Abstract
In prostate cancer, the signals that drive cell proliferation change as tumors progress from castration-sensitive (androgen-dominant) to castration-resistant states. While the mechanisms underlying this change remain uncertain, characterization of common signaling components that regulate both stages of prostate cancer proliferation is important for developing effective treatment strategies. Here, we demonstrate that paxillin, a known cytoplasmic adaptor protein, regulates both androgen- and EGF-induced nuclear signaling. We show that androgen and EGF promoted MAPK-dependent phosphorylation of paxillin, resulting in nuclear translocation of paxillin. We found nuclear paxillin could then associate with androgen-stimulated androgen receptor (AR). This complex bound AR-sensitive promoters, retaining AR within the nucleus and regulating AR-mediated transcription. Nuclear paxillin also complexed with ERK and ELK1, mediating c-FOS and cyclin D1 expression; this was followed by proliferation. Thus, paxillin is a liaison between extranuclear MAPK signaling and nuclear transcription in response to androgens and growth factors, making it a potential regulator of both castration-sensitive and castration-resistant prostate cancer. Accordingly, paxillin was required for normal growth of human prostate cancer cell xenografts, and its expression was elevated in human prostate cancer tissue microarrays. Paxillin is therefore a potential biomarker for prostate cancer proliferation and a possible therapeutic target for prostate cancer treatment.
Introduction
Prostate cancer is associated with significant morbidity and mortality and is the second most common cancer among men worldwide. One feature of prostate cancer progression is its apparent change in androgen responsiveness over time. At diagnosis, locally advanced prostate cancers are treated successfully by prostatectomy, irradiation, and/or anti-androgen therapy, suggesting that tumor cells are dependent on androgens for continued growth and survival. In contrast, despite treatment, many advanced tumors enter a more aggressive castration-resistant state after 18–24 months. This observation suggests fundamental changes in the extracellular triggers and intracellular signaling pathways that regulate proliferation and cell migration (1–3) in aggressive tumors. The reasons for this dramatic transformation in phenotype are not well understood. However, several likely interconnected mechanisms of castration resistance have been proposed. For example, alterations in intraprostatic androgen production or metabolism (“intracrine” androgen production) in castration-resistant prostate cancer (4, 5) could lead to elevated local androgen concentrations in the setting of low serum androgen levels. Alternatively, the abundance or activity of androgen receptors (ARs) and their various transcriptional coregulators might be modified in castration-resistant prostate cancer cells so that they respond to lower concentrations of androgen or other steroids (6). Finally, prostate cancer cells might adopt alternative mechanisms of proliferation that are less dependent on androgens (although still dependent on ARs), but more sensitive to growth factors such as EGF or IGF-1 (6, 7).
Interestingly, others and we have shown that androgen and growth factor signaling are tightly linked in prostate cancer cells (1, 7–11). For example, we demonstrated that androgen binding to extranuclear ARs leads to MMP-mediated release of EGFR ligands (8). Subsequent activation of the EGFR results in MAPK signaling within the cytoplasm, followed by enhanced cellular proliferation. Based on this observation, we propose that androgen-triggered membrane-initiated signaling events in prostate cancer cells are in fact very similar to growth factor–triggered (e.g., EGF) signaling. In both scenarios, receptor tyrosine kinase activation stimulates MAPK signaling, which then regulates expression of AR- and ERK-dependent genes involved in proliferation and migration (8). Thus, crosstalk between extranuclear kinase signaling and transcription appears to be critical for prostate cancer progression regardless of the mitogenic ligand.
How do extranuclear AR and MAPK signaling regulate AR- and ERK-dependent transcription? We previously showed that paxillin (PXN), a protein involved in cytoskeletal remodeling and focal adhesion function, mediates both extra- and intranuclear AR and ERK signaling in prostate cancer cells (8). Specifically, EGFR-induced phosphorylation of PXN on tyrosine residues is required for activation of Raf and downstream MAPK signaling in response to both androgen and EGF stimulation. Some AR- and ERK-regulated gene expressions, and ultimately prostate cancer cell proliferation (8), are then dependent upon subsequent MAPK-dependent phosphorylation of specific serine residues on PXN.
Here, we used in vitro models, xenograft tumor studies, and human tissue microarrays (TMAs) to examine the importance of and the mechanisms by which serine-phosphorylated PXN regulates transcription and proliferation in prostate cancer cells. We found that phosphoserine-PXN (PS-PXN) localized to the nucleus upon MAPK-mediated phosphorylation in both androgen/castration-sensitive (LNCaP) and androgen/castration-insensitive (PC3) prostate cancer cells. Once in the nucleus, PS-PXN both retained androgen-bound AR within the nucleus and complexed with AR on the prostate-specific antigen (PSA) and NKX3-1 promoter to enhance its transcriptional activity. Furthermore, PS-PXN was in fact a general regulator of ERK-mediated nuclear actions regardless of the initial external trigger. In response to EGF, PS-PXN was required for ERK-mediated phosphorylation of E twenty-six–like (ETS-like) transcription factor 1 (ELK1), which in turn regulated expression of the proliferative genes c-FOS and cyclin D1. The relevance of PXN to prostate cancer cell growth in vivo was established by both xenograft studies and PXN overexpression in human prostate cancer tissue samples. Thus, PXN regulates both androgen and growth factor signaling in prostate cancer cells, making it a potential mediator of both castration-sensitive and castration-resistant prostate cancer.
Results
PS-PXN is localized in the nucleus.
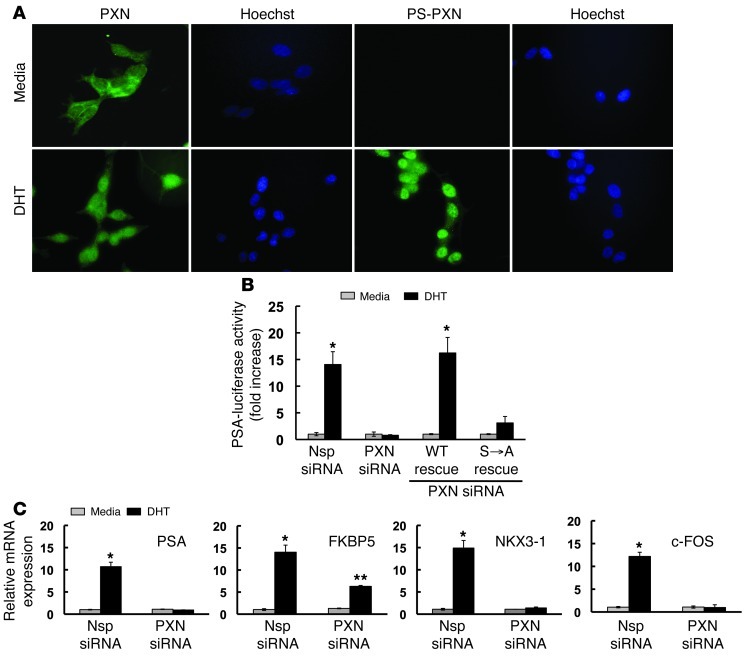
Our previous study demonstrated that the focal adhesion protein PXN was required for androgen-induced expression of prostate-specific antigen (PSA) mRNA in prostate cancer cells, suggesting that PXN might also function within the nucleus (8). To determine whether PXN regulated nuclear processes, we used immunofluorescence to examine subcellular localization of PXN in androgen-sensitive LNCaP prostate cancer cells (Figure 1A). PXN localized primarily in the cytoplasm of serum-starved, unstimulated LNCaP cells (11.4% ± 1.3% of cells contained nuclear PXN). Surprisingly, upon dihydrotestosterone (DHT) stimulation, more PXN was found in the nucleus (73.5% ± 0.6% of cells contained nuclear PXN, P ≤ 0.01 vs. unstimulated cells; Figure 1A). An antibody directed against phosphorylated serine residue 126 of PS-PXN showed that PS-PXN was predominantly nuclear in DHT-treated cells (91.0% ± 3.55% of cells contained mainly nuclear PS-PXN; Figure 1A), indicating that some or all of the nuclear PXN was serine phosphorylated in response to MAPK signaling. Notably, biochemical separation of cytoplasmic and nuclear fractions confirmed the immunofluorescence results (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI62044DS1).
Figure 1. In LNCaP cells, phosphorylated PXN is a nuclear protein that regulates AR-mediated transcription.
(A) PS-PXN localizes to the nucleus. Immunofluorescence of PXN and PS-PXN with medium or DHT (25 nM) treatment (30 minutes). For all immunofluorescence, adjacent Hoechst staining represents the nucleus; experiments were repeated more than 3 times with identical results. Original magnification, ×40. (B) DHT stimulation of the PSA promoter requires PXN. Nsp or PXN siRNA–treated cells were transfected with PSA-luciferase and cytomegalovirus–β-gal plasmid; this was followed by DHT treatment (25 nM; 24 hours). PXN-knockdown cells were transfected with plasmids encoding WT PXN or PXN(S→A). Luciferase activity was normalized to β-gal and represented as fold increase over medium (n = 4). (C) PXN regulates expression of multiple androgen targets. Relative expression of DHT-induced PSA, FKBP5, NKX3-1, and c-FOS mRNAs in Nsp or siRNA-mediated PXN knockdown LNCaP cells. Data are represented as mean ± SEM (n = 3). *P ≤ 0.001; **P ≤ 0.05 relative to Nsp siRNA.
