Abstract
Cardiac conduction system (CCS) disease, which results in disrupted conduction and impaired cardiac rhythm, is common with significant morbidity and mortality. Current treatment options are limited, and rational efforts to develop cell-based and regenerative therapies require knowledge of the molecular networks that establish and maintain CCS function. Recent genome-wide association studies (GWAS) have identified numerous loci associated with adult human CCS function, including TBX5 and SCN5A. We hypothesized that TBX5, a critical developmental transcription factor, regulates transcriptional networks required for mature CCS function. We found that deletion of Tbx5 from the mature murine ventricular conduction system (VCS), including the AV bundle and bundle branches, resulted in severe VCS functional consequences, including loss of fast conduction, arrhythmias, and sudden death. Ventricular contractile function and the VCS fate map remained unchanged in VCS-specific Tbx5 knockouts. However, key mediators of fast conduction, including Nav1.5, which is encoded by Scn5a, and connexin 40 (Cx40), demonstrated Tbx5-dependent expression in the VCS. We identified a TBX5-responsive enhancer downstream of Scn5a sufficient to drive VCS expression in vivo, dependent on canonical T-box binding sites. Our results establish a direct molecular link between Tbx5 and Scn5a and elucidate a hierarchy between human GWAS loci that affects function of the mature VCS, establishing a paradigm for understanding the molecular pathology of CCS disease.
Introduction
The cardiac conduction system (CCS) consists of a network of specialized cardiomyocytes that generate and propagate the electrical impulses that organize cardiac contraction. The CCS is composed of the slowly propagating atrial nodes, including the sinoatrial (SA) and atrioventricular (AV) nodes, and the rapidly propagating ventricular conduction system (VCS), including the AV (His) bundle and right and left bundle branches. The VCS is uniquely adapted for fast conduction in order to rapidly transmit the electrical impulse governing ventricular contraction from the AV node to the ventricular apex. Disorders of the VCS are common, carry significant morbidity, and are poorly understood from a molecular perspective.
The transcriptional networks required to maintain function of the adult CCS are undefined. Our current understanding of the molecular mediators of CCS function stems largely from heritable monogenic disorders and mouse models that have identified a limited number of genes essential for maintaining cardiac rhythm, most of which encode ion channels and their interacting partners (reviewed in ref. 1). Similar approaches have also begun to uncover the transcriptional networks required for CCS development (reviewed in ref. 2). Recent genome-wide association studies (GWAS) have identified loci implicated in ECG interval variation (3–8), providing candidate genes with potentially important roles in CCS function in the general population. Specifically, numerous loci near genes encoding ion channels and transcription factors have been associated with PR and QRS interval variation, reflecting VCS function. Although these data identify a number of candidate loci that may play important roles in VCS function, a challenge for the field is to determine the functional importance of such genomic variation and identify molecular pathways that integrate GWAS loci into a mechanistic understanding of VCS function. Notably, a core set of loci identified consistently by multiple studies has emerged. Specifically, variation near TBX5 and SCN5A has been consistently implicated in VCS function, prioritizing these loci for further functional studies.
TBX5 is a T-box transcription factor that plays a crucial role in heart development (reviewed in ref. 9). Dominant mutations in TBX5 cause Holt-Oram syndrome in humans (10), which is characterized by developmental defects of the upper limb and heart and conduction system disease including age related AV conduction delay. The cardiac phenotype of Holt-Oram disease is largely recapitulated in Tbx5 heterozygous mice (11). Furthermore, numerous GWAS on CCS function have identified genetic variation near TBX5 that associates with PR and/or QRS interval variation (4, 5, 7, 8), which suggests that TBX5 plays a role in CCS function in the general population.
We hypothesized that TBX5 plays an essential role in the mature VCS. Efforts to identify the role of essential genes such as Tbx5 and unveil the transcriptional networks that establish and maintain mature CCS function have been hampered by the lack of CCS-specific in vivo molecular tools. We recently circumvented this hurdle for VCS study in mice by generating a tamoxifen-inducible VCS-specific Cre BAC transgenic mouse line, minKCreERT2 (12). In the present study, we found that removal of Tbx5 from the VCS in Tbx5minKCreERT2 mice resulted in sudden death, slowing of conduction through the VCS, and arrhythmias including spontaneous ventricular tachycardia. TBX5 orchestrated a molecular network required for fast conduction in the VCS, including regulation of the gap junction connexin 40 (Cx40; encoded by Gja5) and the voltage-gated sodium channel Nav1.5 (encoded by Scn5a). Here, we found a direct molecular link between TBX5 and Scn5a via a TBX5-responsive downstream enhancer that was sufficient to direct VCS-specific gene expression. Our results identified a TBX5-Scn5a molecular network essential for function of the mature VCS. The consistent identification of these genes in GWAS on CCS function highlights the importance of this pathway in regulating CCS function.
Results
Decreased survival in adult VCS-specific Tbx5 mutant mice.
The requirement for TBX5 in the mature VCS was tested using a strategy providing normal Tbx5 gene dosage during development, followed by VCS-specific deletion in mature mice. VCS-specific CreERT2 expression from the minKCreERT2 BAC transgene (12) was used to recombine conditional (floxed) Tbx5 alleles (11) in the mature VCS. A tamoxifen administration regime at 6 weeks of age inactivated TBX5 in the mature VCS of Tbx5minKCreERT2 mice. VCS-selective loss of TBX5 expression, as evaluated by immunofluorescence 4 weeks after tamoxifen treatment, was observed in the AV bundle of Tbx5minKCreERT2 adult mice, but not Tbx5fl/fl littermate controls (Figure 1). Consistent with the VCS selectivity of Cre activity in minKCreERT2 mice (12), TBX5 expression was maintained in atrial myocardium of both Tbx5minKCreERT2 and Tbx5fl/fl mice (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI62617DS1).
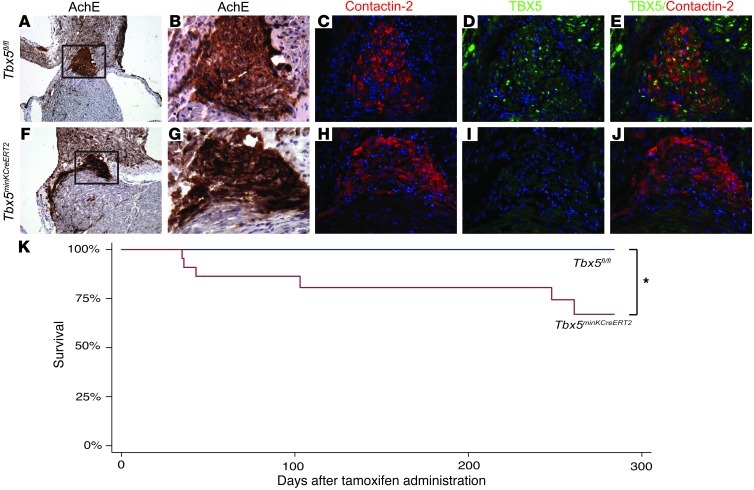
Figure 1. Removal of Tbx5 from the ventricular CCS reduces survival.
(A–J) Tbx5fl/fl and Tbx5minKCreERT2 littermates were administered tamoxifen at 6–7 weeks of age, and TBX5 expression in the VCS was evaluated by immunofluorescence at 10–11 weeks of age. (A–E) Serial sections demonstrating TBX5 expression through the AV bundle (positive for acetylcholinesterase [AchE] and contactin-2)of Tbx5fl/fl mice. (F–J) In contrast, TBX5 was not detected in the AV bundle of Tbx5minKCreERT2 mice. Boxed areas in A and F are shown at higher magnification in B and G. TBX5 and contactin-2 were evaluated on serial sections (merged in E and J). Nuclei were stained with hematoxylin (A, B, F, and G) or DAPI (C–E and H–J). Original magnification, ×10 (A and F), ×40 (B–E and G–J). (K) Tbx5minKCreERT2 mice (n = 22) and Tbx5fl/fl littermates (n = 15) were followed longitudinally after tamoxifen administration at 6–7 weeks of age. Kaplan-Meier survival estimates demonstrated significantly decreased survival after Tbx5 removal. *P < 0.05, log-rank test.
