Abstract
Preadipocytes secrete several WNT family proteins that act through autocrine/paracrine mechanisms to inhibit adipogenesis. The activity of WNT ligands is often decreased by secreted frizzled-related proteins (SFRPs). Sfrp5 is strongly induced during adipocyte differentiation and increases in adipocytes during obesity, presumably to counteract WNT signaling. We tested the hypothesis that obesity-induced Sfrp5 expression promotes the development of new adipocytes by inhibiting endogenous suppressors of adipogenesis. As predicted, mice that lack functional SFRP5 were resistant to diet-induced obesity. However, counter to our hypothesis, we found that adipose tissue of SFRP5-deficient mice had similar numbers of adipocytes, but a reduction in large adipocytes. Transplantation of adipose tissue from SFRP5-deficient mice into leptin receptor–deficient mice indicated that the effects of SFRP5 deficiency are tissue-autonomous. Mitochondrial gene expression was increased in adipose tissue and cultured adipocytes from SFRP5-deficient mice. In adipocytes, lack of SFRP5 stimulated oxidative capacity through increased mitochondrial activity, which was mediated in part by PGC1α and mitochondrial transcription factor A. WNT3a also increased oxygen consumption and the expression of mitochondrial genes. Thus, our findings support a model of adipogenesis in which SFRP5 inhibits WNT signaling to suppress oxidative metabolism and stimulate adipocyte growth during obesity.
Introduction
Obesity is a common disorder that predisposes individuals to type 2 diabetes, atherosclerosis, hypertension, and hyperlipidemia (1). The expansion of white adipose tissue (WAT) during obesity development is initially caused by an increase in adipocyte size, although with time, the total number of adipocytes increases due to preadipocyte differentiation (2, 3). It is well established that the inability to store excess energy in adipose tissue contributes to insulin resistance and metabolic complications (4, 5). Although regulation of adipocyte metabolism and differentiation have been extensively studied (reviewed in refs. 6–10), further understanding of molecular mechanisms influencing adipocyte biology is critical for our understanding and potential treatment of obesity and associated metabolic diseases.
Adipogenesis is the process by which mesenchymal precursor cells differentiate into adipocytes (11–13). It is well established that locally secreted and circulating factors regulate differentiation of precursors to adipocytes and also to alternative mesenchymal cell fates, such as osteoblasts (6). One of the most important regulators of adipogenesis is the WNT/β-catenin signaling pathway (14). Endogenous inhibitors of preadipocyte differentiation include WNT6, WNT10a, and WNT10b (15–18), which are expressed in precursor cells and decline during differentiation. In contrast, WNT5b and WNT4 are transiently induced during adipogenesis and act to promote this process (19, 20). Transgenic mice with expression of Wnt10b in adipose tissues are lean, are resistant to diet-induced and genetic obesity, have improved glucose homeostasis and increased trabecular bone, and are resistant to osteoporosis (21–25). Conversely, Wnt10b–/– mice have a low bone mass phenotype (23) and transient expression of adipocyte markers during regeneration of skeletal myoblasts (26). Thus, numerous studies have demonstrated that WNT ligands are important regulators of mesenchymal cell fate, both in vitro and in vivo (27, 28).
The activity of WNTs is highly regulated by negative extracellular regulators, such as dickkopfs, WNT inhibitory factor 1, and secreted frizzled-related proteins (SFRPs) (27, 29). Of these, SFRPs have been the most extensively studied in the context of adipose tissue (16, 30–32). SFRPs have an N-terminal cysteine-rich domain that is homologous to frizzled proteins, the cell surface receptors for WNT ligands; these proteins also have a C-terminal netrin domain of unknown function (29). The SFRP family consists of 5 members in both human and mouse genomes, with SFRP1, SFRP2, and SFRP5 forming a subfamily based on sequence similarity within the cysteine-rich domain (33). SFRPs are thought to prevent downstream WNT signaling by binding to and sequestering WNT ligands in the extracellular space; however, under some circumstances, SFRPs may stimulate WNT signaling (33).
Sfrp1 expression increases during adipogenesis, is transiently stimulated by a high-fat diet (HFD), and is elevated with mild — but not morbid — obesity (30). In 3T3-L1 preadipocytes, purified recombinant SFRP1 (and SFRP2) or ectopic SFRP1 expression inhibits WNT/β-catenin signaling and stimulates adipogenesis (16, 30). Sfrp5 is also induced during 3T3-L1 adipogenesis and is expressed more highly in isolated adipocytes than in stromal- vascular cells (31, 32, 34). Although 3 groups have reported that Sfrp5 is highly induced with genetic and/or diet-induced obesity (31, 32, 35), another found suppression of Sfrp5 under these conditions (34). Expression of Sfrp5 in WAT was also identified as one of the best a priori predictors of whether genetically identical C57BL/6J mice will gain adiposity when exposed to HFD (32). Taken together, these data suggest that during the progression of obesity, increasing lipid accumulation stimulates SFRP5 — and, to some extent, SFRP1 — to inhibit WNT signaling and thereby promote development of new adipocytes to help store excess energy. Although this is a logical hypothesis, the results of the present study indicated that SFRP5 is not a required regulator of adipocyte development in vivo; instead, our data suggested that SFRP5 and WNT signaling play unexpected roles in regulating mitochondrial oxidative metabolism and growth of adipocytes during obesity.
Results
Sfrp5 mRNA expression is induced during adipogenesis and further increased with obesity.
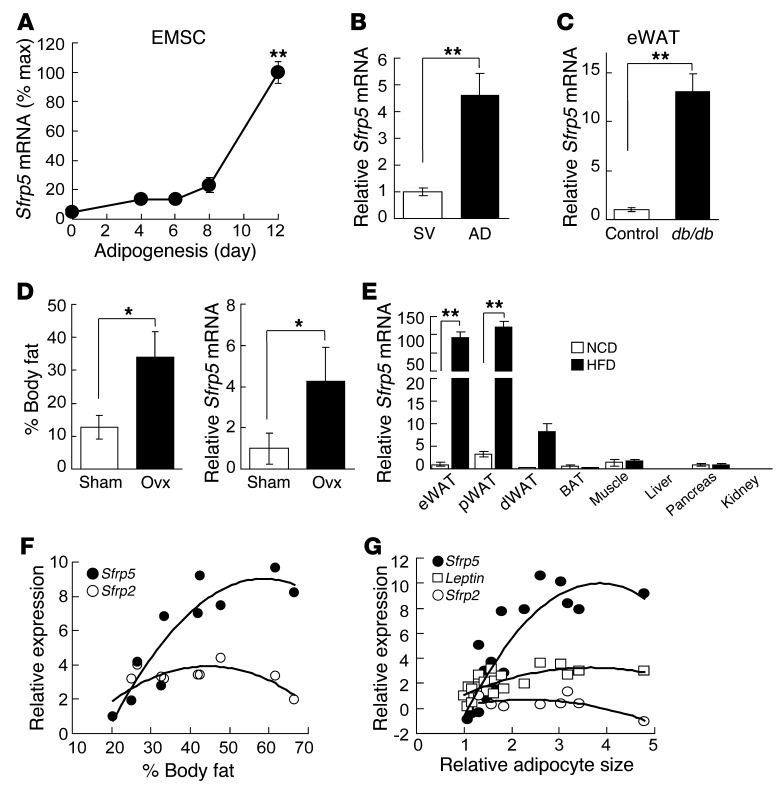
To investigate the role of SFRP5 in WAT biology, we first evaluated Sfrp5 expression during adipogenesis. We found that Sfrp5 mRNA was induced with differentiation of 3T3-L1 preadipocytes (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI63604DS1) and with adipogenesis of ear mesenchymal stem cells (EMSCs) (36) isolated from the outer ears of mice (Figure 1A). Consistent with Sfrp5 induction during preadipocyte differentiation, and in agreement with previous studies (31, 32, 34), expression of Sfrp5 mRNA was markedly higher in the adipocyte fraction than in the stromal-vascular fraction of WAT from lean mice (Figure 1B). Based on Ct values derived from quantitative real-time RT-PCR (qPCR), expression in this context was approximately 10 times higher than in cultured adipocyte models (data not shown). This suggests that Sfrp5 may be expressed relative to adipocyte size, because primary adipocytes are much larger than cultured adipocytes. This hypothesis is supported by our observation that Sfrp5 mRNA expression in adipose tissues was elevated further in a variety of obese models, including leptin receptor–deficient Leprdb/db mice (Figure 1C), leptin-deficient Lepob/ob mice (Supplemental Figure 1B), ovariectomized mice (Figure 1D), hyperphagic Pomc-Tsc1 conditional KO mice (data not shown), or HFD-fed mice (Figure 1E). In addition, Sfrp5 expression was reduced in adipose tissue from Lxrβ–/– mice (Supplemental Figure 1C), in which adipocyte size does not increase with HFD feeding (37). Our data confirmed and extended the work of other investigators (31, 32, 35), but not that of Ouchi, Walsh, and colleagues (34), who report that SFRP5 declines with genetic or dietary obesity. Although Sfrp5 mRNA was expressed at low levels in a number of metabolic tissues, the dramatic induction with diet-induced obesity was specific to WAT depots, with higher expression in visceral WAT and epididymal WAT (eWAT) compared with subcutaneous depots (Figure 1E). Furthermore, Sfrp5 expression in WAT positively correlated with body fat percentage (Figure 1F, Supplemental Figure 1D, and ref. 32) and adipocyte size (Figure 1G). No relationship to adiposity was observed with Sfrp2, which was expressed in preadipocytes and declined during adipogenesis (Figure 1F and Supplemental Figure 1A). Taken together, these data provide compelling evidence that Sfrp5 mRNA is induced during adipogenesis, and that expression closely follows adipocyte growth and hypertrophy, both throughout development and with obesity.
Figure 1. Sfrp5 mRNA is induced during adipogenesis and further increased with obesity.
(A) RNA was isolated from EMSCs during adipogenesis (average Ct at day 12, 27.5). Values were normalized to Tbp mRNA. n = 7–11. (B) Sfrp5 mRNA was higher in adipocytes (AD; average Ct, 24.8) than the stromal vascular fraction (SV; average Ct, 33.0). n = 10. (C) Increased Sfrp5 mRNA in eWAT of 12-week-old male Leprdb/db mice (Ct, ∼24.0) relative to Lepr+/+ controls (Ct, ∼27.5). n = 7 per group. (D) Increased Sfrp5 in WAT after ovariectomy (Ovx). Shown is percent body fat 8 weeks after surgery and relative Sfrp5 mRNA expression in dorsolumbar WAT of ovariectomized (Ct, ∼18.8; n = 5) and sham-operated control (Ct, ∼21.6; n = 6) mice. (E) Beginning at 8 weeks of age, mice were fed NCD or HFD for 12 weeks, and Sfrp5 mRNA was assessed in eWAT (NCD Ct, ∼30.2; HFD Ct, ∼19.91), perirenal WAT (pWAT), dorsolumbar WAT (dWAT), brown adipose tissue (BAT), muscle, liver, pancreas, and kidney. n = 7 per group. (F) Assessment of Sfrp5 and Sfrp2 mRNA in eWAT from mice fed low-fat diet or HFD for 6 months (n = 10 per group) showed a positive correlation between Sfrp5 mRNA expression and percent body fat (R2 = 0.84). (G) Positive correlation between adipocyte size and Sfrp5 mRNA (R2 = 0.78). Leptin (R2 = 0.5141) and Sfrp2 (R2 = 0.1917) are shown for comparison. For A–E, values are mean ± SEM. *P < 0.05; **P < 0.01.
