Abstract
How neural circuit evolution relates to behavioral evolution is not well understood. Here the relationship between neural circuits and behavior is explored with respect to the swimming behaviors of the Nudipleura (Mollusca, Gastropoda, Opithobranchia). Nudipleura is a diverse monophyletic clade of sea slugs among which only a small percentage of species can swim. Swimming falls into a limited number of categories, the most prevalent of which are rhythmic left–right body flexions (LR) and rhythmic dorsal–ventral body flexions (DV). The phylogenetic distribution of these behaviors suggests a high degree of homoplasy. The central pattern generator (CPG) underlying DV swimming has been well characterized in Tritonia diomedea and in Pleurobranchaea californica. The CPG for LR swimming has been elucidated in Melibe leonina and Dendronotus iris, which are more closely related. The CPGs for the categorically distinct DV and LR swimming behaviors consist of nonoverlapping sets of homologous identified neurons, whereas the categorically similar behaviors share some homologous identified neurons, although the exact composition of neurons and synapses in the neural circuits differ. The roles played by homologous identified neurons in categorically distinct behaviors differ. However, homologous identified neurons also play different roles even in the swim CPGs of the two LR swimming species. Individual neurons can be multifunctional within a species. Some of those functions are shared across species, whereas others are not. The pattern of use and reuse of homologous neurons in various forms of swimming and other behaviors further demonstrates that the composition of neural circuits influences the evolution of behaviors.
Keywords: evolvability, neuromodulation, rhythmic movement, species differences, neuroethology
Behavior and neural mechanisms can be considered to represent two different levels of biological organization (1–4). Nevertheless, the evolution of behavior and the evolution of neural circuits underlying behavior are intertwined. For example, it has been suggested that the properties of neural circuits affect the evolvability of behavior; the evolution of particular behaviors could be constrained or promoted by the organization of neural circuits (5–9). Darwin and the early ethologists recognized that behaviors, like anatomical features, are heritable characters that are amenable to a phylogenetic approach (10–13). The use of behavioral traits to determine phylogenies has been validated several times (14–17), and the historical debates about homology and homoplasy of behavior have been thoroughly reviewed (2–4, 15, 17, 18). Examining the neural bases for independently evolved (i.e., homoplastic) behaviors within a clade could provide insight into fundamental aspects of neural circuit organization. However, it is difficult enough to determine the neural basis for behavior in one species. Doing this in several species with quantifiable behaviors is even more challenging.
Studies of the neural bases of swimming behaviors in the Nudipleura (Mollusca, Gastropoda, Opisthobranchia) offer such a possibility. These sea slugs exhibit well differentiated categories of swimming behaviors, and their nervous systems have large individually identifiable neurons, allowing the neural circuitry underlying the swimming behaviors to be determined with cellular precision.
Here we will summarize what is known about the phylogeny of Nudipleura, their swimming behaviors, and the neural circuits underlying swimming. We will also provide data comparing the roles of homologous neurons. We find that neural circuits underlying the behaviors of the same category are composed of overlapping sets of neurons even if they most likely evolved independently. In contrast, neural circuits underlying categorically distinct behaviors use nonoverlapping sets of neurons. Furthermore, homologous neurons can have different functions in different behaviors and even in similar behaviors.
Phylogeny of Nudipleura
The Nudipleura form a monophyletic clade within Opisthobranchia (Gastropoda) that contains two sister clades: Pleurobranchomorpha and Nudibranchia (19–21) (Fig. 1). Molecular evidence suggests that the two sister groups separated approximately 125 Mya (21). Nudibranchia (or, informally, nudibranchs), which are shell-less and have a slug-shaped appearance with “naked gills,” were traditionally classified as their own order. The most recently agreed upon taxonomic classification system for nudibranchs uses unranked clades instead of orders, suborders, and superfamilies (22). There are at least 2,000 to 3,000 identified nudibranch species (23). Studies that used morphological and molecular data support the monophyly of Nudibranchia (19–21, 24–26).
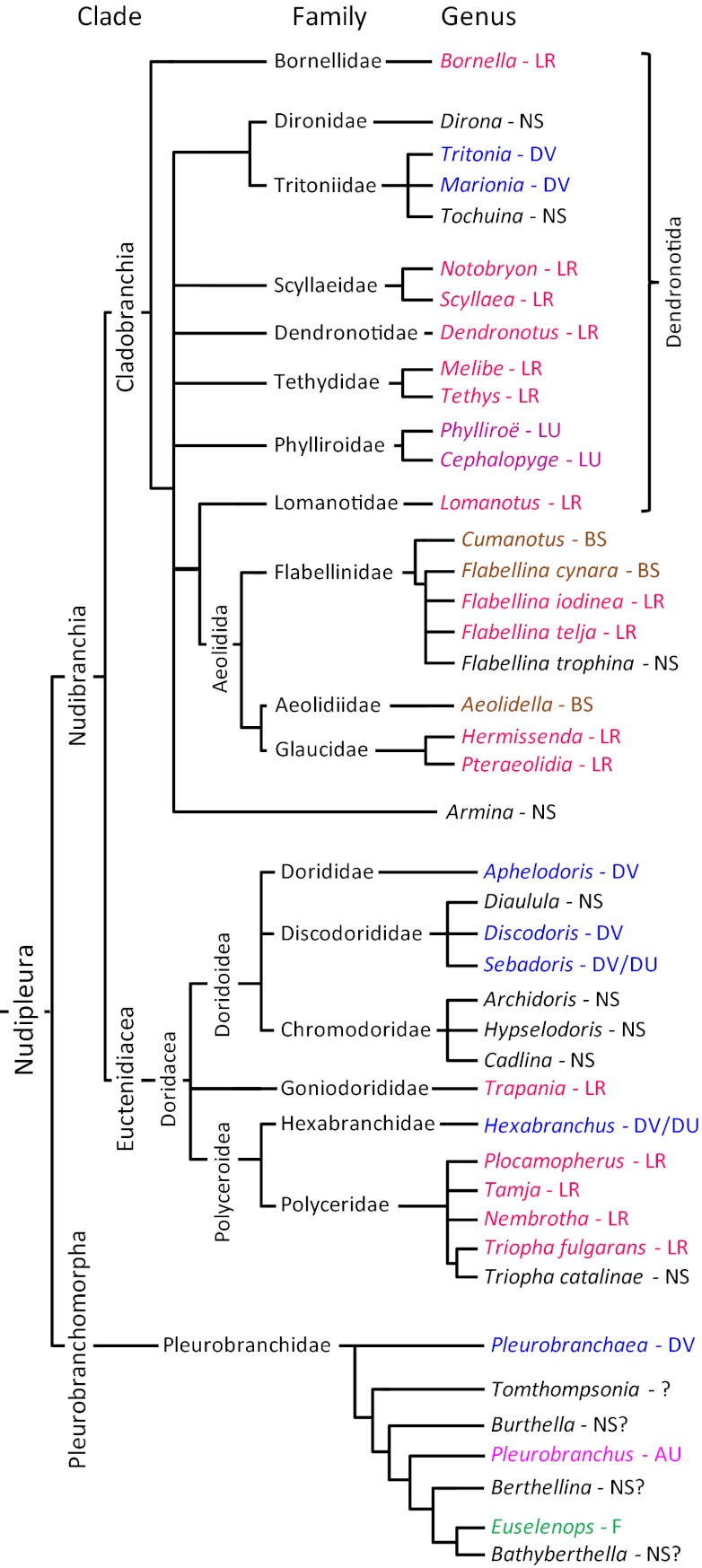
Fig. 1.
An abbreviated phylogeny of the Nudipleura with reference to their behavior. Only the genera of the species listed in Table S1 are shown here unless species differences exist within the genus. The phylogenic relationships are based on refs. 19–21, 25, 26, 28. The references for the behavior are listed in Table S1. Note that this figure represents all the known swimming species and only a tiny fraction of the more than 2,000 species that are not capable of swimming or for which there are no published reports of swimming. LR, left-right flexion; NS, nonswimmer; DV, dorsal-ventral flexion; LU, left-right undulation; BS, breast stroke; DU, dorsal-ventral undulation; AU, asymmetric undulation; F, flapping.