Tbx5minKCreERT2 mice demonstrated sudden death (Figure 1K) as early as 5 weeks after tamoxifen administration.
Adult Tbx5minKCreERT2 mice showed significantly increased mortality relative to adult Tbx5fl/fl mice in longitudinal studies (P = 0.03, log-rank test). These results demonstrated a requirement for TBX5 in the mature VCS and prompted us to investigate the electrophysiologic consequences of VCS-specific Tbx5 knockout.
Loss of fast conduction in adult Tbx5minKCreERT2 mice.
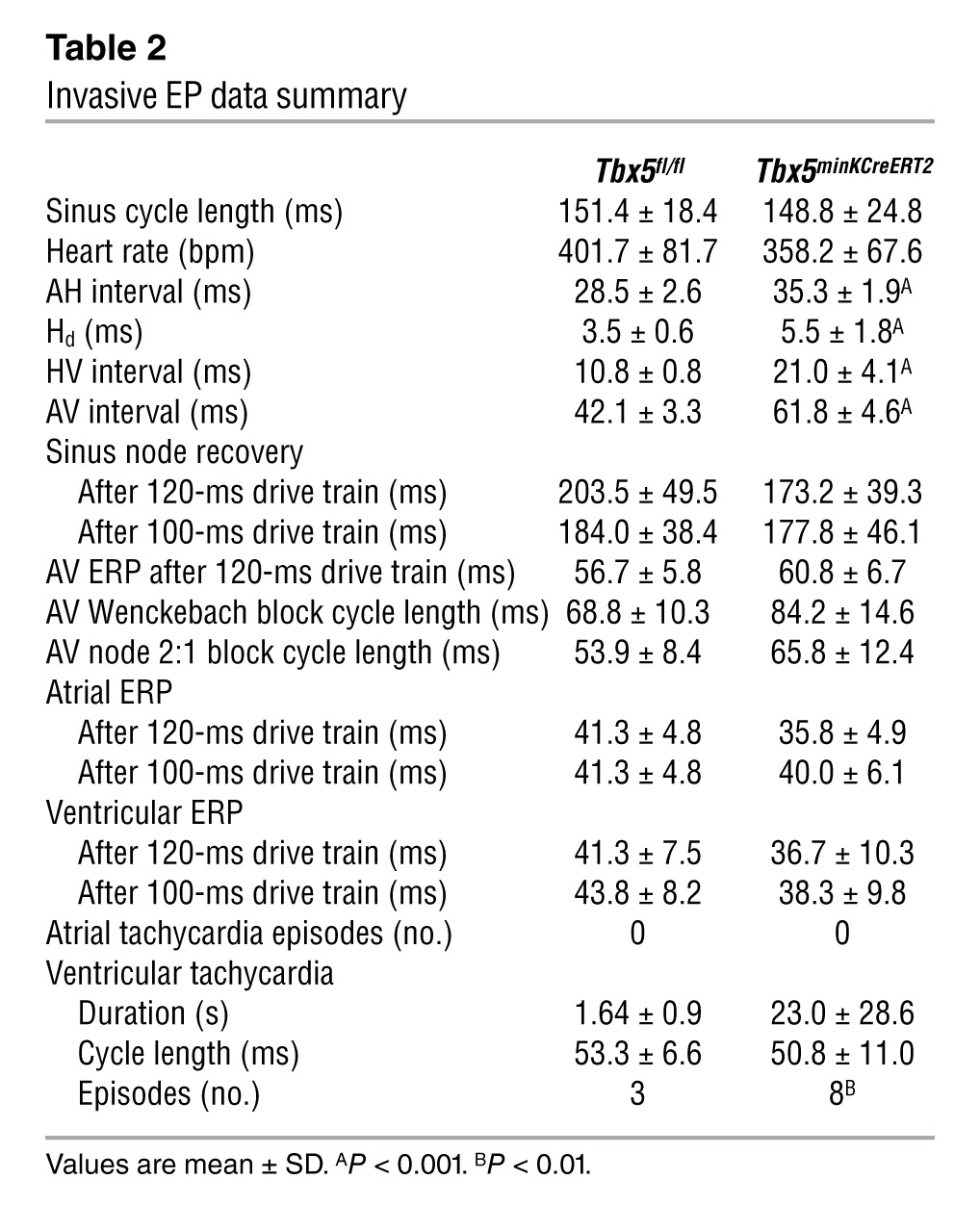
To determine the conduction system effects of VCS-specific Tbx5 removal, we performed conscious, ambulatory telemetry ECG analysis on Tbx5minKCreERT2 animals and littermate controls 4–5 weeks after tamoxifen administration. VCS-specific Tbx5 deletion caused severe conduction slowing. The PR interval, representing the period between atrial and ventricular depolarization, and QRS duration, representing the length of ventricular depolarization and — in the mouse — early repolarization (Figure 2A), were both significantly increased (Figure 2, B and C, and Table 1). To specifically localize the anatomic region of slowed conduction, we performed invasive electrophysiologic (EP) studies. Removal of Tbx5 resulted in moderate prolongation of the atrio-Hisian (AH) interval (representing conduction through the AV node and proximal His bundle), severe widening of the His duration (Hd), and severe prolongation of the Hisioventricular (HV) interval, demonstrating slowed conduction through the His bundle, bundle branches, and Purkinje network (Figure 2, D–F, Tables 1 and 2, and Supplemental Figure 2). Non-VCS function was not altered, with normal heart rate, sinus node, and AV node recovery times and atrial and ventricular effective refractory periods (Table 2). Collectively, these findings indicated an essential role for Tbx5 as a regulator of fast conduction in the VCS.
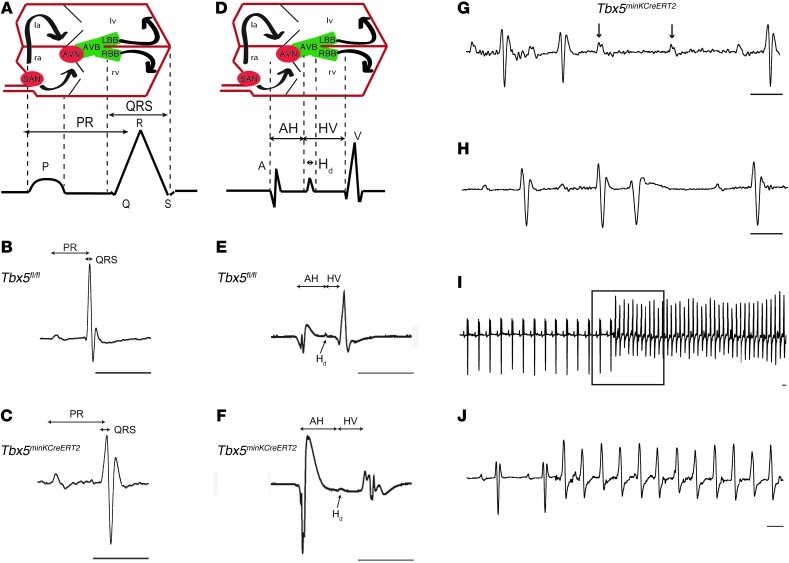
Figure 2. Conduction slowing and arrhythmias after removal of Tbx5 from the ventricular CCS.
