Abstract
Background
Seven genome-wide association studies (GWAS) have been published in AIDS and only associations in the HLA region on chromosome 6 and CXCR6 have passed genome-wide significance.
Methods
We reanalyzed the data from three previously published GWAS, targeting specifically low frequency SNPs (minor allele frequency (MAF)<5%). Two groups composed of 365 slow progressors (SP) and 147 rapid progressors (RP) from Europe and the US were compared with a control group of 1394 seronegative individuals using Eigenstrat corrections.
Results
Of the 8584 SNPs with MAF<5% in cases and controls (Bonferroni threshold=5.8×10−6), four SNPs showed statistical evidence of association with the SP phenotype. The best result was for HCP5 rs2395029 (p=8.54×10−15, OR=3.41) in the HLA locus, in partial linkage disequilibrium with two additional chromosome 6 associations in C6orf48 (p=3.03×10−10, OR=2.9) and NOTCH4 (9.08×10−07, OR=2.32). The fourth association corresponded to rs2072255 located in RICH2 (p=3.30×10−06, OR=0.43) in chromosome 17. Using HCP5 rs2395029 as a covariate, the C6orf48 and NOTCH4 signals disappeared, but the RICH2 signal still remained significant.
Conclusion
Besides the already known chromosome 6 associations, the analysis of low frequency SNPs brought up a new association in the RICH2 gene. Interestingly, RICH2 interacts with BST-2 known to be a major restriction factor for HIV-1 infection. Our study has thus identified a new candidate gene for AIDS molecular etiology and confirms the interest of singling out low frequency SNPs in order to exploit GWAS data.
Keywords: AIDS, HIV-1, genome-wide association study, SNP, disease progression, RICH2
Introduction
In recent years, the development of new DNA technologies has allowed the successful completion of genome-wide association studies (GWAS) and multiple genetic associations were identified in several diseases such as Celiac disease1, Schizophrenia2, or Type 2 diabetes3. In AIDS, several GWAS have been published since 20074–10 and associations passing genome-wide significance have been found solely in chromosome 6 in the region of HLA (HCP54–6, HLA-C4, 5, 11) and in the CXCR6 gene12. The design of genotyping chips tends to rely mainly on common variants with minor allele frequencies (MAF) > 5%. Moreover, the power to detect low allele frequency SNP associations in AIDS is weakened for two complementary reasons: 1. lower frequency means less individuals at stake and thus weaker p values, and 2. AIDS-related genomic cohorts have enrolled fewer patients compared to other pathologies such as chronic kidney disease13 or Type 2 diabetes3. However, low frequency SNPs are predicted to have the potential for greater functional consequences than common alleles and may contribute strongly to genetic susceptibility to common diseases14–16; thus, they constitute very good candidates for genetic association studies. We therefore decided to re-analyze the genome-wide data obtained from the GRIV cohort on slow progression 6 and on rapid progression7, by focusing specifically on low frequency SNPs (MAF < 5 %). In order to increase the power of the study, we also included in our analysis rapid and slow progressors from the Dutch ACS cohort11 and from the American MACS156 group8.
Methods
The case groups
Slow Progressors and Rapid Progressors were gathered from 3 HIV cohorts:
The GRIV cohort
The GRIV (Genomics of Resistance to Immunodeficiency Virus) cohort, established in 1995 in France, is a collection of DNA samples to identify host genes associated with slow-progression and with rapid progression to AIDS17–20. Only Caucasians of European descent living in France were eligible for enrolment to reduce confounding by population substructure. These criteria limit the influence of the ethnic and environmental factors (all subjects live in a similar environment and are infected by HIV-1 subtype B strains) and put an emphasis on the genetic make-up of each individual in determination of rapid (RP) or slow progression (SP) to AIDS. The RP and the SP were included on the basis of the main clinical outcomes, CD4 T-cell count and time to disease progression. SP were defined as asymptomatic HIV-1 infected individuals for more than 8 years, no treatment and a CD4 T-cell count above 500/mm3. The SP group (n=270) was composed of 200 males and 70 females aged at inclusion from 19 to 62 (mean=35). Rapid progressors (RP) were stringently defined as having a two or more CD4 T-cell counts below 300/mm3 less than 3 years after the last seronegative testing. The RP group (n=84) was composed of 72 males and 12 females aged at inclusion from 21 to 55 (median=32). DNA was obtained from fresh peripheral blood mononuclear cells or from EBV-transformed cell lines. All patients provided written informed consent before enrolment in the GRIV genetic association study.
