Abstract
The diverse array of body plans possessed by arthropods is created by generating variations upon a design of repeated segments formed during development, using a relatively small “toolbox” of conserved patterning genes. These attributes make the arthropod body plan a valuable model for elucidating how changes in development create diversity of form. As increasingly specialized segments and appendages evolved in arthropods, the nervous systems of these animals also evolved to control the function of these structures. Although there is a remarkable degree of conservation in neural development both between individual segments in any given species and between the nervous systems of different arthropod groups, the differences that do exist are informative for inferring general principles about the holistic evolution of body plans. This review describes developmental processes controlling neural segmentation and regionalization, highlighting segmentation mechanisms that create both ectodermal and neural segments, as well as recent studies of the role of Hox genes in generating regional specification within the central nervous system. We argue that this system generates a modular design that allows the nervous system to evolve in concert with the body segments and their associated appendages. This information will be useful in future studies of macroevolutionary changes in arthropod body plans, especially in understanding how these transformations can be made in a way that retains the function of appendages during evolutionary transitions in morphology.
The phylum Arthropoda derives its name from the Greek words for “joint” and “foot” (or “leg”), and the remarkable functional diversity of these arthropod appendages has contributed to the notable evolutionary success of this animal group. The basic arthropod body plan consists of serially repeated body segments, with a pair of appendages on most of these segments. Individual segments (or groups of adjacent segments), along with their associated appendages, are often specialized for particular functions (1). These patterns of specialization vary enormously between arthropod species, and this flexible, modular body plan accounts for the superb mobility and specialized feeding modalities that have enabled arthropods to fill a wide variety of terrestrial and aquatic ecological niches. In turn, the great adaptability of arthropod body morphology may be a result of a highly coordinated patterning mechanism that uses a common regulatory network to align regional identity for the ectoderm, mesoderm, and nervous system along the body axis.
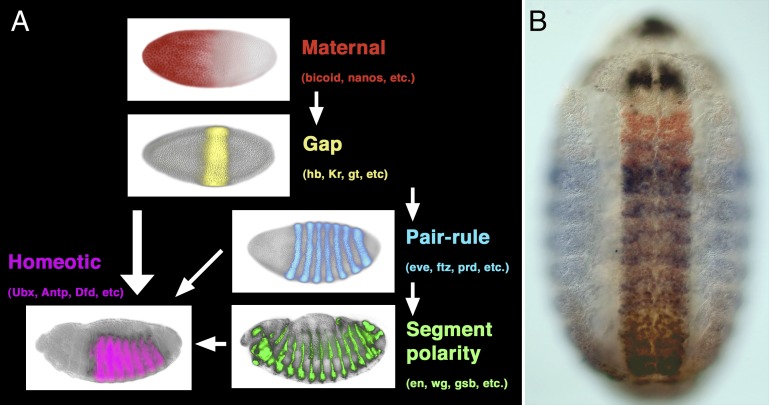
Genetic and molecular studies in the model arthropod, Drosophila melanogaster, have provided us with a detailed understanding of the mechanisms that subdivide the embryo into segments and provide regional identity to these units. The sequential action of maternal gradients and zygotic gap, pair-rule, and segment polarity genes sequentially subdivides the embryos into smaller and smaller units, ultimately organizing the pattern of segmentation. A portion of this segmentation network also regulates the expression of homeotic (Hox) genes, which provide regional identity to the developing segments to make segments distinct from one another (Fig. 1). Altering the expression patterns of these Hox genes leads to the transformation of one or more segments toward the identity of adjacent segments. Subsequent studies revealed a remarkable level of evolutionary conservation of these Hox gene transcription factors, and it appears that Hox genes play a well-conserved role in patterning regional identity along the antero-posterior axis in all bilaterian animals.
Fig. 1.
Early embryo patterning along the antero-posterior axis in Drosophila. (A) Hierarchy of maternal gradients and zygotic gap, pair-rule, and segment polarity genes establishes the repetition of segments, whereas the homeotic genes regionalize the body plan, making segments differ from one another. (B) Protein expression pattern produced by four (of the eight) Hox genes at midembryogenesis. Scr is in black, Antp in red, Ubx in blue, and Abd-B in brown. More intensely stained area in the middle is the central nervous system. Anterior is to the left in A and up in B.