(A–J) Conduction system function in Tbx5fl/fl (B and E) and Tbx5minKCreERT2 (C and F–J) mice was evaluated by ambulatory telemetry (B, C, and G–J) and invasive EP studies (E and F). Electroanatomical correlates of ECG and EP recordings are shown in A and D, respectively. la and ra, left and right atria; AVB, AV bundle; AVN and SAN, AV and SA nodes; LBB and RBB, left and right bundle branches. PR and QRS intervals were prolonged during ambulatory telemetry analysis (representative recordings in B and C), and intracardiac recordings (representative recordings in E and F) demonstrated prolongation of AH interval, Hd, and HV interval. Mobitz type II second-degree AV block (G) occurred exclusively in Tbx5minKCreERT2 mice. PVCs (H) were more common in Tbx5minKCreERT2 mice, and ventricular tachycardia (I and J) was observed exclusively in Tbx5minKCreERT2 mice. Boxed area in I is shown at slower scale in J. Scale bars: 50 ms. Arrows in G represent nonconducted p waves. See Table 1 for quantification of ECG and EP intervals.
Table 1.
Conduction slowing after loss of TBX5 in the VCS
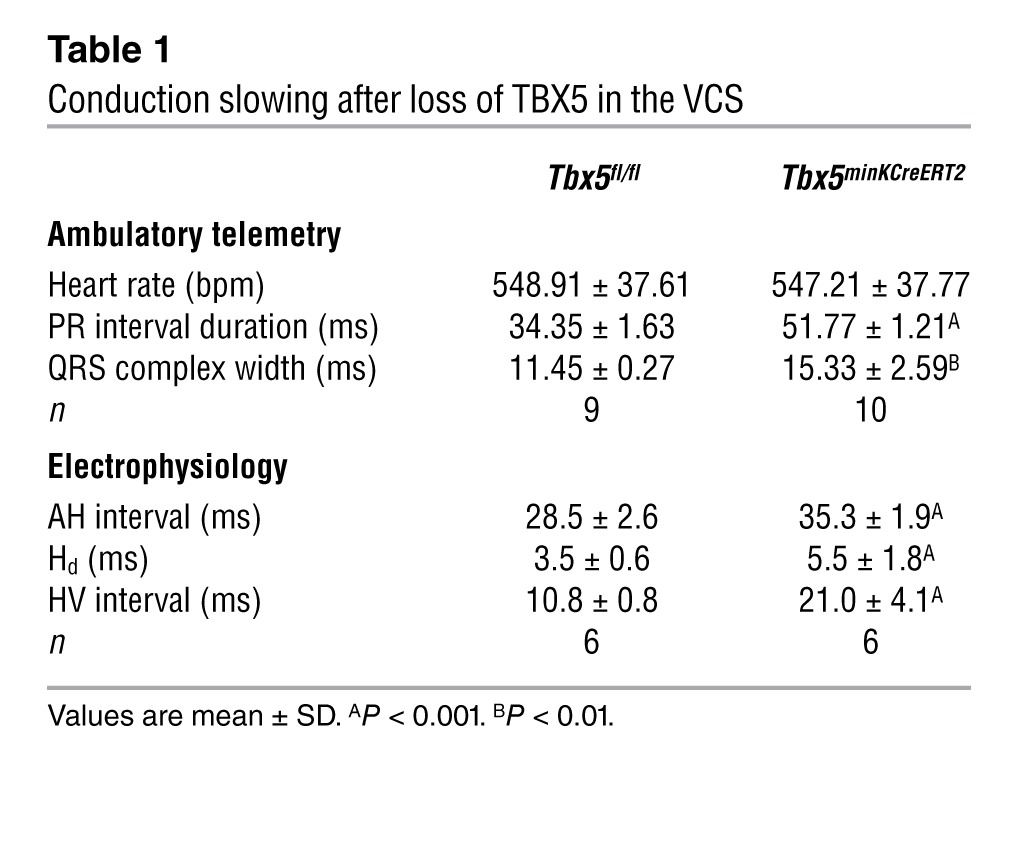
Table 2.
Invasive EP data summary
The autonomic nervous system can play an important role in modulating cardiac conduction. Specifically, activation of the sympathetic nervous system can increase heart rate and contractility and accelerate conduction through the AV node, whereas parasympathetic stimulation via the vagus nerve can act at the SA and AV nodes to decrease heart rate and slow AV conduction. To determine whether slowed conduction in Tbx5minKCreERT2 mice is secondary to increased vagal tone, we administered atropine, a cholinergic antagonist, to tamoxifen-treated Tbx5minKCreERT2 and Tbx5fl/fl mice and evaluated conduction intervals by EP studies. Cholinergic blockade did not result in accelerated conduction in Tbx5minKCreERT2 or Tbx5fl/fl mice (Supplemental Table 1), which suggests that slowed conduction in the absence of Tbx5 in the VCS is not secondary to effects on the autonomic nervous system.
Cardiac arrhythmias in Tbx5minKCreERT2 mice.
Removal of TBX5 from the VCS resulted in significant arrhythmias. Observed rhythm disturbances that occurred exclusively in Tbx5minKCreERT2 mice included Mobitz type II second-degree AV block (Figure 2G), indicative of defects in the His bundle and/or bundle branches (13), and spontaneous ventricular tachycardia (Figure 2, I and J) in ambulatory recordings. Occasional second-degree AV block was observed in both Tbx5fl/fl and Tbx5minKCreERT2 mice (Supplemental Table 2). However, Mobitz type II AV block, a sign of infranodal conduction system disease characterized by one or more dropped QRS complexes without changes in the PR interval, was observed exclusively in Tbx5minKCreERT2 mice. In contrast, AV block in Tbx5fl/fl mice was characterized by a shortened PR interval after the dropped beat, indicative of Wenkebach (Mobitz type I) AV block, considered a benign AV nodal block unlikely to progress to complete AV block. Premature ventricular contractions (PVCs) were also much more common in Tbx5minKCreERT2 than control Tbx5fl/fl mice (Figure 2H), with a maximum of 7 PVCs per 24-hour recording in Tbx5fl/fl mice compared with greater than 100 in 6 of 10 Tbx5minKCreERT2 mice.
Episodes of spontaneous, monomorphic ventricular tachycardia were observed in 3 of 10 Tbx5minKCreERT2 mice versus 0 of 9 littermate controls in ambulatory studies and 1 of 6 Tbx5minKCreERT2 mice versus 0 of 6 littermate controls in EP studies. In addition to the observation of spontaneous tachyarrhythmias in both ambulatory monitoring (Figure 2, I and J) and EP (Supplemental Figure 3) studies, Tbx5minKCreERT2 mice showed significantly increased susceptibility to ventricular tachycardia after burst stimulation in EP studies (Table 2). Episodes of ventricular tachycardia induced by programmed stimulation in Tbx5minKCreERT2 mice resembled those that occurred spontaneously (Figure 2, I and J, and Supplemental Figure 3), but were of shorter duration. In contrast, Tbx5fl/fl mice exhibited only nonsustained episodes of polymorphic ventricular tachycardia after programmed stimulation (Supplemental Figure 4). Furthermore, the Tbx5minKCreERT2 mouse that developed spontaneous ventricular tachycardia during EP studies died suddenly, prior to any electrophysiologic testing. Although we cannot rule out the possibility that nonarrhythmic or bradyarrhythmic causes contributed to the reduced survival of Tbx5minKCreERT2 mice, the observation of sudden death after an episode of spontaneous ventricular tachycardia strongly suggests that ventricular tachycardia may contribute to sudden death after selective removal of Tbx5 from the VCS.
Normal contractile function in Tbx5minKCreERT2 mice.