The ACS cohort
The ACS (Amsterdam Cohort Studies) cohort is composed of 316 HIV-1 homosexual men and 100 HIV-1 drug users. This cohort was established to follow the course of HIV-1 infection using various endpoints related to HIV-1 infection and AIDS. The ACS participants were described in detail previously11.
Since CD4 T-cell count was assessed during routine clinical follow-up, we could extract the ACS SP and RP patients respecting the GRIV criteria (SP: n=36, RP: n=41). The SP and RP status was easily determined among seroconverter subjects since the date of seroconversion was known. We could also extract SP from seroprevalent subjects when the time of seropositivity was known to be higher than 8 years.
The MACS156 group
The MACS156 study comprises a subset of 156 HIV-1 homosexual men enrolled in the MACS (MultiCenter AIDS Cohort Study) cohort, a prospective cohort originally established to investigate the natural history of HIV infection21. This subset of MACS European American participants was chosen to be enriched with the extremes AIDS progression phenotypes 8. The MACS156 participants were described in detail previously8.
Since CD4 T-cell count was assessed during routine clinical follow-up, we could extract the MACS SP and PR respecting the GRIV definition (SP: n=59, RP: n=22). As with the ACS cohort, the SP and the RP were selected from seroconverter subjects. SP were also selected from seroprevalent subjects.
Control groups
Three Caucasian control groups from France, The Netherlands, and the USA were merged and used as a control group.
The D.E.S.I.R. control group
The Data from an Epidemiological Study on Insulin Resistance Syndrome (D.E.S.I.R) program was a 9-year follow-up study designed to clarify the development of the insulin resistance syndrome. Subjects were recruited from 1994 to 1996 from volunteers insured by the French social security system, which offers periodic health examinations free of charge22. This control group comprised 694 participants both non obese and normoglycemic of the D.E.S.I.R. trial, all French and HIV-1 seronegative. It was composed of 281 males and 413 females aged from 30 to 64 years.
Dutch controls (CTR-ACS)
This control group corresponds to 376 Dutch subjects genotyped with HumanHap300 BeadChips23.
Illumina control group
This control group corresponds to Caucasian subjects genotyped with HumanHap300 BeadChips from the Illumina Genotyping Control Database (www.illumina.com). There were 324 individuals.
Genotyping method, quality control
The GRIV cohort, the ACS cohort, and the control groups were genotyped using the Illumina Infinium II HumanHap300 BeadChips (Illumina, San Diego, CA, USA). The genotyping quality was assessed for each group using the BeadStudio software (version 3.1, Illumina). Missing data >2%, minor allele frequency (<1%) and deviations from Hardy-Weinberg equilibrium in the control groups (p<1.0×10−3) were removed during these quality control steps6, 7, 11. The MACS156 group genotype data were obtained through the Affymetrix GeneChip Human Mapping 500K Array (Affymetrix, Santa Clara, CA, USA). Different quality control filters were applied to ensure reliable genotyping data8. For all the cohorts, we removed outliers exhibiting non-Caucasian ancestry, cohort by cohort, using the Eigenstrat method24.
SNP selection
We considered two pooled case groups (SP from GRIV, ACS, and MACS156 group on the one hand, RP from GRIV, ACS, and MACS156 on the other hand) and the pooled control groups (D.E.S.I.R., CTR-ACS, Illumina-CTR). We retained the 8584 SNPs exhibiting a MAF < 5% for the SP-CTR comparison (Bonferroni 5.8×10−6), and 10295 SNPs exhibiting a MAF < 5% for the RP-CTR comparison (Bonferroni 4.8×10−6). It was important to choose SNPs with low frequency in either groups since we were looking for factors either promoting progression (low MAF in SP compared to CTR or low MAF in CTR compared to RP) or preventing progression (low MAF in RP compared to CTR or low MAF in CTR compared to SP). The choice to screen specifically low frequency SNPs stems from two complementary reasons: 1. a biological reason: HIV-1 infection is a multi-factorial disease with several genetic factors impacting disease. Several groups have pointed out that most signals involved in complex diseases should deal with the low frequency variants spectrum 14–16. Indeed, this observation was confirmed in AIDS since the main signal found up to now was in HCP5 with a low frequency variant 4–6, 9. 2. a statistical fact: in our case-control configuration, a low frequency either in the CTR or in the CASE group means systematically a weaker p value for a given Odds Ratio (OR which measures the real biological impact of the SNP). For instance, for an OR of 0.5 in the dominant mode, the p values obtained are: 0.02 with a SP MAF of 2% and a CTR MAF=3.9%, 1.5×10−5 with a SP MAF of 5% and a CTR MAF=9.5%, 9.98×10−8 with a SP MAF of 10% and CTR MAF=18.2%. Moreover, in the Illumina genotyping chips used, SNPs with low MAF are under-represented in genotyping chips compared to SNPs with higher MAF (data not shown). Overall for a biological effect measured by a given OR, SNP associations are thus more difficult to identify in the low frequency spectrum since they exhibit weaker p values by essence and since they are under-represented and thus artificially penalized through global Bonferroni corrections.
