4: S2 cell lysates expressing the indicated combination of MopCS-V5 and HA-Yki constructs were immunoprecipitated and probed with the indicated antibodies.
(C) Physical association between MopWT and Yki. Lanes 1-4: S2 cell lysates expressing the indicated combination of MopWT-V5 and HA-Yki constructs were immunoprecipitated and probed with the indicated antibodies.
(D) Physical association between MopPPxY mutants and wild-type Yki. Lanes 1-4: negative control IPs of S2 cell lysates expressing the indicated Mop-V5 constructs. Lanes 5-8: S2 cell lysates expressing the indicated combination of wild-type Yki and Mop-V5 constructs were precipitated and probed with the indicated antibodies. A diagram of the Mop protein shows the location of the two PPxY1791 and PPxY2823 sites.
(E-G) Phosphorylation-independent repression of Yki-driven overgrowth by Mop. Confocal sections of eye imaginal discs expressing GMR-Gal4, UAS-Yki:V5 (A), GMR-Gal4, UAS-Yki:V53SA(B), GMR-Gal4, UAS-Yki:V53SA + UAS-Mop (C) and stained for V5 (red in E) or Yki (red in F, G) and with nuclei labeled by YOYO-1 iodide (green).
(H-J) YAP-driven overgrowth is sensitive to the genetic dose of mop. Scanning electron micrographs (SEM) of GMR-Gal4, mop−/+ (H), GMR-Gal4/UAS-YAP (I) and GMR-Gal4/UAS-YAP; mop +/− (J) adult eyes.
Mop inhibition of Yki is phosphorylation-independent
A version of Yki carrying Ser-to-Ala mutations in three Wts phosphorylation sites (S111A, S168A, and S250A; UAS-yki:V5S3A) shows enhanced oncogenic activity due to a loss of inhibitory phosphorylation by Wts (Oh and Irvine, 2009). However, expression of Hpo, Wts or Ex, which directly bind the Yki WW domains via their PPxY motifs, are able to suppress YkiS3A phenotypes (Oh et al., 2009). To test whether Mop can also regulate Yki in a phosphorylation-independent manner, UAS-yki:V5 or UAS-yki:V5S3A were co-expressed with Mop (UAS-mop) in the larval eye. As reported, Yki:V5 shows little effect on growth of the larval eye disc while Yki:V5S3A produces enlarged disc with folds of excess tissue (Fig. 5E-F). Expression of Mop resulted in substantial suppression of the Yki:V5S3A growth phenotype (Fig. 5G). This ability of Mop to antagonize a form of Yki that is refractory to SWH-mediated phosphorylation is consistent with a model in which Mop inhibits Yki directly via the WW:PPxY interaction. Significantly, heterozygosity for mop enhances the pro-growth effect of the human Yki homolog Yap in the developing eye (Fig. 5H-J), suggesting that the functional relationship between Yki and Mop-like proteins may be conserved.
mop regulates association of Yki with endosomal compartments
Studies with HD-PTP indicate that the Mop N-terminal Bro1 domain may facilitate interactions with ESCRT endosomal complexes (Doyotte et al., 2008; Ichioka et al., 2007; Odorizzi et al., 2003) and that this underlies the ability of Mop to promote signaling by the EGF and Toll receptors (Huang et al., 2010; Miura et al., 2008). The finding that Mop sequences outside the Bro1 domain bind Yki suggests that Mop may regulate the association of Yki with specific endosome-associated protein complexes, or that Mop physically links Yki to the endosomal trafficking of a receptor-like SWH component such as Fat (rev. in Reddy and Irvine, 2008). To test whether the mopsfvH99 growth phenotype requires signaling downstream of Fat, we expressed an RNAi knockdown transgene to approximated (app), which is required for the overgrowth of mutant fat tissue (Matakatsu and Blair, 2008), in the mopsfvH99/M(3) background. Knockdown of app had no effect on the size of mopsfvH99/M(3) heads, and reciprocally loss of Mop had no discernable effect on Fat levels or the degree of Fat co-localization with the early endosomal marker EEA1 (Fig. S2E-H), indicating that mopsfvH99 may promote growth independent of a defect in Fat signaling.