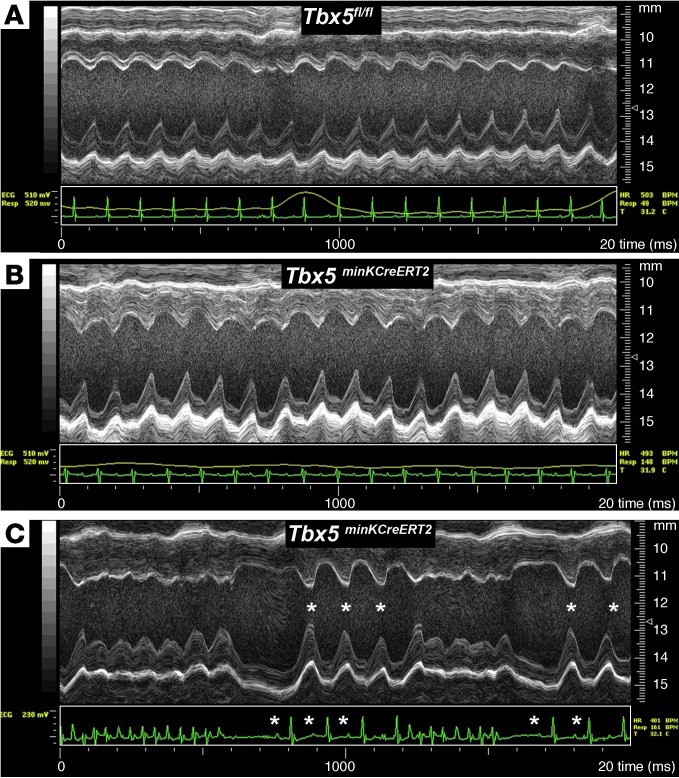
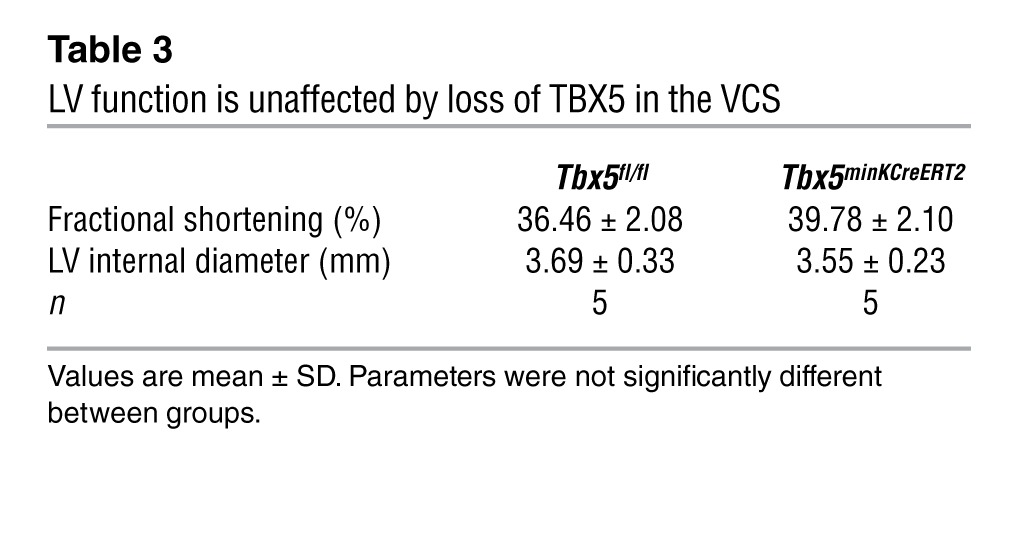
To distinguish between a primary conduction system abnormality and secondary conduction system defects caused by primary contractile dysfunction, we assessed cardiac contractility via echocardiography. Transthoracic echocardiography demonstrated that LV function was indistinguishable between Tbx5fl/fl and Tbx5minKCreERT2 mice (Figure 3, A and B, and Table 3). Furthermore, the mutant mice demonstrated immediate recovery of normal cardiac function after episodes of spontaneous ventricular tachycardia (Figure 3C). Together, these data indicate that the conduction defects observed in Tbx5minKCreERT2 mice did not derive from a secondary consequence of myocardial dysfunction.
Figure 3. Normal cardiac function after loss of TBX5 in the VCS.
(A and B) Cardiac function, assessed by M-mode echocardiography, of Tbx5fl/fl controls (A) and Tbx5minKCreERT2 littermates (B) administered tamoxifen at 6–7 weeks of age and studied 4–5 weeks later in sinus rhythm. No functional difference between mutant and control mice was detected. (C) A Tbx5minKCreERT2 animal with episodic spontaneous ventricular tachycardia demonstrated rapid recovery of ventricular function during sinus beats (asterisks) that followed tachycardic episodes.
Table 3.
LV function is unaffected by loss of TBX5 in the VCS
Tbx5 is not required for survival of VCS cells.
Based on the known requirement for Tbx5 for cell survival in other contexts (14, 15), we hypothesized that the conduction abnormalities in Tbx5minKCreERT2 mice may be caused by decreased survival of VCS cells. However, the VCS fate map was not affected by deletion of Tbx5. Simultaneously deleting Tbx5 from the VCS and marking cells with the Cre-dependent lacZ reporter ROSA-26R (R26RminKCreERT2/+Tbx5minKCreERT2 mice) generated a fate map indistinguishable from that generated in the presence of Tbx5 (R26RminKCreERT2/+Tbx5+/+ mice) (Figure 4). This result demonstrated that the conduction defects in Tbx5minKCreERT2 mice could not be attributed to loss of VCS cells.
Figure 4. Tbx5 removal does not affect cell survival in the VCS.
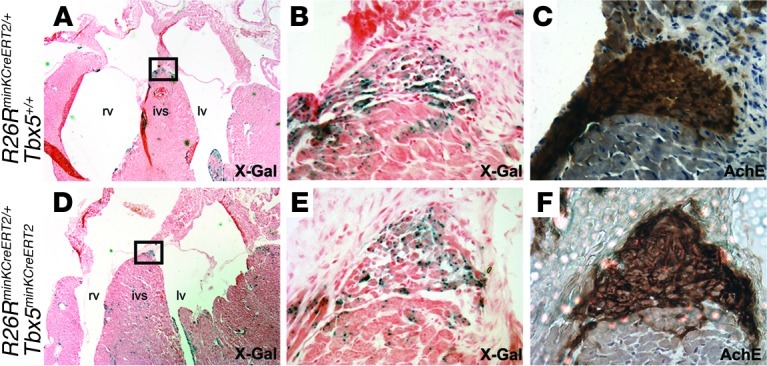
X-Gal– (A, B, D, and E) and acetylcholinesterase-stained (C and F) serial sections through the AV bundle of R26RminKCreERT2/+Tbx5+/+ (A–C) and R26RminKCreERT2/+Tbx5minKCreERT2 (D–F) animals administered tamoxifen showed similar distribution of cells labeling the VCS fate map, which demonstrated that Tbx5 was not required for VCS cell survival. ivs, interventricular septum. Boxed regions in A and D are shown at higher magnification in B and E. Original magnification, ×2.5 (A and D), ×40 (B, C, E, and F).
Tbx5 is required for VCS expression of Cx40 and Nav1.5 to modulate fast conduction.
We investigated the hypothesis that TBX5 is required for a functional molecular pathway mediating fast VCS conduction. We analyzed the molecular basis for loss of fast conduction in Tbx5minKCreERT2 mice by examining expression of known mediators of fast conduction in the VCS of Tbx5minKCreERT2 mutant and Tbx5fl/fl control hearts. Fast conduction in the VCS requires a high degree of cell-cell electrical coupling and rapid depolarization (16), which are substantially mediated by Cx40 and Nav1.5, respectively. We found that expression of Cx40 and Nav1.5 was drastically reduced in the VCS after removal of Tbx5 (Figure 5). The proximal AV bundle and distal AV bundle/bundle branches were identified by their anatomic location, acetylcholinesterase activity (12, 17–19), and contactin-2 expression (20). Tbx5fl/fl controls demonstrated high Cx40 and Nav1.5 expression throughout the molecularly defined VCS, whereas Tbx5minKCreERT2 animals demonstrated dramatic reductions in Cx40 and Nav1.5 VCS expression. In keeping with the results shown in Figure 1, TBX5 was expressed in the AV bundle and bundle branches of Tbx5fl/fl mice, but not Tbx5minKCreERT2 littermates (Figure 5). Consistent with the specificity of Cre activity in minKCreERT2 BAC transgenic mice (12) and the hypothesis that slowed conduction is caused by the primary effects of TBX5 loss in the VCS, Cx40 and Nav1.5 expression were not altered outside of the VCS (Supplemental Figure 5).