Within Nudibranchia, there are two monophyletic clades (19): Euctenidiacea (Anthobranchia) (27, 28) and Cladobranchia (25). Euctenidiacea includes Doridacea, which is larger than Cladobranchia, subdividing into 25 families (28). Within Cladobranchia, Bornellidae forms a sister group to the other subclades (25). Aeolidida is a monophyletic clade with Lomanotidae as a sister group (25). What was traditionally called Dendronotida forms a paraphyletic grouping. A recent study was unable to include the nudibranch Melibe in Cladobranchia because of a 12-bp deletion in its genome (25). However, its natural affinity with Tethys in terms of shared derived characteristics strongly suggests that it belongs in Cladobranchia, as we have indicated in Fig. 1. There are several additional unresolved relations in Nudibranchia, most notably in Dendronotida and Doridacea. Consideration of locomotor behavior and neural circuits may help resolve these relations.
Categories of Locomotor Behavior
Crawling is the primary form of locomotion for all Nudipleura (29–31). The majority of species crawl via mucociliary locomotion; cilia on the bottom of the foot beat and propel the animal over a surface of secreted mucus. The speed of crawling is affected by efferent serotonergic and peptidergic neurons that control the ciliary beat frequency (30–32). Some species also use muscular crawling, which relies on waves of contraction or extension and contraction of the foot. Crawling is a trait shared with most Opisthobranchia and is therefore plesiomorphic to the Nudipleura. Only three nudibranch species do not crawl because they are truly pelagic: Phylliroë atlantica, Phylliroë bucephala, and Cephalopyge trematoides (33). This is also true for gastropods in general; there are ∼40,000 marine gastropod species but only approximately 150 are pelagic (33).
In addition to crawling, a limited number of benthic species can also swim (34). We classify swimming in the Nudipleura into seven general categories: (i) left–right flexion (LR), (ii) dorsal–ventral flexion (DV), (iii) left–right undulation (LU), (iv) dorsal–ventral undulation (DU), (v) asymmetric undulation (AU), (vi) breaststroke (BS), and (vii) flapping (F) (Table S1).
LR swimming is characterized by the flattening of the body in the sagittal plane and repeated left–right bending near the midpoint of the body axis with the head and tail coming together laterally (Fig. 2A). This movement propels the animal through the water. Some animals, such as Melibe leonina, exhibit foot-first directionality, presumably because the dorsal cerata create drag. Other animals, such as Tambja eliora, proceed headfirst, with the tail lagging slightly, causing the body to take on an “S” form (34). Animals in the genus Plocamopherus typically have a dorsal crest at the posterior end of the body that may act as a paddle and cause the head to proceed the tail (35).
Fig. 2.
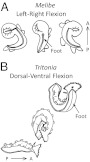
Two examples of swimming behaviors. (A) LR swimming exhibited by M. leonina. The ventral side of the animal is shown with the mouth at the top of the image. During swimming, the foot is narrowed to a strip and the animal rhythmically flexes its body leftward and rightward, bending at a point midway along the body axis. (B) DV swimming exhibited by T. diomedea. The animal starts on the substrate, shown at the bottom with its head to the right. It launches with a ventral flexion, where the head and tail meet under the foot. Then, it flexes so that the head and tail meet above the dorsal body surface. The foot is flattened and expanded to the width of the body. A, anterior; P, posterior.
Plocamopherus ceylonicus (35, 36) and Plocamopherus maderae (37) swim with LR flexions when dislodged from a substrate or disturbed in some way. Tambja appears to use LR swimming as an escape response; contact with the predacious nudibranch Roboastra will elicit swimming in Tambja (34, 38). LR swimming in Melibe and Dendronotus iris can be initiated in response to loss of contact with the substrate or in response to the touch of a predatory sea star (39, 40). Melibe may also swim seasonally to disperse (41). The flexion cycle period for Melibe and Dendronotus is approximately 3 s, and swim bouts can last many minutes (39, 40).
As its name suggests, Bornella anguilla swims with an eel-like movement caused by waves of muscular contraction (42). Therefore, unlike other members of its genus, it is classified as an LU swimmer. LU swimming, which otherwise is found mostly in pelagic species, may be a further refinement of LR swimming.
DV swimming involves the animal flattening its body in the horizontal plane and repeatedly bending such that the tail and head meet in alternation above and below the midpoint of the body (Fig. 2B). Tritonia diomedea and Pleurobranchaea californica are two examples of DV swimmers that have been extensively studied (43–46). Swim bouts for Tritonia and Pleurobranchaea last less than 1 min and are triggered by contact with a predatory sea star or in the laboratory by high salt solutions or electric shock (47). The flexion cycle period under natural conditions is 5 to 10 s in Tritonia (48) and 3 to 6 s in Pleurobranchaea (49).
DU swimming, like DV swimming, involves movement in dorsal and ventral directions, but here there are progressive symmetric waves of body wall or mantle muscular contraction. The Spanish Dancer, Hexabranchus sanguineus, and other members of that genus are famous for their flamboyant swimming behavior (34, 50, 51). Hexabranchus swimming differs in several ways from the DV swimming of Tritonia and Pleurobranchaea; in addition to the symmetrical undulation of the lateral fringes of the mantle, it has a shorter flexion cycle period (2–4 s), swim bouts occur spontaneously, and swimming can last for long periods of time.
F swimming is similar to DV swimming in that the movement is bilaterally symmetric and dorsal–ventral in orientation, but instead of the head and tail meeting, the lateral edges of the mantle or foot rise and fall. F swimming is much more common in Opisthobranchia outside of the Nudipleura, such as Clione limacina (52) and many species of Aplysia (53, 54).
AU and BS are less common forms of locomotion. AU is characteristic of Pleurobranchus membranaceus (55) in which the animal swims upside down using its mantle as a passive keel while producing alternating muscular waves along its foot. BS involves the use of appendages including cerata and tentacles to stroke the water in a manner similar to a human swimmer’s movements. Only four nudibranch species have been described as exhibiting this type of behavior (Table S1).
Phylogenetic Distribution of Swimming Behaviors
As noted earlier, we have been unable to find reports of swimming by 97% to 98% of nudibranch species and approximately half the major subfamilies in the Pleurobranchomorpha clade. However, this does not mean they are not capable of swimming. Some species swim only as a high threshold escape response. Still, it is highly probable that the vast majority of the Nudipleura cannot and do not swim. This discussion is limited to species for which the type of swimming has been reported or for which swimming has been explicitly tested and shown not to occur.
LR swimming is by far the most prevalent of the six modes of swimming exhibited by nudibranchs: of the 60 nudibranch species documented to swim in the scientific literature, 40 species use LR or LU (Table S1). These 40 species are phylogenetically disparate, encompassing species in Doridacea and Cladobranchia (Fig. 1). Within the latter, there are LR swimmers in Aeolidoidea and Dendronotoidea. In Doridacea, all but one of the LR swimmers is in the family Polyceridae. There are no LR swimmers in the Pleurobranchomorpha or, to our knowledge, in any other Opisthobranch clade. This suggests that LR swimming is a derived characteristic of the nudibranch clade.
Unlike LR swimming, DV swimming is found in Nudibranchia and in Pleurobranchomorpha (Fig. 1). DV swimming is not present outside of Nudipleura and is therefore likely to be a synapomorphy of this clade. However, it is not widely displayed within Nudibranchia, appearing in just one family of Dendronotida (Tritoniidae) and in three families of Doridacea (Discodorididae, Dorididae, and Hexibranchidae). Discodorididae and Hexibranchidae also exhibit dorsal ventral undulations (i.e., DU).