Since PS-PXN was primarily nuclear, we postulated that PS-PXN might regulate PSA mRNA expression by modulating AR-mediated transcription. We therefore examined the role of PS-PXN in regulating AR-induced activation of the PSA promoter (Figure 1B). We transfected a luciferase construct driven by the PSA promoter into LNCaP prostate cancer cells and measured DHT-triggered luciferase activity. While DHT induced PSA promoter activity in cells transfected with control (Nsp) siRNA, knockdown of endogenous PXN (PXN siRNA) abrogated DHT-induced PSA promoter activity (Figure 1B). Reexpression of WT PXN in knockdown cells restored DHT-induced PSA promoter activity (WT Rescue; Figure 1B). In contrast, expression of a PXN mutant lacking the MAPK-dependent serine phosphorylation sites (serines 83, 126, and 130), PXN(S→A) failed to rescue DHT-induced PSA promoter activity (S→A rescue; Figure 1B). This result confirms that MAPK-dependent phosphorylation of PXN is required for AR-mediated activation of the PSA promoter.
We next examined the role of PXN in the expression of multiple androgen targets using quantitative PCR (qPCR) (Figure 1C). Androgen-induced expression of PSA, FKBP5, and NKX3-1 mRNAs was significantly inhibited after siRNA-mediated PXN knockdown in LNCaP cells (Figure 1C) without affecting total AR expression (Supplemental Figure 2A). These results suggest that PXN is a broad regulator of AR-mediated transcription. Furthermore, androgen-triggered upregulation of c-FOS mRNA, a proliferative signal requiring ERK signaling but not AR-mediated transcription, also required PXN (Figure 1C). This result demonstrates that, in addition to directly regulating AR-mediated transcription, PXN also regulates indirect effects of membrane-initiated extranuclear AR signaling on transcription (in this case via AR-triggered ERK signaling). PXN expression was confirmed by Western blot (Supplemental Figure 3A).
PXN regulates nuclear AR localization.
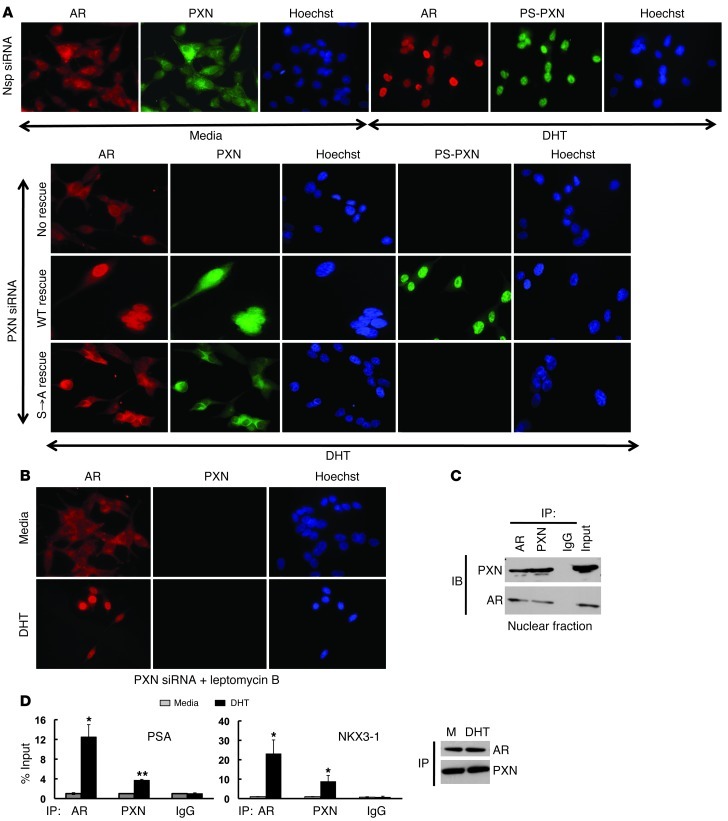
Since PS-PXN is localized in the nucleus and required for AR-mediated expression of several androgen-induced mRNAs, we postulated that PS-PXN might mediate AR’s nuclear localization in response to androgen. Therefore, we examined colocalization of PXN or PS-PXN with AR in LNCaP cells (Figure 2A). In serum-starved cells (Nsp siRNA-medium; Figure 2A), AR and total PXN were primarily cytoplasmic (14.7% ± 1.1% and 8.1% ± 0.7% of cells contained nuclear AR and PXN, respectively). Upon DHT stimulation, both AR and PS-PXN colocalized to the nucleus (97.1 ± 2.1% and 93.5 ± 2.3% of cells contained predominantly nuclear AR and PS-PXN, respectively). However, after PXN knockdown (PXN siRNA–no rescue; Figure 2A), AR nuclear localization was markedly reduced in response to DHT (14.1% ± 0.6% of cells contained nuclear AR, P ≤ 0.05 vs. nonspecific [Nsp] siRNA). Reexpression of WT PXN rescued DHT-induced AR nuclear localization (WT rescue; 89.1% of cells contained nuclear AR; Figure 2A). However, reexpression of PXN(S→A) lacking the MAPK-dependent phosphorylation sites did not rescue DHT-induced AR nuclear localization (S→A rescue; 18.75% ± 1.6% of cells contained nuclear AR, P ≤ 0.05 vs. Nsp siRNA or WT rescue; Figure 2A), suggesting that PS-PXN is required for AR nuclear localization. Importantly, PXN(S→A) is expressed in these cells but, as expected, is not phosphorylated in response to DHT. Furthermore, subcellular localizations of PXN, PS-PXN, and AR were confirmed with nuclear and cytoplasmic isolation followed by Western blot (Supplemental Figure 1A).
Figure 2. In LNCaP cells, phosphorylated PXN maintains DHT-triggered AR nuclear localization and binds to promoter DNA.
(A) PXN is required for AR nuclear localization. Immunofluorescence of AR (red), PXN (green), or PS-PXN (green) in cells treated with Nsp or PXN siRNA, followed by medium or DHT (25 nM; 30 minutes). PXN-knockdown cells were transfected with plasmids expressing WT PXN or PXN(S→A). (B) PXN retains AR within the nucleus. Cells were treated with PXN siRNA, then leptomycin B (15 ng/ml), followed by medium or DHT (25 nM; 30 minutes) stimulation. Original magnification, ×40. (C) PXN and AR form a complex in the nucleus. AR or PXN was precipitated from nuclear extracts of DHT-treated LNCaP cells (IP), followed by immunoblotting (IB) (n = 3 with identical results). IgG represents precipitation with Nsp antibody. (D) AR and PXN bind to the PSA and NKX3-1 promoters. Cells were starved overnight; this was followed by medium or DHT (25 nM; 45 minutes) treatment and chromatin IP using antibodies against the indicated proteins (IP). IgG represents Nsp antibody. Values are represented as percentage input (mean ± SEM, n = 3). *P ≤ 0.001; **P ≤ 0.05 relative to medium.
PS-PXN could be regulating AR nuclear localization by 1 of 2 mechanisms. First, PS-PXN may directly mediate AR translocation to the nucleus in response to DHT. Alternatively, DHT might be sufficient to promote AR nuclear localization, while PS-PXN functions to retain AR within the nucleus. To differentiate between these 2 possibilities, we examined PXN’s role in mediating subcellular localization of AR in LNCaP cells treated with leptomycin B, which blocks nuclear export by inhibiting CRM1/exportin (Figure 2B). Under these conditions, DHT promoted AR accumulation in the nucleus (89.6% ± 0.5% of cells) relative to untreated cells (2.8% ± 0.5% of cells, P ≤ 0.01), even in the absence of detectable PXN, indicating that PS-PXN most likely regulates AR nuclear retention rather than nuclear entry.
PXN and AR complex in the nucleus and on the PSA and NKX3-1 promoters.
To determine whether AR and PXN formed a complex in the nucleus, we performed co-IP assays (Figure 2C) in nuclear extracts from LNCaP cells treated with DHT. AR and PXN coprecipitated regardless of the precipitating antibody, confirming that they form a complex in the nucleus (Figure 2C). Surprisingly, AR and PXN were also in close proximity on the PSA and NKX3-1, but not FKBP5, promoters, as ChIP assays revealed both AR and PXN associated with the PSA and NKX3-1 promoters in a DHT-dependent fashion (Figure 2D). To determine whether AR and PXN are associated at the same time on the PSA promoter, we performed ChIP analysis in LNCaP cells with an antibody against PXN followed by re-ChIP with an antibody to AR. Indeed, we generated DHT-dependent PCR products from the PSA promoter after the consecutive precipitations (Supplemental Figure 2B), confirming that, in addition to regulating AR retention in the nucleus, PXN may coregulate AR-mediated transcription in response to DHT.