Characterization of Sfrp5Q27stop mutant mice.
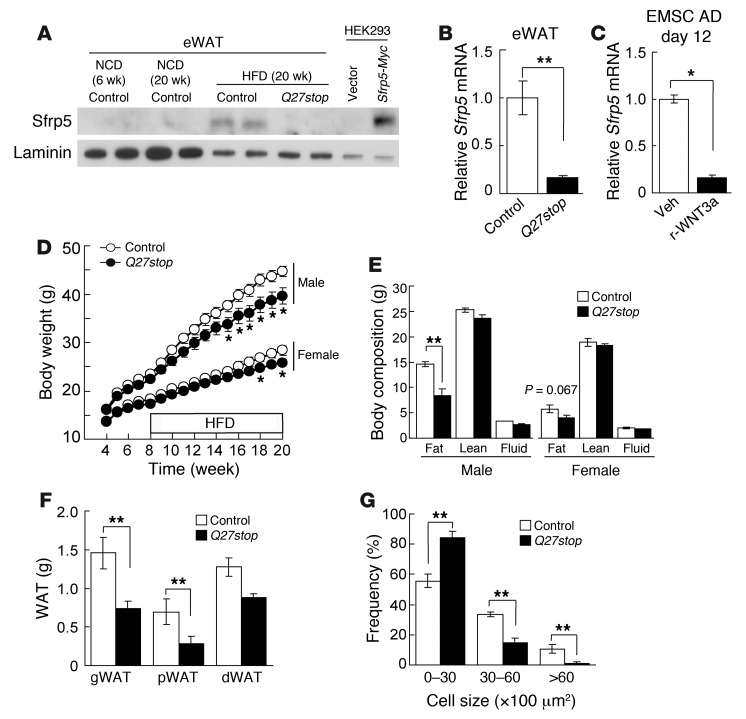
To evaluate the role of Sfrp5 in adipose biology and obesity, we obtained Sfrp5Q27stop mice generated by N-ethyl-N-nitrosourea mutagenesis (38, 39). In these mice, a single base pair mutation results in a premature stop codon at glutamine 27 that is predicted to result in a nonfunctional allele (Supplemental Figure 2A). To test this prediction, we used immunoblotting to analyze Sfrp5 expression in WAT of control or Sfrp5Q27stop mice. In control mice, SFRP5 was detectable only after 20 weeks of HFD feeding (Figure 2A), consistent with the elevated Sfrp5 transcript levels observed under these conditions (Figure 1A). In contrast, SFRP5 was not detectable in HFD-fed Sfrp5Q27stop mice using antisera directed to either the N terminus (Figure 2A) or an internal antigen (data not shown). In addition, expression of Sfrp5 mRNA was decreased in adipose tissues and the adipocyte fraction of mutant mice (Figure 2B and Supplemental Figure 2B), which suggests that expression of Sfrp5 mRNA is maintained through a positive feedback mechanism. The well-known function of SFRPs as WNT inhibitors (29) suggests that Sfrp5Q27stop mice may have elevated WNT signaling, which might contribute to suppression of Sfrp5 mRNA. Consistent with this possibility, addition of recombinant WNT3a to cultured EMSC adipocytes rapidly suppressed Sfrp5 mRNA (Figure 2C). Finally, in light of the functional redundancy that exists for Sfrp1, Sfrp2, and Sfrp5 during development (40), we further evaluated expression of other Sfrp family members in Sfrp5Q27stop mice. This revealed a compensatory increase in Sfrp1, but not in Sfrp2, Sfrp3, or Sfrp4, in adipose tissue of mutant animals (Supplemental Figure 2C). Taken together, these data indicate that Sfrp5Q27stop mice have reduced expression of Sfrp5 mRNA and complete deficiency of SFRP5 protein, with potential compensatory upregulation of Sfrp1.
Figure 2. Characterization of Sfrp5Q27stop mutant mice.
(A) SFRP5 protein was detected in eWAT of HFD-fed control mice, but not eWAT of NCD-fed control or Sfrp5Q27stop mice. Mice were fed HFD for 12 weeks, starting at 8 weeks of age. HEK 293T cells transfected with Sfrp5-Myc fusion construct served as a positive control. (B and C) Decreased Sfrp5 mRNA in Sfrp5Q27stop eWAT might be caused by elevated Wnt signaling. (B) Sfrp5 mRNA expression in eWAT from HFD-fed control or Sfrp5Q27stop mice, normalized to Tbp mRNA and expressed relative to controls (n = 17 per group). (C) EMSC adipocytes (day 12) were treated with vehicle or recombinant WNT3a (100 ng/ml) for 48 hours. Sfrp5 mRNA expression was normalized to Tbp and expressed relative to vehicle. (D) Sfrp5Q27stop mice resisted diet-induced weight gain. Body weight in mice from 4 to 20 weeks of age (n = 14–19). HFD was started at 8 weeks of age. (E) Decreased fat mass in Sfrp5Q27stop mice at 20 weeks of age (n = 8). (F) Reduced weight of WAT depots in female Sfrp5Q27stop mice at 20 weeks of age (n = 6–7). (G) Decreased weight of adipose tissue in Sfrp5Q27stop mice was due to reduced adipocyte size. H&E-stained gWAT samples from 20-week-old female mice were evaluated by histomorphometry, and the frequency of adipocyte sizes was plotted. For B–G, data are average ± SEM. *P < 0.05; **P < 0.01.
SFRP5 is required for adipocyte hypertrophy, but not hyperplasia, during obesity.
The phenotypes of control and Sfrp5Q27stop mice were indistinguishable when fed normal chow diet (NCD), as assessed by total body weight, tissue and organ weights, and body composition (data not shown); this may relate to the low levels of Sfrp5 mRNA and SFRP5 protein expression observed under this condition (Figure 1 and Figure 2A). Thus, we sought to investigate the role of SFRP5 by challenging Sfrp5Q27stop mice with HFD. Compared with controls, Sfrp5Q27stop mice resisted HFD-induced weight gain, with males demonstrating a stronger phenotype than females (Figure 2D). Notably, no change in body weight was observed in approximately 40% of cohorts (data not shown). Although some cohorts were statistically underpowered, we speculate that incomplete phenotypic penetrance of Sfrp5Q27stop mice was due to differential compensation by Sfrp1 and/or other factors. Investigation of body composition revealed that the differences in total body weight were due to reduced fat mass in Sfrp5Q27stop mice (Figure 2, E and F), with more dramatic reductions in those depots that showed the highest Sfrp5 expression (Figure 1E).
We and others have established that several WNTs, including WNT10b, are endogenous repressors of adipogenesis (15, 16, 18, 41–43), and enforced expression of Wnt10b in adipose tissues combats the development of dietary and genetic obesity (21, 22). We thus hypothesized that, as adipocytes reach their capacity to store lipid, they secrete SFRP5 to act in a paracrine manner to inhibit these WNTs and thereby recruit new adipocytes. To test this hypothesis, we performed histological analyses on eWAT from control and Sfrp5Q27stop mice that had been challenged with HFD. Unexpectedly, we found that the total number of adipocytes within the adipose depot was similar between genotypes (Supplemental Figure 2D). However, the number of large adipocytes was significantly reduced in Sfrp5Q27stop mice (Figure 2G and Supplemental Figure 2D), accounting for the decreased eWAT mass in these animals. The observed reduction in lipid storage in adipocytes of Sfrp5Q27stop mice suggests that SFRP5 and WNT signaling have unanticipated effects on local or whole-body energy metabolism. Paradoxically, the phenotype of Sfrp5Q27stop mice was opposite to that described by Ouchi et al. (34), who found that Sfrp5–/– mice gain more weight than controls due to adipocyte hypertrophy.
Loss of SFRP5 results in reduced leptin and mild improvements in glucose tolerance and insulin sensitivity.
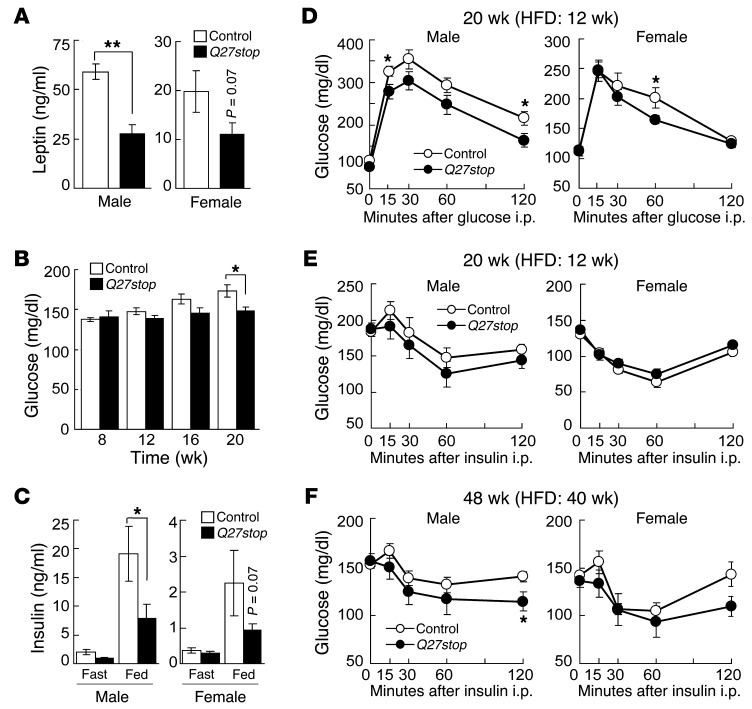
We next measured metabolic variables, such as circulating leptin, glucose, and insulin concentrations, to determine whether these were altered in HFD-fed Sfrp5Q27stop mice. Consistent with the reduced adiposity we observed (Figure 2), serum leptin concentrations in male Sfrp5Q27stop mice were significantly lower than in controls (Figure 3A). In addition, we monitored blood glucose concentrations in ad libitum–fed mice at 4-week intervals, starting with the first HFD feeding at 8 weeks of age. No significant differences in circulating glucose in either genotype were observed through 16 weeks of age (Figure 3B and Supplemental Figure 3A); however, by 20 weeks, glucose concentrations of male Sfrp5Q27stop mice were lower than those of control mice (Figure 3B). In addition, male Sfrp5Q27stop mice were resistant to HFD-induced hyperinsulinemia, a trend also observed in female Sfrp5Q27stop mice (Figure 3C).