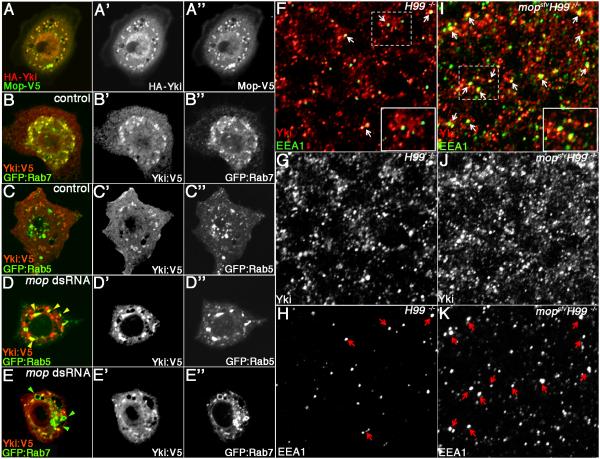
The effect of mopIR on Yki protein (see Fig. S2I-J) led us to examine the relationship between Mop and the cytoplasmic pool of Yki more closely. In imaginal discs and S2 cells, Mop is normally found in cytoplasmic puncta adjacent to Rab5-positive endosomes and partially co-localizes with Rab7-positive endosomes (Huang et al., 2010; Miura et al., 2008). To test whether the physical interaction of Mop and Yki proteins is accompanied by a co-localization of the proteins to specific cytoplasmic structures, HA-Yki and Mop-V5 were visualized in cultured S2 cells. HA-Yki and Mop-V5 substantially co-localize to discrete puncta in the cytoplasm (Fig. 6A-A”). To identity these Yki-containing structures, Yki:V5 was co-expressed with either GFP-Rab7 or GFP-Rab5 to mark late and early endosomes respectively. A majority of Yki:V5 protein localizes to Rab7-containing late endosomes in S2 cells (Fig. 6B-B”). This parallels the reported partial location of Mop to Rab7-endosomes (Miura et al., 2008) and indicates that Mop and Yki partially colocalize at these structures. By contrast, Yki:V5 is largely excluded from Rab5-containing early endosomes (Fig. 6C-C”). To test whether depletion of mop affects these patterns, Yki:V5 was expressed with either GFP-Rab5 or GFP-Rab7 in cells treated with mop dsRNA. As reported (Miura et al., 2008), depletion of mop by RNA interference caused an enlargement of GFP-Rab7 endosomes (Fig, 6E-E”). These enlarged mop-knockdown GFP-Rab7 endosomes are depleted for Yki:V5 relative to untreated S2 cells (green arrowheads, Fig. 6E-E”), and in parallel, Yki:V5 appears in GFP-Rab5 endosomes (yellow arrowheads, Fig. 6D-D”). This endosomal redistribution of exogenous Yki correlates with an increase in overall levels of the protein and increased expression of the Yki target ex as measured by qRT-PCR (Fig. S3A,C). A similar rise in endogenous Yki levels is observed upon mop knockdown in S2 cells, and this correlates with elevated expression of ex (Fig. S3B-C). Moreover, when expressed together with HA-Yki, Mop can retard Yki-driven transcriptional upregulation of ex (Fig. S3C).
Figure 6. Mop Controls the Endosomal Localization of Yki.
(A-C) Confocal images of S2 cells expressing HA-Yki (A-A” (red)) together with Mop-V5 (A-A”(green)), or Yki:V5 (B-B”, C-C”) together with GFP:Rab7 (B-B”) or GFP:Rab5 (C-C”).
(D-E) Confocal images of mop dsRNA-treated S2 cells expressing Yki:V5 and either GFP:Rab5 (D-D”) or GFP:Rab7 (E-E’). In mop dsRNA-treated S2 cells, a portion of Yki:V5 is re-localized to large Rab5-positive endosomes (yellow arrowheads in (D)) and is largely excluded from enlarged Rab7-positive endosomes (green arrowheads in (E)).
(F-K) Confocal sections of H99 (F-H) and mopsfvH99 (I-K) mutant eye imaginal disc tissue, immunostained for endogenous Yki (red) and early endosome antigen-1 (EEA1 (green)). Yki/EEA1 double-positive endosomes are marked by white arrows in (F) and (I). Red arrows denote slightly enlarged EEA1 endosomes in mop,H99 cells (H vs. K) that also colocalize with Yki. Dashed boxes correspond to insets. Images were collected at 100× magnification.