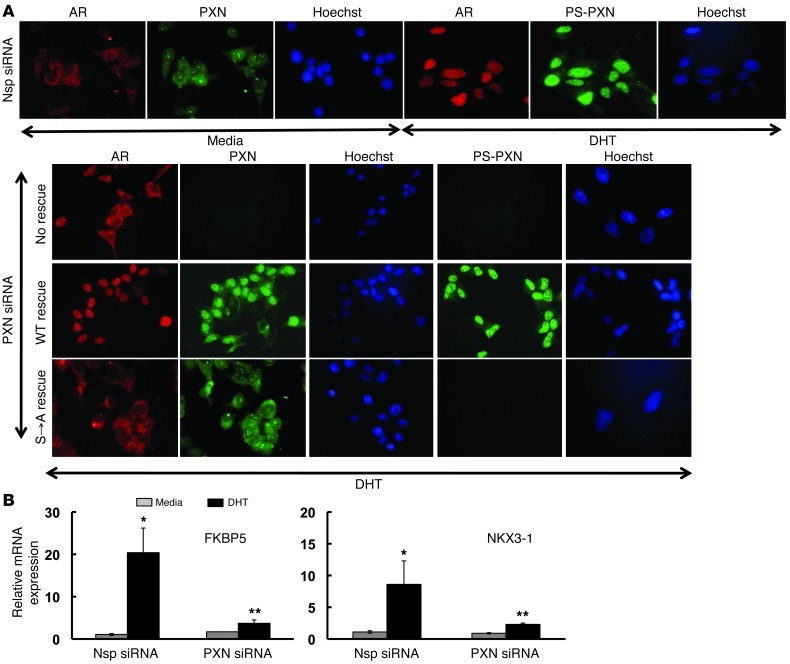
Importantly, we performed the same experiments in an AR-positive, “castration-resistant” prostate cancer cell line (C4-2). Similar to LNCaP cells, PS-PXN regulated AR nuclear localization (Figure 3A) as well as DHT-induced FKBP5 and NKX3-1 mRNA expression in C4-2 cells (Figure 3B), suggesting that PXN may be a common regulator of AR actions in prostate cancer. We confirmed subcellular localization of AR and PXN in C4-2 cells by Western blot (Supplemental Figure 1B).
Figure 3. In C4-2 cells, PXN regulates AR nuclear localization and androgen-induced mRNA expression.
(A) PXN is required for AR nuclear localization. Immunofluorescence of AR (red), PXN (green), or PS-PXN (green) in cells treated with Nsp or PXN siRNA, followed by treatment with medium or DHT (25 nM; 30 minutes). PXN-knockdown cells were transfected with plasmids expressing WT PXN or PXN(S→A). Original magnification, ×40. (B) PXN regulates androgen-induced gene expression. Relative expression of DHT-induced FKBP5 and NKX3-1 mRNAs by qPCR in Nsp or siRNA-mediated PXN-knockdown in C4-2 cells. Data are represented as mean ± SEM (n = 3). *P ≤ 0.001; **P ≤ 0.05 relative to Nsp siRNA.
PXN regulates AR nuclear localization in primary mouse granulosa cells.
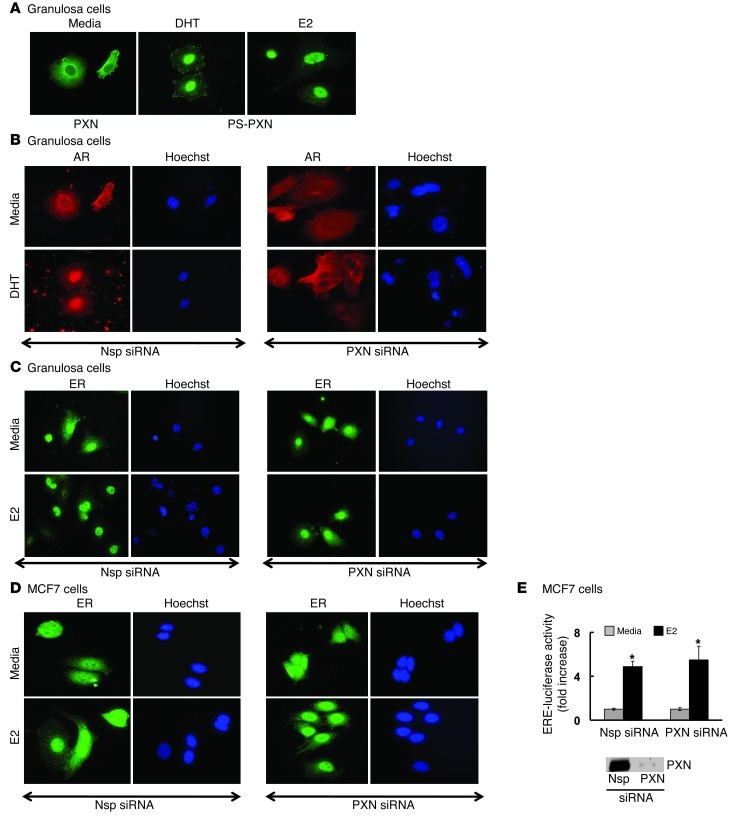
Since PXN was necessary for DHT-induced AR nuclear localization in prostate cancer cells, we determined whether it functioned similarly in AR-dependent cells that were neither prostate nor tumor cell lines. Therefore, we examined primary mouse granulosa cells (GCs), where AR plays an important role in ovarian function (12, 13). Interestingly, as seen in prostate cancer cell lines, both PXN (Figure 4A) and AR (Figure 4B) were primarily cytoplasmic under basal conditions in primary GCs (1.8% ± 0.3% and 12.9% ± 0.7% of cells with nuclear PXN and AR, respectively). As expected, PS-PXN (89.4% ± 1.3% of cells; Figure 4A) and AR (98.2% ± 0.3% cells; Figure 4B) became exclusively nuclear with DHT stimulation. Furthermore, as in prostate cancer cells, knockdown of PXN (PXN siRNA) expression in primary GCs abrogated nuclear localization of AR (Figure 4B) in response to DHT (24.05% ± 1.4% of cells with nuclear AR) compared with Nsp siRNA (98.2 ± 0.3% of cells with nuclear AR, P ≤ 0.05). This result suggests that PXN may be a general regulator of DHT-induced AR nuclear retention regardless of cell type.
Figure 4. PXN specifically regulates AR nuclear localization in primary GCs.
Primary GCs from C57BL/6J mouse ovaries were treated with DHT or estradiol (25 nM) for 30 minutes. (A) Immunofluorescence studies (n = 3 experiments with identical results) showed that, under basal conditions (medium), PXN is predominantly cytoplasmic. Under DHT or estradiol (E2) stimulation, PS-PXN is primarily nuclear. Adjacent Hoechst staining represents the nucleus. (B and C) Immunofluorescence studies (n = 3 experiments with identical results) of primary GCs treated with Nsp or PXN-specific siRNA. PXN ablation prevents DHT-induced nuclear translocation of AR (B), but has no effect on nuclear localization of ERα in medium or estradiol-treated cells (C). (D) Immunofluorescence studies (n = 3 experiments with identical results) in MCF7 breast cancer cells showing siRNA-mediated knockdown of PXN has no effect on ERα nuclear localization in medium or estradiol-treated cells. Original magnification, ×40. (E) PXN is not required for ERE-mediated transcription. Nsp or PXN-specific siRNA treated MCF7 cells were transiently transfected with ERE reporter luciferase construct plus cytomegalovirus–β-gal plasmid. ERE-luciferase activity was normalized to β-gal expression and data represented as fold increase with respect to medium treatment (mean ± SEM, n = 3). *P ≤ 0.001 relative to medium. The blot on the right shows PXN knockdown.
PXN has been suggested as mediating nongenomic estrogen signaling in breast cancer cells (14). We therefore tested PXN’s role in regulating ERα nuclear localization in mouse primary GCs (Figure 4C). ERα was expressed primarily in the nucleus both in the absence (80% ± 1.7% of cells) and presence (93.6% ± 0.3% of cells) of estradiol. Although estradiol triggered PS-PXN nuclear expression (98.4% ± 1.4% of cells; Figure 4A), knockdown of PXN expression by siRNA had no effect on ERα subcellular localization (Figure 4C). Similar studies in MCF7 breast cancer cells also showed that PXN knockdown had no effect on ERα nuclear localization (Figure 4D). Importantly, PXN knockdown did not reduce estrogen-induced luciferase activity in MCF7 cells expressing an estrogen response element–luciferase (ERE-luciferase) construct (Figure 4E). Therefore, PXN exerts selective effects on steroid hormone–regulated transcription, being critical for AR- but not ERα-mediated nuclear retention and transcriptional activation.