Evolution of Swimming Behaviors
There are a number of possible scenarios that could account for the phylogenetic distribution of swimming behaviors among the Nudipleura. Considering the extreme rarity of swimming, it is possible, maybe even likely, that swimming evolved on multiple occasions from nonswimming species. The repeated gain of a function such as rhythmic movement could suggest that there is a predisposition toward these behaviors. The repeated appearance of LR and DV swimming may simply indicate that these two basic movements are the most likely to occur in a slug-shaped body with few appendages. When appendages such as moveable cerata are present, they have been repeatedly used for BS swimming. In the absence of such appendages, the only means of swimming are with LR-like or DV-like movements.
Given the presence of swimming across the phylogeny, it is possible that, rather than evolving independently many times from nonswimmers, swimming behaviors were repeatedly lost. Although this may lead to more transformations, it may be easier to lose a character than to gain one, as has been seen in other systems (56–60).
For the moment, we will only consider the possible evolutionary scenarios that include transformations from one swimming state to another and ignore nonswimmers. It is generally the case that members of the same genus and often the same family exhibit the same form of swimming (Table S1), allowing us to group them together (Fig. 3). Here we will consider potential scenarios involving just the evolution of DV and LR swimming. It is possible that the ancestral species was able to swim by using DV and LR movements. However, this seems unlikely because there are no extant species that exhibit both these behaviors. It is also unlikely that the ancestral state was LR swimming because of its absence in Pleurobranchomorpha.
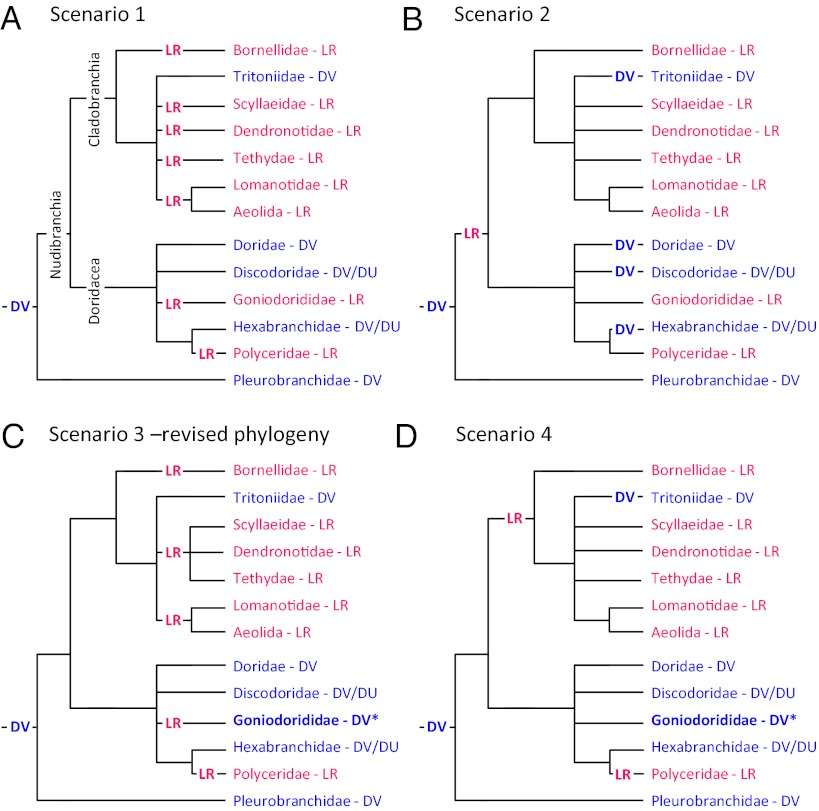
Fig. 3.
Possible evolutionary scenarios explaining the phylogenetic distribution of swimming behaviors. Just the families of the DV and LR swimming animals are shown. (A) In scenario 1, DV swimming is a synapomorphy of the Nudipleura that was lost and replaced six times by LR swimming. (B) In scenario 2, LR swimming is a synapomorphy of the Nudibranchia. DV swimming then reappears four times in different nudibranch lineages. (C) For scenario 3, the phylogenetic tree of Dendronotida is altered to group LR swimmers together. Goniodorididae (asterisk), which includes T. velox, is switched from LR to DV (as discussed in the text). This reduces the number of transitions to LR from six in scenario 1 to four. (D) Scenario 4 is similar to scenario 2, with Goniodorididae (asterisk) switched to DV. This represents the most parsimonious explanation if DV swimming is ancestral, with just three transitions from the basal DV state.
Consider scenario 1 (Fig. 3A) in which DV swimming arose once at the base of the Nudipleura and LR swimming evolved independently several times. In this scenario, DV swimming behaviors in Pleurobranchomorpha, Doridacea, and Cladobranchia are homologous because they are shared by a common ancestor. Scenario 1 would also suggest that LR swimming evolved independently as many as seven times. Because of the unresolved branches in the phylogeny, there may be fewer switches in phenotype than this. In scenario 2 (Fig. 3B), LR swimming evolved once in the Nudibranchia and DV swimming reevolved independently as many as four times. Again, the number of homoplastic events could be lower if the bifurcations in the phylogeny were better resolved.
The phylogenetic distribution of the swimming behavior suggests a resolution to the Dendronotida phylogeny, with Tritoniidae branching off separately from the LR swimmers. This would reduce the number of homoplastic events in Cladobranchia according to scenario 1 from five to three (scenario 3; Fig. 3C).
The phylogenetic distribution of the behavior also calls into question the accuracy of a report about the behavior of Trapania velox. Outside of the family Polyceridae, T. velox (family: Goniodorididae) is the only doridacean reported to swim with left–right flexions. Farmer (34) categorized T. velox as an LR swimmer based on a previous report by Cockerell (61), who described T. velox as being, “very active when swimming with an undulating motion on the surface of the water.” However, there is no indication as to the plane of movement. Farmer (34) reported working with this rare species and being unsuccessful at making it swim, and was thus unable to provide any additional information. We were unable to find any other reports of its behavior. If T. velox is reclassified as a DV swimmer, it would further decrease the number of homoplastic events in scenario 1 from seven to four (Fig. 3C). Thus, examining the phylogenetic distribution of behavior makes a prediction about the behavior of this rare species.
Redefining T. velox as a DV swimmer also suggests a fourth scenario (Fig. 3D), whereby LR swimming arose independently in Cladobranchia and Polyceridae. This would also involve reevolution of DV swimming in Tritoniidae. Scenario 4 would therefore be the most parsimonious explanation for the phylogenetic distribution of swimming behaviors if one does not take into account the hundreds of nonswimming species.
Neural Circuits Underlying Swimming
With our potential scenarios about the homology and homoplasy of swimming behaviors, it is now of interest to compare the neural mechanisms for these behaviors. The neural activity that underlies rhythmic DV and LR movements originates from central pattern generator (CPG) circuits (62). These swim CPGs are composed of neurons whose anatomical and physiological properties allow them to be individually identifiable from animal to animal within a species. The same sets of characteristics can be used to identify homologous neurons in other species (63). This allows the composition of neural circuits and the roles of homologous neurons to be compared across species. The neural circuits underlying swimming have been determined in two DV swimmers [T. diomedea (44) and P. californica (45, 64)] and two LR swimmers [M. leonina (40, 65) and D. iris (40)]. We can now begin to compare neural circuits underlying behaviors of animals to address phylogenetic and functional hypotheses.
DV Swim CPGs.