Figure 3. Loss of SFRP5 results in reduced leptin and mild improvements in glucose tolerance and insulin sensitivity.
(A) Lower serum leptin in Sfrp5Q27stop mice. Animals were fed HFD for 16 weeks. n = 8 (male); 13–15 (female). (B) Blood glucose and (C) insulin concentrations in mice fed HFD ad libitum (n = 14). Insulin was determined in 12-hour fasted or ad libitum HFD-fed mice at 20 weeks of age. n = 10–13 (male); 20 (female). (D) Glucose tolerance in 20-week-old mice fed HFD. n = 14 (male); 14–15 (female). (E and F) Insulin tolerance test after a 3-hour fast. Mice were evaluated at (E) 20 (n = 14 [male]; 18–19 [female]) or (F) 48 (n = 8) weeks of age. For A–F, data are average ± SEM. *P < 0.05; **P < 0.01.
Ouchi et al. (34) suggest that Sfrp5–/– mice fed an obesogenic diet develop insulin resistance. To determine whether glucose homeostasis was altered in Sfrp5Q27stop mice, we performed glucose tolerance tests and insulin tolerance tests on HFD-fed mice. Extensive experimentation revealed modestly improved glucose tolerance in male Sfrp5Q27stop animals, with significant reductions in circulating glucose observed at some time points after glucose administration to male and female mice (Figure 3D). Although glucose concentrations after insulin injection of male Sfrp5Q27stop mice tended to be reduced, there was no statistical difference between genotypes in insulin sensitivity in either sex (Figure 3E). We also performed an experiment in which mice received HFD for 40 weeks, similar to the previously reported time frame of dietary treatment in which glucose intolerance was observed (34); again, male Sfrp5Q27stop mice had a weak tendency to improved glucose homeostasis (Figure 3F and Supplemental Figure 3B), but this metabolic phenotype was mild.
To create a model in which we could investigate the potential relationship between SFRP5 and insulin sensitivity in vitro, we isolated EMSCs from control and Sfrp5Q27stop mice and differentiated them into adipocytes. As expected for a gene expressed late in adipocyte differentiation, loss of Sfrp5 did not influence adipogenesis, nor did it influence expression of common adipocyte markers such as PPARγ or FABP4 (Supplemental Figure 3, C and D). However, Sfrp5Q27stop adipocytes had reduced expression of Sfrp5 mRNA and a compensatory increase in Sfrp1 (Supplemental Figure 3E), consistent with our above-described observations in adipose tissue (Figure 2B and Supplemental Figure 2C). Under our cell culture conditions, loss of SFRP5 did not influence signaling events implicated in insulin resistance, including phosphorylation of Ser307-IRS1 or JNK (Supplemental Figure 3D), a target of noncanonical WNT signaling (44, 45). In contrast, both of these signaling proteins were phosphorylated in response to lipopolysaccharide, as expected (46, 47). In contrast to Ouchi et al., who reported that WNT5a stimulates JNK phosphorylation in adipocytes and that SFRP5 binds to and antagonizes WNT5a (34), we found no evidence that recombinant WNT5a stimulates phosphorylation of JNK or IRS1 in either control or Sfrp5Q27stop EMSC adipocytes (Supplemental Figure 3F). To further explore the potential role of SFRP5 in modulation of WNT5a, we overexpressed Sfrp5 in 3T3-L1 adipocytes; however, we did not observe differences in phosphorylation of JNK or IRS1 with WNT5a, in either the absence or the presence of overexpressed SFRP5 (Supplemental Figure 3G). Finally, we measured insulin-stimulated glucose uptake in the above-described primary or immortalized adipocyte models and found that glucose uptake was not influenced by either loss or overexpression of SFRP5 (data not shown). Taken together, these data provided no support for the hypothesis that WNT5a or SFRP5 regulates insulin sensitivity of adipocytes.
SFRP5 deficiency does not detectably alter energy balance.
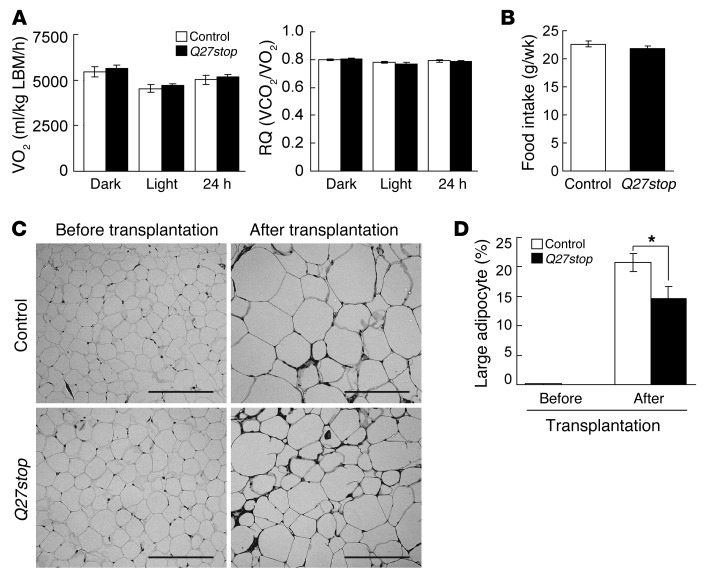
We observed that SFRP5 was required for adipocyte growth under obesogenic conditions (Figure 2). To evaluate the effects of SFRP5 deficiency on whole-body energy balance, we measured metabolic rate in control and Sfrp5Q27stop mice using the Comprehensive Lab Animal Monitoring System (CLAMS). Despite a decrease in fat mass in Sfrp5Q27stop mice, differences in oxygen consumption and respiratory exchange ratio were not detectable between genotypes (Figure 4A and Supplemental Figure 4A). Differences in food intake were also not observed (Figure 4B). As SFRP5 deficiency did not cause detectable changes in whole-body energy balance, small, insensible alterations in food intake and/or energy expenditure over a period of many weeks likely cause the resistance to obesity observed in Sfrp5Q27stop mice.
Figure 4. SFRP5 regulates adipocyte size during obesity in a tissue-autonomous manner.
(A) No difference in oxygen consumption (VO2) or respiratory quotient (RQ) in Sfrp5Q27stop mice. Male control and Sfrp5Q27stop mice (n = 8 each) were evaluated by CLAMS for 3 days. Oxygen consumption was normalized to lean body mass (LBM). (B) Weekly food intake in control and Sfrp5Q27stop mice (n = 9) was similar between genotypes. (C) Adipocyte hypertrophy in Sfrp5Q27stop mice was tissue-autonomous. eWAT from control or Sfrp5Q27stop donors (n = 6 each) was transplanted subcutaneously into Leprdb/db recipients (n = 3) at 4 weeks of age. 10 weeks after transplantation, donor tissues were excised and subjected to morphological analysis. Representative H&E staining of gWAT before and after transplantation into Leprdb/db recipient mice is shown. Scale bars: 200 μm. (D) Reduced frequency of large adipocytes (>8,000 μm2) in Sfrp5Q27stop WAT after transplantation. For A, B, and D, values are mean ± SEM. *P < 0.01.
SFRP5 is a secreted protein tightly associated with the extracellular matrix.
SFRPs are generally believed to be secreted proteins that act through autocrine and paracrine mechanisms; however, the recent report by Ouchi et al. suggests that SFRP5 is detectable in culture media and serum, at least when overexpressed (34). Because adipose tissue appeared to be the major source of SFRP5 in obese mice (Figure 1E), it was important to establish whether SFRP5 influences energy metabolism in distal tissues and organs through an endocrine mechanism, similar to leptin or adiponectin, or whether its effects are exclusively local. To address these questions, we transiently transfected 293T cells with either empty vector or a vector containing a Sfrp5-Myc fusion construct; we also used the above-described 3T3-L1 cells, which stably overexpress Myc-tagged SFRP5 protein. SFRP5 was easily detected in whole-cell lysates and extracellular matrix fractions for 293T cells and 3T3-L1 adipocytes. In contrast, SFRP5 was detected in conditioned media of 293T cells only with long film exposures and very sensitive enhanced chemiluminescence, and could not be detected in 3T3-L1 adipocyte media even under these conditions (Supplemental Figure 4, B and C). However, treatment of 293T cells or 3T3-L1 adipocytes with heparin stimulated release of some SFRP5 from the extracellular matrix into the media (Supplemental Figure 4, B and C), consistent with previous reports identifying SFRPs as heparin-binding proteins (48–50).
Effects of SFRP5 on adipocyte growth are adipose tissue–autonomous.
The results of our investigations as to whether SFRP5 is secreted in a soluble form suggested that the effects of this protein on metabolism are likely restricted to adipose tissue, which was the major source of SFRP5 in obese animals (Figure 1E). Therefore, to investigate whether alterations in adipocyte size in Sfrp5Q27stop mice are tissue-autonomous, we transplanted eWAT from control and Sfrp5Q27stop mice subcutaneously into Leprdb/db recipient mice at 4 weeks of age (Supplemental Figure 4D). At 10 weeks after transplantation, the donor tissue was excised (Supplemental Figure 4E) and subjected to histomorphological analysis. Substantial adipocyte growth occurred within the obesogenic environment of the Leprdb/db recipient mice (Figure 4, C and D); however, the proportion of large adipocytes in the Sfrp5Q27stop transplant was reduced approximately 30% compared with that of controls (Figure 4, C and D), consistent with results obtained in WAT of Sfrp5Q27stop mice. Taken together, these data indicate that SFRP5 acts through a tissue-autonomous mechanism to regulate adipocyte size during obesity.
SFRP5 regulates mitochondria number.