Statistical analysis
We performed a case-control analysis by comparing either the SP group (n=365) or the RP group (n=147) consisting in GRIV, ACS, and MACS patients with the control group consisting of D.E.S.I.R, CTR-ACS, and Illumina control individuals (n=1394). The statistical analysis was performed by a logistic regression (with SNPtest software25) in the dominant mode, taking into account stratification by adding the 2 first Eigenstrat PC axes as covariates using EIGENSOFT. Testing for association under the dominant model was appropriate since we lacked power to test for associations under the recessive model and additionnaly, in this context, the dominant model is identical to the additive mode. For each SNP passing the Bonferroni threshold, we recomputed the regression by adding the HCP5 SNP (rs2395029) as a covariate to check for non-independence due to linkage disequilibrium (with SNPtest software25).
SNP imputation
Using SNPtest Impute software 25 it was possible to impute untyped SNPs of the MACS156 study subjects, absent from the the Affymetrix GeneChip Human Mapping 500K Array (Affymetrix, Santa Clara, CA, USA) and present in the Illumina HumanHap300 BeadChips (Illumina, San Diego, CA, USA). They were imputed using the HapMap release 21 phased data for the European population (CEU) as panel of reference (http://www.hapmap.org). Only the SNPs imputed with high reliability (imputation quality score25 P>0.9) were retained.
Internal replication
After comparing the total SP group (combined from GRIV, ACS, and MACS 156) with the total control group (combined from D.E.S.I.R, CTR-ACS, and CTR_Illumina), we also performed an individual analysis of each group GRIV, ACS, and MACS156 for the four SNPs passing the Bonferroni threshold. For GRIV, we checked the result obtained in our previous GWAS6. For ACS, we tested the association between the four SNPs and times to time to AIDS93 by linear regression. For MACS156 group, we also tested the association between the SNPs and time to AIDS93 by linear regression.
Linkage disequilibrium (LD)
For each SNP exhibiting a significant association, we looked for the other SNPs in linkage disequilibrium (r2≥0.9) in the HapMap population of Western European ancestry (CEU, HapMap data Release 21a/phase II January 2007, on NCBI B35 assembly, dbSNP125, http://www.hapmap.org) in order to identify the genes possibly tracked by the SNP associations. A SNP was assigned to a gene if it was located within the gene or in the 2kb flanking regions (potential regulatory sequence), otherwise it was considered intergenic.
Bioinformatics exploration
To further explore the associations observed, we tried to identify putative modifications in mRNA expression as shown in Genevar26 and Dixon27 databases, in splicing (FastSNP28, http://fastsnp.ibms.sinica.edu.tw/pages/input_CandidateGeneSearch.jsp/), in polyadenylation (polyAH, http://linux1.softberry.com/berry.phtml?topic=polyah&group=programs&subgroup=promoter and polyApred, http://www.imtech.res.in/raghava/polyapred/submission.html), or in transcription factor binding sites (SignalScan, http://www-bimas.cit.nih.gov/molbio/signal/, TESS, http://www.cbil.upenn.edu/cgi-bin/tess/tess?RQ=WELCOME, and TFSearch, http://www.cbrc.jp/research/db/TFSEARCH.html, derived from TRANSFAC database). We used the Genecards database to look for the tissues and organs expressing the proteins (GeneCards29, http://www.genecards.org).