Whereas Hox genes have provided developmental biologists with an outstanding example of a deeply conserved mechanism of pattern formation, changes in these genes have also been implicated in the evolutionary process that has led to the diversification of body plans both between and within animal phyla. For example, comparisons of Hox gene expression and function within the various groups of arthropods led to a number of hypotheses regarding the possible role of these genes in the evolution of the arthropod body plan (reviewed in ref. 2). Indeed, work on Hox genes played a key role in the renaissance of evolutionary developmental biology (“evo-devo”) during the past 30 years.
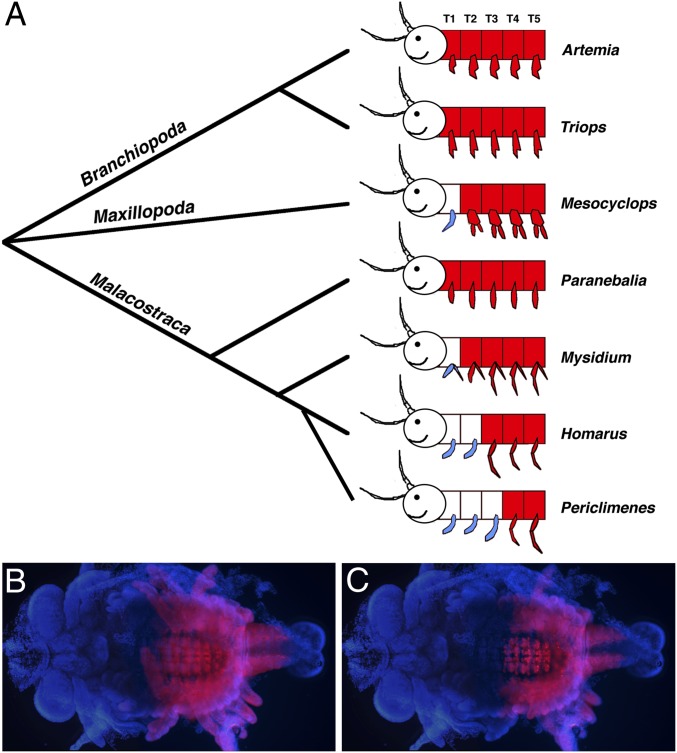
One example of the potential role of Hox genes in morphological evolution comes from work on Ultrabithorax (Ubx) in crustaceans. In this case, changes in the expression pattern of Ubx are associated with the evolution of a specific type of appendage, known as a maxilliped, which is a jaw-like feeding appendage that is part of the anterior thorax. Depending on the species, crustaceans possess anywhere from zero to three pairs of maxillipeds, and, as illustrated in Fig. 2, the point of transition from maxilliped-bearing segments to more posterior thoracic-type segments (usually used for locomotion) is correlated with the boundary of Ubx expression (3). This relationship between Ubx and the evolution of body patterning was moved beyond correlation with functional studies in the amphipod crustacean, Parhyale hawaiensis. A combination of misexpression and knockdown experiments revealed that the number of maxillipeds could be increased or reduced by knocking down or misexpressing Ubx, respectively, in Parhyale (4, 5). The change in the number of maxillipeds was not due to a change in the total number of appendages, but rather due to homeotic transformations altering the relative ratio of different appendage types, resembling the general pattern of differences seen between existing crustacean species.
Fig. 2.
Correspondence between Ubx expression and the transition from feeding to locomotory appendages along the antero-posterior axis during crustacean evolution. (A) Ubx expression is shown in red, maxillipeds are shown in blue, and anterior is to the left. The anterior boundary of Ubx expression in various crustacean species corresponds to the transition point from feeding to locomotory appendages. The head appendages of Mn, MxI, and MxII are not shown, but would also be classified as feeding (jaw) appendages (adapted from ref. 3). (B and C) Ubx protein expression (in red) in a marble crayfish embryo focused to highlight expression in the appendages (B) and the nervous system (C). In this species, there are maxillipeds in T1–T3, and Ubx expression begins at T4 in both the ectoderm and the nervous system.