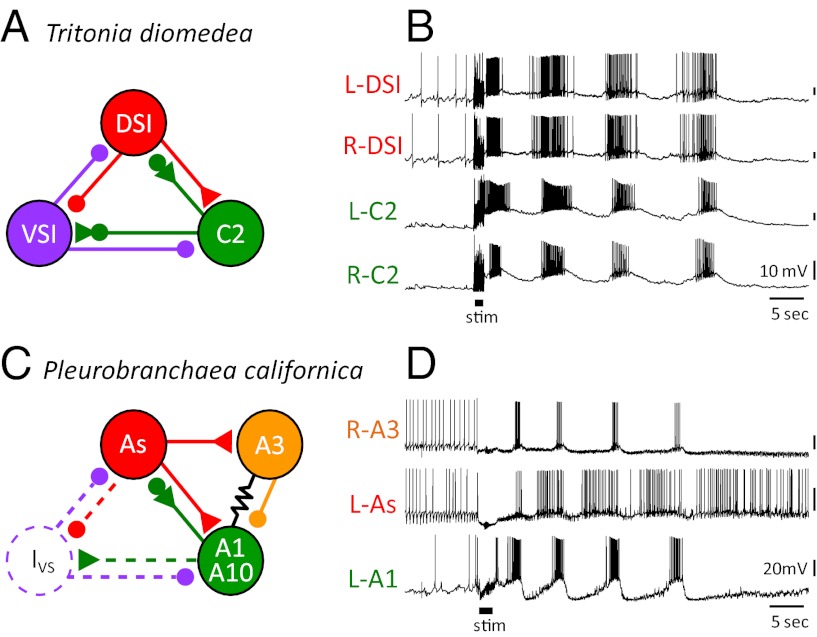
The neural basis for DV swimming was first studied in T. diomedea (43, 66–69). The swim CPG consists of just three neuron types (Fig. 4A). On each side of the brain, there are three dorsal swim interneurons (DSIs), one ventral swim interneuron (VSI), and one cerebral interneuron 2 (C2), for a total of 10 neurons (44, 47). The DSIs initiate the dorsal flexion cycle in which C2 participates. C2 then excites VSI, which inhibits DSI and C2 and elicits the ventral phase of the movement. As would be expected for a DV swimmer, the contralateral counterparts for each neuron fire in relative synchrony (Fig. 4B).
Fig. 4.
The neural circuits and swim motor patters for the DV swimmers Tritonia and Pleurobranchaea. (A) The Tritonia swim CPG consists of three neuron types: DSI, C2, and VSI. (B) Simultaneous intracellular microelectrode recordings show that two contralateral DSIs fire bursts of action potentials in phase with each other and slightly ahead of the two C2s. VSI (not recorded here) fires action potentials in the interburst interval. The motor pattern is initiated by electrical stimulation of a body wall nerve (stim). (C) The Pleurobranchaea swim CPG contains five types of neurons (64). The As neurons are homologues of the DSIs. A1 is homologous to C2. A10 is strongly electrically coupled to A1 and, for simplicity, is shown together with it. A3 is not found in Tritonia. The Ivs neuron has not been found, but has been postulated to exist based on recordings of inhibitory postsynaptic potentials in other neurons. (D) Simultaneous intracellular recordings from an A3, As, and A1. The As neuron leads the A1 neuron just as DSI leads C2. The swim motor pattern is initiated by electrical stimulation of a body wall nerve (stim). In A and C, the small filled circles represent inhibitory synapses, the triangles are excitatory synapses and combinations are mixed inhibition and excitation. The resistor symbol represents electrical synapses. B and D are previously unpublished recordings.
The neurons comprising the CPG for DV swimming in P. californica include DSI and C2 homologues called As and A1, respectively (49, 64). The connectivity and activity of these homologues is similar in both species (Fig. 4 C and D). The homologue of the Tritonia VSI has not been identified in Pleurobranchaea, although there is synaptic input to As and A1 during the ventral phase of the motor pattern that may arise from such a neuron (i.e., Ivs neuron) (64). Alternatively, ventral phase synaptic input may arise from a neuron that is not homologous to VSI, but serves a similar role.
There are also Pleurobranchaea swim CPG neurons (A3 and A10) that have not been identified in Tritonia. Despite more than 40 y of electrophysiological study concentrated in the area where the A3 and A10 somata would be, no neurons with equivalent synaptic connectivity or activity have been found in Tritonia. Thus, either these neurons do not exist in Tritonia or they cannot be recognized with electrophysiological criteria.
With the information available about the swim CPGs in Tritonia and Pleurobranchaea, we can currently say that some homologous neurons are used for similar functions in distantly related species. This result is compatible with any of the phylogenetic scenarios (Fig. 3). If DV swimming is homologous (scenarios 1 or 3; Fig. 3 A and C), the similarities in the DV swim CPGs in Tritonia and Pleurobranchaea could be a result of their homology and the potential differences in the swim CPGs could represent divergence of the circuit architecture. The differences in the swim CPGs may just as readily reflect independent evolutionary paths (scenarios 2 or 4; Fig. 3 B and D), which might suggest a predisposition to use certain neurons to produce these behaviors.
LR Swim CPGs.
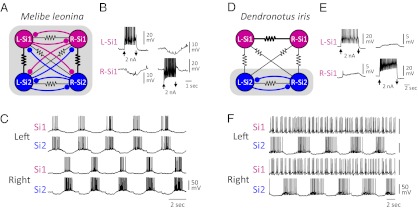
The LR swim CPG was first described in M. leonina (65, 70). The published circuit consists of a pair of bilaterally represented neurons: swim interneuron 1 (Si1) and swim interneuron 2 (Si2; Fig. 5A). Based on their anatomy and neurochemistry, these neurons are not homologous to any of the Tritonia or Pleurobranchaea swim CPG neurons.
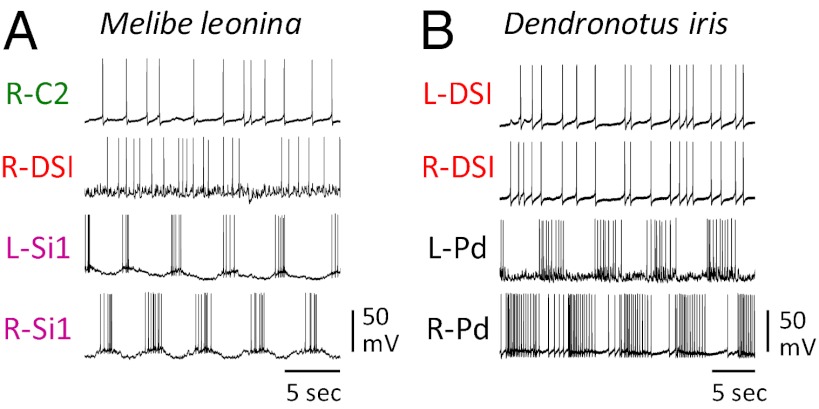
Fig. 5.
Neural circuitry and swim motor pattern for the LR swimmers Melibe and Dendronotus. (A) In the Melibe swim CPG (65), there are two bilaterally represented neurons Si1 and Si2 that are mutually inhibitory across the midline and exhibit strong electrical coupling ipsilaterally (as indicated by thicker resistor symbol). (B) Depolarization of one Si1 by injecting 2nA of current into it hyperpolarizes the contralateral counterpart. (C) The Melibe swim motor pattern consists of ipsilateral synchrony and alternation with the contralateral side. (D) In Dendronotus, the inhibitory connections to and from Si1 are absent, and the electrical coupling between the contralateral Si1 pair dominates (40). (E) Depolarization of an Si1 with 2nA current injection depolarizes the contralateral counterpart. (F) In the Dendronotus swim motor pattern, the left and right Si2 fire alternating bursts of action potentials, but the Si1s fire irregularly. In A and D, the shaded boxes represent the functional CPGs. All recordings are previously unpublished.
In the Melibe swim CPG, each neuron reciprocally inhibits the two contralateral counterparts (Fig. 5B). There is also strong electrical coupling between the ipsilateral Si1 and Si2, causing them to fire in phase with each other and 180° out of phase with the contralateral pair (Fig. 5C). This bursting pattern drives the left–right alternations of the swimming behavior (71).