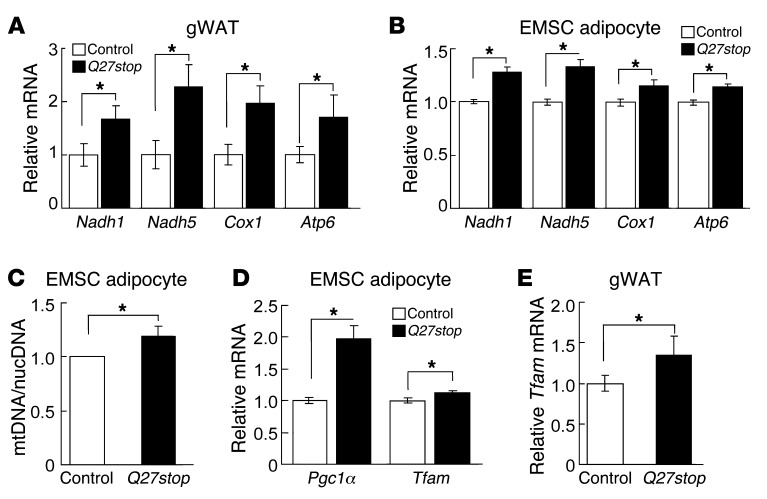
To obtain a global and unbiased perspective on genes that may be regulated by SFRP5, we performed gene profiling of eWAT from HFD-fed control and Sfrp5Q27stop mice. To identify primary effects of SFRP5 ablation, rather than effects secondary to altered adiposity, we profiled mice with similar body masses between genotypes. Affymetrix microarray data were analyzed using Ingenuity Pathway Analyses to identify relevant biological networks or pathways (Supplemental Figure 5A). Gene profiling results indicated an upregulation of genes involved in mitochondrial oxidative phosphorylation (termed OXPHOS complex genes) in Sfrp5Q27stop mice, which raises the possibility that SFRP5 deficiency protects against obesity by increasing mitochondrial oxidative phosphorylation in adipocytes. qPCR analysis confirmed that RNAs encoding mitochondrial OXPHOS complex proteins, such as NADH dehydrogenase subunit 1 (Nadh1), NADH dehydrogenase subunit 5 (Nadh5), cytochrome c oxidase subunit I (Cox1), and ATP synthase FO subunit 6 (Atp6), were increased in the Sfrp5Q27stop samples evaluated by gene profiling (Supplemental Figure 5B) as well as in eWAT from an independent cohort of HFD-fed mice (Figure 5A). These effects of SFRP5 deficiency were cell-autonomous and specific to adipocytes, since we also observed elevated mitochondrial gene expression in cultured adipocytes (Figure 5B), but not in EMSC precursors (data not shown), from Sfrp5Q27stop mice. To determine whether elevated expression of these mitochondrial RNAs was due to increased number of mitochondria, we tested the ratio of mitochondrial DNA (mtDNA) to nuclear DNA (nucDNA) by qPCR in the cultured adipocytes. Although a significant increase in mitochondrial number was not always observed in Sfrp5Q27stop adipocytes, when all 6 independent experiments were taken together, the number of mitochondria increased approximately 20% (Figure 5C), consistent with the observed increase in mitochondrial gene expression (Figure 5B). These data suggest that Sfrp5Q27stop mice may restrain adipocyte growth under obesogenic environmental conditions due to increased number of mitochondria.
Figure 5. SFRP5 regulates mitochondrial biogenesis.
(A) Increased mitochondrial OXPHOS complex genes in gWAT of Sfrp5Q27stop mice. In gWAT from female mice fed HFD for 12 weeks, mRNA for Nadh1, Nadh5, Cox1, and Atp6 was measured by qPCR. Values were normalized to Hprt mRNA and expressed relative to control mice (n = 22 per genotype). (B) Increased mitochondrial OXPHOS complex genes in Sfrp5Q27stop EMSC adipocytes. Values were normalized to Tbp mRNA and expressed relative to control adipocytes (n = 6–9 independent experiments). (C) Sfrp5Q27stop adipocytes have more mitochondria. mtDNA/nucDNA ratio was assessed by qPCR in EMSC adipocytes (day 12). mtDNA values were normalized to nuclear-encoded Gcg gene (n = 6 independent experiments). (D) Elevated Pgc1α and Tfam mRNA in Sfrp5Q27stop day-12 EMSC adipocytes. (E) Increased Tfam mRNA in gWAT from Sfrp5Q27stop mice. Sfrp5Q27stop female mice fed HFD for 12 weeks were measured as in A. For B–D, data are mean ± SEM relative to control values (assigned as 1) within each independent experiment. For A and E, values are mean ± SEM for all samples. *P < 0.05.
To evaluate the mechanism whereby SFRP5 deficiency increases mitochondrial copy number, we evaluated established mechanisms for increased biogenesis (51). Although differences in nuclear respiratory factor 1 (Nrf1), Nrf2, and Errγ mRNAs in cultured adipocytes derived from Sfrp5Q27stop mice were not observed (data not shown), the elevated expression of PPARγ coactivator 1α (Pgc1α) (Figure 5D) was suggestive of a potential mechanism for increased mitochondrial biogenesis. This idea was supported by the increased expression of mitochondrial transcription factor A (Tfam) observed in both differentiated adipocytes and WAT from Sfrp5Q27stop mice (Figure 5, D and E), as Tfam is a well-established target of PGC1α that regulates both mitochondrial transcription and mtDNA copy number (52). Taken together, our results suggest that the increase in SFRP5 with obesity facilitates lipid storage by reducing mitochondrial biogenesis.
Elevated adipocyte respiration in SFRP5-deficient adipocytes is due to mitochondria number and functionality.
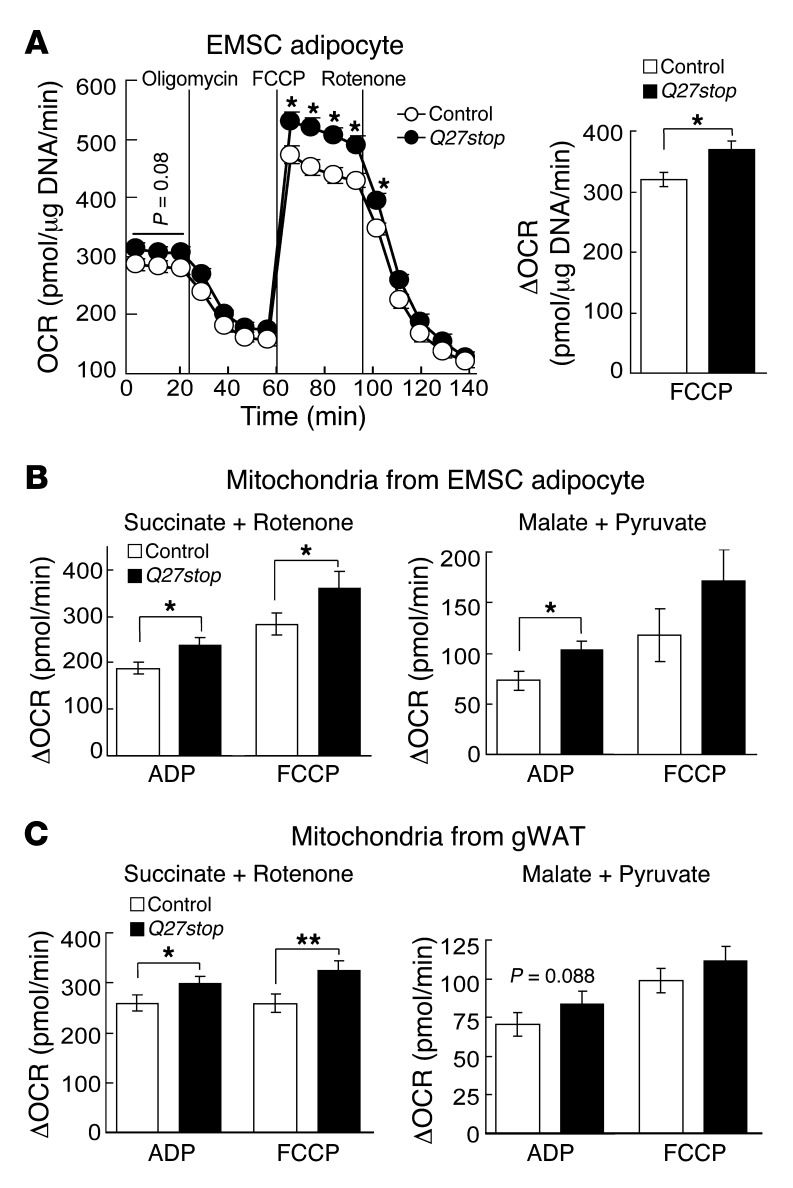
Although white adipocytes have less oxidative capacity than do brown adipocytes, several reports have demonstrated the importance of white adipocyte mitochondria for adipogenesis and development of adiposity (53, 54). To further investigate whether loss of SFRP5 affects adipocyte oxidative capacity, we used a Seahorse XF Analyzer to measure oxygen consumption rate (OCR) of differentiated adipocytes and mitochondria isolated from cultured adipocytes or adipose tissue, both under basal conditions and in response to oligomycin, FCCP, and rotenone. Although basal OCR in Sfrp5Q27stop EMSC adipocytes tended to be higher than in control adipocytes, this difference did not reach statistical significance (Figure 6A). However, both maximal OCR (after injection of the uncoupling agent FCCP) and respiratory capacity were significantly higher in Sfrp5Q27stop EMSC adipocytes (Figure 6A), indicative of greater maximal aerobic capacity in Sfrp5Q27stop than control adipocytes. These observations are consistent with SFRP5 deficiency causing increased mitochondrial number, but they could also result from increased mitochondrial functionality.
Figure 6. SFRP5 regulates adipocyte respiration and mitochondrial function.
(A) Higher oxidative capacity in Sfrp5Q27stop EMSC adipocytes. Basal OCR and effects of oligomycin, FCCP, and rotenone are demonstrated. The difference between maximal OCR after FCCP and minimum OCR after oligomycin (ΔOCR) is also shown. Values (mean ± SEM) are normalized to DNA (n = 10 independent experiments). (B and C) Increased difference between maximal OCR after ADP or FCCP and average OCR after oligomycin (ΔOCR) in mitochondria isolated from (B) adipocytes and (C) gWAT of Sfrp5Q27stop mice fed HFD for 12 weeks. Succinate (2.5 mM) and rotenone (1 μM) were used to assay complex II–dependent respiration; malate (2.5 mM) and pyruvate (2.5 mM) were used to assay complex I–dependent respiration (n = 5 independent experiments). Data are mean ± SEM. *P < 0.05; **P < 0.01.
To determine whether increased respiratory capacity in EMSCs from Sfrp5Q27stop mice is exclusively caused by increased mitochondrial number, we first measured OCR in isolated mitochondria from EMSC adipocytes. Interestingly, maximal consumption of oxygen in mitochondria from Sfrp5Q27stop adipocytes was higher than that of controls (Figure 6B), despite similar numbers of mitochondria included in the assay for each genotype. These results were observed for functionality of both complex II (succinate plus rotenone) and complex I (malate plus pyruvate) (Figure 6B and Supplemental Figure 6B). We then extended this analysis to mitochondria isolated from eWAT. Mitochondria from HFD-fed Sfrp5Q27stop mice had elevated OCR, with improved functionality for complex II and a trend for complex I (Figure 6C and Supplemental Figure 6B). These data suggest that increased respiratory capacity in Sfrp5Q27stop EMSC adipocytes is due to increased mitochondria biogenesis as well as improved aerobic capacity per mitochondrion.