Figure 5. Decreased Cx40 and Nav1.5 expression in the VCS after removal of TBX5.
The proximal (A–J) and distal (K–T) AV bundle was identified by acetycholinesterase activity (A, B, F, G, K, L, P, and Q) and contactin-2 expression (C, D, H, I, M, N, R, and S) on serial sections from Tbx5fl/fl and Tbx5minKCreERT2 hearts. Whereas the contactin-2–positive AV bundle expressed high levels of Cx40, Nav1.5, and TBX5 in Tbx5fl/fl mice, their expression was drastically reduced in that of Tbx5minKCreERT2 mice. (C, D, H, I, M, N, R, and S) Dual-color immunofluorescence for Cx40 or Nav1.5 and contactin-2 is shown. Contactin-2 expression in E, J, O, and T is from sections adjacent to those stained for TBX5; contactin-2 and TBX5 antibodies were both raised in goat, preventing dual-color immunofluorescence on the same section. Nuclei were stained with hematoxylin (A, B, F, G, K, L, P, and Q) or DAPI (blue; C–E, H–J, M–O, and R–T). Boxed regions in A, F, K, and P are shown at higher magnification in B, G, L, and Q. Original magnification, ×10 (A, F, K, and P); ×40 (B–E, G–J, L–O, and P–T).
TBX5-dependent activation of an Scn5a enhancer.
We hypothesized that TBX5 directly drives a molecular network required for fast VCS conduction via direct regulation of Gja5 and Scn5a, and focused our efforts on Scn5a, as no direct regulators of Scn5a in the VCS have been identified; Gja5 is a known TBX5 target in the embryo (11). We bioinformatically interrogated the Scn5a locus to identify potential TBX5-responsive enhancers using the overlap of 4 independent data sets: (a) evolutionary conservation (21); (b) ChIP-seq studies identifying TBX5 binding sites in the atrial cardiomyocyte HL-1 cell line (22); (c) p300 ChIP-seq peaks to mark active enhancers, both in vitro and in vivo (22, 23); and (d) bioinformatic predictions of cardiac enhancers (24). Within the genomic region including the Scn5a locus and complete upstream and downstream intergenic regions, a single region approximately 15 kb downstream of Scn5a demonstrated overlap in all 4 data sets (Figure 6A). We hypothesized that this genomic region represents a TBX5-dependent Scn5a enhancer.
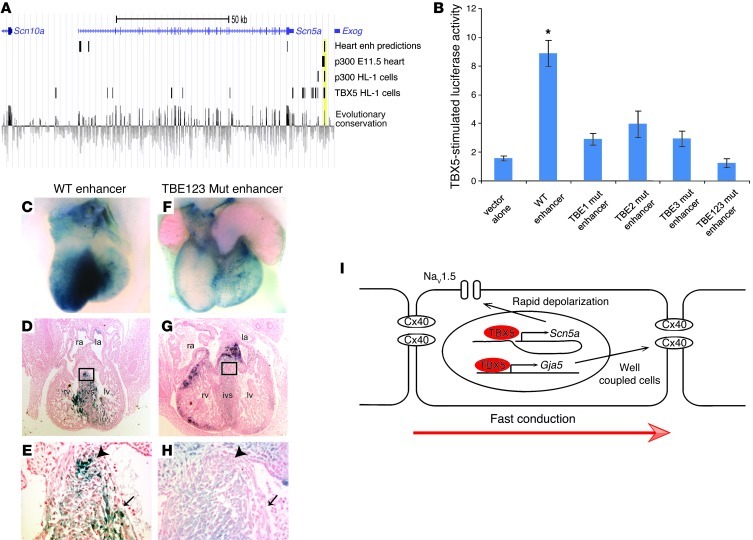
Figure 6. TBX5 directly regulates an enhancer downstream of Scn5a.
(A) Bioinformatic identification of a candidate enhancer downstream of Scn5a. We used previously reported data sets to identify potential TBX-responsive enhancers: bioinformatic predictions of cardiac enhancers (24); p300 ChIP-seq peaks to mark active enhancers in the E11.5 heart (23); ChIP-seq studies identifying both p300 and TBX5 binding sites in the atrial cardiomyocyte HL-1 cell line (22); and evolutionary conservation, as assessed by genomic evolutionary rate profiling score (21). A region demonstrating overlap in all 4 data sets, approximately 15 kb downstream of Scn5a, is shaded yellow. (B) The WT candidate enhancer demonstrated robust TBX5-mediated activation in dual luciferase reporter assays in HEK-293T cells. Luciferase activity was blunted by single mutation of any of 3 conserved T-box elements (TBE1 mut, TBE2 mut, or TBE3 mut) and eliminated by mutation of all 3 T-box elements (TBE123 mut). *P < 0.05 versus all other groups; n ≥ 3; mean ± SEM. (C–H) VCS of transient transgenic embryos, analyzed at E13.5. Whereas the WT enhancer reproducibly drove lacZ expression from a minimal promoter (C–E), mutation of the T-box elements in the enhancer resulted in blunted and regionally variable expression (F–H). Shown are representative posterior (C and F) and sagittal section (D and G) views of X-Gal staining. Higher-magnification views of boxed regions in D and G demonstrated X-gal expression (E) or its absence (H) in the developing AV bundle (arrowhead) and bundle branches (arrow). Note that, in contrast to the weak, non-CCS lacZ expression in F, the more robustly stained heart in G demonstrated ectopic expression in the endocardial cushions and compact myocardium. (I) Model for the role of TBX5 in driving fast conduction in the VCS via direct regulation of Scn5a and Gja5. Original magnification, ×4 (D and G); ×40 (E and H).
We tested the ability of this putative enhancer to activate transcription in a TBX5-dependent manner. TBX5 dramatically upregulated enhancer-dependent luciferase reporter expression from this genomic region (chr9:119,378,051–119,379,479; NCBI build 37/mm9) in vitro (Figure 6B). This TBX5-responsive enhancer contains 3 conserved T-box binding sites (CAGGTGTGAGCC, chr9:119,379,020–119,379,031; TGGGGTGTGGAG, chr9:119,379,008–119,378,997; GGAGGTGTGAAT, chr9:119,378,918–119,378,929). The core of the TBX5 binding motif is GTG (22, 25, 26), a sequence conserved in each of the 3 TBX5 binding sites present in the defined enhancer. Individual mutation of single TBX5 binding sites from GTG to AAA at the core of each of the 3 conserved TBX5 consensus motifs significantly decreased TBX5-mediated activation of the enhancer in vitro, and mutation of all 3 TBX5 binding sites completely abolished TBX5-dependent enhancer activation (Figure 6B).
TBX5-responsive activity in vitro identified this enhancer as a candidate VCS enhancer. We tested whether this enhancer was sufficient for VCS-specific expression in vivo. The WT enhancer proved sufficient to drive in vivo VCS expression of lacZ from a minimal promoter in 13 of 16 transgenic embryos (Figure 6, C–E). Specifically, the enhancer reproducibly drove robust lacZ expression in the VCS, including the AV bundle and bundle branches as well as the dorsal wall of the atria, in an overall pattern closely resembling native Scn5a expression (27, 28). The VCS-specific activity of the Scn5a enhancer was T-box dependent: VCS-specific lacZ expression was severely diminished by mutation of the 3 conserved T-box sites in the enhancer. Cardiac lacZ expression in T-box mutant transgenic embryos was weak and variable, with only 3 of 12 transgenic embryos demonstrating detectable VCS lacZ expression (Figure 6, F–H, and Supplemental Figure 6). These results indicate that TBX5 directly regulates an enhancer downstream of Scn5a sufficient for patterning VCS-specific gene expression.