Homologues of the Melibe Si1 and Si2 were identified in D. iris based on anatomical, neurochemical, and electrophysiological features (40). However, there are important differences in the neural circuit formed by these neurons (Fig. 5D). Although the contralateral Si2 neurons reciprocally inhibit each other, Si1 does not inhibit or receive inhibition from either contralateral neuron. Instead, Si1 exhibits strong electrical coupling to its contralateral counterpart (Fig. 5E). During a swim motor pattern, the contralateral Si2 neurons fire bursts of action potentials in alternation, but the Si1 pair fire irregularly (Fig. 5F). Thus, whereas both Si1 and Si2 are members of the LR swim CPG in Melibe, only Si2 is in Dendronotus.
If LR swimming in Melibe and Dendronotus is homologous, as would be expected from scenarios 2, 3, or 4 (Fig. 3 B–D), this would be an example in which the neural mechanisms diverged while the behavior stayed the same. However, it could be the case that the differences in neural mechanism reflect a different evolutionary origin for LR swimming in Melibe and Dendronotus as in scenario 1 (Fig. 3A).
Functions of DV Swim CPG Neurons in Other Species
DSI and C2 homologues can be recognized by using neuroanatomical and neurochemical criteria, allowing them to be identified in species that are not DV swimmers (Table 1). The DSIs are serotonergic (72, 73) and have a characteristic axon projection pattern (67). They have been identified in 10 different genera, including two opisthobranchs outside of the Nudipleura (74). Electrophysiological traits of the DSI homologues show little correlation with the type of behavior produced by the species (74). C2 has been identified based on peptide immunoreactivity and characteristic morphology in five genera within the Nudipleura (75). Thus, these DV swim CPG neurons are present regardless of the animal’s mode of locomotion. This suggests that the swimming CPGs were built upon previously existing neural circuits, coopting existing neurons for new functions.
Table 1.
Homologous neurons identified in different species with different behaviors
| Nudipleura |
||||
| Neuron | DV swimmers | LR swimmers | Nonswimmers | Other Opisthobranchia |
| DSI | Tritonia (76) | Melibe (74) | Armina (74) | Aplysia (78–81) |
| Pleurobranchaea (64) | Dendronotus (74) | Triopha (74) | Clione (82, 83) | |
| Hermissenda (77) | Tochina (74) | |||
| C2 | Tritonia (76, 84) | Melibe (75) | ||
| Pleurobranchaea (49) | Hermissenda (75) | |||
| Flabellina (75) | ||||
The DV swim CPG neurons are not members of the LR swim CPGs. The DSI and C2 homologs in Melibe are not rhythmically active in phase with the motor pattern (Fig. 6A), nor are the DSI homologues rhythmically active during the Dendronotus swim motor pattern (Fig. 6B). Thus, categorically distinct behaviors are produced by CPGs containing nonoverlapping sets of neurons.
Fig. 6.
Homologues of the Tritonia DV swim CPG neurons are not rhythmically active during LR swim motor patterns. (A) In Melibe, the C2 and DSI homologues do not display any rhythmic bursting in phase with the swim motor pattern reflected in the alternating firing pattern of the left and right Si. (B) In Dendronotus, a contralateral pair of DSI homologues exhibit synchronous irregular spiking that shows no relation to the ongoing LR swim motor pattern displayed by two contralateral pedal motor neurons (L-Pd and R-Pd). All recordings are previously unpublished.
It was shown that the DSI homologues in Melibe do have an effect on the production of the swim motor pattern; they can initiate a motor pattern in a quiescent preparation and hyperpolarization can temporarily halt an ongoing motor pattern (85). In contrast to Tritonia, in which the DSIs are an integral part of the DV swim CPG, in Melibe, they act as extrinsic modulators. Thus, the functions of homologous neurons differ in species with different behaviors.
The DSIs are not dedicated to one function even within a species. In Pleurobranchaea, the DSI homologues synapse onto serotonergic neurons that increase ciliary beating and thereby increase the speed of crawling (86). In Tritonia, DSI accelerates crawling through synapses onto the efferent peptidergic pedal neuron Pd5, which in turn increases cilia beat frequency (87). DSI homologues in the nonswimming Tochuina tetraquetra and Triopha catalinae also monosynaptically excite homologues of Pd5 and presumably increasing the speed of crawling (74). In Hermissenda, which produces LR flexions, the DSI homologues do not increase ciliary beating, but instead excite motor neurons that cause contraction of the anterior foot (77). In the more distantly related opisthobranch, Aplysia californica, DSI homologues also initiate muscular crawling (78). Whereas, in the pelagic opisthobranch, C. limacina, the DSI homologues increase the frequency of parapodial “wing” flapping and excite motor neurons that innervate the wings (83, 88). Thus, the DSI homologues share common functions in controlling the foot and/or locomotion.
The C2 and DSI homologues have additional roles outside of locomotion. In Pleurobranchaea, the C2 homologue (A1) suppresses feeding through its connections to feeding-related interneurons (49). In contrast, the DSI homologues (As) have the opposite effect by exciting a number of feeding interneurons (86). This is a shared function with other opisthobranchs such as A. californica, in which the DSI homologues (CC9-10) help excite one of the same feeding interneurons as in Pleurobranchaea, the metacerebral cell (78). Thus, individual neurons are multifunctional. Some functions are shared across species, whereas other functions are particular to some species.
Conclusions
A phylogenetic analysis of the neural basis for swimming in the Nudipleura has revealed several interesting aspects about the evolution of behavior. First, the basic building blocks of neural circuits, namely the neurons, are shared across diverse species. For example, DSI homologues are found across Opisthobranchia. Second, neurons, which are multifunctional within a species, appear to take on additional functions over the course of evolution. For instance, the DSI homologues are involved in several behaviors in various species, including generating DV swimming or enhancing other types of locomotion such as enhancing LR swimming or wing flapping. They also accelerate crawling and promote feeding. It is reasonable to expect that highly interconnected interneurons would not be dedicated to a single function, but would dynamically interact with many neurons involved in a variety of different behaviors.
This comparative analysis has also revealed that species with categorically similar behaviors such as the two DV swimmers, Tritonia and Pleurobranchaea, or the two LR swimmers, Melibe and Dendronotus, have overlapping sets of neurons in the swim CPG circuits. In contrast, the CPGs underlying categorically distinct behaviors consist of nonoverlapping sets of neurons. However, even in species that exhibit similar behaviors such as Melibe and Dendronotus, the CPG circuits can differ in neuronal and synaptic composition. Thus, although behavior itself is not a predictor of its underlying neural mechanism, it is a good first approximation.
We do not understand why the circuits in Melibe and Dendronotus differ. There could be functional reasons; perhaps Si1, which is not rhythmically active in Dendronotus, has an additional function that is incompatible with swimming in that species. There may also be phylogenetic reasons; perhaps Melibe and Dendronotus independently evolved swim CPGs and came up with different circuit organizations. Whatever the reason, the results show that analogous behaviors can be generated by circuits with different circuit architectures. Recent work in invertebrates has shown that there can be variability in neural circuits that is not reflected in the performance of the behavior even across individuals within a species (89, 90).
There is a great degree of behavioral homoplasy. Although scenario 4 (Fig. 3D) may be the most parsimonious explanation for the phylogenetic distribution of the swimming behaviors, it should be kept in mind that only approximately 2% to 3% of nudibranch species have been reported to swim. Therefore, there is probably even more behavioral homoplasy than any of the scenarios in Fig. 3 indicate. It is conceivable that swimming arose independently in each family where it is found, 16 times in all (Fig. 1 and Table S1).
Given that Tritonia and Pleurobranchaea are very distantly related within the Nudipleura clade, it is even more likely that they independently evolved DV swim CPGs. If so, the incorporation of DSI and C2 homologues into such a circuit represents parallel evolution, whereby homologous structures independently came to have similar functions (91–94). This has been suggested for other systems as well. For example, homologous brain nuclei appear to be involved in vocal learning in lineages of birds that evolved song independently (95, 96). Similarly, interaural coincidence detection circuits arose independently in the brainstem nuclei of birds and mammals (97). Finally, the appearance of similar cortical areas are correlates with the independent evolution of precision hand control in primates (98), suggesting that constraints in cortical organization led to the evolution of similar neural mechanisms underlying dexterity (99).