We next examined whether mop loss in imaginal disc tissue affects endosomal distribution of endogenous Yki. In control H99 clones, Yki localizes to punctate cytoplasmic structures that occasionally co-localize with early endosomal antigen-1 (EEA1)-positive early endosomes (Fig. 6F-H). In mopsfvH99 mutant tissue, EEA1-positive endosomes are more abundant, and tend to be larger than in control cells (red arrows in Fig. 6H,K). Yki also accumulates in punctate structures in mopsfvH99 that substantially overlap with enlarged EEA1-positive endosomes (Fig. 6I-K). The extent of Yki:EEA1 co-localization was quantitated by counting the percentage of EEA1-positive endosomes of all sizes that are also positive for Yki. In H99 control clones the percentage of EEA1/Yki-positive endosomes is 19±2% (complied from 10 clones containing a total of 380 EEA1-positive endosomes). In mopsfvH99 tissue this approximately doubles to 38±3% (compiled from 10 clones containing a total of 584 EEA1-positive endosomes). To determine whether this enrichment of Yki in EEA1 endosomes is accompanied by changes in localization of Yki on late endosomes, the association of Yki with Rab7-positive late endosomes was examined in H99 and mopsfvH99 clones. In H99 control clones, Yki localizes to structures that are adjacent to, but do not substantially overlap with Rab7 late endosomes (Fig. S4E-E”). mopsfvH99 clones contain enlarged Rab7-positive endosomes that exclude the increased pool of cytoplasmic endogenous Yki (Fig. S4D-D”). Thus, mop loss in imaginal disc tissue leads to an increase in the early-endosome associated pool of cytoplasmic Yki.
Mop restricts ectopic ex but not diap1 transcription in imaginal discs
Yki lacks DNA binding activity but has been shown to activate transcription of the diap1 promoter via the DNA binding factor and TEAD homolog Scalloped (Sd) (Goulev et al., 2008; Wu et al., 2008; Zhang et al., 2008). Interestingly, qRT-PCR analysis shows that mopsfvH99 mutant discs elevate expression of ex mRNA transcripts but not diap1 transcripts, while wts mutant cells elevate levels of both ex and diap1 transcripts (Fig. 7B). In addition, DIAP1 protein is not elevated in mopsfvH99 mutant clones (Fig. 7A-A’) but is readily detected at elevated levels in wts mutant eye disc clones (Fig. S4A-A’). diap1-lacZ (th-Z) reporter expression is also unaffected in mopsfvH99 mutant eye clones marked by up-regulated Crb (Fig. S4B-B’). The lack of an effect on diap1 indicates that Mop may be required to regulate the expression of some SWH targets and not others. To test this, MARCM analysis was used to analyze SWH target-gene induction in wts mutant clones over-expressing Mop. As previously reported (Hamaratoglu et al., 2006), ex-Z and th-Z are strongly upregulated in wts mutant clones (Fig. 7C-C’,D-D’). Over-expression of wild-type Mop rescues the effect of wts loss on ex-Z (Fig. 7E-E’) but does not block elevated th-Z expression (Fig. 7F-6F’). The catalytic-site mutant form of MopCS (Miura et al., 2008) was also able to efficiently suppress ectopic ex-Z levels in wts clones (Fig. 7G-G’). These data suggest that Mop acts to restrict Yki-driven upregulation of ex transcription but not diap1 transcription, and that it does so through a mechanism that does not require Mop catalytic tyrosine phosphatase activity.
Discussion
Here we describe a screening strategy to identify mutations in Drosophila that require a synergistic block in cell death in order to promote tissue overgrowth. Using this approach we have identified the endosomal protein Myopic, which is the Drosophila homolog of the candidate mammalian tumor suppressor HD-PTP, as a regulator of the SWH growth inhibitory pathway. Through multiple approaches, we demonstrate that Mop regulates Yki activity via a mechanism involving direct binding and modulation of Yki endosomal association.
This study defines a pool of cytoplasmic Yki that binds Mop and colocalizes with it on endosomes. Data from discs and cultured cells indicate Mop controls endosomal association of this pool of Yki and that a positive correlation exists between Yki colocalization with EEA1-positive early endosomes, and Yki levels and activity. A growing body of genetic and molecular data support a role for endosomes as key signaling centers for signal transduction pathways that influence the nuclear translocation of latent cytoplasmic transcription factors (Birtwistle and Kholodenko, 2009; Bokel et al., 2006; Devergne et al., 2007; Di Guglielmo et al., 2003; Fortini and Bilder, 2009; rev. in Miaczynska et al., 2004; Murphy et al., 2009; e.g. Taelman et al., 2010). For example, the activated c-Met receptor associates with the STAT3 transcription factor on EEA1-positive endosomes prior to STAT3 nuclear accumulation, and c-Met delivery to a perinuclear endosomal compartment is necessary to sustain nuclear STAT3 (Kermorgant and Parker, 2008). The enrichment of Yki on EEA1 endosomes and activation of a subset of Yki nuclear targets in mop mutant cells suggests that Yki, perhaps in association with receptor complexes, may take a similar route to the nucleus. Intriguingly, microtubule-regulated perinuclear transport of Merlin (Mer) controls nucleocytoplasmic shuttling of Yki (Bensenor et al., 2010). The direct link between Mer transport and Yki shuttling is not clear. However, as Mer can control internalization of transmembrane receptors (rev. in McClatchey and Fehon, 2009), perinuclear transport of Mer might in turn modulate endosomal internalization and transit of Yki:receptor complexes en route to the nucleus.