PXN modulates ERK-mediated transcription.
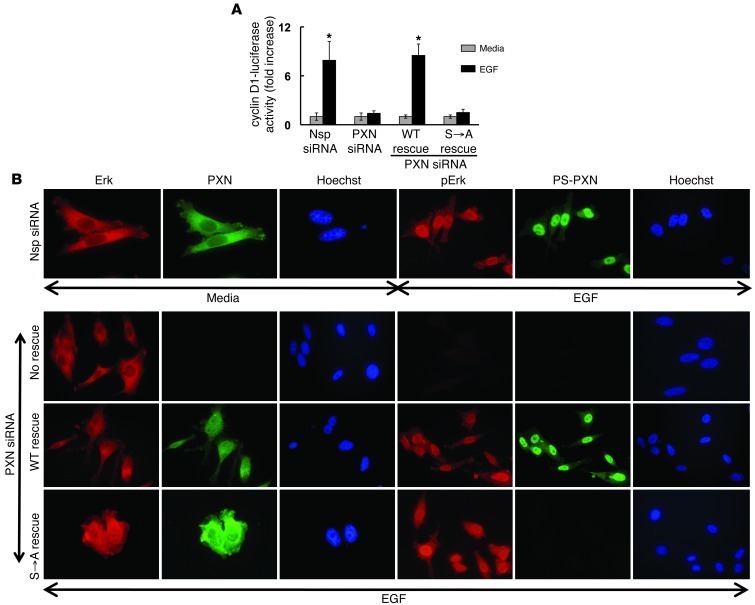
DHT-induced activation of extranuclear ARs leads to transactivation of the EGFR and subsequent MAPK activation (8, 10, 14). Furthermore, nuclear PS-PXN functions downstream of ERK to regulate AR-mediated transcription (Figure 1). Since PXN functions downstream of the EGFR and ERK in response to DHT, we reasoned that PXN might in fact be a general regulator of ERK signaling in response to EGF, independent of androgens. We therefore used a cyclin D1 promoter–luciferase construct to determine whether PXN is required for EGF-induced cyclin D1 promoter activity (Figure 5A). Knockdown of PXN expression by siRNA abrogated EGF-induced cyclin D1 promoter activity. Reexpression of WT PXN, but not PXN(S→A), rescued EGF-induced cyclin D1 promoter activity (Figure 5A). These results suggest that, as with DHT-induced PSA promoter activity, ERK-dependent phosphorylation of PXN is required for EGF-induced cyclin D1 promoter activity. Confirmation of PXN knockdown and rescue in PC3 cells were determined by Western blot (Supplemental Figure 3B).
Figure 5. PXN modulates ERK-mediated transcription independent of androgens.
(A) PXN is required for EGF-induced cyclin D1 promoter activity. Nsp/PXN-specific siRNA-treated PC3 cells were transiently transfected with cyclin D1 promoter-luciferase plus cytomegalovirus–β-gal plasmid, followed by EGF stimulation (20 ng/ml; 24 hours). PXN-knockdown cells were transfected with plasmids expressing WT PXN or PXN lacking the MAPK-targeted serines (S→A). Cyclin D1 promoter-luciferase activity was normalized to β-gal expression and data represented as fold increase with respect to medium (mean ± SEM, n = 3). *P ≤ 0.001. (B) Nuclear localization of ERK is independent of PS-PXN. Immunofluorescence of ERK (red), PXN (green), p-ERK (green), or PS-PXN (green) in PC3 cells treated with Nsp siRNA or PXN siRNA, followed by medium or EGF (20 ng/ml; 30 minutes) stimulation. PXN-knockdown cells were transfected with plasmids expressing WT PXN or PXN lacking the MAPK-targeted serine (S→A). Adjacent Hoechst staining represents the nucleus (n = 5 experiments, all with identical results). Original magnification, ×40.
Since nuclear PS-PXN is required for AR nuclear localization, we determined whether PXN might also regulate ERK nuclear localization (Figure 5B). While ERK and PXN (Nsp siRNA–medium; Figure 5B) were predominantly cytoplasmic in serum-starved PC3 cells (7.6% ± 0.9% and 2.9% ± 0.3% of cells contained nuclear ERK and PXN, respectively), EGF induced (Nsp siRNA–EGF; Figure 5B) nuclear expression of both phosphorylated ERK (p-ERK) and PS-PXN (89.7% ± 1.4% and 95.4% ± 2.2% of cells for p-ERK and nuclear PS-PXN, respectively). Upon siRNA-mediated ablation of PXN, ERK was no longer activated/phosphorylated and did not translocate into the nucleus in response to EGF (PXN siRNA–no rescue; Figure 5B). Interestingly, reexpression of either WT PXN or PXN(S→A) (WT and S→A rescue; Figure 5B) restored nuclear localization of p-ERK in response to EGF stimulation (98.2% ± 0.2% and 85.1% ± 0.6%, respectively). Therefore, unlike DHT-induced AR trafficking, EGF-induced nuclear localization of ERK requires PXN but not PS-PXN. We confirmed subcellular localization of ERK and PXN by Western blot (Supplemental Figure 1C).
PS-PXN complexes with ERK in the nucleus to regulate c-FOS mRNA expression.
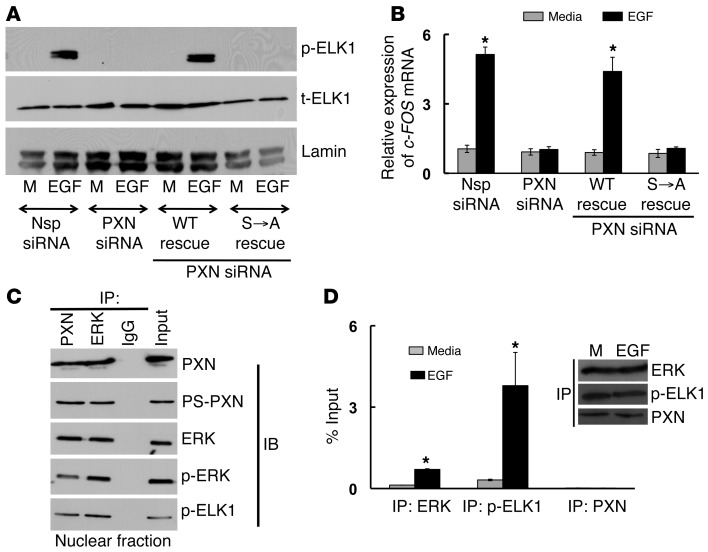
Although MAPK-dependent phosphorylation of PXN is not required for ERK nuclear localization, it is still necessary to activate the cyclin D1 promoter (Figure 5A). ELK1 is an ERK nuclear target that mediates transcription (15). For example, phosphorylated ELK1 (p-ELK1) increases expression of the early response gene c-FOS, which in turn regulates expression of cyclin D1 and other genes (16). In PC3 cells, knockdown of PXN eliminated EGF-induced phosphorylation of ELK1 (Figure 6A) as well as c-FOS mRNA expression (Figure 6B), both of which could be rescued upon reexpression of WT PXN but not PXN(S→A). Therefore, MAPK-dependent phosphorylation of PXN is required for ERK-mediated phosphorylation of ELK1 and subsequent ELK1 actions on the c-FOS promoter. Co-IP studies from nuclear extracts of EGF stimulated PC3 cells, demonstrated a complex among PS-PXN, p-ERK, and p-ELK1, regardless of whether PXN or ERK was initially precipitated (Figure 6C). Furthermore, ChIP revealed that both p-ELK1 and ERK associated with the c-FOS promoter in an EGF-dependent fashion (Figure 6D). However, unlike PXN regulation of AR actions, PXN did not bind to the c-FOS promoter in EGF-stimulated PC3 cells (Figure 6D). Together, these observations suggest that, although PS-PXN is not required for ERK nuclear localization and does not directly bind to the c-FOS promoter, PS-PXN is still required for ERK-mediated actions in the nucleus, perhaps functioning as a nuclear scaffold to facilitate formation of an ERK and ELK1 complex. Once together, ERK-mediated phosphorylation of ELK1 induces the expression of c-FOS, which triggers expression of cyclin D1 and subsequent cell proliferation.
Figure 6. PS-PXN regulates c-FOS mRNA expression in the nucleus by complexing with ERK and ELK1 to allow ERK-mediated activation of ELK1.