If homologous neurons are repeatedly incorporated into neural circuits for analogous behaviors, it suggests that these neurons may be part of a more readily achievable state for swimming. Thus, the nervous system may affect the evolvability of behavior because some configurations of existing neurons could be more robust than others. The concept of evolvability first arose from genetics (100, 101), but has since been applied to nervous systems (5, 7–9). Exploring the aspects of neural organization that lead to repeated evolution of particular behaviors will point to the factors that are most important for behavioral output.
Supplementary Material
Acknowledgments
We thank Arianna Tamvacakis for feedback on the manuscript. This work was supported by National Science Foundation Integrative Organismal Systems Grants 0814411, 1120950, and 1011476.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution VI: Brain and Behavior,” held January 19–21, 2012, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at www.nasonline.org/evolution_vi.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201877109/-/DCSupplemental.
References
- 1.Striedter GF, Northcutt RG. Biological hierarchies and the concept of homology. Brain Behav Evol. 1991;38:177–189. doi: 10.1159/000114387. [DOI] [PubMed] [Google Scholar]
- 2.Rendall D, Di Fiore A. Homoplasy, homology, and the perceived special status of behavior in evolution. J Hum Evol. 2007;52:504–521. doi: 10.1016/j.jhevol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Lauder GV. Homology, form, and function. In: Hall BK, editor. Homology: The Hierachical Basis of Comparative Biology. San Diego: Academic; 1994. pp. 151–196. [Google Scholar]
- 4.Lauder GV. Homology, analogy, and the evolution of behavior. In: Nitecki MH, Kitchell JA, editors. Evolution of Animal Behavior. New York: Oxford Univ Press; 1986. pp. 9–40. [Google Scholar]
- 5.Katz PS. Neural mechanisms underlying the evolvability of behaviour. Philos Trans R Soc Lond B Biol Sci. 2011;366:2086–2099. doi: 10.1098/rstb.2010.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson BA, et al. Brain evolution triggers increased diversification of electric fishes. Science. 2011;332:583–586. doi: 10.1126/science.1201524. [DOI] [PubMed] [Google Scholar]
- 7.Bendesky A, Bargmann CI. Genetic contributions to behavioural diversity at the gene-environment interface. Nat Rev Genet. 2011;12:809–820. doi: 10.1038/nrg3065. [DOI] [PubMed] [Google Scholar]
- 8.Airey DC, Castillo-Juarez H, Casella G, Pollak EJ, DeVoogd TJ. Variation in the volume of zebra finch song control nuclei is heritable: developmental and evolutionary implications. Proc Biol Sci. 2000;267:2099–2104. doi: 10.1098/rspb.2000.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto K, Vernier P. The evolution of dopamine systems in chordates. Front Neuroanat. 2011;5:21. doi: 10.3389/fnana.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darwin C. The Origin of Species by Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray; 1876. [Google Scholar]
- 11.Heinroth O. Beiträge zur Biologie, namentlich Ethologie und Psychologie der Anatiden. Verhanalung des V Internationalen Ornithologen Kongresses, Berlin, 1910. 1911. pp. 589–702.
- 12.Whitman CO. Animal Behavior. Biological Lectures of the Marine Biological Laboratory. Woods Hole, MA: Woods Hole Marine Biological Laboratory; 1899. [Google Scholar]
- 13.Lorenz K. The Foundations of Ethology. New York: Springer-Verlag; 1981. [Google Scholar]
- 14.De Queiroz A, Wimberger PH. The usefulness of behavior for phylogeny estimation: Levels of homoplasy in behavioral and morphological characters. Evolution. 1993;47:46–60. doi: 10.1111/j.1558-5646.1993.tb01198.x. [DOI] [PubMed] [Google Scholar]
- 15.Wenzel JW. Behavioral homology and phylogeny. Annu Rev Ecol Syst. 1992;23:361–381. [Google Scholar]
- 16.Stuart AE, Hunter FF, Currie DC. Using behavioural characters in phylogeny reconstruction. Ethol Ecol Evol. 2002;14:129–139. [Google Scholar]
- 17.Proctor HC. Measures of homoplasy. In: Sanderson MJ, Hufford L, editors. Homoplasy: The Recurrence of Similarity in Evolution. San Diego: Academic; 1996. pp. 131–149. [Google Scholar]
- 18.Foster SA, Cresko WA, Johnson KP, Tlusty MU, Willmott HE. In: Homoplasy: The Recurrence of Similarity in Evolution. Sanderson MJ, Hufford L, editors. San Diego: Academic; 1996. pp. 245–269. [Google Scholar]
- 19.Waegele H, Willan RC. Phylogeny of the nudibranchia. Zool J Linn Soc. 2000;130:83–181. [Google Scholar]
- 20.Wollscheid-Lengeling E, Boore J, Brown W, Waegele H. The phylogeny of Nudibranchia (Opisthobranchia, Gastropoda, Mollusca) reconstructed by three molecular markers. Org Divers Evol. 2001;1:241–256. [Google Scholar]
- 21.Göbbeler K, Klussmann-Kolb A. Out of Antarctica?—new insights into the phylogeny and biogeography of the Pleurobranchomorpha (Mollusca, Gastropoda) Mol Phylogenet Evol. 2010;55:996–1007. doi: 10.1016/j.ympev.2009.11.027. [DOI] [PubMed] [Google Scholar]
- 22.Bouchet P, et al. Classification and nomenclator of gastropod families. Malacologia. 2005;47:1–368. [Google Scholar]
- 23.Behrens DW. Nudibranch Behavior. Jacksonville, FL: New World Publications; 2005. [Google Scholar]
- 24.Dinapoli A, Klussmann-Kolb A. The long way to diversity—phylogeny and evolution of the Heterobranchia (Mollusca: Gastropoda) Mol Phylogenet Evol. 2010;55:60–76. doi: 10.1016/j.ympev.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 25.Pola M, Gosliner TM. The first molecular phylogeny of cladobranchian opisthobranchs (Mollusca, Gastropoda, Nudibranchia) Mol Phylogenet Evol. 2010;56:931–941. doi: 10.1016/j.ympev.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Vonnemann V, Schrodl M, Klussmann-Kolb A, Wagele H. Reconstruction of the phylogeny of the Opisthobranchia (Mollusca: Gastropoda) by means of 18S and 28S rRNA gene sequences. J Molluscan Stud. 2005;71:113–125. [Google Scholar]
- 27.Valdes A. A phylogenetic analysis and systematic revision of the cryptobranch dorids (Mollusca, Nudibranchia, Anthobranchia) Zool. J. Linn. Soc. Lond. 2003;136:535–636. [Google Scholar]
- 28.Thollesson M. Phylogenetic analysis of dorid nudibranchs (Gastropoda: Doridacea) using the mitochondrial 16S rRNA gene. J Molluscan Stud. 1999;65:335–353. [Google Scholar]
- 29.Chase R. Behavior and Its Neural Control in Gastropod Molluscs. New York: Oxford Univ Press; 2002. [Google Scholar]
- 30.Audesirk G, McCaman RE, Willows AOD. The role of serotonin in the control of pedal ciliary activity by identified neurons in Tritonia diomedia. Comp Biochem Physiol. 1979;62C:87–91. doi: 10.1016/0306-4492(79)90104-7. [DOI] [PubMed] [Google Scholar]
- 31.Audesirk G. Central neuronal control of cilia in Tritonia diamedia. Nature. 1978;272:541–543. doi: 10.1038/272541a0. [DOI] [PubMed] [Google Scholar]