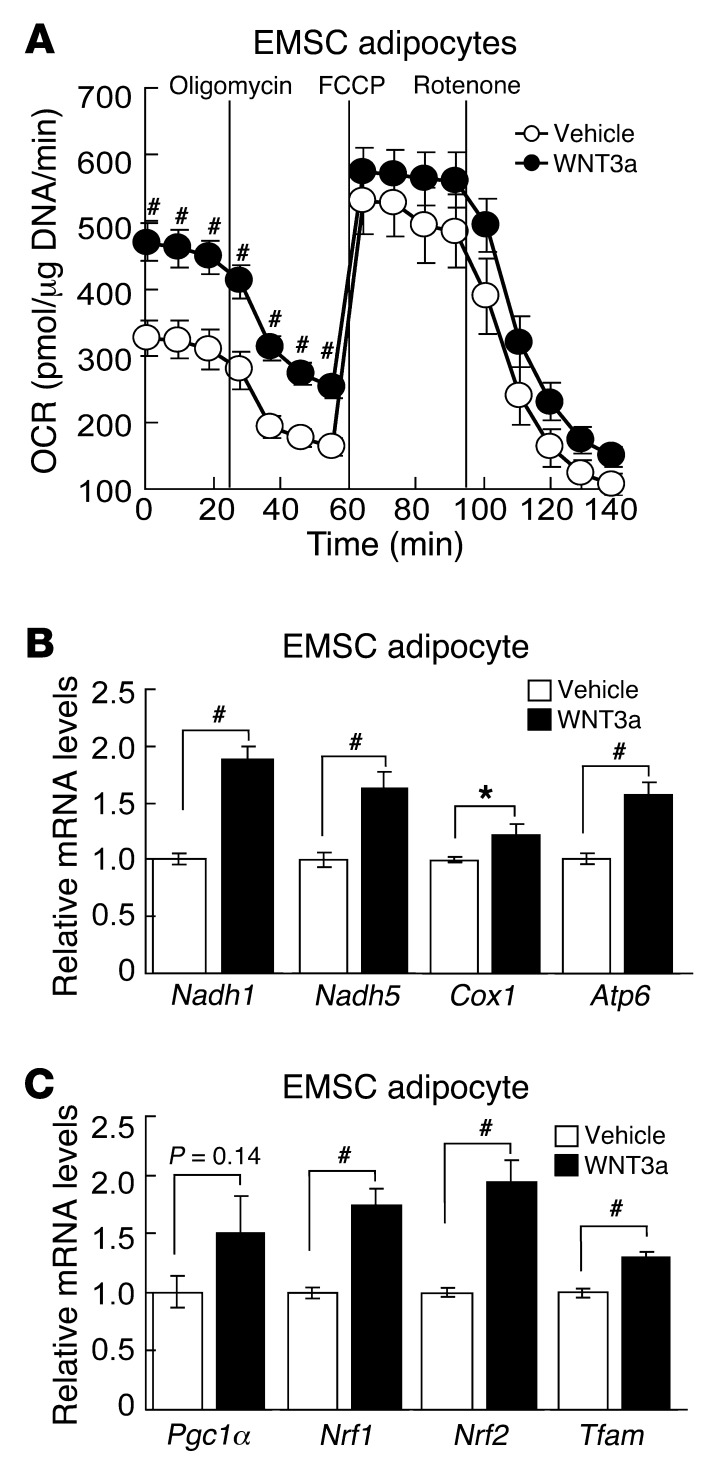
WNT3a stimulates mitochondrial respiration and gene expression.
Although SFRPs are well known to inhibit WNT signaling, they also influence differentiation and other cellular functions through a number of other mechanisms, such as inhibition of bone morphogenetic proteins (33, 55). To evaluate whether SFRP5 deficiency influences mitochondrial biology by increasing WNT signaling, we treated control EMSC adipocytes with recombinant WNT3a for 48 hours. We observed an increase in basal OCR with WNT3a treatment (Figure 7A). In addition, we evaluated a number of genes involved in mitochondrial biogenesis and function and found that WNT3a strongly induced expression of Nadh1, Nadh2, Cox1, and Atp6 (Figure 7B), as observed in Sfrp5Q27stop EMSC adipocytes and adipose tissue (Figure 5). Next, we examined whether mitochondrial biogenesis markers are also regulated by WNT3a treatment. Similar to our results in Sfrp5Q27stop EMSC adipocytes (Figure 5), WNT3a induced expression of both Pgc1α and Tfam (Figure 7C); however, unlike in Sfrp5Q27stop EMSC adipocytes, WNT3a also elevated expression of Nrf1 and Nrf2. These experiments revealed some of the complexity in signaling and effects of WNT ligands in adipocytes and were consistent with the hypothesis that enhanced mitochondrial biogenesis and function in Sfrp5Q27stop mice is caused by increased WNT signaling.
Figure 7. WNT3a stimulates mitochondrial respiration and gene expression.
(A) Control EMSC adipocytes were treated with WNT3a (100 ng/ml) for 48 hours. Basal OCR and effects of oligomycin, FCCP, and rotenone are shown (representative of 5 independent experiments). (B and C) WNT3a induced mRNA expression of (B) mitochondrial genes (Nadh1, Nadh5, Cox1, and Atp6) and (C) nuclear-encoded regulators (Pgc1α, Nrf1, Nrf2, and Tfam). Transcript expression was assessed by qPCR after exposure to WNT3a (100 ng/ml) for 48 hours. Values (normalized to Tbp mRNA) are expressed relative to control mice (n = 3 independent experiments). *P < 0.05; #P < 0.01.
Discussion
The literature regarding the regulation of SFRP5 with obesity shows conflicting data. We found here that Sfrp5 mRNA was strongly induced in adipose tissues of 5 murine models of obesity, with increased SFRP5 protein levels also observed with diet-induced obesity (Figure 1 and Supplemental Figure 1). Our data are consistent with several prior publications (31, 32, 35) reporting elevated Sfrp5 mRNA with obesity. However, Ouchi et al. reported very different findings from the previous literature, although the authors did not note the discrepancy (34). They report that, in C57BL/6J mice fed an obesogenic diet (35.8% fat and 36.8% carbohydrate), expression of Sfrp5 is induced after 12 weeks, but is suppressed relative to controls after 24 weeks of the high-fat/high-sucrose diet. The basis for this incongruity is unclear, but is unlikely to be the extent of obesity or duration of feeding, since these were similar with our present study and others (31, 32). Perhaps the answer lies in the specific dietary content: Ouchi et al. uniquely used Bio-Serv diet no. F1850 (34), whereas we and others (31) used Research Diets D12451; however, the calories from fat and carbohydrate, and contributions from lard and sucrose, appear similar between these 2 diets. Ouchi et al. also reported that Sfrp5 expression in WAT is reduced in ZDF-Leprfa/fa rats and in Lepob/ob mice (34); although the literature shows no comparable data for SFRP5 expression in rat WAT, these Lepob/ob data are again discordant with ours and others’ (31). The Lepob/ob mice in our experiment were 12 weeks of age, and those of Lagathu et al. (31) were 5 and 16 weeks of age; thus, it is formally possible that the expression pattern changes dramatically between these ages and 20 weeks of age, as evaluated by Ouchi et al. (34). However, the growth chart from JAX labs for B6.V-Lepob/J mice (56) indicates that obesity is well developed by 8 weeks, and only incremental increases in body weight occur after 16 weeks. Thus, the preponderance of evidence indicates that expression of Sfrp5 is induced during adipogenesis, is higher in isolated adipocytes than in the stromal-vascular fraction of WAT from lean animals, and increases dramatically with obesity development.
Although further study is required to establish the mechanistic basis for elevated SFRP5 in adipose tissue of obese mice, hints as to the mechanism come from our data that Sfrp5Q27stop mice had not only premature termination of SFRP5 protein (Figure 2A, Supplemental Figure 2A, and ref. 39), but also reduced Sfrp5 mRNA expression (Figure 2B). This observation suggests a positive feedback loop in which expression of SFRP5 inhibits WNT signaling, further increasing Sfrp5 expression. This idea is supported by our demonstration that WNT3a suppressed Sfrp5 mRNA expression in adipocytes (Figure 2C). We also observed that WNT3a increased Sfrp1 mRNA expression (data not shown), which is consistent with the upregulation of Sfrp1 observed in adipose tissue and EMSC adipocytes from Sfrp5Q27stop mice (Supplemental Figure 2C and Supplemental Figure 3E). Therefore, this regulation may contribute to the incomplete penetrance of SFRP5 deficiency on resistance to HFD-induced obesity. In addition, knockdown of Sfrp1 in adipocytes resulted in a slight increase in Sfrp5 and dramatic induction of Sfrp2 (data not shown), which supports the genetic evidence for functional redundancy among this subfamily of SFRPs during early development of mice (40, 57).
Elegant work from the Kozak lab on variation in Sfrp5 gene expression in genetically identical C57BL/6J mice fed HFD suggests that expression of SFRP5 is regulated by epigenetic mechanisms (32). This idea is supported by the cancer literature, which indicates that methylation and inactivation of SFRP5 is associated with ovarian, gastric, breast, and renal cancers as well as myeloid leukemia (58–62). However, Sfrp5 upregulation is absent in 3T3-L1 adipocytes treated with a demethylating agent (35). Additionally, obesity does not affect methylation of specific CpG sites in the Sfrp5 promoter (35), although it is possible that regulation is from distal enhancers that remain to be evaluated.
The finding of Ouchi et al. that SFRP5 can be systemically delivered (34) is intriguing in light of prior work indicating that SFRPs act through autocrine and paracrine mechanisms (63). Our work with overexpressed SFRP5 in 293T and 3T3-L1 cells demonstrated that, under these conditions, SFRP5 was easily detected in whole-cell lysates and extracellular matrix fractions and that significant detection in media was possible only when SFRP5 was competed off the extracellular matrix using heparin. These biochemical characteristics of SFRP5 are consistent with properties of other SFRPs (48–50). Compelling experiments in Xenopus indicate that SFRPs readily diffuse within extracellular matrix of the developing embryo, which extends the gradient and signaling ranges of WNTs and BMPs (63). SFRP5 or other family members have not been detected in proteomic analysis of serum proteins (64, 65); however, a recent report suggests that circulating human SFRP5 can be detected by ELISA and that circulating concentrations are not influenced by obesity, but increase with calorie restriction of very obese individuals (66). Although adipose tissue is an endocrine organ that secretes many factors influencing satiety, insulin resistance, and metabolism (67, 68), the available evidence suggests that metabolic effects of SFRP5 are mediated through actions restricted to adipose tissue.
Our present data demonstrated that Sfrp5Q27stop mice were resistant to diet-induced obesity due to impaired adipocyte growth. In contrast, Ouchi et al. found that Sfrp5–/– mice fed HFD gain additional fat mass and show increased adipocyte size (34). In accordance with these basic observations, Sfrp5Q27stop mice had a mild improvement in glucose tolerance (Figure 3), whereas Sfrp5–/– mice developed severe glucose intolerance and hepatic steatosis (34). The proposed mechanism for this observation is that SFRP5 is required to neutralize WNT5a, which is elevated with obesity and acts in adipose tissue to activate JNK1, a well-known cause of metabolic dysfunction and macrophage activation. Our experiments confirmed the observation of Ouchi et al. that Wnt5a is slightly elevated with obesity (data not shown and ref. 34), which had previously been reported by Koza et al. (32). However, our data did not support the elevated WNT5a/SFRP5 ratio reported by Ouchi et al. (34), given the dramatic increase in SFRP5 we observed (Figure 1 and Supplemental Figure 1).