Genetic data show that exogenous Mop is sufficient to restrict ectopic expression of the Yki-target ex but not diap1 and that loss of endogenous Mop upregulates a set of Yki targets that do not include diap1. Mop thus appears to define a regulatory step in determining outputs of the SWH pathway, perhaps as part of the endosomal sorting process. Trafficking of transmembrane proteins down alternate endosomal routes contributes to the activation of different nuclear programs in the Notch, Jak/STAT and Akt pathways (Hori et al., 2004; Kermorgant and Parker, 2008; Schenck et al., 2008). Similarly, association of Yki-containing complexes with different endosomal compartments may shift Yki nuclear output, perhaps by bringing Yki into contact with post-translational modifiers or binding partners that affect its ability to activate its suite of target promoters. Further studies will be required to establish whether loss of Mop indeed alters Yki post-translational modification or the assembly of Yki transcriptional complexes.
In the context of SWH signaling, the differential effect of mop loss on ex and diap1 expression place Mop within the growth-regulatory arm of the SWH network. Differential effects on the growth and apoptotic outputs of the SWH pathway is also a feature of mutations in ex and mer, which preferentially drive Yki-dependent clonal growth or anti-apoptotic signals respectively (Pellock et al., 2007) and whose combined mutant phenotypes are more severe than those of single mutants (Hamaratoglu et al., 2006; Maitra et al., 2006). The synergy between ex and mop alleles on IOC number extends this model and supports the hypothesis that Ex is downstream of wts in growth control but upstream of wts in apoptotic control (Badouel et al., 2009; Hamaratoglu et al., 2006).
mop mutant cells undergo high rates of caspase-dependent apoptosis in developing eye and wing imaginal discs (this study and Miura et al., 2008). It is probable that this apoptosis is not caused by an effect on diap1 expression but rather a requirement for Mop in additional pro-survival mechanisms. Knockdown of vertebrate HD-PTP/PTPN23 elevates levels of tyrosine phosphorylated focal adhesion kinase (FAK) (Castiglioni et al., 2007), which is implicated in cell migration and integrin-mediated survival signals (Mitra and Schlaepfer, 2006). Mop facilitates trafficking of the EGFR receptor into late endosomal compartments and promotes Ras/MAPK signaling downstream of EGFR in the developing retina (Miura et al., 2008). Because the Ras/MAPK module is required to restrain cell death pathways (e.g. Bergmann et al., 1998; Kurada and White, 1998), reduced EGFR-dependent signaling seems likely to contribute to a subset of the apoptotic phenotype of mop mutant cells.
The Mop:Yki interaction involves a WW:PPxY interaction mechanism shared by the SWH proteins Ex, Wts, and Hpo that can bind Yki directly and regulate its activity independent of S168 phosphorylation status (Badouel et al., 2009; Oh et al., 2009). Mop represses growth driven by the YkiS3A mutant, indicating that its repressive mechanism is not dependent on Wts kinase activity. As Mop controls the distribution of Yki across endosomal compartments, the paired Bro1 and PPxY domains in Mop could function as a bridge between Yki-containing SWH signaling complexes in the cytoplasm and complexes on the outer membrane of endosomes such as ESCRTs. These complexes could be fairly static or they could assemble and disassemble in response to specific signals. The fact that Mop, Ex, Hpo and Wts share a WW:PPxY binding mechanism suggests these proteins might compete for Yki binding in the cytosol, or that Mop acts as an endocytic scaffolding factor in a ‘hand-off’ mechanism from the upstream components Ex, Hpo, and Wts. Indeed understanding the dynamics and composition of the Mop:Yki complex is a significant question going forward. Intriguingly loss of the Lgl kinase, which regulates cell polarity and membrane compartmentalization (Bilder, 2004), elevates Yki activity by mislocalizing Hpo and the SWH component RASSF in the cytoplasm of disc cells (Grzeschik et al., 2010), suggesting that Hpo and RASSF proteins participate in dynamic and localized interactions in the cytoplasm that are important for their Yki-regulatory function.