- 32.Willows AOD, Pavlova GA, Phillips NE. Modulation of ciliary beat frequency by neuropeptides from identified molluscan neurons. J Exp Biol. 1997;200:1433–1439. doi: 10.1242/jeb.200.10.1433. [DOI] [PubMed] [Google Scholar]
- 33.Lalli CM, Gilmer RW. Pelagic Snails. Stanford, CA: Stanford Univ Press; 1989. [Google Scholar]
- 34.Farmer WM. Swimming gastropods (Opisthobranchia and Prosobranchia) Veliger. 1970;13:73–89. [Google Scholar]
- 35.Rudman WB, Darvell BW. Opisthobranch molluscs of Hong Kong: Part 1. Goniodorididae, Onchidorididae, Triophidae, Gymnodorididae, Chromodorididae (Nudibranchia) Asian Marine Biology. 1990;7:31–79. [Google Scholar]
- 36.Marshall JG, Willan RC. Nudibranchs of Heron Island, Great Barrier Reef - a survey of the Opisthobranchia (sea slugs) of Heron and Wistari Reefs. Leiden, The Netherlands: Backhuys; 1999. [Google Scholar]
- 37.Lowe RT. Description of a new dorsibranchiate gasteropod discovered at Madeira. Proc Zool Soc Lond. 1842;10:51–53. [Google Scholar]
- 38.Pola M, Cervera JL, Gosliner TM. Description of two new phanerobranch nembrothid species (Nudibranchia: Polyceridae: Doridacea) J Mar Biol Assoc U K. 2006;86:403–409. [Google Scholar]
- 39.Lawrence KA, Watson WH., 3rd Swimming behavior of the nudibranch Melibe leonina. Biol Bull. 2002;203:144–151. doi: 10.2307/1543383. [DOI] [PubMed] [Google Scholar]
- 40.Sakurai A, Newcomb JM, Lillvis JL, Katz PS. Different roles for homologous interneurons in species exhibiting similar rhythmic behaviors. Curr Biol. 2011;21:1036–1043. doi: 10.1016/j.cub.2011.04.040. [DOI] [PubMed] [Google Scholar]
- 41.Mills CE. In: Reproduction and Development of Marine Invertebrates. Wilson Jr WH, Stricker SA, Shinn GL., editors. Baltimore: Johns Hopkins Univ Press; 1994. pp. 313–319. [Google Scholar]
- 42.Johnson S. A new Indo-West pacific species of the dendronotacean nudibranch Bornella (Mollusca: Opisthobranchia) with anguilliform swimming behaviour. Micronesica. 1984;19:17–26. [Google Scholar]
- 43.Willows AO. Behavioral acts elicited by stimulation of single, identifiable brain cells. Science. 1967;157:570–574. doi: 10.1126/science.157.3788.570. [DOI] [PubMed] [Google Scholar]
- 44.Katz PS. Tritonia swim network. Scholarpedia. 2009;4:3638. [Google Scholar]
- 45.Gillette R, Jing J. The role of the escape swim motor network in the organization of behavioral hierarchy and arousal in Pleurobranchaea. Am Zool. 2001;41:983–992. [Google Scholar]
- 46.Davis WJ, Mpitsos GJ. Behavioral choice and habituation in the marine mollusk Pleurobranchaea californica MacFarland (Gastropoda, Opisthobranchia) Z Vgl Physiol. 1971;75:207–232. [Google Scholar]
- 47.Katz PS. In: Handbook of Microcircuits. Shepherd G, Grillner S, editors. New York: Oxford Univ Press; 2010. pp. 443–449. [Google Scholar]
- 48.Hume RI, Getting PA, Del Beccaro MA. Motor organization of Tritonia swimming. I. Quantitative analysis of swim behavior and flexion neuron firing patterns. J Neurophysiol. 1982;47:60–74. doi: 10.1152/jn.1982.47.1.60. [DOI] [PubMed] [Google Scholar]
- 49.Jing J, Gillette R. Neuronal elements that mediate escape swimming and suppress feeding behavior in the predatory sea slug Pleurobranchaea. J Neurophysiol. 1995;74:1900–1910. doi: 10.1152/jn.1995.74.5.1900. [DOI] [PubMed] [Google Scholar]
- 50.Edmunds M. On the swimming and defensive response of Hexabranchus marginatus (Mollusca, Nudibranchia) J. Linnean Soc. 1968;47:425–429. [Google Scholar]
- 51.Gohar HAF, Soliman GN. The biology and development of Hexabranchus sanguineus (Rüpp. and Leuck.) (Gastropoda, Nudibranchiata) Publ Mar Biol Sta Ghardaqa (Red Sea) 1963;12:219–247. [Google Scholar]
- 52.Arshavsky YuI, et al. Control of locomotion in marine mollusc Clione limacina VI. Activity of isolated neurons of pedal ganglia. Exp Brain Res. 1986;63:106–112. doi: 10.1007/BF00235652. [DOI] [PubMed] [Google Scholar]
- 53.Donovan DA, Pennings SC, Carefoot TH. Swimming in the sea hare Aplysia brasiliana: Cost of transport, parapodial morphometry, and swimming behavior. J Exp Mar Biol Ecol. 2006;328:76–86. [Google Scholar]
- 54.Bebbington A, Hughes GM. Locomotion in Aplysia (Gastropoda, Opisthobranchia) J Molluscan Stud. 1973;40:399–405. [Google Scholar]
- 55.Thompson TE, Slinn DJ. On the biology of the opisthobranch Pleurobranchus membranaceus. J Mar Biol Assoc U K. 1959;38:507–524. [Google Scholar]
- 56.Moczek AP, Cruickshank TE, Shelby A. When ontogeny reveals what phylogeny hides: Gain and loss of horns during development and evolution of horned beetles. Evolution. 2006;60:2329–2341. [PubMed] [Google Scholar]
- 57.Duboué ER, Keene AC, Borowsky RL. Evolutionary convergence on sleep loss in cavefish populations. Curr Biol. 2011;21:671–676. doi: 10.1016/j.cub.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 58.Harshman J, et al. Phylogenomic evidence for multiple losses of flight in ratite birds. Proc Natl Acad Sci USA. 2008;105:13462–13467. doi: 10.1073/pnas.0803242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiens JJ, Kuczynski CA, Duellman WE, Reeder TW. Loss and re-evolution of complex life cycles in marsupial frogs: Does ancestral trait reconstruction mislead? Evolution. 2007;61:1886–1899. doi: 10.1111/j.1558-5646.2007.00159.x. [DOI] [PubMed] [Google Scholar]
- 60.Whiting MF, Bradler S, Maxwell T. Loss and recovery of wings in stick insects. Nature. 2003;421:264–267. doi: 10.1038/nature01313. [DOI] [PubMed] [Google Scholar]
- 61.Cockerell TDA. Three new nudibranchs from California. J Malacol. 1901;8:85–87. [Google Scholar]
- 62.Delcomyn F. Neural basis of rhythmic behavior in animals. Science. 1980;210:492–498. doi: 10.1126/science.7423199. [DOI] [PubMed] [Google Scholar]
- 63.Croll RP. Identified neurons and cellular homologies. In: Ali MA, editor. Nervous Systems in Invertebrates. New York: Plenum; 1987. pp. 41–59. [Google Scholar]
- 64.Jing J, Gillette R. Central pattern generator for escape swimming in the notaspid sea slug Pleurobranchaea californica. J Neurophysiol. 1999;81:654–667. doi: 10.1152/jn.1999.81.2.654. [DOI] [PubMed] [Google Scholar]
- 65.Thompson S, Watson WH., 3rd Central pattern generator for swimming in Melibe. J Exp Biol. 2005;208:1347–1361. doi: 10.1242/jeb.01500. [DOI] [PubMed] [Google Scholar]
- 66.Dorsett DA, Willows AOD, Hoyle G. Centrally generated nerve impulse sequences determining swimming behavior in Tritonia. Nature. 1969;224:711–712. [Google Scholar]
- 67.Getting PA, Lennard PR, Hume RI. Central pattern generator mediating swimming in Tritonia. I. Identification and synaptic interactions. J Neurophysiol. 1980;44:151–164. doi: 10.1152/jn.1980.44.1.151. [DOI] [PubMed] [Google Scholar]
- 68.Getting PA. Mechanisms of pattern generation underlying swimming in Tritonia. I. Neuronal network formed by monosynaptic connections. J Neurophysiol. 1981;46:65–79. doi: 10.1152/jn.1981.46.1.65. [DOI] [PubMed] [Google Scholar]
- 69.Getting PA. Neural control of swimming in Tritonia. In: Roberts A, Roberts BL, editors. Symposia of the Society for Experimental Biology, No. 37, Neural Origin of Rhythmic Movements, New York: Cambridge Univ Press; 1983. pp. 89–128. [PubMed] [Google Scholar]