Whereas these experiments result in the striking transformation of appendage morphology that mimics evolutionary transitions, reservations about the relevance of such Hox-mediated transformation to the natural process of evolution still remain. Such radical morphological transformations in a single step are probably unlikely to be adaptive, but it is reasonable to consider that gradual morphological changes would simply require incremental changes in the patterns and levels of Hox gene expression. Indeed, some crustaceans, such as mysids, possess appendages of intermediate morphology (between a standard maxilliped and a swimming leg) that are associated with intermediate levels and mosaic patterns of Ubx expression (3). Thus, it is likely that microevolutionary changes in Hox gene regulation could occur over time to lead to macroevolutionary changes in morphology.
A more important consideration is that even gradual transformation during evolution must occur in such a way that the appendage and associated segment remain functional and useful to the organism at each point in the transition. For this to happen, more than just the external morphology of the appendage needs to be altered. Coordinated changes must also be made in the musculature and nervous system associated with the transforming appendage. It is reasonable to assume that the segment must evolve as a whole, with coordination between the ectoderm, mesoderm, and nervous system. We suggest that the Hox gene system functions in arthropods in a manner that facilitates such a coordinated transformation. Our purpose here is to review the manner in which the nervous system is patterned in arthropods, highlighting first that the same system used for ectodermal segmentation, particularly at the level of segment polarity genes, contributes to generating the segmental organization of the nervous system, and second, that Hox genes play a major role in the regionalization of the nervous system just as they do for the ectoderm. Most of the data come from studies in Drosophila, but comparative studies have helped to define properties that are generally conserved in neural patterning across the phylum. In conclusion, we argue that the manner in which Hox genes function in the nervous system provides a mechanism to coordinate the different parts of the segment during evolutionary transitions.
Neurogenesis in Arthropods
In arthropods, neurogenesis takes place within a broad ventral domain called the ventral neuroectoderm (VNE), which is competent to form both ectoderm and neural precursor cells. In the VNE of insects, groups of four to eight cells within each hemisegment are recruited into a proneural fate by the achaete-scute complex, and stochastic interactions mediated by Delta-Notch signaling specify one of these cells to become a neural stem cell, called a neuroblast (NB), and the remaining cells become epidermal (6, 7).The specified NB then delaminates from the surrounding epithelium (Fig. 3A) and undergoes several rounds of asymmetric division perpendicular to the epithelium, thereby generating a column of cells called ganglion mother cells. Each ganglion mother cell divides once, symmetrically, to produce either two postmitotic neurons or two postmitotic glial cells (7, 8). The lineage resulting from each NB is invariant.
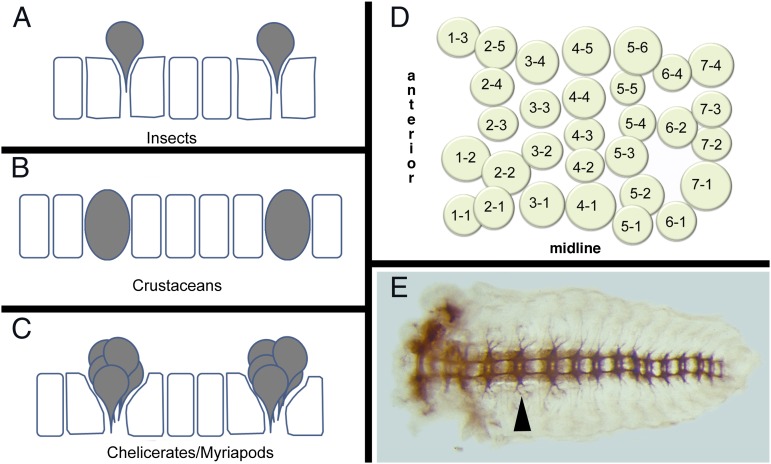
Fig. 3.