(A and B) PS-PXN is essential for EGF-induced phosphorylation of ELK1 and c-FOS mRNA expression. Immunoblot (A) and qPCR (B) analysis in PC3 cells (treated with Nsp or PXN-specific siRNA) showing EGF-induced (20 ng/ml; 30 minutes) (A) phosphorylation of ELK1 and (B) c-FOS mRNA expression. PXN-knockdown cells were also transfected with plasmids expressing WT PXN or PXN lacking the MAPK-targeted serines (S→A). Data are represented as mean ± SEM (n = 3). *P ≤ 0.005 relative to medium. (C) PS-PXN forms a complex with ERK and ELK1 in the nucleus. IP assays were performed in nuclear extracts of LNCaP cells treated with EGF (20 ng/ml, 24 hours) (n = 3 experiments with identical results). Either PXN, PS-PXN, ERK, p-ERK, or p-ELK1 were precipitated, followed by immunoblotting as indicated. (D) ERK and ELK1, but not PXN, bind to the c-FOS promoter in EGF-stimulated PC3 cells. PC3 cells were serum starved overnight, followed by treatment with medium or EGF (20 ng/ml) for 24 hours, before chromatin IP. The occupancy of ERK, p-ELK1, and PXN on c-FOS promoter was examined. Values are presented as percentage input (mean ± SEM, n = 3). *P ≤ 0.005 with respect to medium. Representative immunoblot shows equal amounts of ERK, p-ELK1, and PXN were IP from medium (M) and EGF-treated samples.
Knockdown of PXN inhibits in vivo growth of human prostate cancer xenografts.
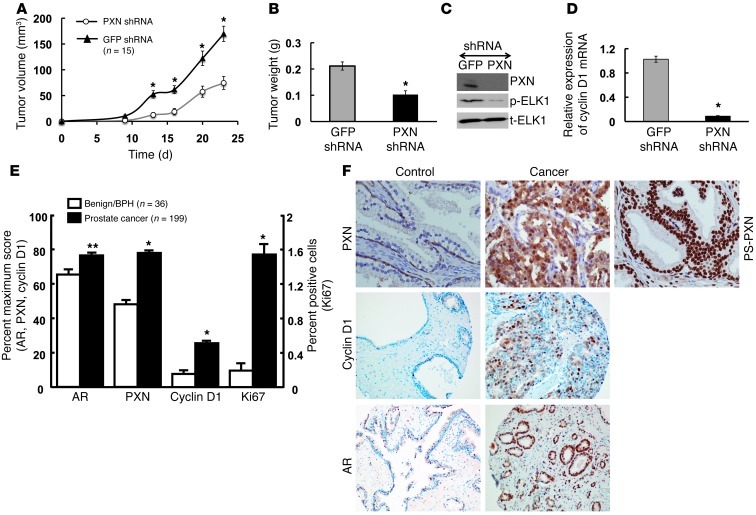
In vitro studies showed that PXN was essential for PC3 cell proliferation, migration, and invasion (8). To determine whether PXN is similarly required for prostate tumor cell growth in vivo, we stably knocked down PXN expression in PC3 cells using a lentiviral shRNA system and compared the growth of these cells to PC3 cells infected with Nsp (GFP) shRNA in immunocompromised mice. PXN knockdown significantly reduced tumor formation when compared with control PC3 cells infected with Nsp shRNA. The volume (Figure 7A) and weight (Figure 7B) of the tumors derived from PC3 cells transfected with PXN shRNA were 40%–45% lower compared with those of control groups. Notably, levels of PXN and p-ELK protein (Figure 7C) as well as cyclin D1 mRNA (Figure 7D) were markedly reduced in the PXN shRNA–treated tumors relative to control (GFP) shRNA-treated mice at the end of the experiment. These observations confirm the importance of the PXN-mediated signaling pathway for prostate cancer cell proliferation in vivo.
Figure 7. PXN knockdown inhibits prostate cancer growth and tumor development, and PXN expression is upregulated in prostate cancer patient samples.
(A–D) PXN knockdown in PC3 cells suppresses tumor formation in xenografts. Nude mice (n = 15/group) were injected subcutaneously with 0.2 ml (1.5 × 106) PC3 cells infected with PXN shRNA or GFP shRNA-negative control. (A) The growth curve of tumors bearing PC3 cell xenografts. (B) The tumor weights of prostate cancer cell xenografts. *P ≤ 0.005 comparing PXN shRNA and GFP shRNA. (C and D) ELK1 phosphorylation and cyclin D1 expression are lower in xenograft tumors lacking PXN. Protein levels of PXN, p-ELK1, and t-ELK1 (C) as well as mRNA levels of cyclin D1 (D) in excised tumors that were treated with GFP or PXN shRNA. (E and F) PXN levels and downstream effectors are upregulated in prostate cancer TMAs. TMA sections were subjected to immunohistochemistry and PXN, AR, cyclin D1, and Ki67 expression measured by scoring intensity and cell number. (E) Data represented as percentage of maximum score for AR, PXN, and cyclin D1 and as percentage of positive cells for Ki67. *P ≤ 0.0001; **P ≤ 0.001 relative to control or BPH samples. All data are represented as mean ± SEM (n = 15). (F) Representative TMA sections showing PXN, cyclin D1, and AR expression in normal/BPH and cancer samples and PS-PXN expression in cancer samples (note that PS-PXN expression was not statistically different in benign versus cancerous prostate).
PXN expression is upregulated in human prostate cancer patient samples.
Given that PXN and other downstream proliferative signaling molecules are important for prostate cancer cell proliferation both in vitro (8) and in vivo (Figure 7, A–D), we postulated that these proteins might be overexpressed in human prostate cancer tissues. Using a human TMA of human prostate biopsies that contained 199 malignant samples and 36 normal or benign prostate hyperplasia (BPH) samples, we determined the abundance of PXN by immunohistochemistry (Figure 7, E and F). Immunohistochemistry revealed primarily cytoplasmic staining of PXN in epithelial cells (Figure 7F), with a significantly higher degree of intensity and cell number scoring in the malignant (cancer) versus normal and/or BPH samples (Figure 7E). Furthermore, as shown by immunofluorescence in prostate cancer cell lines, immunohistochemistry with an antibody directed against PS-PXN revealed almost exclusive nuclear localization of PS-PXN in both benign and tumor tissues (representative photo in Figure 7F), though, unlike in total PXN, scoring of intensity and cell number did not reveal a statistically significant difference in PS-PXN expression (not shown). Notably, AR, cyclin D1, and Ki67 expression in these same TMAs were all significantly higher in the malignant (cancer) versus normal and/or BPH samples (Figure 7, E and F), confirming that the PXN-regulated signaling/proliferative pathway is upregulated in prostate tumors. Together, the xenograft studies showing the importance of PXN and associated signals for prostate cancer cell growth in vivo and the TMA data demonstrating increased expression of PXN and associated signals in human prostate cancers suggest a significant role of PXN in prostate cancer progression.
Discussion
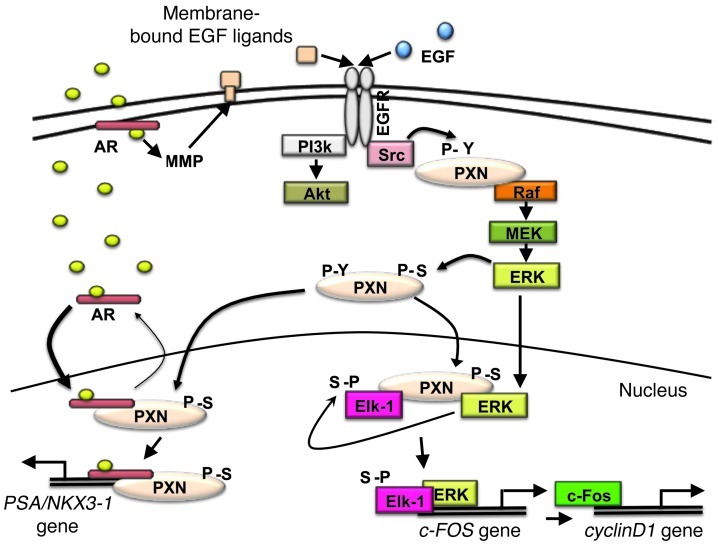
Prostate cancer cell proliferation requires stimulation from many sources. These sources include androgens and growth factors at early stages, with an emphasis on the former. However, in later castration-resistant stages, the character of androgen dependence seems to change, and prostate cancer seems to proliferate effectively in response to other signals, with perhaps more emphasis on growth factors. Here, we demonstrate that PXN is a critical regulator of both androgen- and growth factor–mediated proliferation in prostate cancer cell lines, making it a potential mediator of both androgen/castration-sensitive and castration-resistant prostate cancer. We find that PXN regulates both androgen- and growth factor–mediated cell proliferation by modulating ERK signaling and subsequent downstream effects (Figure 8).
Figure 8. PXN mediates extranuclear and nuclear AR and ERK signaling.