- 70.Watson WH, Lawrence KA, Newcomb JM. Neuroethology of Melibe leonina swimming behavior. Am Zool. 2001;41:1026–1035. [Google Scholar]
- 71.Watson WH, 3rd, Newcomb JM, Thompson S. Neural correlates of swimming behavior in Melibe leonina. Biol Bull. 2002;203:152–160. doi: 10.2307/1543384. [DOI] [PubMed] [Google Scholar]
- 72.Katz PS, Getting PA, Frost WN. Dynamic neuromodulation of synaptic strength intrinsic to a central pattern generator circuit. Nature. 1994;367:729–731. doi: 10.1038/367729a0. [DOI] [PubMed] [Google Scholar]
- 73.McClellan AD, Brown GD, Getting PA. Modulation of swimming in Tritonia: Excitatory and inhibitory effects of serotonin. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1994;174:257–266. doi: 10.1007/BF00193792. [DOI] [PubMed] [Google Scholar]
- 74.Newcomb JM, Katz PS. Homologues of serotonergic central pattern generator neurons in related nudibranch molluscs with divergent behaviors. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2007;193:425–443. doi: 10.1007/s00359-006-0196-4. [DOI] [PubMed] [Google Scholar]
- 75.Lillvis JL, Gunaratne CA, Katz PS. Neurochemical and neuroanatomical identification of central pattern generator neuron homologues in Nudipleura molluscs. PLoS ONE. 2012;7:e31737. doi: 10.1371/journal.pone.0031737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Getting PA. Neuronal organization of escape swimming in Tritonia. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1977;121:325–342. [Google Scholar]
- 77.Tian LM, Kawai R, Crow T. Serotonin-immunoreactive CPT interneurons in Hermissenda: Identification of sensory input and motor projections. J Neurophysiol. 2006;96:327–335. doi: 10.1152/jn.00035.2006. [DOI] [PubMed] [Google Scholar]
- 78.Jing J, Vilim FS, Cropper EC, Weiss KR. Neural analog of arousal: Persistent conditional activation of a feeding modulator by serotonergic initiators of locomotion. J Neurosci. 2008;28:12349–12361. doi: 10.1523/JNEUROSCI.3855-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xin YP, Koester J, Jing J, Weiss KR, Kupfermann I. Cerebral-abdominal interganglionic coordinating neurons in Aplysia. J Neurophysiol. 2001;85:174–186. doi: 10.1152/jn.2001.85.1.174. [DOI] [PubMed] [Google Scholar]
- 80.Wright WG, Jones K, Sharp P, Maynard B. Widespread anatomical projections of the serotonergic modulatory neuron, CB1, in Aplysia. Invert Neurosci. 1995;1:173–183. doi: 10.1007/BF02331914. [DOI] [PubMed] [Google Scholar]
- 81.Mackey SL, Kandel ER, Hawkins RD. Identified serotonergic neurons LCB1 and RCB1 in the cerebral ganglia of Aplysia produce presynaptic facilitation of siphon sensory neurons. J Neurosci. 1989;9:4227–4235. doi: 10.1523/JNEUROSCI.09-12-04227.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Panchin YV, Popova LB, Deliagina TG, Orlovsky GN, Arshavsky YI. Control of locomotion in marine mollusk Clione limacina. VIII. Cerebropedal neurons. J Neurophysiol. 1995;73:1912–1923. doi: 10.1152/jn.1995.73.5.1912. [DOI] [PubMed] [Google Scholar]
- 83.Satterlie RA, Norekian TP. Serotonergic modulation of swimming speed in the pteropod mollusc Clione limacina. III. Cerebral neurons. J Exp Biol. 1995;198:917–930. doi: 10.1242/jeb.198.4.917. [DOI] [PubMed] [Google Scholar]
- 84.Taghert PH, Willows AOD. Control of a fixed action pattern by single, central neurons in the marine mollusk, Tritonia diomedea. J Comp Physiol. 1978;123:253–259. [Google Scholar]
- 85.Newcomb JM, Katz PS. Different functions for homologous serotonergic interneurons and serotonin in species-specific rhythmic behaviours. Proc Biol Sci. 2009;276:99–108. doi: 10.1098/rspb.2008.0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jing J, Gillette R. Escape swim network interneurons have diverse roles in behavioral switching and putative arousal in Pleurobranchaea. J Neurophysiol. 2000;83:1346–1355. doi: 10.1152/jn.2000.83.3.1346. [DOI] [PubMed] [Google Scholar]
- 87.Popescu IR, Frost WN. Highly dissimilar behaviors mediated by a multifunctional network in the marine mollusk Tritonia diomedea. J Neurosci. 2002;22:1985–1993. doi: 10.1523/JNEUROSCI.22-05-01985.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arshavsky YI, Deliagina TG, Orlovsky GN, Panchin YV, Popova LB. Interneurones mediating the escape reaction of the marine mollusc Clione limacina. J Exp Biol. 1992;164:307–314. [Google Scholar]
- 89.Roffman RC, Norris BJ, Calabrese RL. Animal-to-animal variability of connection strength in the leech heartbeat central pattern generator. J Neurophysiol. 2011;107:1681–1693. doi: 10.1152/jn.00903.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goaillard JM, Taylor AL, Schulz DJ, Marder E. Functional consequences of animal-to-animal variation in circuit parameters. Nat Neurosci. 2009;12:1424–1430. doi: 10.1038/nn.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hoekstra HE, Price T. Evolution. Parallel evolution is in the genes. Science. 2004;303:1779–1781. doi: 10.1126/science.1096413. [DOI] [PubMed] [Google Scholar]
- 92.Scotland RW. What is parallelism? Evol Dev. 2011;13:214–227. doi: 10.1111/j.1525-142X.2011.00471.x. [DOI] [PubMed] [Google Scholar]
- 93.Wake DB, Wake MH, Specht CD. Homoplasy: From detecting pattern to determining process and mechanism of evolution. Science. 2011;331:1032–1035. doi: 10.1126/science.1188545. [DOI] [PubMed] [Google Scholar]
- 94.Sanderson MJ, Hufford L. Homoplasy: The Recurrence of Similarity in Evolution. San Diego: Academic; 1996. [Google Scholar]
- 95.Hara E, Rivas MV, Ward JM, Okanoya K, Jarvis ED. Convergent differential regulation of parvalbumin in the brains of vocal learners. PLoS ONE. 2012;7:e29457. doi: 10.1371/journal.pone.0029457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Feenders G, et al. Molecular mapping of movement-associated areas in the avian brain: A motor theory for vocal learning origin. PLoS ONE. 2008;3:e1768. doi: 10.1371/journal.pone.0001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schnupp JW, Carr CE. On hearing with more than one ear: Lessons from evolution. Nat Neurosci. 2009;12:692–697. doi: 10.1038/nn.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Padberg J, et al. Parallel evolution of cortical areas involved in skilled hand use. J Neurosci. 2007;27:10106–10115. doi: 10.1523/JNEUROSCI.2632-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Krubitzer L. In search of a unifying theory of complex brain evolution. Ann N Y Acad Sci. 2009;1156:44–67. doi: 10.1111/j.1749-6632.2009.04421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Masel J, Trotter MV. Robustness and evolvability. Trends Genet. 2010;26:406–414. doi: 10.1016/j.tig.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kirschner M, Gerhart J. Evolvability. Proc Natl Acad Sci USA. 1998;95:8420–8427. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.