Discussion
The function of the VCS is rapid conduction of electrical impulses from the AV node to the apical ventricular myocardium to establish coordinated ventricular contraction with apex-to-base polarity. The consequences of VCS failure have long been apparent in the clinical setting and include AV block, bundle branch/interventricular conduction defects, and ventricular tachycardia, all of which carry significant morbidity and mortality (29–32). To our knowledge, it was not previously possible to evaluate the specific role for TBX5 within the conduction system, given its broad cardiac expression in the adult heart, its requirement during cardiac development, and the structural heart defects frequently associated with Tbx5 haploinsufficiency (11, 33). By selectively removing TBX5 from the adult VCS in this study, we identified a role for TBX5 in the mature VCS of structurally normal hearts. Removal of TBX5 from the mature VCS resulted in a significant increase in mortality accompanied by arrhythmias, including ventricular tachycardia, and a dramatic slowing of conduction through the VCS. Slowed VCS conduction was manifest as AV block with dramatic increases in Hd and HV interval as well as a prolonged QRS interval, indicative of interventricular conduction delay. We further demonstrated that loss of fast conduction in the VCS was not secondary to loss of contractile function or loss of VCS cells, but rather was associated with reductions in Cx40 and Nav1.5 expression in the VCS. TBX5 directly regulated Nav1.5 expression via an enhancer downstream of the Scn5a locus that possessed T-box element–dependent VCS-specific expression in vivo. Our results are the first to our knowledge to establish a transcriptional pathway required for function of the mature VCS, and further establish Tbx5minKCreERT2 mice as a model for the pathogenesis of VCS disease.
Based on the present findings, we propose a model whereby Tbx5 serves as an essential regulator of VCS function and is required for Cx40 and Nav1.5 expression in the VCS (Figure 6I). Cx40, a high-conductance gap junction expressed in the atria and VCS, mediates tight cell-cell coupling (34), and Gja5 knockout mice demonstrate functional VCS slowing similar to, but less severe than, that of Tbx5minKCreERT2 mice (18, 35–41). Neither spontaneous ventricular tachycardia nor sudden death (both of which were observed in our Tbx5minKCreERT2 mice) have been reported in Gja5 knockout mice. Our observation that TBX5 was required for expression of Cx40 in the adult VCS extends previous work identifying Gja5 as a TBX5 target in the embryonic heart (11).
Rapid depolarization in the VCS is mediated by the voltage-gated sodium channel Nav1.5 (28, 38). Mutations in SCN5A, as well as alterations in its expression levels, are associated with numerous human cardiac disease conditions, yet regulation of its conduction system expression has been unknown (42). Scn5a haploinsufficiency in mice causes conduction slowing in the VCS (38) and, on occasion, spontaneous ventricular arrhythmias, including ventricular tachycardia (37). Furthermore, GWAS have consistently linked SNPs near SCN5A to variation in PR and QRS intervals, which suggests that genetic variation at this locus may exert substantial influence on VCS function in the general population. Given that cell-cell electrical coupling and excitability are key parameters governing conduction velocity, we hypothesize that the reductions in Cx40 and Nav1.5 expression after removal of Tbx5 are responsible for a key component of the observed conduction slowing.
Recent CCS GWAS have uncovered numerous loci linked to cardiac conduction function in the general population (3–8). An ongoing challenge is to identify the source of functional variation tagged by GWAS. TBX5, TBX3, NKX2-5, and SCN5A have all been implicated in PR and/or QRS interval variation (4–8). TBX5 is in close genomic proximity to TBX3, a related transcription factor expressed throughout the central CCS (43, 44). SNPs have been identified in cardiac conduction GWAS upstream of TBX3 that correlate with PR and QRS interval variation (5, 8) TBX3 is a potent transcriptional repressor capable of repressing Scn5a, as well as Gja1 (encoding Cx43) and Gja5, and is essential for development and function of the SA and AV nodes as well as the VCS (45–47). TBX3 may thereby contribute to repression of Gja5 and Scn5a in the VCS after Tbx5 removal in Tbx5minKCreERT2 mice, possibly by acting directly on the downstream Scn5a enhancer identified in this study. Perinatal removal of Nkx2-5 results in loss of Nav1.5 in the ventricles, whereas Nav1.5 expression is preserved in the atria and VCS (48), areas with high levels of Tbx5 expression. These data suggest that a balance of TBX5, TBX3, and NKX2-5 activities is essential for regional regulation of Nav1.5, with a strict requirement for TBX5 for maximal Nav1.5 expression in the VCS. Furthermore, GWAS have identified genetic variations downstream, in the distal introns, and in the 3′ untranslated region of SCN5A that correlate with PR and/or QRS interval variation (4, 5, 8). The enhancer identified in the present study is in linkage disequilibrium with this region. An exciting possibility is that the reported GWAS SNPs tag functional enhancer variants that influence SCN5A expression by disrupting the TBX5-responsive conduction system enhancer identified in the current study.
We here identified a molecular link between TBX5 and Scn5a, a gene implicated in one of the few existing mouse models of spontaneous ventricular tachyarrhythmia, Scn5a+/– mice (37, 49). Future studies will determine the degree to which this molecular pathway mediates the pathogenesis of these lethal arrhythmias. Given the essential role for Nav1.5 in cardiac conduction (38), the consequences of SCN5A mutations for human disease (50), and the consistent association of the SCN5A locus with variation in VCS function in diverse populations (4–8), understanding the regulation of SCN5A — particularly in the VCS — is of crucial importance. This study identified a direct molecular pathway between TBX5 and Scn5a, demonstrated that CCS GWAS anticipate a molecular pathway required for normal CCS function, and established a molecular paradigm for understanding the pathology of VCS disease.
Methods
Transgenic mice.
Tbx5minKCreERT2, Tbx5fl/fl, and ROSA-26R-LacZ mice have all been previously described (11, 12, 51). Mice were maintained on a mixed genetic background, and littermates were used as controls in all experiments except for the fate map studies shown in Figure 4, in which age-matched controls were used because of the prohibitive number of mice that would be necessary to generate littermate controls. Tamoxifen was administered at a dose of 0.167 mg/g body weight for 5 consecutive days by oral gavage at 6–7 weeks of age, as previously described (12).
Telemetry ECG analysis.
10- to 12-week-old mice were anesthetized with isofluorane, and telemetry transmitters (ETA-F10; DSI) were implanted in the back with leads tunneled to the right upper and left lower thorax, as previously described (52). Heart rate and PR and QRS intervals were calculated using Ponemah Physiology Platform (DSI) from 24-hour recordings.
Electrophysiology studies.
Detailed protocols for in vivo electrophysiology studies have been previously described (53). Briefly, 10- to 12-week-old mice were anesthetized with pentobarbital (33 mg/kg i.p.), and a 1.1-Fr octapolar electrode catheter (EPR-800; Millar) was advanced via a right jugular venous cut-down to record right atrial, His bundle, and RV potentials and to perform programmed electrical stimulation. See Supplemental Methods for details on atropine administration.
Echocardiography studies.
Transthoracic echocardiography in mice was performed under inhaled isoflurane anesthesia, delivered via nose cone. Chest hairs were removed with a topical depilatory agent. Limb leads were attached for electrocardiogram gating, and animals were imaged in the left lateral decubitus position with a VisualSonics Vevo 770 machine using a 30-MHz high-frequency transducer. Body temperature was maintained using a heated imaging platform and warming lamps. 2-dimensional images were recorded in parasternal long- and short-axis projections, with guided M-mode recordings at the midventricular level in both views. LV cavity size and percent fractional shortening were measured in at least 3 beats from each projection and averaged. M-mode measurements were used to determine LV chamber dimensions and percent LV fractional shortening, the latter calculated as ([LVIDd – LVIDs]/LVIDd), where LVIDd and LVIDs are LV internal diameter in diastole and systole, respectively).
Acetylcholinesterase and β-galactosidase activity.
Acetylcholinesterase and β-galactosidase activity were assayed on 7- to 10-μM fresh-frozen cryosections, as described previously (12).
Analysis of protein expression.