Results
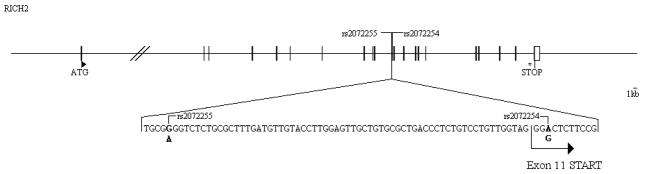
For the 8584 SNPs with a MAF < 5 %, we tested a total of 365 SP patients and compared them with a control cohort of 1394 seronegative individuals (from France, the Netherlands, and the US, see methods). Four signals passed the Bonferroni threshold in the dominant mode, three in chromosome 6 and one in chromosome 17 (Table 1). Not unexpectently, the best result was obtained for the well-replicated HCP5 rs2395029 (p=8,54×10−15). The two other associations found in chromosome 6 were for rs9368699 in C6orf48 (p= 3,03×10−10) and rs8192591 in NOTCH4 (p=9,08×10−07). These genes are in partial linkage disequilibrium with HCP5-rs2395029 (resp. r2=0.68, r2=0.57). The fourth association corresponded to the chromosome 17 SNP rs2072255 located in the RICH2 gene (p=3.30×10−6). This SNP is in full LD (r2=1) with rs2072254 corresponding to a synonymous change (Gly188Gly) of RICH2 (Figure 1 and see map, Supplemental Digital Content 1). The fours SNPs corresponded to the higher end of the MAF distribution of the SNP studied (see histogram, Supplemental Digital Content 2), which is logical since larger numbers of subjects lead to stronger p values.
Table 1.
Four SNPs passed the Bonferroni threshold after comparing slow progressors against controls on low frequency SNPs (MAF < 5%). For each SNP, the chromosome, the minor allele A1, the minor allele frequency (MAF), the frequency of the group in the dominant mode (used for association tests), the p value in the dominant mode, the odds ratio (OR), and the genes concerned (SNP and/or SNP(s) in LD with r2≥0.9 in Caucasians HapMap data) are provided. A SNP was assigned to a gene if it was located in the gene or within the 2kb flanking regions.
| SNP | chr | A1 | A2 | MAFSP | MAFCTR | Frequency dominant mode: A1A1 & A1A2 |
PSP-CTR | OR | Gene(s)/LD | |
|---|---|---|---|---|---|---|---|---|---|---|
| SPALL | CTRALL | |||||||||
| rs2395029 | 6 | C | A | 10.19% | 3.41% | 20.38% | 6.81% | 8.54×10−15 | 3.41 CI 95%: 2.4–4.8 | HCP5, intergenic, MICB |
| rs9368699 | 6 | G | A | 9.20% | 3.48% | 18.13% | 6.96% | 3.03×10−10 | 2.9 CI 95%: 2.04–4.16 | C6orf48, RDBP, TNXB |
| rs8192591 | 6 | T | C | 6.83% | 3.15% | 13.63% | 6.30% | 9.08×10−7 | 2.32 CI 95%: 1.56– 3.41 | NOTCH4 GPSM3 |
| rs2072255 | 17 | A | G | 4.79% | 10.04% | 9.04% | 18.87% | 3.30×10−6 | 0.43 CI 95%: 0.28–0.63 | RICH2 |
Figure 1.
Genetic map of the RICH2 gene region. Exons and UnTranslated Regions are symbolized by full and empty rectangles, respectively. The positions of the ATG and STOP codons are indicated by a triangle (▸) and by an asterisk (*), respectively. The 2 SNPs rs2072255 (intronic) and rs2072254 (exonic) are represented (r2=1) and a zoom with the genetic sequences is provided.
In order to evaluate the role of LD in these associations, we recomputed the p-values using the HCP5 SNP (rs2395029) as covariate. The two chromosome 6 SNPs were not significant in the adjusted analysis (p=0.78 for rs9368699 in C6orf48, p=0.31 for rs8192591 in NOTCH4), but the association remained statistically robust for the chromosome 17 SNP after controlling for the HCP5 SNP, p=1.82×10−6 for rs2072255 in RICH2. In line with this computation, we found that the rs2072255 frequency was not significantly different between the SP elite controllers12 and among the SP non elite controllers 12 (p=0.7).