The Sfrp5Q27stop mutation, while generated by chemical mutagenesis, was backcrossed to the wild-type parental strain (C57BL/6J) for about 20 generations. Thus, only very closely linked secondary mutations would not have been eliminated. Sfrp5 was chosen for this functional study on the basis of its expression profile and close connection to adipocyte biology. Thus, the phenotype revealed was also expected; this would not be the case for a hypothetical second mutation, which would effectively have been chosen at random. To further address this possibility, we examined the 11 genes contained within the 400 kb surrounding Sfrp5; of these, only 3 showed a moderate level of expression in adipose tissue, based on publicly available tissue arrays. However, to our knowledge, there is no published literature on any of the genes related to adipose tissue. Thus, although a confounding second mutation cannot be excluded without sequencing the genome, it appears to be unlikely. Excision of the first exon of Sfrp5 to create the Sfrp5–/– line used by Ouchi et al. (34), as first described by Satoh (40), is not expected to directly interfere with expression of non-coding RNAs or other proteins, although it is conceivable that transcription factors binding to the exon could serve as enhancers for distal genes. An independent Sfrp5–/– mouse line has been created by Leaf et al. (57); however, these mice unfortunately no longer exist. Further studies using adipose-specific knockout of Sfrp5 will be required to shed light on the discrepancy between the phenotypes described herein for Sfrp5Q27stop mice and the Sfrp5–/– mouse line used by Ouchi et al. (34).
Mitochondrial biogenesis and remodeling is important for adipocyte differentiation and function (53, 54, 69), and to our knowledge, regulation of these processes by WNT signaling has not been previously reported in this context. We observed that WNT3a stimulated mitochondrial biogenesis and oxygen consumption in adipocytes. These effects appeared to be similar to those observed in C2C12 cells, in which WNT3a stimulates mitochondrial biogenesis through stabilization of β-catenin and induction of IRS1 and MYC (70). However, the mechanism in adipocytes is unlikely to involve the β-catenin pathway, since β-catenin is suppressed during adipogenesis and targeted for ubiquitin-mediated turnover by PPARγ (15, 42). Furthermore, we did not observe increases in either IRS1 or MYC in WNT3a-treated adipocytes (data not shown). Instead, the mechanism in adipocytes appeared to be through induction of PGC1α, TFAM, and potentially other regulators of mitochondrial biogenesis and function, such as NRF1 and NRF2 (Figures 5 and 7), although these latter factors were not elevated in Sfrp5Q27stop mice. This mechanism is reminiscent of prior reports regarding the effects of WNT3a-conditioned media on osteogenesis (71), but contrary to our previous observations in brown adipocytes (72). Whereas PGC1α is well known for regulated induction of UCP1 (73), under no circumstances did we observe elevated UCP1 in our studies (data not shown). In addition to effects on mitochondrial biogenesis, increased maximum oxygen capacity was also observed in isolated mitochondria of Sfrp5Q27stop mice (Figure 6), which indicates that WNT signaling also causes functional changes to this organelle. In summary, inhibition of WNT signaling by SFRP5 was required to suppress oxidative metabolism and stimulate maximal adipocyte growth during obesity. Thus, SFRP5 deficiency in adipose tissue elevated WNT signaling to stimulate mitochondrial biogenesis and function, thereby resulting in mice resistant to adipocyte growth under obesogenic conditions.
Methods
Further information can be found in Supplemental Methods.
Animals and animal care.
Sfrp5Q27stop mice were generated by N-ethyl-N-nitrosourea mutagenesis, in which a C79T mutation created a premature stop codon at glutamine 27 (38, 39), and the mutation was back-bred about 20 generations onto the C57BL/6 background. C57BL/6, Leprdb/db, and Lepob/ob mice were obtained from The Jackson Laboratory. Lxrβ–/– mice were as described previously (37). The following diets were used: NCD (catalog no. 5001, LabDiet; PMI Nutrition International); low-fat diet (10% calories from fat, catalog no. D12450B; Research Diets); HFD (45% calories from fat, catalog no. D12451; Research Diets Inc.). C57BL/6 mice were subjected to ovariectomy or sham operation at 3 months of age, as described previously (23). All mice were housed on a 12-hour light/12-hour dark cycle in the Unit for Laboratory Animal Medicine at the University of Michigan, with free access to water and diet.
Animal measurements.
Blood glucose levels were determined using an automated blood glucose reader (Accu-Check; Roche). Serum insulin and leptin levels were measured by ELISA (Crystal Chem Inc.). Glucose tolerance tests were performed on mice that were fasted overnight (12 hours). Blood was collected immediately before as well as 15, 30, 60, and 120 minutes after i.p. injection of glucose (1 g/kg body weight). For insulin tolerance tests, mice were fasted for 3 hours and then injected with 0.5 U/kg body weight of human insulin (Novolin R; Novo Nordisk). Body fat, lean mass, and free fluid were measured in conscious animals using an NMR analyzer (Minispec LF90II; Bruker Optics). Oxygen consumption, carbon dioxide production, spontaneous motor activity, and food intake were measured using CLAMS (Columbus Instruments International).
Analysis of adipocyte morphology.
Ovarian adipose tissue from 20-week-old females was divided into 4 pieces (proximal side of ovary to distal), then fixed in 4% paraformaldehyde and paraffin embedded. For each piece, 3 independent sections spaced by 100 μm were stained with H&E, and 6–7 pictures were taken for each section. We counted more than 1,500 adipocytes per piece of WAT, resulting in a total of 6,700–8,900 adipocytes counted per mouse. Quantification of adipocyte size was done with ImageJ software (NIH). All images were converted into binary files using a unique threshold value that separated positively labeled cells from background.
Adipose tissue transplantation.
Gonadal WAT (gWAT) was excised from control and Sfrp5Q27stop donor mice and transplanted subcutaneously into Leprdb/db recipient mice at 4 weeks of age. To control for host differences in hyperphagia, vasculature formation, or other variables, 100-mg pieces of tissue from 2 control and 2 Sfrp5Q27stop mice were transplanted into the same Leprdb/db recipient. After 10 weeks, the transplanted tissue was harvested and fixed in 4% paraformaldehyde. H&E-stained sections were subjected to morphometric analysis as described above.
Stromal-vascular and adipocyte fractionation.
Fractionation of WAT into stromal-vascular cells and adipocytes was performed as previously described (22, 74).
Cell culture.
3T3-L1 adipogenesis was as described previously (75). Cells that had been confluent for 2 days (assigned as day 0) were treated with 10% FBS with 0.5 mM methylisobutylxanthine, 1 μM dexamethasone, and 1 μg/ml insulin. On day 2, cells were fed 1 μg/ml insulin in 10% FBS, and on day 4 and every 2 days thereafter, cells were fed with 10% FBS. Lipid accumulation in adipocytes was visualized by staining with Oil Red-O (76, 77). EMSCs, isolated from control and Sfrp5Q27stop mice as previously described (36), were maintained in 5% CO2 and DMEM/F12 1:1 media (Gibco; Invitrogen) supplemented with 15% FBS (Atlas Biologicals), Primocin as antibiotics (InVivoGen), and 10 ng/ml recombinant bFGF (PeproTech). To induce adipocyte differentiation, recombinant bFGF was removed and replaced with 10% FBS with 0.5 mM methylisobutylxanthine, 1 μM dexamethasone, 5 μg/ml insulin, and 5 μM troglitazone. On day 2, cells were fed 5 μg/ml insulin plus 5 μM troglitazone. On day 4 and every 2 days thereafter, cells were fed with 15% FBS.
Immunoblot analysis.
Tissue or cell extracts were immunoblotted with antibodies specific for SFRP5 (SARP3 E-19; Santa Cruz Biotechnology); laminin (Novus Biologicals); p-JNK, JNK, IRS1, p-S6 (Ser240/244), S6, p-AKT (Thr308), AKT, MYC, and β-catenin (Cell Signaling); FABP4 (R&D Systems); PPARγ (Santa Cruz Biotechnology); and α-tubulin (Sigma-Aldrich). p-IRS1 (Ser307) antibody was provided by L. Rui (University of Michigan). For SFRP5 blots, we prepared concentrated adipose tissue lysates using StrataClean Resin according to the manufacturer’s protocol (Agilent Technologies).
Plasmids.
Sfrp5 was amplified from a mouse eye cDNA library using forward and reverse primers containing 5′ EcoR1 and 3′ Xho1 restriction sites, respectively. For the Sfrp5-Myc fusion construct, the Myc tag sequence was inserted into the reverse primer so as to be in-frame at the C terminus of SFRP5 protein. The resulting amplicons were cloned into the EcoR1 and Xho1 sites of pcDNA3.1+ (Invitrogen) to generate pcDNA3.1+ (Sfrp5-Myc) constructs. For subsequent stable infection into 3T3-L1 preadipocytes, Sfrp5-Myc was subcloned into the pMSCVneo retroviral vector (Clontech Laboratories) using the EcoR1 and Xho1 sites.
Reagents.
Recombinant murine WNT3a, WNT5a, and WNT5b were purchased from R&D Systems and each used at a concentration of 100 ng/ml. Oligomycin, FCCP, and rotenone were from Enzo Life Sciences. Adenosine diphosphate, succinate, malate, pyruvate, and antimycin A were purchased from Sigma-Aldrich.
mRNA quantification by RT-PCR.
Total RNA was prepared from frozen tissue or cells by using RNA Stat60 according to the manufacturer’s protocol (Tel-Test Inc.). Genomic DNA was removed from total RNA using DNA-free Dnase 1 (Ambion Inc.). Total RNA was reverse transcribed with random hexamers (TaqMan Reverse Transcription kit; Applied Biosystems). qPCR was performed using the MyiQ real-time PCR detection system with SYBR green reagents (Bio-Rad Laboratories). Primers for qPCR were as follows: Sfrp5 sense, 5′-GTTTCCCTTGGCCCGAGAT-3′; Sfrp5 antisense, 5′-CACCGCGATGCAGAGGTC-3′; Sfrp1 sense, 5′-GCCACAACGTGGGCTACAA-3′; Sfrp1 antisense, 5′-ACCTCTGCCATGGTCTCGTG-3′; Sfrp2 sense, 5′-CCTCGCTAGTAGCGACCACCT-3′; Sfrp2 antisense, 5′-GTTTTGCAGGCTTCACACACC-3′; Sfrp3 sense, 5′-ATTTGGTGTTCTGTACCCTG-3′; Sfrp3 antisense, 5′-CGTTTCCTCATAAATGCTTC-3′; Sfrp4 sense, 5′-AGAAGGTCCATACAGTGGGAAG-3′; Sfrp4 antisense, 5′-GTTACTGCGACTGGTGCGA-3′; Pgc1α sense, 5′-GTAGGCCCAGGTACGACAGC-3′; Pgc1α antisense, 5′-GCTCTTTGCGGTATTCATCCC-3′; Nadh1 sense, 5′-CACTCCTCGTCCCCATTCTA-3′; Nadh1 antisense, 5′-ATGCCGTATGGACCAACAAT-3′; Nadh5 sense, 5′-TCTCACCCAAAACGACATCA-3′; Nadh5 antisense, 5′-TTGAAGAATGCGTGGGTACA-3′; Cox1 sense, 5′-GAGAGGCCTTTGCTTCAAAAA-3′; Cox1 antisense, 5′-AGGTTGGTTCCTCGAATGTG-3′; Atp6 sense, 5′-AGGATTCCCAATCGTTGTAGCC-3′; Atp6 antisense, 5′-CCTTTTGGTGTGTGGATTAGCA-3′. Other primers were described previously: Nrf2 and Tfam (78) as well as TATA box–binding protein (Tbp), hypoxanthine phosphoribosyltransferase 1 (Hprt), and 18S (76, 79).
qPCR to estimate mtDNA.