Androgen binds ARs at or near the plasma membrane, which transactivates the EGFR via MMP-mediated release of EGFR ligands. EGFR can also be activated directly by EGF. This activates Src, which phosphorylates PXN on tyrosine residues, enabling Raf to activate MEK/ERK. ERK then mediates phosphorylation of PXN at serine residues (though not necessarily directly), and PS-PXN enters the nucleus. Meanwhile, androgen-bound AR also translocates to the nucleus. In the nucleus, PS-PXN interacts with androgen-bound AR to retain it in the nucleus. PS-PXN then associates with or near the AR on the PSA/NKX3-1 promoter to help promote AR-driven transcription (left). Activated ERK also enters the nucleus, where PS-PXN, p-ERK, and ELK1 form a complex and ERK phosphorylates ELK1, which induces c-FOS expression. The latter then activates cyclin D1 promoter activity/cyclin D1 expression (right), thereby promoting cell proliferation.
First, PXN is required for receptor tyrosine kinase–mediated activation of ERK. We previously showed that EGFR activation (either directly by EGF or indirectly by DHT through MMP-mediated release of EGFR ligands) leads to phosphorylation of tyrosine residues on PXN, most likely by Src (8). This tyrosine phosphorylation of PXN is required for downstream Raf, MEK, and ERK activation, with PXN potentially serving as a scaffold that holds this signalosome together (8, 17, 18).
Second, PXN functions downstream of ERK as an effector of nuclear signaling. As described previously, the MAPK pathway regulates phosphorylation of PXN at serine residues, most likely while PXN resides within the cytoplasm (8). Once PXN is phosphorylated, however, we find that PS-PXN enters the nucleus, demonstrating that PXN is not only an important cytoplasmic signaling molecule, but also a critical nuclear signaling molecule. In fact, previous studies showed that PXN shuttles between the cytoplasm and the nucleus in COS and HeLa cells (19), although how this shuttling is mediated and its biological relevance/significance were not addressed. Our results provide evidence that PXN nuclear localization requires MAPK-dependent serine phosphorylation and that nuclear PS-PXN serves many functions once it enters the nucleus. Of note, although a core nuclear export signal (NES) within the LD4 motif of PXN has been reported (19), no nuclear localization signal has been identified.
Third, once in the nucleus, PS-PXN regulates several transcriptional responses (Figures 1 and 6). For example, PXN forms a complex with ligand-bound AR, enhancing its retention within the nucleus and thereby facilitating AR transcriptional responses (e.g., PSA, NKX3-1, and FKBP5). Furthermore, PXN binds to the PSA and NKX3-1 promoters along with the AR, suggesting that PXN might be a transcriptional coregulator of the AR. In contrast, we did not find PXN to associate with AR on the FKBP5 promoter. However, transcriptional regulation of FKBP5 by AR is different than that from PSA. The promoter region of FKBP5 consists of several distal androgen response elements (AREs) that are important enhancers (20, 21), which might have led to our missing the appropriate PXN-binding site. The ability of PXN to regulate AR-mediated transcription is consistent with earlier studies in CV-1 and prostate cancer cells overexpressing transfected PXN and AR, where PXN potentiated AR-mediated activation of MMTV-luciferase (22) and PSA promoter-luciferase constructs (23).
Importantly, our studies demonstrate that PXN regulates AR nuclear localization, not only in prostate cancer cells, but also in primary mouse GCs, suggesting that PXN is a common regulator of both membrane-initiated and genomic androgen signaling in multiple cell types. In contrast to the AR, PXN had no effect on ERα nuclear localization in the same primary GCs and in MCF7 breast cancer cells. Previously, PXN was implicated as a regulator of nongenomic estrogen effects in breast cancer cells (24), although transcription was not examined (25). Our data confirm that estradiol rapidly triggers serine phosphorylation of PXN and subsequent nuclear localization in granulosa and breast cancer cells. However, we find that PXN is not required for ERα-mediated activation of an ERE-luciferase construct in MCF7 cells. Together, these data indicate that PXN has different effects on AR- versus ERα-mediated signaling and suggest that extranuclear ERK signaling might be more important for transcription by AR versus ERα.
Fourth, PXN modulates more than just AR-mediated transcription. Independent of steroids or steroid receptors, PXN regulates cytoplasmic ERK activation and subsequent nuclear ERK-dependent transcription (Figure 7), providing evidence that PXN is in fact a broad regulator of ERK actions inside the nucleus. ERK is known to translocate to the nucleus and regulate gene expression (26). Our study provides insight into the intranuclear mechanism of ERK-regulated gene expression, demonstrating the importance of nuclear PS-PXN in mediating these effects. Although EGF-induced MAPK activation requires cytoplasmic PXN (8), translocation of activated ERK to the nucleus does not require subsequent MAPK-dependent phosphorylation of PXN. However, PS-PXN is still required for nuclear ERK actions, in which ERK, PS-PXN, and ELK1 form a complex that permits ERK-mediated phosphorylation of ELK1. Activated ELK1 and ERK, but not PXN, then bind to the c-FOS promoter to induce c-FOS expression (Figure 7). As reported (15, 16), c-FOS can then activate cyclin D1 promoter activity and cyclin D1 expression, thereby promoting cell proliferation. Thus, while the cytoplasmic scaffolding properties of PXN, specifically with regard to ERK signaling, are well established, our data demonstrate that PXN serves a similar function in the nucleus, forming a platform that enables ERK to interact with and phosphorylate ELK1. Given the general importance of ERK, the physiological significance of PXN regulation of ERK actions may extend beyond cancer and in fact apply to normal development and function of many tissues. Accordingly, global knockout of PXN in mice causes embryonic lethality (27), confirming PXN’s physiological importance.
Finally, our in vivo xenograft and TMA studies reinforce our in vitro results and establish PXN’s potential significance in prostate cancer progression. Knockdown of PXN expression in prostate cancer cells leads to decreased and slower xenograft tumor growth, demonstrating the importance of PXN in prostate cancer cell proliferation in vivo. Furthermore, TMA studies demonstrate increased PXN expression in prostate tumor samples, consistent with previous work demonstrating elevated PXN levels in premalignant lung lesions (28, 29) and colorectal tumors (30). Most importantly, our results show that the impact of PXN on cyclin D1 expression, ELK1 phosphorylation, and proliferative index (Ki67) is conserved between in vitro and in vivo studies with prostate cancer cell lines as well as in human TMAs, confirming the importance of PXN-mediated signaling for prostate cancer progression. Of note, under the conditions of our TMA studies, although we saw a trend of increased PS-PXN expression in tumors versus benign prostate (not shown), the difference did not reach statistical significance. This is likely due to the combination of PS-PXN concentrating in the small nuclear volume and the high affinity of our antibody to PS-PXN leading to a cluster of high scores that made discerning significant differences difficult. Further examination of larger data sets may reveal statistically significant difference in PS-PXN expression between cancerous and benign prostate as well as between different Gleason grades of prostate cancer.
In summary, our model (Figure 8) highlights 2 major concepts regarding PXN’s role in ERK and AR signaling: (a) PXN has functions beyond focal adhesion signaling and acts as a liaison between extranuclear kinase signaling and intranuclear transcription; and (b) just as PXN serves as a signaling scaffold molecule in the cytoplasm, it functions as a signaling scaffold molecule in the nucleus. The data here are consistent with PXN being a critical mediator of kinase-regulated transcription and subsequent prostate cancer cell proliferation in both castration-sensitive and castration-resistant prostate cancer, thereby establishing PXN as a potential biomarker for prostate cancer proliferation and an attractive therapeutic target for prostate cancer treatment.
Methods
Cell culture.
LNCaP, C4-2, PC3, and MCF7 cells were obtained from ATCC and cultured in RPMI 1640 (Invitrogen) containing 10% FBS and 1% penicillin-streptomycin. Primary GCs isolated from C57BL/6J mouse ovaries were cultured in DMEM/F-12 containing 5% FBS, 1% penicillin-streptomycin, and 2.5 μg/ml fungizone for 48 hours (31, 32); this was followed by serum starvation for 4 hours and then DHT or estradiol (25 nM) treatment for 30 minutes.
PXN knockdown (siRNA) and rescue experiments.
siRNA experiments were performed (8) using nontargeting siRNA pool or human PXN siRNA ON-TARGET plus SMARTpool containing 4 siRNAs targeting PXN mRNA according to the manufacturer’s instructions (Thermo Fisher Scientific). After 72 hours, cells were treated overnight with serum-free, phenol red–free RPMI 1640 and stimulated with or without DHT (25 nM, LNCaP cells) or EGF (20 ng/ml, PC3 cells).
For rescue experiments, after siRNA, cells were transfected (FuGENE 6; Roche) with WT PXN or serine mutated PXN(S→A) (serines 83, 126, 130 converted to alanines), as described (8). After 24 hours, cells were treated overnight with serum-free, phenol red–free RPMI 1640 followed by stimulation with or without DHT (25 nM) or EGF (20 ng/ml) for 30 minutes or 24 hours. For leptomycin B experiments, cells were preincubated with 15 ng/μl of leptomycin B for 30 minutes prior to medium/DHT treatment (30 minutes).
Western blots.