Immunofluorescence studies were performed on 7-μM fresh-frozen cryosections as described previously (12), using the following antibodies: rabbit polyclonal anti-Cx40 (1:250 dilution; catalog no. 36-4900; Zymed), rabbit polyclonal anti-Nav1.5 (1:200 dilution; catalog no. ASC-005; Alomone Laboratories), goat polyclonal anti-TBX5 (1:250 dilution; catalog no. sc-17866; Santa Cruz), goat polyclonal anti–contactin-2 (1:50 dilution; catalog no. AF1714; R&D), donkey anti-rabbit IgG Alexa Fluor 488 (Invitrogen), and donkey anti-goat IgG Alexa Fluor 594 (Invitrogen). Images shown are representative of sampling a minimum of 6 animals of each genotype at 5–10 levels per heart through the AV bundle and proximal bundle branches. See Supplemental Methods for Western blotting methods.
Cell culture studies.
The Scn5a 3′ enhancer (chr9:119378051-119379479; NCBI build 37/mm9) was cloned from BAC RP23-103G4 (Invitrogen) into the XhoI and BglII sites of a pGL3 Basic vector (Promega) that was modified to include a minimal TK promoter between the BglII and HindIII sites. Conserved TBX5 binding elements, identified using ECR Browser (54) and rVISTA (54) software, had the following genomic coordinates: TBE1, chr9:119,379,020–119,379,031; TBE2, chr9:119,378,997–119,379,008; TBE3, chr9:119,378,918–119,378,929. Site-directed mutagenesis was performed using the QuikChange Lightning kit (Agilent). TBX5 was cloned from a mouse atrial cDNA library into pcDNA3.1 hygro (Invitrogen). All constructs were sequence verified (see Supplemental Table 3 for primer sequences).
Transient transfections were performed in HEK-293T cells using FuGene HD (Promega) according to the manufacturer’s instructions. The day before transfection, 2 × 105 cells/well were plated in 12-well plates in growth medium (D-MEM plus 10% FBS and l-glutamine). The following day, each well was transfected with 600 ng luciferase reporter, 400 ng TBX5 or empty pcDNA vector, and 1 ng pRL-CMV using a 3:1 FuGene HD/DNA ratio. Growth medium was replaced 24 hours after transfection, cells were harvested 48 hours after transfection, and luciferase activity was assayed using the Promega Dual Luciferase Reporter kit according to the manufacturer’s instructions. All transfections were performed in duplicate and repeated in a minimum of 3 independent replicates.
Transient transgenic experiments.
The Scn5a enhancer and T-box element (TBE) mutant enhancer were subcloneæd from the pGL3 reporter vector into the XhoI and PstI sites of the Hsp68-LacZ transgenic reporter vector (55). The enhancer-Hsp68-LacZ fragment was digested with XhoI and NotI, gel purified, and used to create transient transgenic embryos at the University of Chicago Transgenic Core Facility. Embryos were harvested at E13.5, stained with X-Gal overnight at 37°C as previously described (56), embedded in paraffin, sectioned at 5 μM, and counterstained with Nuclear Fast Red.
Statistics.
Kaplan-Meier survival estimates were created using the STATA software package and tested for significance using the log-rank test. Dual luciferase reporter assays were analyzed in SPSS using 1-way ANOVA and Games-Howell post-hoc testing. 24-hour ECG intervals and EP intervals were calculated as described above and analyzed using Student’s t test (2-tailed). A P value less than 0.05 was considered significant.
Study approval.
All animal experiments were conducted in accordance with national and institutional guidelines and were approved by the Institutional Animal Care and Use Committees of the University of Chicago and University of Pennsylvania.
Supplementary Material
Acknowledgments
This study was supported by grants from the NIH (HL092153 and HL114010 to I.P. Moskowitz; HL105734 to V.V. Patel; HL092443 to EMM; HL097587 to J.P. Fahrenbach; HL098565 to G.H. Kim; K.J. Schillinger supported by HL007843) and the American Heart Association (10PRE2670012 to D.E. Arnolds; 11IRG4930008 to V.V. Patel).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(7):2509–2518. doi:10.1172/JCI62617.
References
- 1.Priori SG. The fifteen years of discoveries that shaped molecular electrophysiology: time for appraisal. Circ Res. 2010;107(4):451–456. doi: 10.1161/CIRCRESAHA.110.226811. [DOI] [PubMed] [Google Scholar]
- 2.Hatcher CJ, Basson CT. Specification of the cardiac conduction system by transcription factors. Circ Res. 2009;105(7):620–630. doi: 10.1161/CIRCRESAHA.109.204123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers JC, et al. Genetic variation in SCN10A influences cardiac conduction. Nat Genet. 2010;42(2):149–152. doi: 10.1038/ng.516. [DOI] [PubMed] [Google Scholar]
- 4.Holm H, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42(2):117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 5.Pfeufer A, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42(2):153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith JG, et al. Genome-wide association study of electrocardiographic conduction measures in an isolated founder population: Kosrae. Heart Rhythm. 2009;6(5):634–641. doi: 10.1016/j.hrthm.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith JG, et al. Genome-wide association studies of the PR interval in African Americans. PLoS Genet. 2011;7(2):e1001304. doi: 10.1371/journal.pgen.1001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sotoodehnia N, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet. 2010;42(12):1068–1076. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoogaars WM, Barnett P, Moorman AF, Christoffels VM. T-box factors determine cardiac design. Cell Mol Life Sci. 2007;64(6):646–660. doi: 10.1007/s00018-007-6518-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basson CT, et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15(1):30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 11.Bruneau BG, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106(6):709–721. doi: 10.1016/S0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 12.Arnolds DE, Moskowitz IP. Inducible recombination in the cardiac conduction system of mink:CreERT(2) BAC transgenic mice. Genesis. 2011;49(11):878–884. doi: 10.1002/dvg.20759. [DOI] [PubMed] [Google Scholar]
- 13.Vijayaraman P, Ellenbogen KA. Hurst’s The Heart. 2008. Bradyarrhythmias and pacemakers. In: Fuster V, O’Rourke RA, Walsh RA, Poole–Wilson P, eds. pp. 1020–1054. New York, New York, USA: McGraw Hill; [Google Scholar]
- 14.Goetz SC, Brown DD, Conlon FL. TBX5 is required for embryonic cardiac cell cycle progression. Development. 2006;133(13):2575–2584. doi: 10.1242/dev.02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He ML, et al. Induction of apoptosis and inhibition of cell growth by developmental regulator hTBX5. Biochem Biophys Res Commun. 2002;297(2):185–192. doi: 10.1016/S0006-291X(02)02142-3. [DOI] [PubMed] [Google Scholar]
- 16.Kléber AG, Rudy Y. Basic mechanisms of cardiac impulse propagation and associated arrhythmias. Physiol Rev. 2004;84(2):431–488. doi: 10.1152/physrev.00025.2003. [DOI] [PubMed] [Google Scholar]
- 17.Lamers WH, te Kortschot A, Los JA, Moorman AF. Acetylcholinesterase in prenatal rat heart: a marker for the early development of the cardiac conductive tissue? Anat Rec. 1987;217(4):361–370. doi: 10.1002/ar.1092170407. [DOI] [PubMed] [Google Scholar]
- 18.Simon AM, Goodenough DA, Paul DL. Mice lacking connexin40 have cardiac conduction abnormalities characteristic of atrioventricular block and bundle branch block. Curr Biol. 1998;8(5):295–298. doi: 10.1016/S0960-9822(98)70113-7. [DOI] [PubMed] [Google Scholar]
- 19.van Veen TA, et al. Discontinuous conduction in mouse bundle branches is caused by bundle-branch architecture. Circulation. 2005;112(15):2235–2244. doi: 10.1161/CIRCULATIONAHA.105.547893. [DOI] [PubMed] [Google Scholar]