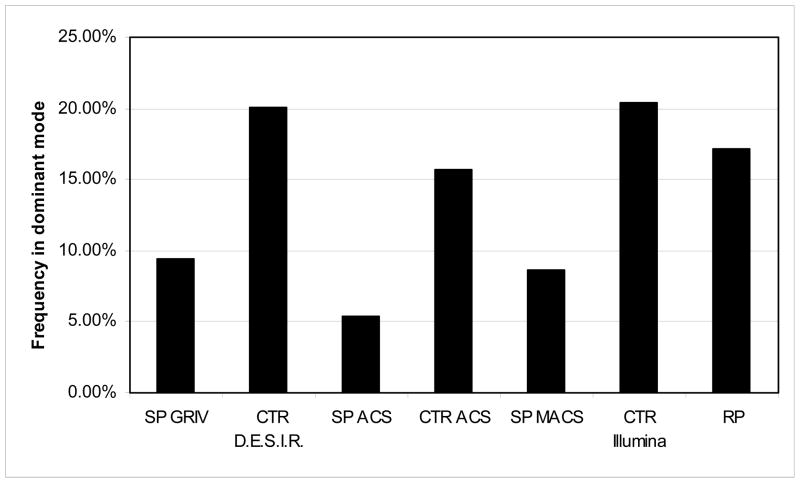
The subjects carrying the rs2072255-A allele were 9.04% in the SP group (Table 1), 18.87% in the control group, and 17.1% in the RP group (this excludes the hypothesis of an effect on infection). Interestingly these frequencies were consistent within each of the three SP groups as well as within each of the three control groups (Figure 2). Moreover, the positive signal for association of rs2072255 was confirmed in the individual cohorts GRIV, ACS, and MACS156 study: the comparison of the NP with D.E.S.I.R controls in GRIV led to p= 8.1×10−5, the analysis of ACS by linear regression led to p=0.05, and the analysis of the MACS156 group by linear regression led also to p=0.05. A table summarizing the results in the different cohorts is provided in supplemental digital content 3. Finally, the rs2072254 RICH2 exonic SNP (in LD with the rs2072255) is located in a splicing site according to FastSNP28 (Figure 1).
Figure 2.
Frequency of individuals carrying the rs2072255-A variant (dominant mode) in the various groups: SP GRIV (n=270), SP ACS (n=31), SP MACS (n=59), the D.E.S.I.R. control group (n=694). CTR-ACS (n=376), the Illumina CTR (n=324), and the RP group (n=147). The frequencies within each of the GRIV RP, ACS RP, and MACS156 study RP groups was similar.
When comparing the 10295 selected SNPs between the 147 RP patients with the 1394 seronegative control group, no signal passed the Bonferroni threshold.
Discussion
We decided to reanalyze previous GWAS data on AIDS cohorts by focusing specifically on low frequency SNPs (MAF<5%). For that, we combined rapid and slow progressors from three international cohorts from France (GRIV), Netherlands (ACS), and US (MACS156 study) totalling 365 SP and 147 RP, who were compared with 1394 controls (seronegative individuals). No association was found when comparing the RP group with the CTR group. This was not a surprise since the RP group comprised only 147 individuals and for a MAF of 5% in the CTR, one needed to get a quite strong biological effect (OR>2.8) to pass the Bonferroni threshold. Four SNPs passed the Bonferroni threshold when comparing the SP group with the CTR group. Among them, three are in chromosome 6 and were previously found significant by several studies: rs2395029 in HCP54–6, 8, 9, 11, rs9368699 in C6orf485, 6, rs8192591 in NOTCH45. NOTCH4 is an interesting candidate gene due to its role in immune regulation and the NOTCH4 rs8192591 corresponds to a non-synonymous Gly534Ser protein variant. This association was found independent from the HCP5 rs2395029 by Fellay et al.5, however the signal disappeared in our own study when using HCP5 rs2395029 as covariate. A possible explanation for this discrepancy could be the use of viral load as an endpoint in the study by Fellay et al.5 while we used here a progression phenotype.