Western blots were performed as described (33). Primary antibodies used were as follows: rabbit polyclonal PXN and mouse monoclonal anti-AR (441) at 1:1,000 (Santa Cruz Biotechnology Inc.); rabbit polyclonal PXN–phospho-S126 (Invitrogen); rabbit polyclonal PXN-phospho-S83 (ECM Biosciences); rabbit polyclonal p44/42-ERK1/2, phospho-p44/42-ERK1/2 (T202/Y204), phospho-ELK1 (Ser383), and lamin at 1:1,000 and GAPDH at 1:5,000 (Cell Signaling Technology).
Nuclear and cytoplasmic fractionation.
Separation of nuclear and cytoplasmic fractions was performed using NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific) according to the manufacturer’s protocol. Expression of GAPDH and lamin were used as cytoplasmic and nuclear controls, respectively.
ChIP.
LNCaP and PC3 cells were serum starved overnight, then treated with DHT (25 nM) or EGF (20 ng/ml) for 45 minutes or 24 hours, respectively, before ChIP with MAGnify Chromatin Immunoprecipitation System (Invitrogen) according to the manufacturer’s instructions. Chromatin fragments were immunoprecipitated with Dynabeads coupled with anti-PXN or anti-AR (Santa Cruz Biotechnology Inc.) antibodies for LNCaP cells and anti-PXN (Santa Cruz Biotechnology Inc.), anti-ERK1/2, or anti–p-ELK1 (Cell Signaling Technology) antibodies for PC3 cells. For controls, cells were pretreated with rabbit IgG, and qPCR was performed using EXPRESS SYBR GreenER qPCR SuperMixes (Invitrogen) with primers designed for PSA promoter (34), NKX3-1 (35), FKBP5 (21), c-FOS promoter (SimpleChIP human c-FOS promoter primers; Cell Signaling Technology), and negative control regions. Amplification efficiency of the qPCR reaction was calculated using 10-fold serial dilutions of input DNA, and quantification of precipitated DNA was analyzed by the ΔΔCt method and normalized to input controls. Input values were obtained from samples treated in the same way, but with no IP step.
ChIP–re-ChIP assay.
LNCaP cells were serum starved overnight; this was followed by treatment with medium or DHT (25 nM) for 45 minutes. Thereafter, ChIP analysis followed by re-ChIP was performed with Re-ChIP-IT Magnetic ChIP Kit (Active Motif) according to the manufacturer’s instructions. Chromatin fragments were immunoprecipitated first with anti-PXN antibody; this was followed by reimmunoprecipitation with anti-AR antibody (Santa Cruz Biotechnology Inc.). IgG and no-antibody IP were used as negative controls. Processed DNA samples were used for qPCR evaluation using EXPRESS SYBR GreenER qPCR SuperMixes.
Xenograft studies.
Recombinant lentivirus expressing an shRNA directed against human PXN (NM_002859.2) was prepared from 293T cells (36) by the University of Pittsburgh Cancer Institute Lentiviral Core Facility. PC-3 cells grown on 6-well plates were infected with 106 virus particles in the presence of 8 μg/ml polybrene overnight at 37°C and pooled clones selected in 2 μg/ml puromycin.
NOD.CB17-Prkdcscid/J mice (15 per group) were injected subcutaneously into flanks with 0.2 ml (1.5 × 106 cells) PC-3 cells transfected with PXN shRNA or GFP shRNA–negative control. Tumor volumes were measured every 3 days. Mice were sacrificed at 4 weeks and tumor weights measured. Expression of PXN, p-ELK1, and total ELK1 (t-ELK1) were confirmed by Western blot, while cyclin D1 mRNA expression was measured by qPCR.
Transcription (luciferase) assay.
Transcription assays were performed as described (37, 38). Briefly, LNCaP, PC3, or MCF7 cells were treated with Nsp or PXN siRNA for 72 hours. Thereafter cells were transfected with reporter luciferase constructs containing the PSA promoter (LNCaP cells), the cyclin D1 promoter (PC3 cells; promoter from Karen Knudsen, Thomas Jefferson University, Philadelphia, Pennsylvania, USA), or ERE (for MCF7 cells) plus a cytomegalovirus–β-gal plasmid in medium containing 5% charcoal-stripped fetal bovine serum using FuGENE 6 (Roche) according to the manufacturer’s protocol. For rescue experiments, WT and serine mutated PXN(S→A) were transfected. The next day, cells were treated with and without DHT (25 nM, LNCaP cells), EGF (20 ng/ml, PC3 cells), or estradiol (50 nM, MCF7 cells) for 24 hours and extracts analyzed for luciferase and β-gal expression using the Promega Luciferase Assay System (Promega Corp.) and PerkinElmer Galacto-Star Kit. PSA-luciferase activity was normalized to β-gal expression and data represented as fold increase with respect to medium. To confirm PXN knockdown and rescue, cell lysates were subjected to Western blot analysis.
IP.
IP experiments were done with nuclear fraction (nuclear and cytoplasmic fractionation) using protein G–agarose beads (Millipore) according to the manufacturer’s instructions. Antibodies used for IP were as follows: mouse monoclonal AR (no. 441), p-ERK1/2 (no. 12D4), PXN (no. 177), and IgG (control) antibodies (Santa Cruz Biotechnology Inc).
Immunofluorescence.
Cells were grown in poly-D-lysine precoated glass bottom dishes (MatTech Corp.). Cells were treated with PXN siRNA and rescued with WT or serine mutated PXN(S→A) followed by serum starvation and stimulation with DHT or EGF for 30 minutes. Cells were fixed in 4% paraformaldehyde and dual immunofluorescence microscopy performed (39). Cells were stained with rabbit polyclonal PXN (1:100) and mouse monoclonal AR (1:50) (Santa Cruz Biotechnology Inc.); rabbit polyclonal PXN-phospho-S126 (1:50; Invitrogen); rabbit polyclonal PXN-phospho-S83 (1:50; ECM Biosciences); rabbit polyclonal p44/42-ERK1/2 (1:100), phospho-p44/42-ERK1/2 (T202/Y204) (1:100; Cell Signaling Technology), and ERα (1:10). Binding of primary antibody was detected by reporter IgG conjugated to Texas Red (anti-mouse IgG) and fluorescein (anti-rabbit IgG) (Vector Laboratories). For all immunofluorescence studies, cells were counted from 3 random fields/experiment (each experiment was performed at least 3 times) and cytoplasmic versus nuclear localization of protein of interest scored. Data are represented as percentage of cells.
TMAs.
Tumors were sampled from radical prostatectomy specimens from 199 prostate cancer patients with localized disease and 36 normal or BPH patients. All patient samples were obtained from the New York University Cooperative Prostate Cancer Tissue Resource. An intermediate-density TMA was assembled using a semi-automated tissue puncher/array (Beecher Instruments) (40). Four core samples of prostate cancer tissue were acquired per case. TMA sections were cut at 4-μm–thick intervals. Every tenth section was stained with H&E to verify histology. PXN, AR, cyclin D1, Ki67, and PS-PXN expression were manually scored by 2 uropathologists. The score for each core was assigned a number between 0 and 8 using a distribution–intensity scoring system (41). Four replicates of each TMA slide were averaged.
Statistics.
Unless otherwise indicated, data are presented as mean ± SEM. Data for the transcription assays, immunofluorescence scoring, real-time PCR, ChIP, and re-ChiP experiments as well as for the xenograft and TMA studies were analyzed using Students’s t test. A value of P ≤ 0.05 was considered significant.
Study approval.
All animal studies reported were approved by the University Committee on Animal Resources at the University of Rochester Medical Center, which is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. All human tissues were collected with institutional review board approval, and patients gave informed consent.
Supplementary Material
Acknowledgments
We thank Anindita Biswas for technical assistance with the generation of paxillin mutants. This work was supported by NIH grant DK59913 (to S.R. Hammes).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(7):2469–2481. doi:10.1172/JCI62044.