Arthropod neurogenesis. (A–C) Process of neuroblast formation in insects (A) and crustaceans (B) and precursor clusters in myriapods and chelicerates (C). Individual neuroblasts delaminate inward in insects, whereas they remain in the epithelia in crustaceans. In myriapods and chelicerates, instead of neuroblasts, clusters of cells move in to form neurons, although their arrangement is reminiscent of the neuroblast pattern seen in insects and crustaceans (adapted from ref. 39). (D) Map of neuroblasts in a Drosophila hemisegment. The 30 neuroblasts are arranged in seven rows along the antero-posterior axis. (E) Nervous system (axon staining) of a grasshopper embryo showing the segmental arrangement of neural ganglia that is coincident with the segmental arrangement of the body ectoderm. Arrowhead points to the ganglion in the T2 segment.
Neuroblast formation and proliferation to form neurons and glia in malacostracan crustaceans are similar to those in insects, with some notable exceptions. Crustacean NBs remain within the VNE and do not delaminate from the epithelium (Fig. 3B) and NB specification appears to involve an invariant lineage pattern (9), as opposed to the inductive system seen in insects.
Neurogenesis in both myriapods and chelicerates is fundamentally different from that seen in insects and crustaceans. Rather than specifying a single stem cell that buds off multiple neurons and glia, an entire cluster of cells is recruited into a neural fate (Fig. 3C) (10–12). Each cell within this cluster invaginates from the VNE, forming a conspicuous layer of cells beneath the presumptive ectoderm. In centipedes and spiders, each cluster consists of 5–9 cells (10, 12). In the millipede, clusters of up to 11 cells are observed (11), which, due to the greater number of cells, are arranged in a grapelike as opposed to planar configuration and are less apically constricted than in spiders and chelicerates. In myriapods, each cell within the invaginated cluster divides equally, resulting in a column of cells within the embryo. However, spider clusters appear to proliferate preferentially within the apical presumptive ectoderm layer, before invagination (13). Cell lineage tracing experiments have yet to be performed in chelicerates and myriapods to determine the relationships between neurons and glia within each cluster.
Despite these differences in the manner in which neural precursor cells form, across all arthropods each hemisegment generates ∼30 NBs (insects and crustaceans) or clusters of precursors (in the case of chelicerates and myriapods) arranged in a stereotyped configuration of seven rows, with a characteristic number of NBs per row (Fig. 3D) (8, 10–12, 14). This configuration is serially repeated between segments and, at least for insects and crustaceans, is important for inferring NB homology between segments of the same animal and between segments of different animals (15).
Specification of Neuroblast Identity Creates Segmental Neuromeres
In Drosophila, the NB array described above is arranged in a segmentally repeated pattern from the outset because of the action of the segmentation network that patterns all ectodermal derivatives. Indeed, much of the specification of the individual NBs occurs before, or just after, their delamination from the ectodermal layer. Detailed studies of the function of segment polarity genes reveal that this level of the segmentation hierarchy acts to pattern the NBs in a manner similar to its role in patterning the overlying ectoderm, although in a few cases it is possible to separate the function of segment polarity genes for patterning the neuroblasts vs. the ectoderm (16, 17).
Once specified, individual neuroblasts generate specific lineages of identified motor neurons, interneurons, and glial cells. Lineage specification involves the sequential expression of genes such as hunchback, Kruppel, castor, and PDM in ganglion mother cells (reviewed in ref. 18) and cell–cell interactions between ganglion mother cell progeny. This process results in a specific and highly reproducible arrangement of ∼600 neurons and glial cells within each segment of the nervous system. Many of these neurons are uniquely identifiable on the basis of morphological criteria such as cell body position and patterns of axonal projection, as well as on the basis of molecular criteria such as patterns of transcription factor and neurotransmitter expression (and some sets of glia are also uniquely identifiable on the basis of cell body position and trancription factor expression). By the midway point of Drosophila embryogenesis, the segmental organization of both the ectoderm (with associated appendage primordia) and the underlying central nervous system is clearly visible, and the same is true during the development of all arthropods. In most arthropods, the neural segments (neuromeres) condense into structures known as ganglia (Fig. 3E), and these ganglia remain located within their respective body segments (Drosophila is a notable exception in which the ganglia fuse and move anteriorly, but remain appropriately connected by nerves to their segments of origin).