DNA was prepared from frozen tissue or cells using Gentra Puregene Kits including RNase A treatment (Qiagen). mtDNA copy number per nuclear genome in EMSC adipocytes was quantified as described previously (80). PCR primers for amplification of nucleotides 314–489 of the mitochondrial genome were as follows: sense, 5′-AACGCGGTCATACGATTAAC-3′; antisense, 5′-CCCAGTTTGGGTCTTAGCTG-3′. PCR primers for amplification of the mouse nuclear-encoded Gcg gene were as follows: sense, 5′-CAGGGCATCTCAGACC-3′; antisense, 5′-GCTATTGGAAAGCCTCTTGC-3′. qPCR was performed using the MyiQ real-time PCR detection system as described above, using separate tubes for mtDNA and nucDNA amplification.
Isolation of mitochondria from adipose tissue and cultured EMSC adipocytes.
Isolation of mitochondria was essentially as described previously (81). Briefly, adipose tissues were removed after euthanasia and rinsed twice in PBS plus EDTA. Tissues were then homogenized using a Dounce homogenizer and centrifuged at 800 g for 10 minutes at 4°C. The supernatant was then centrifuged for 15 minutes at 9,000 g at 4°C, and the pellet washed with ice-cold buffer (81). After centrifugation at 8,000 g for 10 minutes at 4°C, the pellet containing mitochondria was resuspended for analyses. Mitochondria isolation from EMSC adipocytes was as described above, except that a glass–Teflon potter was used for homogenization (81).
Adipocyte and mitochondria respiration.
Measurement of oxygen consumption in both EMSC adipocytes and isolated mitochondria was performed using the XF24 Extracellular Flux Analyzer according to the manufacturer’s instructions (Seahorse Bioscience). A sheet of EMSC adipocytes was mechanically stripped from the original cell culture plate and cut to fit XF24 Islet Capture Microplates. A capture screen was inserted to maintain the sheet of adipocytes at the bottom of the well. Oxygen consumption rate was calculated using the AKOS technique and normalized to DNA. Isolated mitochondria (5 μg) from EMSC adipocytes or adipose tissue from mice fed HFD for 12 weeks were seeded to XF24 Capture Microplates, and oxygen consumption rate was assayed as described above.
Microarray analysis.
Gene profiling data are available from GEO ( http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE37514. See Supplemental Methods for details.
Statistics.
All data are presented as mean ± SEM and were analyzed by 2-tailed Student’s t test or ANOVA. Differences were considered significant for P values less than 0.05.
Study approval.
Animal experiments were conducted following protocols approved by the University of Michigan University Committee on the Use and Care of Animals (UCUCA).
Supplementary Material
Acknowledgments
This work was supported by NIH grants DK51563 and DK62876 to O.A. MacDougald. H. Mori was supported by a mentor-based postdoctoral fellowship from the American Diabetes Association. W.P. Cawthorn is supported by a Postdoctoral Research Fellowship from the Royal Commission for the Exhibition of 1851 (United Kingdom). I. Gerin was “chargé de recherches” of the Belgian FNRS. This work used Animal Phenotyping and Morphology Core Services supported by NIH grants DK089503 and P60DK020572, respectively. The authors thank Janet Hoff, Colin McCoin, Kathy Feig, Nathan Qi, Sydney Bridges, Angela Tucker, Katie Gee, Katherine Overmyer, Nathan Kanner, Seema Dhindaw, Joe Washburn, and Sam Langberg for technical assistance; Subramaniam Pennathur and Anuradha Vivekanandan for mass spectrometry; and members of the MacDougald lab for helpful discussions and assistance.
Footnotes
Conflict of interest: O.A. MacDougald owns stock in Merck and Express Scripts. V. Krishnan owns stock in and is an employee of Eli Lilly and Co. V.S. Susulic owns stock in Johnson and Johnson and is an employee of Centocor. K.A. Longo is an employee of Proteostasis Therapeutics.
Citation for this article: J Clin Invest. 2012;122(7):2405–2416. doi:10.1172/JCI63604.
See the related Commentary beginning on page 2349.
References
- 1.Ode KL, Frohnert BI, Nathan BM. Identification and treatment of metabolic complications in pediatric obesity. Rev Endocr Metab Disord. 2009;10(3):167–188. doi: 10.1007/s11154-009-9115-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faust IM, Johnson PR, Stern JS, Hirsch J. Diet-induced adipocyte number increase in adult rats: a new model of obesity. Am J Physiol. 1978;235(3):E279–E296. doi: 10.1152/ajpendo.1978.235.3.E279. [DOI] [PubMed] [Google Scholar]
- 3.Jo J, et al. Hypertrophy and/or hyperplasia: dynamics of adipose tissue growth. PLoS Comput Biol. 2009;5(3):e1000324. doi: 10.1371/journal.pcbi.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan CY, Vidal-Puig A. Adipose tissue expandability: the metabolic problems of obesity may arise from the inability to become more obese. Biochem Soc Trans. 2008;36(pt 5):935–940. doi: 10.1042/BST0360935. [DOI] [PubMed] [Google Scholar]
- 5.Virtue S, Vidal-Puig A. It’s not how fat you are, it’s what you do with it that counts. PLoS Biol. 2008;6(9):e237. doi: 10.1371/journal.pbio.0060237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7(12):885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 7.Lefterova MI, Lazar MA. New developments in adipogenesis. Trends Endocrinol Metab. 2009;20(3):107–114. doi: 10.1016/j.tem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 8. Farmer SR. Regulation of PPARgamma activity during adipogenesis. Int J Obes (Lond). 2005. 29 suppl 1: S13 S16 10.1038/sj.ijo.0802907 [DOI] [PubMed] [Google Scholar]
- 9.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 10.Marshall S. Role of insulin, adipocyte hormones, and nutrient-sensing pathways in regulating fuel metabolism and energy homeostasis: a nutritional perspective of diabetes, obesity, and cancer. Sci STKE. 2006;2006(346):re7. doi: 10.1126/stke.3462006re7. [DOI] [PubMed] [Google Scholar]
- 11.Cawthorn WP, Scheller EL, Macdougald OA. Adipose tissue stem cells: the great WAT hope [published online ahead of print March 12, 2012]. Trends Endocrinol Metab. doi: 10.1016/j.tem.2012.01.003. doi:10.1016/j.tem.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cawthorn WP, Scheller EL, MacDougald OA. Adipose tissue stem cells meet preadipocyte commitment: going back to the future. J Lipid Res. 2012;53(2):227–246. doi: 10.1194/jlr.R021089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacDougald OA, Lane MD. When precursors are also regulators. Curr Biol. 1995;5(6):618–621. doi: 10.1016/S0960-9822(95)00125-4. [DOI] [PubMed] [Google Scholar]
- 14.Prestwich TC, MacDougald OA. Wnt/beta-catenin signaling in adipogenesis and metabolism. Curr Opin Cell Biol. 2007;19(6):612–617. doi: 10.1016/j.ceb.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross SE, et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289(5481):950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 16.Bennett CN, et al. Regulation of Wnt signaling during adipogenesis. J Biol Chem. 2002;277(34):30998–31004. doi: 10.1074/jbc.M204527200. [DOI] [PubMed] [Google Scholar]
- 17.Bennett CN, Hodge CL, MacDougald OA, Schwartz J. Role of Wnt10b and C/EBPalpha in spontaneous adipogenesis of 243 cells. Biochem Biophys Res Commun. 2003;302(1):12–16. doi: 10.1016/S0006-291X(03)00092-5. [DOI] [PubMed] [Google Scholar]
- 18.Cawthorn WP, et al. Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a beta-catenin-dependent mechanism. Bone. 2012;50(2):477–489. doi: 10.1016/j.bone.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanazawa A, et al. Association of the gene encoding wingless-type mammary tumor virus integration-site family member 5B (WNT5B) with type 2 diabetes. Am J Hum Genet. 2004;75(5):832–843. doi: 10.1086/425340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishizuka M, Koyanagi A, Osada S, Imagawa M. Wnt4 and Wnt5a promote adipocyte differentiation. FEBS Lett. 2008;582(21–22):3201–3205. doi: 10.1016/j.febslet.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Longo KA, et al. Wnt10b inhibits development of white and brown adipose tissues. J Biol Chem. 2004;279(34):35503–35509. doi: 10.1074/jbc.M402937200. [DOI] [PubMed] [Google Scholar]
- 22.Wright WS, et al. Wnt10b inhibits obesity in ob/ob and agouti mice. Diabetes. 2007;56(2):295–303. doi: 10.2337/db06-1339. [DOI] [PubMed] [Google Scholar]
- 23.Bennett CN, et al. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci U S A. 2005;102(9):3324–3329. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett CN, et al. Wnt10b increases postnatal bone formation by enhancing osteoblast differentiation. J Bone Miner Res. 2007;22(12):1924–1932. doi: 10.1359/jbmr.070810. [DOI] [PubMed] [Google Scholar]
- 25.Aslanidi G, et al. Ectopic expression of Wnt10b decreases adiposity and improves glucose homeostasis in obese rats. Am J Physiol Endocrinol Metab. 2007;293(3):E726–736. doi: 10.1152/ajpendo.00248.2007. [DOI] [PubMed] [Google Scholar]
- 26.Vertino AM, et al. Wnt10b deficiency promotes coexpression of myogenic and adipogenic programs in myoblasts. Mol Biol Cell. 2005;16(4):2039–2048. doi: 10.1091/mbc.E04-08-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krishnan V, Bryant HU, Macdougald OA. Regulation of bone mass by Wnt signaling. J Clin Invest. 2006;116(5):1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennell JA, O’Leary EE, Gummow BM, Hammer GD, MacDougald OA. T-cell factor 4N (TCF-4N), a novel isoform of mouse TCF-4, synergizes with beta-catenin to coactivate C/EBPalpha and steroidogenic factor 1 transcription factors. Mol Cell Biol. 2003;23(15):5366–5375. doi: 10.1128/MCB.23.15.5366-5375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116(pt 13):2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 30.Lagathu C, et al. Secreted frizzled-related protein 1 regulates adipose tissue expansion and is dysregulated in severe obesity. Int J Obes (Lond). 2010;34(12):1695–1705. doi: 10.1038/ijo.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagathu C, et al. Dact1, a nutritionally regulated preadipocyte gene, controls adipogenesis by coordinating the Wnt/beta-catenin signaling network. Diabetes. 2009;58(3):609–619. doi: 10.2337/db08-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koza RA, et al. Changes in gene expression foreshadow diet-induced obesity in genetically identical mice. PLoS Genet. 2006;2(5):e81. doi: 10.1371/journal.pgen.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J Cell Sci. 2008;121(pt 6):737–746. doi: 10.1242/jcs.026096. [DOI] [PubMed] [Google Scholar]