References
- 1.Zhu ML, Kyprianou N. Androgen receptor and growth factor signaling cross-talk in prostate cancer cells. Endocr Relat Cancer. 2008;15(4):841–849. doi: 10.1677/ERC-08-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharifi N, McPhaul MJ, Auchus RJ. “Getting from here to there”—mechanisms and limitations to the activation of the androgen receptor in castration-resistant prostate cancer. J Investig Med. 2010;58(8):938–944. doi: 10.231/JIM.0b013e3181ff6bb8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Traish AM, Morgentaler A. Epidermal growth factor receptor expression escapes androgen regulation in prostate cancer: a potential molecular switch for tumour growth. Br J Cancer. 2009;101(12):1949–1956. doi: 10.1038/sj.bjc.6605376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montgomery RB, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 2008;68(11):4447–4454. doi: 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaoka M, Hara T, Kusaka M. Overcoming persistent dependency on androgen signaling after progression to castration-resistant prostate cancer. Clin Cancer Res. 2010;16(17):4319–4324. doi: 10.1158/1078-0432.CCR-10-0255. [DOI] [PubMed] [Google Scholar]
- 6.Bennett NC, Gardiner RA, Hooper JD, Johnson DW, Gobe GC. Molecular cell biology of androgen receptor signalling. Int J Biochem Cell Biol. 2010;42(6):813–827. doi: 10.1016/j.biocel.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 7.Culig Z, et al. Androgen receptor activation in prostatic tumor cell lines by insulin-like growth factor-I, keratinocyte growth factor, and epidermal growth factor. Cancer Res. 1994;54(20):5474–5478. [PubMed] [Google Scholar]
- 8.Sen A, O’Malley K, Wang Z, Raj GV, Defranco DB, Hammes SR. Paxillin regulates androgen—and epidermal growth factor—induced MAPK signaling and cell proliferation in prostate cancer cells. J Biol Chem. 2010;285(37):28787–28795. doi: 10.1074/jbc.M110.134064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sen A, Prizant H, Hammes SR. Understanding extranuclear (nongenomic) androgen signaling: what a frog oocyte can tell us about human biology. Steroids. 2011;76(9):822–828. doi: 10.1016/j.steroids.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Migliaccio A, et al. Crosstalk between EGFR and extranuclear steroid receptors. Ann N Y Acad Sci. 2006;1089:194–200. doi: 10.1196/annals.1386.006. [DOI] [PubMed] [Google Scholar]
- 11.Unni E, et al. Changes in androgen receptor nongenotropic signaling correlate with transition of LNCaP cells to androgen independence. Cancer Res. 2004;64(19):7156–7168. doi: 10.1158/0008-5472.CAN-04-1121. [DOI] [PubMed] [Google Scholar]
- 12.Sen A, Hammes SR. Androgens: they don’t just make a man out of you. Expert Rev Obstet Gynecol. 2011;6(1):23–36. doi: 10.1586/eog.10.73. [DOI] [Google Scholar]
- 13.Sen A, Hammes SR. Granulosa cell-specific androgen receptors are critical regulators of ovarian development and function. Mol Endocrinol. 2010;24(7):1393–1403. doi: 10.1210/me.2010-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auricchio F, Migliaccio A, Castoria G. Sex-steroid hormones and EGF signalling in breast and prostate cancer cells: targeting the association of Src with steroid receptors. Steroids. 2008;73(9–10):880–884. doi: 10.1016/j.steroids.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 15.Zhang HM, et al. Mitogen-induced recruitment of ERK and MSK to SRE promoter complexes by ternary complex factor Elk-1. Nucleic Acids Res. 2008;36(8):2594–2607. doi: 10.1093/nar/gkn099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein EA, Assoian RK. Transcriptional regulation of the cyclin D1 gene at a glance. J Cell Sci. 2008;121(pt 23):3853–3857. doi: 10.1242/jcs.039131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown MC, Turner CE. Paxillin: adapting to change. Physiol Rev. 2004;84(4):1315–1339. doi: 10.1152/physrev.00002.2004. [DOI] [PubMed] [Google Scholar]
- 18.Deakin NO, Turner CE. Paxillin comes of age. J Cell Sci. 2008;121(pt 15):2435–2444. doi: 10.1242/jcs.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong JM, Lau LS, Ng YW, Lim L, Manser E. Paxillin nuclear-cytoplasmic localization is regulated by phosphorylation of the LD4 motif: evidence that nuclear paxillin promotes cell proliferation. Biochem J. 2009;418(1):173–184. doi: 10.1042/BJ20080170. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Lou Z, Wang L. The role of FKBP5 in cancer aetiology and chemoresistance. Br J Cancer. 2011;104(1):19–23. doi: 10.1038/sj.bjc.6606014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makkonen H, Kauhanen M, Paakinaho V, Jaaskelainen T, Palvimo JJ. Long-range activation of FKBP51 transcription by the androgen receptor via distal intronic enhancers. Nucleic Acids Res. 2009;37(12):4135–4148. doi: 10.1093/nar/gkp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasai M, Guerrero-Santoro J, Friedman R, Leman ES, Getzenberg RH, DeFranco DB. The Group 3 LIM domain protein paxillin potentiates androgen receptor transactivation in prostate cancer cell lines. Cancer Res. 2003;63(16):4927–4935. [PubMed] [Google Scholar]
- 23.Obinata D, et al. ARFGAP3, an androgen target gene, promotes prostate cancer cell proliferation and migration. Int J Cancer. 2012;130(10):2240–2248. doi: 10.1002/ijc.26224. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, et al. Estrogen stimulation of cell migration involves multiple signaling pathway interactions. Endocrinology. 2010;151(11):5146–5156. doi: 10.1210/en.2009-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almeida M, Han L, Ambrogini E, Bartell SM, Manolagas SC. Oxidative stress stimulates apoptosis and activates NF-kappaB in osteoblastic cells via a PKCbeta/p66shc signaling cascade: counter regulation by estrogens or androgens. Mol Endocrinol. 2010;24(10):2030–2037. doi: 10.1210/me.2010-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madak-Erdogan Z, Lupien M, Stossi F, Brown M, Katzenellenbogen BS. Genomic collaboration of estrogen receptor alpha and extracellular signal-regulated kinase 2 in regulating gene and proliferation programs. Mol Cell Biol. 2011;31(1):226–236. doi: 10.1128/MCB.00821-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagel M, et al. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol Cell Biol. 2002;22(3):901–915. doi: 10.1128/MCB.22.3.901-915.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackinnon AC, et al. Paxillin expression and amplification in early lung lesions of high-risk patients, lung adenocarcinoma and metastatic disease. J Clin Pathol. 2011;64(1):16–24. doi: 10.1136/jcp.2010.075853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jagadeeswaran R, et al. Paxillin is a target for somatic mutations in lung cancer: implications for cell growth and invasion. Cancer Res. 2008;68(1):132–142. doi: 10.1158/0008-5472.CAN-07-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, et al. Identification and functional characterization of paxillin as a target of protein tyrosine phosphatase receptor T. Proc Natl Acad Sci U S A. 2010;107(6):2592–2597. doi: 10.1073/pnas.0914884107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peluso JJ, Pappalardo A. Progesterone regulates granulosa cell viability through a protein kinase G-dependent mechanism that may involve 14-3-3sigma. Biol Reprod. 2004;71(6):1870–1878. doi: 10.1095/biolreprod.104.031716. [DOI] [PubMed] [Google Scholar]
- 32.Hernandez Gifford JA, Hunzicker-Dunn ME, Nilson JH. Conditional deletion of beta-catenin mediated by Amhr2cre in mice causes female infertility. Biol Reprod. 2009;80(6):1282–1292. doi: 10.1095/biolreprod.108.072280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evaul K, Hammes SR. Cross-talk between G protein-coupled and epidermal growth factor receptors regulates gonadotropin-mediated steroidogenesis in Leydig cells. J Biol Chem. 2008;283(41):27525–27533. doi: 10.1074/jbc.M803867200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonit M, et al. Hormone depletion-insensitivity of prostate cancer cells is supported by the AR without binding to classical response elements. Mol Endocrinol. 2011;25(4):621–634. doi: 10.1210/me.2010-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan PY, Chang CW, Chng KR, Wansa KD, Sung WK, Cheung E. Integration of regulatory networks by NKX3-1 promotes androgen-dependent prostate cancer survival. Mol Cell Biol. 2012;32(2):399–414. doi: 10.1128/MCB.05958-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubinson DA, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33(3):401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 37.Lutz LB, Jamnongjit M, Yang WH, Jahani D, Gill A, Hammes SR. Selective modulation of genomic and nongenomic androgen responses by androgen receptor ligands. Mol Endocrinol. 2003;17(6):1106–1116. doi: 10.1210/me.2003-0032. [DOI] [PubMed] [Google Scholar]
- 38.Evaul K, Jamnongjit M, Bhagavath B, Hammes SR. Testosterone and progesterone rapidly attenuate plasma membrane Gbetagamma-mediated signaling in Xenopus laevis oocytes by signaling through classical steroid receptors. Mol Endocrinol. 2007;21(1):186–196. doi: 10.1210/me.2006-0301. [DOI] [PubMed] [Google Scholar]
- 39.Razandi M, Pedram A, Rubin T, Levin ER. PGE2 and PGI2 inhibit ET-1 secretion from endothelial cells by stimulating particulate guanylate cyclase. Am J Physiol. 1996;270(4 pt 2):H1342–H1349. doi: 10.1152/ajpheart.1996.270.4.H1342. [DOI] [PubMed] [Google Scholar]
- 40.Matysiak BE, et al. Simple, inexpensive method for automating tissue microarray production provides enhanced microarray reproducibility. Appl Immunohistochem Mol Morphol. 2003;11(3):269–273. doi: 10.1097/00129039-200309000-00011. [DOI] [PubMed] [Google Scholar]
- 41.Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17(5):1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.