Neuromeres Show Distinct Segment-Specific Properties Under Hox Gene Control
Just as with the ectoderm, the individual neural segments are not equivalent. Whereas serially homologous neuroblasts of the thorax and abdomen generally produce the same progeny in each segment, there are at least seven lineages that show differences between segments, and it is the expression of Hox genes within the nervous system that controls these regional differences (19–22). During neurogenesis, Hox genes control NB lineage character by specifying cell number (by regulating both proliferation and apoptosis), cell type (specifying different types of neurons), and neural wiring (regulating axonogenesis). These differences ultimately give rise to segment-specific neural networks. Below, we focus on three individual NB lineages (Fig. 2) to demonstrate how Hox genes control NB fate at various stages throughout neurogenesis.
NB 1-1.
Each neuroblast 1-1 (neuroblast occupying the first column and the first row) in the thoracic segments generates 8–14 cells, but NB 1-1s in the abdominal segments generate only 5–6 cells. In addition, all thoracic NB 1-1 progeny are neurons, whereas the abdominal NB 1-1 produces both neurons and glia (14, 23). This thoracic vs. abdominal neuromere fate difference for NB 1-1 is specified before NB delamination by Ultrabithorax (Ubx) and abdominal A (abdA); and these Hox genes are sufficient to induce an abdominal NB 1-1 fate when misexpressed in the thorax (24).
NB 6-4.
In the embryonic thorax, the NB 6-4 lineage generates neurons and glial cells, whereas in the abdomen, the NB 6-4 lineage produces only glial cells (25). In the thoracic lineage, the absence of abd-A and Abd-B allows CycE to be expressed before the first division of the NB. CycE localizes to one daughter cell via asymmetric division of the neuroblast, which marks it for a neural fate; the absence of CycE in the other daughter cell promotes a glial fate. In the abdomen, abd-A and Abd-B directly repress CycE, and the NB divides symmetrically to produce only glial cells (20).
NB 7-3.
In embryonic segments of the labium and T3 to A8, the NB 7-3–generated motor neuron GW undergoes apoptosis, whereas in T1 and T2, GW is preserved (26). The segments in which GW survives correspond to the expression domain of Antp. Rogulja-Ortmann et al. (22) demonstrate that an early antagonistic interaction between Antp and Ubx regulates the survival of GW during late embryogenesis. Antp is required for the survival of GW, whereas Ubx promotes apoptosis in this cell. In T3, where both Antp and Ubx are expressed, Ubx is strongly up-regulated in late embryogenesis and counteracts the survival signal of Antp, resulting in GW apoptosis. The GW motor neuron of the labial segment never receives the Antp survival signal and thus undergoes apoptosis. The Ubx-directed apoptosis of GW is likely mediated by the proapoptotic gene reaper.
Toward the end of embryogenesis, neuroblast division ceases, and the majority of the NBs in abdominal segments undergo apoptosis, whereas in the thorax, very few NBs apoptose (26, 27). During the larval stage, neurogenesis begins once again as the quiescent neuroblasts begin dividing again (and are now known as postembryonic neuroblasts) (28). The number of postembryonic neuroblasts (pNB) in each hemisegment varies along the antero-posterior axis, with ∼23 pNBs in the thorax and only 3 pNBs in the central abdomen (29). These region-specific differences between homologous pNBs reflect the greater sensory and motor complexity of the adult thorax relative to the abdomen, and again these differences are due to the activity of Hox genes. For example, the three abdominal pNBs transiently express abd-A during proliferation, which limits the number of cells they produce (29).