- 20.Pallante BA, et al. Contactin-2 expression in the cardiac Purkinje fiber network. Circ Arrhythm Electrophysiol. 2010;3(2):186–194. doi: 10.1161/CIRCEP.109.928820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol. 2010;6(12):e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci U S A. 2011;108(14):5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blow MJ, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42(9):806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narlikar L, et al. Genome-wide discovery of human heart enhancers. Genome Res. 2010;20(3):381–392. doi: 10.1101/gr.098657.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh TK, Packham EA, Bonser AJ, Robinson TE, Cross SJ, Brook JD. Characterization of the TBX5 binding site and analysis of mutations that cause Holt-Oram syndrome. Hum Mol Genet. 2001;10(18):1983–1994. doi: 10.1093/hmg/10.18.1983. [DOI] [PubMed] [Google Scholar]
- 26.Mori AD, et al. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev Biol. 2006;297(2):566–586. doi: 10.1016/j.ydbio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 27.Dominguez JN, de la Rosa A, Navarro F, Franco D, Aranega AE. Tissue distribution and subcellular localization of the cardiac sodium channel during mouse heart development. Cardiovasc Res. 2008;78(1):45–52. doi: 10.1093/cvr/cvm118. [DOI] [PubMed] [Google Scholar]
- 28.Remme CA, et al. The cardiac sodium channel displays differential distribution in the conduction system and transmural heterogeneity in the murine ventricular myocardium. Basic Res Cardiol. 2009;104(5):511–522. doi: 10.1007/s00395-009-0012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng S, et al. Long-term outcomes in individuals with prolonged PR interval or first-degree atrioventricular block. JAMA. 2009;301(24):2571–2577. doi: 10.1001/jama.2009.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheinman MM. Role of the His-Purkinje system in the genesis of cardiac arrhythmia. Heart Rhythm. 2009;6(7):1050–1058. doi: 10.1016/j.hrthm.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 31.Schneider JF, Thomas HE, Jr, McNamara PM, Kannel WB. Clinical-electrocardiographic correlates of newly acquired left bundle branch block: the Framingham Study. Am J Cardiol. 1985;55(11):1332–1338. doi: 10.1016/0002-9149(85)90499-0. [DOI] [PubMed] [Google Scholar]
- 32.Huikuri HV, Castellanos A, Myerburg RJ. Sudden death due to cardiac arrhythmias. N Engl J Med. 2001;345(20):1473–1482. doi: 10.1056/NEJMra000650. [DOI] [PubMed] [Google Scholar]
- 33.Basson CT, et al. The clinical and genetic spectrum of the Holt-Oram syndrome (heart-hand syndrome). N Engl J Med. 1994;330(13):885–891. doi: 10.1056/NEJM199403313301302. [DOI] [PubMed] [Google Scholar]
- 34.van Veen AA, van Rijen HV, Opthof T. Cardiac gap junction channels: modulation of expression and channel properties. Cardiovasc Res. 2001;51(2):217–229. doi: 10.1016/S0008-6363(01)00324-8. [DOI] [PubMed] [Google Scholar]
- 35.Bevilacqua LM, et al. A targeted disruption in connexin40 leads to distinct atrioventricular conduction defects. J Interv Card Electrophysiol. 2000;4(3):459–467. doi: 10.1023/A:1009800328836. [DOI] [PubMed] [Google Scholar]
- 36.Hagendorff A, Schumacher B, Kirchhoff S, Lüderitz B, Willecke K. Conduction disturbances and increased atrial vulnerability in Connexin40-deficient mice analyzed by transesophageal stimulation. Circulation. 1999;99(11):1508–1515. doi: 10.1161/01.CIR.99.11.1508. [DOI] [PubMed] [Google Scholar]
- 37.Leoni AL, et al. Variable Na(v)1.5 protein expression from the wild-type allele correlates with the penetrance of cardiac conduction disease in the Scn5a(+/–) mouse model. PLoS One. 2010;5(2):e9298. doi: 10.1371/journal.pone.0009298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papadatos GA, et al. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc Natl Acad Sci U S A. 2002;99(9):6210–6215. doi: 10.1073/pnas.082121299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamaddon HS, Vaidya D, Simon AM, Paul DL, Jalife J, Morley GE. High-resolution optical mapping of the right bundle branch in connexin40 knockout mice reveals slow conduction in the specialized conduction system. Circ Res. 2000;87(10):929–936. doi: 10.1161/01.RES.87.10.929. [DOI] [PubMed] [Google Scholar]
- 40.VanderBrink BA, et al. Connexin40-deficient mice exhibit atrioventricular nodal and infra-Hisian conduction abnormalities. J Cardiovasc Electrophysiol. 2000;11(11):1270–1276. doi: 10.1046/j.1540-8167.2000.01270.x. [DOI] [PubMed] [Google Scholar]
- 41.Verheule S, van Batenburg CA, Coenjaerts FE, Kirchhoff S, Willecke K, Jongsma HJ. Cardiac conduction abnormalities in mice lacking the gap junction protein connexin40. J Cardiovasc Electrophysiol. 1999;10(10):1380–1389. doi: 10.1111/j.1540-8167.1999.tb00194.x. [DOI] [PubMed] [Google Scholar]
- 42.Rook MB, Evers MM, Vos MA, Bierhuizen MF. Biology of cardiac sodium channel Nav1.5 expression. Cardiovasc Res. 2012;93(1):12–23. doi: 10.1093/cvr/cvr252. [DOI] [PubMed] [Google Scholar]
- 43.Hoogaars WM, et al. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res. 2004;62(3):489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 44.Sizarov A, Devalla HD, Anderson RH, Passier R, Christoffels VM, Moorman AF. Molecular analysis of patterning of conduction tissues in the developing human heart. Circ Arrhythm Electrophysiol. 2011;4(4):532–542. doi: 10.1161/CIRCEP.111.963421. [DOI] [PubMed] [Google Scholar]
- 45.Hoogaars WM, et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21(9):1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frank DU, et al. Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc Natl Acad Sci U S A. 2012;109(3):E154–E163. doi: 10.1073/pnas.1115165109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bakker ML, et al. T-box transcription factor TBX3 reprograms mature cardiac myocytes into pacemaker-like cells. Cardiovasc Res. 2012;94(3):439–449. doi: 10.1093/cvr/cvs120. [DOI] [PubMed] [Google Scholar]
- 48.Briggs LE, et al. Perinatal loss of Nkx2-5 results in rapid conduction and contraction defects. Circ Res. 2008;103(6):580–590. doi: 10.1161/CIRCRESAHA.108.171835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stengl M. Experimental models of spontaneous ventricular arrhythmias and of sudden cardiac death. Physiol Res. 2010. 59 suppl 1: S25 S31 [DOI] [PubMed] [Google Scholar]
- 50.Tfelt-Hansen J, Winkel BG, Grunnet M, Jespersen T. Inherited cardiac diseases caused by mutations in the Nav1.5 sodium channel. J Cardiovasc Electrophysiol. 2010;21(1):107–115. doi: 10.1111/j.1540-8167.2009.01633.x. [DOI] [PubMed] [Google Scholar]
- 51.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 52.Wheeler MT, Allikian MJ, Heydemann A, Hadhazy M, Zarnegar S, McNally EM. Smooth muscle cell-extrinsic vascular spasm arises from cardiomyocyte degeneration in sarcoglycan-deficient cardiomyopathy. J Clin Invest. 2004;113(5):668–675. doi: 10.1172/JCI20410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu F, et al. Histone-deacetylase inhibition reverses atrial arrhythmia inducibility and fibrosis in cardiac hypertrophy independent of angiotensin. J Mol Cell Cardiol. 2008;45(6):715–723. doi: 10.1016/j.yjmcc.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32(Web Server issue):W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105(4):707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- 56.Hoffmann AD, Peterson MA, Friedland-Little JM, Anderson SA, Moskowitz IP. sonic hedgehog is required in pulmonary endoderm for atrial septation. Development. 2009;136(10):1761–1770. doi: 10.1242/dev.034157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.