The fourth signal identified in the present study is new and corresponds to rs2072255 in the chromosome 17 RICH2 gene. The RICH2 gene encodes a Rho-type GTPase activator composed of 818 amino acid (89,247 kDa) with an intracellular localisation. It is expressed highly in the brain, and at a basal level in several tissues notably in the lymph nodes29. A recent study has shown that RICH2 is a part of the physical link between BST-2 and the actin cytoskeleton and prevents the internalisation of BST-230. RICH2 could thus contribute to the externalisation of BST-2 which prevents HIV-1 virion budding and release31. This is a counteraction of HIV-1 Vpu known to favor internalization and degradation of BST-231. The rs2072255-A RICH2 allele favors progression to AIDS since 18.87% of the CTR carry the variant while only 9.04% of the SP carry it (Table 1, Figure 2). Interestingly, rs2072255 is in total LD with the SNP rs2072254 located in a splicing site of RICH2 as suggested by FastSNP (Figure 2). If the rs2072254-G minor allele alters mRNA splicing, it could lead to a down-modulation of RICH2 and thus explain a diminished effect of BST-2 against HIV-1 production.
The identification of three chromosome 6 signals already confirmed by several studies shows the relevance of targeting specifically low frequency SNPs in GWAS. The genetic and biological data regarding the RICH2 signal are also quite compelling and provide a new relevant candidate gene to explore the molecular etiology of HIV-1 pathogenesis. Further genetic and experimental studies will be needed to confirm and understand the effect of RICH2 in AIDS pathogenesis.
Supplementary Material
Acknowledgments
Sources of support: This work was supported by Agence Nationale de Recherche sur le SIDA (ANRS), Sidaction, Fondation de France, Innovation 2007 program of Conservatoire National des Arts et Métiers (CNAM), AIDS Cancer Vaccine Development Foundation, Neovacs SA, Vaxconsulting. Sophie Limou benefits from a fellowship from the French Ministry of Education, Technology and Research and Sigrid Le Clerc benefits from a fellowship of ANRS. The Amsterdam Cohort Studies on HIV infection and AIDS, a collaboration between the Amsterdam Health Service, the Academic Medical Center of the University of Amsterdam, Sanquin Research, and the University Medical Center Utrecht, are part of the Netherlands HIV Monitoring Foundation and financially supported by the Netherlands National Institute for Public Health and the Environment. The authors acknowledge funding from the Netherlands Organization for Scientific Research (TOP, registration number 9120.6046). The MACS is funded by the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute. UO1-AI-35042, UL1-RR025005 (GCRC), UO1-AI-35043, UO1-AI-35039, UO1-AI-35040, UO1-AI-35041. This project has been funded in part with federal funds from the National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
The authors are grateful to all the patients and medical staff who have kindly collaborated with the GRIV project. Data in this manuscript were collected by the Multicenter AIDS Cohort Study (MACS) with centers (Principal Investigators) at The Johns Hopkins Bloomberg School of Public Health (Joseph B. Margolick, Lisa P. Jacobson), Howard Brown Health Center, Feinberg School of Medicine, Northwestern University, and Cook County Bureau of Health Services (John P. Phair, Steven M. Wolinsky), University of California, Los Angeles (Roger Detels), and University of Pittsburgh (Charles R. Rinaldo). Website located at http://www.statepi.jhsph.edu/macs/macs.html. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References
- 1.Dubois PC, Trynka G, Franke L, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010 Apr;42(4):295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Donovan MC, Craddock N, Norton N, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008 Sep;40(9):1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 3.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008 May;40(5):638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007 Aug 17;317(5840):944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fellay J, Ge D, Shianna KV, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009 Dec;5(12):e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Limou S, Le Clerc S, Coulonges C, et al. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02) J Infect Dis. 2009 Feb 1;199(3):419–426. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- 7.Le Clerc S, Limou S, Coulonges C, et al. Genomewide association study of a rapid progression cohort identifies new susceptibility alleles for AIDS (ANRS Genomewide Association Study 03) J Infect Dis. 2009 Oct 15;200(8):1194–1201. doi: 10.1086/605892. [DOI] [PubMed] [Google Scholar]