The human HD-PTP/PTPN23 gene resides in a region of the genome (3p21.3) associated with loss-of-heterozygosity (LOH) in greater than 90% of small cell (SCLC) and non-small cell (NSCLC) lung cancers (Braga et al., 2002; Kok et al., 1997; Szeles et al., 1997; Toyooka et al., 2000). Yap protein is predominantly nuclear in a subset of primary NSCLC samples, promotes cell proliferation and invasion in NSCLC cell lines, and its expression correlates with poor prognosis in NSCLC patients (Wang et al., 2010). Thus mutations that deregulate Yap levels and activity are predicted to promote the inappropriate growth and invasiveness of lung epithelial cells. The mechanism of growth suppression by HD-PTP is not known, but its ability to suppress colony formation of human renal cancer cells is independent of catalytic PTPase activity (Gingras et al., 2009) in much the same way regulation of Yki by Mop does not require PTPase activity. Although HD-PTP lacks a canonical PPxY motif, genetic data indicate that Mop retains the ability to inhibit Yap activity in the Drosophila eye. The extent to which HD-PTP binds Yap or Taz has yet to be examined, but if the relationship between the orthologous Drosophila proteins is conserved in vertebrates, this link to Yki/Yap may contribute to growth regulatory roles of vertebrate HD-PTP proteins in development and disease.
Experimental Procedures
Drosophila Genetics
Alleles used: ex697 (ex-Z) (Boedigheimer et al., 1993), thj5c8 (th-Z) (Hay et al., 1995), ykiB5 (Huang et al., 2005), wtsX1 (Xu et al., 1995) and H99 (White et al., 1994). Transgenes used: UAS-Myc-Wts1 (Jia et al., 2003), UAS-Ex (Boedigheimer et al., 1997), UAS-Mop and UAS-MopCS (Miura et al., 2008), UAS-Yki:V5 (Oh and Irvine, 2009), UAS-mopIR (Vienna Drosophila RNAi Collection), bantam-GFP (Brennecke et al., 2003).
Immunofluoresence
Imaginal discs and S2 cells were fixed and stained following standard procedures. Antibodies used: mouse anti-Dlg (1:20, DSHB), mouse anti-BrdU (1:50, Becton Dickinson), mouse anti-Cyclin E (1:5, H. Richardson), mouse anti-Cyclin A (1:200, DSHB), guinea pig anti-Expanded (1:10,000, R. Fehon), rat anti-Crb (1:500, U. Tepass), mouse anti-Wg (1:800, DSHB), mouse anti-β-galactosidase (1:1000, Promega), mouse anti-V5 (1:200, Invitrogen) and mouse anti-HA High Affinity (1:100, Roche), mouse anti-Yki (1:100, Zhi-Chun Lai), rabbit anti-EEA1 (1:250, Thermo-Scientific), mouse anti-DIAP1 (1:400, B. Hay)
Clonal Cell Counts and Cell Cycle Analysis
For FACS analysis, H99/M(3) and mopsfvH99/M(3) wing discs were dissociated in PBS Trypsin-EDTA and stained with 20 μM DRAQ-5 (Biostatus Limited). Data were acquired on a Becton Dickinson LSR II flow cytometer and analyzed with FloJo Software. Clonal cell count data was generated by producing heat-shock H99 and mopsfvH99 wing clones 48hr AED. At 96 AED wing discs were fixed and stained with DRAQ5 and the number of nuclei per clone and twin-spot pair was counted.
Quantitative RT-PCR
Total RNA was extracted from larval eye discs using TRizol (Invitrogen) and purified using RNeasy Mini kit (Qiagen). Superscript II RT and random primers (Invitrogen) were used to produce cDNAs. Exon-specific primers were used with SYBR Green I Master (Roche) to perform qPCR reactions using a Roche LightCycler 480. All reactions were performed in triplicate and the relative amount of diap1 and ex mRNA was normalized to β-tubulin transcript.
Cell culture
Drosophila S2 cells were maintained at 25°C in Schneider’s Drosophila medium (Gibco) supplemented with 10% heat inactivated fetal bovine serum (Gibco). Proteins were induced with 0.35 mM CuSO4 overnight. To establish a stable cell line of pMK33-GSNTAP-MopCS, S2 cells were transfected using Effectene transfection reagent (Qiagen). After 48 hours of incubation with the transfection reagent, cells were maintained in complete media with 300 μg/ml hygromycin (Sigma). mop dsRNA was generated using the T7 RiboMAX system (Promega) using the following primers: 5′-T7-tgcccacattaccgagttatcg-3′ and 5′-T7-tttccgctattggtttgctgac-3′. mop dsRNA was transfected into cells using Cellfectin (Invitrogen) and cells were incubated for 48 hours, followed by transfection with the following constructs: 1) pAc5.1-Yki:V5, 2) UAS-GFP:Rab5, 3) UAS-GFP:Rab7, 4) pMT-Gal4, followed by another 48 hour incubation and induction with .5 mM CuSO4 for 4 hours.