- 34.Ouchi N, et al. Sfrp5 is an anti-inflammatory adipokine that modulates metabolic dysfunction in obesity. Science. 2010;329(5990):454–457. doi: 10.1126/science.1188280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okada Y, Sakaue H, Nagare T, Kasuga M. Diet-induced up-regulation of gene expression in adipocytes without changes in DNA methylation. Kobe J Med Sci. 2009;54(5):E241–E249. [PubMed] [Google Scholar]
- 36.Rim JS, Mynatt RL, Gawronska-Kozak B. Mesenchymal stem cells from the outer ear: a novel adult stem cell model system for the study of adipogenesis. FASEB J. 2005;19(9):1205–1207. doi: 10.1096/fj.04-3204fje. [DOI] [PubMed] [Google Scholar]
- 37.Gerin I, et al. LXRbeta is required for adipocyte growth, glucose homeostasis, and beta cell function. J Biol Chem. 2005;280(24):23024–23031. doi: 10.1074/jbc.M412564200. [DOI] [PubMed] [Google Scholar]
- 38.Quwailid MM, et al. A gene-driven ENU-based approach to generating an allelic series in any gene. Mamm Genome. 2004;15(8):585–591. doi: 10.1007/s00335-004-2379-z. [DOI] [PubMed] [Google Scholar]
- 39.Cox S, Smith L, Bogani D, Cheeseman M, Siggers P, Greenfield A. Sexually dimorphic expression of secreted frizzled-related (SFRP) genes in the developing mouse Mullerian duct. Mol Reprod Dev. 2006;73(8):1008–1016. doi: 10.1002/mrd.20507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satoh W, Matsuyama M, Takemura H, Aizawa S, Shimono A. Sfrp1, Sfrp2, and Sfrp5 regulate the Wnt/beta-catenin and the planar cell polarity pathways during early trunk formation in mouse. Genesis. 2008;46(2):92–103. doi: 10.1002/dvg.20369. [DOI] [PubMed] [Google Scholar]
- 41.Tseng YH, et al. Gene-expression in brown preadipocytes predicts differentiation: role of insulin receptor substrates and necdin. Nat Cell Biol. 2005;7(6):601–611. doi: 10.1038/ncb1259. [DOI] [PubMed] [Google Scholar]
- 42.Moldes M, et al. Peroxisome-proliferator-activated receptor gamma suppresses Wnt/beta-catenin signalling during adipogenesis. Biochem J. 2003;376(pt 3):607–613. doi: 10.1042/BJ20030426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villanueva CJ, et al. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab. 2011;13(4):413–427. doi: 10.1016/j.cmet.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell. 2003;5(3):367–377. doi: 10.1016/S1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 45.Yamanaka H, et al. JNK functions in the non-canonical Wnt pathway to regulate convergent extension movements in vertebrates. EMBO Rep. 2002;3(1):69–75. doi: 10.1093/embo-reports/kvf008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andreasen AS, Kelly M, Berg RM, Moller K, Pedersen BK. Type 2 diabetes is associated with altered NF-kappaB DNA binding activity, JNK phosphorylation, and AMPK phosphorylation in skeletal muscle after LPS. PLoS One. 2011;6(9):e23999. doi: 10.1371/journal.pone.0023999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aguirre V, Werner ED, Giraud J, Lee YH, Shoelson SE, White MF. Phosphorylation of Ser307 in insulin receptor substrate-1 blocks interactions with the insulin receptor and inhibits insulin action. J Biol Chem. 2002;277(2):1531–1537. doi: 10.1074/jbc.M101521200. [DOI] [PubMed] [Google Scholar]
- 48.Finch P, et al. Purification and molecular cloning of a secreted, frizzled-related antagonist of Wnt action. Proc Natl Acad Sci U S A. 1997;94(13):6770–6775. doi: 10.1073/pnas.94.13.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uren A, et al. Secreted frizzled-related protein-1 binds directly to Wingless and is a biphasic modulator of Wnt signaling. J Biol Chem. 2000;275(6):4374–4382. doi: 10.1074/jbc.275.6.4374. [DOI] [PubMed] [Google Scholar]
- 50.Zhong X, et al. Regulation of secreted Frizzled-related protein-1 by heparin. J Biol Chem. 2007;282(28):20523–20533. doi: 10.1074/jbc.M609096200. [DOI] [PubMed] [Google Scholar]
- 51.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88(2):611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 52.Ekstrand MI, et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13(9):935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 53.Wilson-Fritch L, et al. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J Clin Invest. 2004;114(9):1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson-Fritch L, et al. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol. 2003;23(3):1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mii Y, Taira M. Secreted Wnt “inhibitors” are not just inhibitors: regulation of extracellular Wnt by secreted Frizzled-related proteins. Dev Growth Differ. 2011;53(8):911–923. doi: 10.1111/j.1440-169X.2011.01299.x. [DOI] [PubMed] [Google Scholar]
- 56. Body weight information. The Jackson Laboratory Web site. http://jaxmice.jax.org/support/weight/000632.html . Accessed May 7, 2012.
- 57.Leaf I, Tennessen J, Mukhopadhyay M, Westphal H, Shawlot W. Sfrp5 is not essential for axis formation in the mouse. Genesis. 2006;44(12):573–578. doi: 10.1002/dvg.20248. [DOI] [PubMed] [Google Scholar]
- 58.Veeck J, et al. Epigenetic inactivation of the secreted frizzled-related protein-5 (SFRP5) gene in human breast cancer is associated with unfavorable prognosis. Carcinogenesis. 2008;29(5):991–998. doi: 10.1093/carcin/bgn076. [DOI] [PubMed] [Google Scholar]
- 59.Zhao C, Bu X, Zhang N, Wang W. Downregulation of SFRP5 expression and its inverse correlation with those of MMP-7 and MT1-MMP in gastric cancer. BMC Cancer. 2009;9:224. doi: 10.1186/1471-2407-9-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Su HY, et al. Epigenetic silencing of SFRP5 is related to malignant phenotype and chemoresistance of ovarian cancer through Wnt signaling pathway. Int J Cancer. 2010;127(3):555–567. doi: 10.1002/ijc.25083. [DOI] [PubMed] [Google Scholar]
- 61.Hou HA, et al. Distinct association between aberrant methylation of Wnt inhibitors and genetic alterations in acute myeloid leukaemia. Br J Cancer. 2011;105(12):1927–1933. doi: 10.1038/bjc.2011.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kinoshita T, Nomoto S, Kodera Y, Koike M, Fujiwara M, Nakao A. Decreased expression and aberrant hypermethylation of the SFRP genes in human gastric cancer. Hepatogastroenterology. 2011;58(107–108):1051–1056. [PubMed] [Google Scholar]
- 63.Mii Y, Taira M. Secreted Frizzled-related proteins enhance the diffusion of Wnt ligands and expand their signalling range. Development. 2009;136(24):4083–4088. doi: 10.1242/dev.032524. [DOI] [PubMed] [Google Scholar]
- 64.Anderson NL, et al. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol Cell Proteomics. 2004;3(4):311–326. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Adkins JN, et al. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol Cell Proteomics. 2002;1(12):947–955. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- 66.Schulte DM, et al. Pro-inflammatory wnt5a and anti-inflammatory sFRP5 are differentially regulated by nutritional factors in obese human subjects. PLoS One. 2012;7(2):e32437. doi: 10.1371/journal.pone.0032437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.MacDougald OA, Burant CF. The rapidly expanding family of adipokines. Cell Metab. 2007;6(3):159–161. doi: 10.1016/j.cmet.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 68.Deng Y, Scherer PE. Adipokines as novel biomarkers and regulators of the metabolic syndrome. Ann N Y Acad Sci. 2010;1212:E1–E19. doi: 10.1111/j.1749-6632.2010.05875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Pauw A, Tejerina S, Raes M, Keijer J, Arnould T. Mitochondrial (dys)function in adipocyte (de)differentiation and systemic metabolic alterations. Am J Pathol. 2009;175(3):927–939. doi: 10.2353/ajpath.2009.081155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoon JC, Ng A, Kim BH, Bianco A, Xavier RJ, Elledge SJ. Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes Dev. 2010;24(14):1507–1518. doi: 10.1101/gad.1924910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.An JH, et al. Enhanced mitochondrial biogenesis contributes to Wnt induced osteoblastic differentiation of C3H10T1/2 cells. Bone. 2010;47(1):140–150. doi: 10.1016/j.bone.2010.04.593. [DOI] [PubMed] [Google Scholar]
- 72.Kang S, et al. Effects of Wnt signaling on brown adipocyte differentiation and metabolism mediated by PGC-1alpha. Mol Cell Biol. 2005;25(4):1272–1282. doi: 10.1128/MCB.25.4.1272-1282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3(5):333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 74.Rodbell M. Metabolism of isolated fat cells I. effects of hormones on glucose metabolism and lipolysis. J Biol Chem. 1964;239:375–380. [PubMed] [Google Scholar]
- 75.Hemati N, Ross SE, Erickson RL, Groblewski GE, MacDougald OA. Signaling pathways through which insulin regulates CCAAT/enhancer binding protein α (C/EBPα) phosphorylation and gene expression in 3T3-L1 adipocytes: Correlation with GLUT4 gene expression. J Biol Chem. 1997;272(41):25913–25919. doi: 10.1074/jbc.272.41.25913. [DOI] [PubMed] [Google Scholar]
- 76.Gerin I, Bommer GT, Lidell ME, Cederberg A, Enerback S, Macdougald OA. On the role of FOX transcription factors in adipocyte differentiation and insulin-stimulated glucose uptake. J Biol Chem. 2009;284(16):10755–10763. doi: 10.1074/jbc.M809115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Erickson RL, Hemati N, Ross SE, MacDougald OA. p300 coactivates the adipogenic transcription factor CCAAT/enhancer- binding protein alpha. J Biol Chem. 2001;276(19):16348–16355. doi: 10.1074/jbc.m100128200. [DOI] [PubMed] [Google Scholar]
- 78.Tseng YH, et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454(7207):1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mori H, et al. Critical role for hypothalamic mTOR activity in energy balance. Cell Metab. 2009;9(4):362–374. doi: 10.1016/j.cmet.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang H, et al. Replication of murine mitochondrial DNA following irradiation. Adv Exp Med Biol. 2009;645:43–48. doi: 10.1007/978-0-387-85998-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2007;2(2):287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.