- 8.Herbeck JT, Gottlieb GS, Winkler CA, et al. Multistage genomewide association study identifies a locus at 1q41 associated with rate of HIV-1 disease progression to clinical AIDS. J Infect Dis. 2010 Feb 15;201(4):618–626. doi: 10.1086/649842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalmasso C, Carpentier W, Meyer L, et al. Distinct genetic loci control plasma HIV-RNA and cellular HIV-DNA levels in HIV-1 infection: the ANRS Genome Wide Association 01 study. PLoS One. 2008;3(12):e3907. doi: 10.1371/journal.pone.0003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelak K, Goldstein DB, Walley NM, et al. Host determinants of HIV-1 control in African Americans. J Infect Dis. 2010 Apr 15;201(8):1141–1149. doi: 10.1086/651382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Manen D, Kootstra NA, Boeser-Nunnink B, Handulle MA, van’t Wout AB, Schuitemaker H. Association of HLA-C and HCP5 gene regions with the clinical course of HIV-1 infection. Aids. 2009 Jan 2;23(1):19–28. doi: 10.1097/QAD.0b013e32831db247. [DOI] [PubMed] [Google Scholar]
- 12.Limou S, Coulonges C, Herbeck JT, et al. Multi-Cohort Genetic Association Study Reveals CXCR6 as a New Chemokine Receptor Involved in AIDS Long-Term Non-Progression. The Journal of infectious diseases. 2010 doi: 10.1086/655782. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kottgen A, Pattaro C, Boger CA, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010 May;42(5):376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007 Apr;80(4):727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorlov IP, Gorlova OY, Sunyaev SR, Spitz MR, Amos CI. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am J Hum Genet. 2008 Jan;82(1):100–112. doi: 10.1016/j.ajhg.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008 Jun;40(6):695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flores-Villanueva PO, Hendel H, Caillat-Zucman S, et al. Associations of MHC ancestral haplotypes with resistance/susceptibility to AIDS disease development. J Immunol. 2003 Feb 15;170(4):1925–1929. doi: 10.4049/jimmunol.170.4.1925. [DOI] [PubMed] [Google Scholar]
- 18.Hendel H, Caillat-Zucman S, Lebuanec H, et al. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J Immunol. 1999 Jun 1;162(11):6942–6946. [PubMed] [Google Scholar]
- 19.Rappaport J, Cho YY, Hendel H, Schwartz EJ, Schachter F, Zagury JF. 32 bp CCR-5 gene deletion and resistance to fast progression in HIV-1 infected heterozygotes. Lancet. 1997 Mar 29;349(9056):922–923. doi: 10.1016/S0140-6736(05)62697-9. [DOI] [PubMed] [Google Scholar]
- 20.Winkler CA, Hendel H, Carrington M, et al. Dominant effects of CCR2-CCR5 haplotypes in HIV-1 disease progression. J Acquir Immune Defic Syndr. 2004 Dec 1;37(4):1534–1538. doi: 10.1097/01.qai.0000127353.01578.63. [DOI] [PubMed] [Google Scholar]
- 21.Kaslow RA, Ostrow DG, Detels R, Phair JP, Polk BF, Rinaldo CR., Jr The Multicenter AIDS Cohort Study: rationale, organization, and selected characteristics of the participants. Am J Epidemiol. 1987 Aug;126(2):310–318. doi: 10.1093/aje/126.2.310. [DOI] [PubMed] [Google Scholar]
- 22.Balkau B. An epidemiologic survey from a network of French Health Examination Centres, (D.E.S.I.R.): epidemiologic data on the insulin resistance syndrome. Rev Epidemiol Sante Publique. 1996 Aug;44(4):373–375. [PubMed] [Google Scholar]
- 23.van Es MA, Veldink JH, Saris CG, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet. 2009 Oct;41(10):1083–1087. doi: 10.1038/ng.442. [DOI] [PubMed] [Google Scholar]
- 24.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006 Aug;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 25.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007 Jul;39(7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 26.Ge D, Zhang K, Need AC, et al. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 2008 Apr;18(4):640–643. doi: 10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dixon AL, Liang L, Moffatt MF, et al. A genome-wide association study of global gene expression. Nat Genet. 2007 Oct;39(10):1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 28.Yuan HY, Chiou JJ, Tseng WH, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W635–641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 1997 Apr;13(4):163. doi: 10.1016/s0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- 30.Rollason R, Korolchuk V, Hamilton C, Jepson M, Banting G. A CD317/tetherin-RICH2 complex plays a critical role in the organization of the subapical actin cytoskeleton in polarized epithelial cells. J Cell Biol. 2009 Mar 9;184(5):721–736. doi: 10.1083/jcb.200804154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tokarev A, Skasko M, Fitzpatrick K, Guatelli J. Antiviral activity of the interferon-induced cellular protein BST-2/tetherin. AIDS Res Hum Retroviruses. 2009 Dec;25(12):1197–1210. doi: 10.1089/aid.2009.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.