Immunoprecipitation
Cell extracts were lysed in lysis buffer (LB: 50 mM Tris pH 7.5, 125 mM NaCl, 5% glycerol, 0.2% IGEPAL, 1.5 mM MgCl2, 1 mM DTT, 25 mM NaF, 1 mM Na3VO4, 1 mM EDTA and Complete protease inhibitor (Roche) and lysates were incubated with anti-HA affinity beads (Sigma) for 2 h at 4°C, followed by extensive washes. Protein complexes were eluted with SDS sample buffer, separated on SDS protein gels, transferred onto Immun-Blot PVDF membranes (Bio-Rad) and probed with mouse anti-HA, or mouse anti-V5 antibodies (Sigma).
Supplementary Material
Acknowledgements
We apologize to those who could not be cited due to space constraints. We thank D.J. Pan, G. Halder, K. Irvine, K. Harvey, H. McNeill, Z-C. Lai, J. Jiang, Y.Chinchore, R. Fehon, J. Treisman, T. Xu, B. Hay, and M. Frolov for gifts of reagents and fly stocks. We acknowledge the Vienna Drosophila RNAi Collection, the Harvard TRiP collection, the Bloomington Drosophila Stock Center, and the Developmental Studies Hybridoma Bank (DSHB) for providing fly stocks and antibodies. We thank members of the Moberg and Veraksa laboratories for helpful comments and discussion, the Taplin Mass Spectrometry Facility (Harvard Univ.) for peptide analysis, and the Robert P. Apkarian Integrated Electron Microscopy Core for SEM images. This work was supported by NIH CA1123368 to KHM and NSF 0640700 to AV.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asano M, Nevins JR, Wharton RP. Ectopic E2F expression induces S phase and apoptosis in Drosophila imaginal discs. Genes Dev. 1996;10:1422–1432. doi: 10.1101/gad.10.11.1422. [DOI] [PubMed] [Google Scholar]
- Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le Bihan T, McNeill H. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Bensenor LB, Barlan K, Rice SE, Fehon RG, Gelfand VI. Microtubule-mediated transport of the tumor-suppressor protein Merlin and its mutants. Proc Natl Acad Sci U S A. 2010;107:7311–7316. doi: 10.1073/pnas.0907389107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann A, Agapite J, McCall K, Steller H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95:331–341. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- Bilder D. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18:1909–1925. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- Birtwistle MR, Kholodenko BN. Endocytosis and signalling: a meeting with mathematics. Mol Oncol. 2009;3:308–320. doi: 10.1016/j.molonc.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedigheimer M, Bryant P, Laughon A. Expanded, a negative regulator of cell proliferation in Drosophila, shows homology to the NF2 tumor suppressor. Mech Dev. 1993;44:83–84. doi: 10.1016/0925-4773(93)90058-6. [DOI] [PubMed] [Google Scholar]
- Boedigheimer MJ, Nguyen KP, Bryant PJ. Expanded functions in the apical cell domain to regulate the growth rate of imaginal discs. Dev Genet. 1997;20:103–110. doi: 10.1002/(SICI)1520-6408(1997)20:2<103::AID-DVG3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Bokel C, Schwabedissen A, Entchev E, Renaud O, Gonzalez-Gaitan M. Sara endosomes and the maintenance of Dpp signaling levels across mitosis. Science. 2006;314:1135–1139. doi: 10.1126/science.1132524. [DOI] [PubMed] [Google Scholar]
- Braga E, Senchenko V, Bazov I, Loginov W, Liu J, Ermilova V, Kazubskaya T, Garkavtseva R, Mazurenko N, Kisseljov F, et al. Critical tumor-suppressor gene regions on chromosome 3P in major human epithelial malignancies: allelotyping and quantitative real-time PCR. Int J Cancer. 2002;100:534–541. doi: 10.1002/ijc.10511. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam Encodes a Developmentally Regulated microRNA that Controls Cell Proliferation and Regulates the Proapoptotic Gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Brumby AM, Richardson HE. Using Drosophila melanogaster to map human cancer pathways. Nat Rev Cancer. 2005;5:626–639. doi: 10.1038/nrc1671. [DOI] [PubMed] [Google Scholar]
- Castiglioni S, Maier JA, Mariotti M. The tyrosine phosphatase HD-PTP: A novel player in endothelial migration. Biochem Biophys Res Commun. 2007;364:534–539. doi: 10.1016/j.bbrc.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci U S A. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi C, Zhu H, Han M, Zhuang Y, Wu X, Xu T. Disruption of lysosome function promotes tumor growth and metastasis in Drosophila. J Biol Chem. 2010;285:21817–21823. doi: 10.1074/jbc.M110.131714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Devergne O, Ghiglione C, Noselli S. The endocytic control of JAK/STAT signalling in Drosophila. J Cell Sci. 2007;120:3457–3464. doi: 10.1242/jcs.005926. [DOI] [PubMed] [Google Scholar]
- Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat Cell Biol. 2003;5:410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyotte A, Mironov A, McKenzie E, Woodman P. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc Natl Acad Sci U S A. 2008;105:6308–6313. doi: 10.1073/pnas.0707601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Bilder D. Endocytic regulation of Notch signaling. Curr Opin Genet Dev. 2009;19:323–328. doi: 10.1016/j.gde.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevet A, Polesello C, Blight K, Robertson F, Collinson LM, Pichaud F, Tapon N. The Hippo pathway regulates apical-domain size independently of its growth-control function. J Cell Sci. 2009;122:2360–2370. doi: 10.1242/jcs.041806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras MC, Zhang YL, Kharitidi D, Barr AJ, Knapp S, Tremblay ML, Pause A. HD-PTP is a catalytically inactive tyrosine phosphatase due to a conserved divergence in its phosphatase domain. PLoS One. 2009;4:e5105. doi: 10.1371/journal.pone.0005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F, Gajewski K, Sansores-Garcia L, Morrison C, Tao C, Halder G. The Hippo tumor-suppressor pathway regulates apical-domain size in parallel to tissue growth. J Cell Sci. 2009;122:2351–2359. doi: 10.1242/jcs.046482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Hori K, Fostier M, Ito M, Fuwa TJ, Go MJ, Okano H, Baron M, Matsuno K. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development. 2004;131:5527–5537. doi: 10.1242/dev.01448. [DOI] [PubMed] [Google Scholar]
- Huang HR, Chen ZJ, Kunes S, Chang GD, Maniatis T. Endocytic pathway is required for Drosophila Toll innate immune signaling. Proc Natl Acad Sci U S A. 2010;107:8322–8327. doi: 10.1073/pnas.1004031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Ichioka F, Takaya E, Suzuki H, Kajigaya S, Buchman VL, Shibata H, Maki M. HD-PTP and Alix share some membrane-traffic related proteins that interact with their Bro1 domains or proline-rich regions. Arch Biochem Biophys. 2007;457:142–149. doi: 10.1016/j.abb.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, Halder G. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- Kermorgant S, Parker PJ. Receptor trafficking controls weak signal delivery: a strategy used by c-Met for STAT3 nuclear accumulation. J Cell Biol. 2008;182:855–863. doi: 10.1083/jcb.200806076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok K, Naylor SL, Buys CH. Deletions of the short arm of chromosome 3 in solid tumors and the search for suppressor genes. Adv Cancer Res. 1997;71:27–92. doi: 10.1016/s0065-230x(08)60096-2. [DOI] [PubMed] [Google Scholar]
- Kurada P, White K. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell. 1998;95:319–329. doi: 10.1016/s0092-8674(00)81764-x. [DOI] [PubMed] [Google Scholar]
- Kyriakakis P, Tipping M, Abed L, Veraksa A. Tandem affinity purification in Drosophila: the advantages of the GS-TAP system. Fly (Austin) 2008;2:229–235. doi: 10.4161/fly.6669. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Levy D, Adamovich Y, Reuven N, Shaul Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell. 2008;29:350–361. doi: 10.1016/j.molcel.2007.12.022. [DOI] [PubMed] [Google Scholar]
- Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, Pan D. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci U S A. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra S, Kulikauskas RM, Gavilan H, Fehon RG. The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr Biol. 2006;16:702–709. doi: 10.1016/j.cub.2006.02.063. [DOI] [PubMed] [Google Scholar]
- Matakatsu H, Blair SS. The DHHC palmitoyltransferase approximated regulates Fat signaling and Dachs localization and activity. Curr Biol. 2008;18:1390–1395. doi: 10.1016/j.cub.2008.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchey AI, Fehon RG. Merlin and the ERM proteins--regulators of receptor distribution and signaling at the cell cortex. Trends Cell Biol. 2009;19:198–206. doi: 10.1016/j.tcb.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaczynska M, Pelkmans L, Zerial M. Not just a sink: endosomes in control of signal transduction. Curr Opin Cell Biol. 2004;16:400–406. doi: 10.1016/j.ceb.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Mitra SK, Schlaepfer DD. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol. 2006;18:516–523. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Miura GI, Roignant JY, Wassef M, Treisman JE. Myopic acts in the endocytic pathway to enhance signaling by the Drosophila EGF receptor. Development. 2008;135:1913–1922. doi: 10.1242/dev.017202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JE, Padilla BE, Hasdemir B, Cottrell GS, Bunnett NW. Endosomes: a legitimate platform for the signaling train. Proc Natl Acad Sci U S A. 2009;106:17615–17622. doi: 10.1073/pnas.0906541106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Nicholson SC, Gilbert MM, Nicolay BN, Frolov MV, Moberg KH. The archipelago Tumor Suppressor Gene Limits Rb/E2F-Regulated Apoptosis in Developing Drosophila Tissues. Curr Biol. 2009 doi: 10.1016/j.cub.2009.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G, Katzmann DJ, Babst M, Audhya A, Emr SD. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J Cell Sci. 2003;116:1893–1903. doi: 10.1242/jcs.00395. [DOI] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–1927. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. Cooperative regulation of growth by Yorkie and Mad through bantam. Dev Cell. 2011;20:109–122. doi: 10.1016/j.devcel.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Reddy BV, Irvine KD. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev Biol. 2009;335:188–197. doi: 10.1016/j.ydbio.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini RA, Xu T. A genetic screen in Drosophila for metastatic behavior. Science. 2003;302:1227–1231. doi: 10.1126/science.1088474. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Pellock BJ, Buff E, White K, Hariharan IK. The Drosophila tumor suppressors Expanded and Merlin differentially regulate cell cycle exit, apoptosis, and Wingless signaling. Dev Biol. 2007;304:102–115. doi: 10.1016/j.ydbio.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HW, Slattery M, Mann RS. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23:2307–2319. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BV, Irvine KD. The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development. 2008;135:2827–2838. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- Ren F, Zhang L, Jiang J. Hippo signaling regulates Yorkie nuclear localization and activity through 14-3-3 dependent and independent mechanisms. Dev Biol. 2010;337:303–312. doi: 10.1016/j.ydbio.2009.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A, Goto-Silva L, Collinet C, Rhinn M, Giner A, Habermann B, Brand M, Zerial M. The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell. 2008;133:486–497. doi: 10.1016/j.cell.2008.02.044. [DOI] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K, Ciampa PJ, Brook A, Dyson N. A genetic screen for modifiers of E2F in Drosophila melanogaster. Genetics. 1999;153:275–287. doi: 10.1093/genetics/153.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano S, Monti O, Pediconi N, Baccarini A, Fontemaggi G, Lapi E, Mantovani F, Damalas A, Citro G, Sacchi A, et al. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA Damage. Mol Cell. 2005;18:447–459. doi: 10.1016/j.molcel.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Szeles A, Yang Y, Sandlund AM, Kholodnyuk I, Kiss H, Kost-Alimova M, Zabarovsky ER, Stanbridge E, Klein G, Imreh S. Human/mouse microcell hybrid based elimination test reduces the putative tumor suppressor region at 3p21.3 to 1.6 cM. Genes Chromosomes Cancer. 1997;20:329–336. [PubMed] [Google Scholar]
- Taelman VF, Dobrowolski R, Plouhinec JL, Fuentealba LC, Vorwald PP, Gumper I, Sabatini DD, De Robertis EM. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell. 2010;143:1136–1148. doi: 10.1016/j.cell.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia VE, Nicolaescu E, McDonald CB, Musi V, Oka T, Inayoshi Y, Satteson AC, Mazack V, Humbert J, Gaffney CJ, et al. Y65C missense mutation in the WW domain of the Golabi-Ito-Hall syndrome protein PQBP1 affects its binding activity and deregulates pre-mRNA splicing. J Biol Chem. 2010;285:19391–19401. doi: 10.1074/jbc.M109.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DCR, Schiripo TA, Haber DA, Hariharan IK. salvador Promotes Both Cell Cycle Exit and Apoptosis in Drosophila and Is Mutated in Human Cancer Cell Lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Toyooka S, Ouchida M, Jitsumori Y, Tsukuda K, Sakai A, Nakamura A, Shimizu N, Shimizu K. HD-PTP: A novel protein tyrosine phosphatase gene on human chromosome 3p21.3. Biochem Biophys Res Commun. 2000;278:671–678. doi: 10.1006/bbrc.2000.3870. [DOI] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Veraksa A, Bauer A, Artavanis-Tsakonas S. Analyzing protein complexes in Drosophila with tandem affinity purification-mass spectrometry. Dev Dyn. 2005;232:827–834. doi: 10.1002/dvdy.20272. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dong Q, Zhang Q, Li Z, Wang E, Qiu X. Overexpression of yes-associated protein contributes to progression and poor prognosis of non-small-cell lung cancer. Cancer Sci. 2010;101:1279–1285. doi: 10.1111/j.1349-7006.2010.01511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. Embo J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Pattern formation in the Drosophila retina. In: Bate M, Arias A. Martinez, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; Plainview, New York: 1993. pp. 1277–1325. [Google Scholar]
- Wu M, Pastor-Pareja JC, Xu T. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature. 2010;463:545–548. doi: 10.1038/nature08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. Embo J. 2004;23:790–799. doi: 10.1038/sj.emboj.7600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.