Regionalized Diversity of Motor Circuits
An important function of the nervous system is to control locomotion, which is achieved through a complex network of sensory neurons, interneurons, and motor neurons. The evolution of arthropods from a worm-like body plan to one with multijointed appendages implies the evolution of a more sophisticated nervous system with segment-specific innervation of individual muscles within the proximodistal axis of the appendages. To organize a series of muscle activations and coordinated movement, each motor neuron must develop a unique identity, extend axons to corresponding muscle targets, and grow proper dendritic trees that connect to sensory and interneurons. Regionalized locomotion is therefore supported by specialized functional networks that emerge during development. Here we discuss studies that show that the regulation by Hox genes of segment-specific neuronal patterning leads to specialized motor control.
Morphological diversity among segmental units of the nervous system is critical for proper axonal targeting and the formation of functional neuromuscular networks. This regionalized diversity is achieved, in part, by the selective cell death and survival of progenitor cells (as described above) and differentiated motor neurons. The regulation of apoptosis has become increasingly refined throughout evolution, and the key roles Hox genes play in the selective death and survival of neurons support their utility in the evolution of neuronal diversification along the antero-posterior axis (30).
The antagonistic effects of Ubx and Antp regulate the survival of two differentiated motor neurons, GW and MNa, in late stages of Drosophila neurogenesis. Antp prevents cell death by blocking reaper- and grim-mediated apoptosis, whereas Ubx, which is strongly up-regulated in the CNS at a late point in development, activates reaper-dependent cell death and executes apoptosis by counteracting the function of Antp (22). The segment-specific levels of Ubx and Antp may therefore enable the refinement of circuitry via the selective paring of motor neurons.
Hox genes may further specify neuronal morphology along the antero-posterior axis by influencing the selective removal of mature neurons. Whereas developmental apoptosis typically occurs immediately after cell birth in Drosophila and other invertebrates, dMP2 and MP1 motor neurons undergo apoptosis only after axonal extension and the guidance of follower neurons has occurred. The MP1 pioneer neuron originates from the ventral midline after gastrulation and forms part of the CNS midline, whereas MP2 (progenitor of dMP2) originates from the ventral neuroectoderm and forms part of the lateral CNS. Postmitotic apoptosis of dMP2 and MP1 takes place only in anterior segments, and the selective survival in posterior segments A6–A8 is mediated by the differential expression of Abd-B, which cell autonomously represses the cell death activators reaper and grim (30).
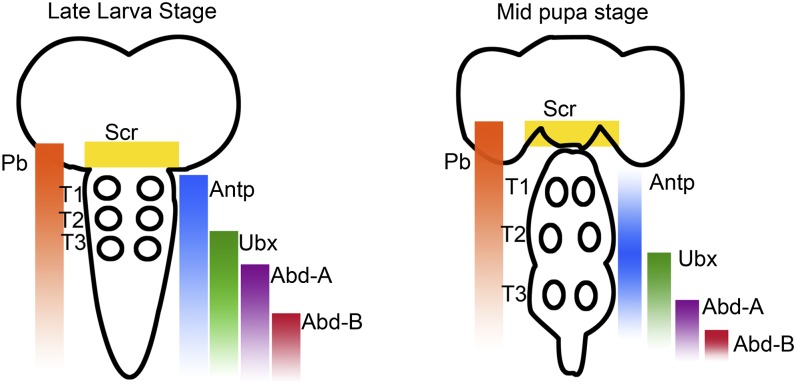
In the leg-bearing segments of Drosophila, motor neurons arise in segment-specific patterns during embryonic and postembryonic neurogenesis (31, 32). In each of these segments, ∼50 motor neurons arise from at least 11 independent lineages, but the majority of these motor neurons derive from only 2 lineages, referred to as Lin A and Lin B. Lin A motor neurons innervate the distal muscles, the femur and the tibia, whereas Lin B innervates the more proximal leg segments, coxa, trochanter, and femur. In addition to their critical role in motor neuron survival and specification during early development, Baek (33) proposed that Hox genes, and the Hox cofactors homothorax (hth) and extradentical (exd), influence axon and dendritic targeting. Pb, Antp, Ubx, hth, and exd are differentially expressed during late larval and midpupal stages in adult leg Drosophila motor neurons within the CNS (see Fig. 4 for expression patterns of Hox genes within the larval and pupal CNS). When the expression of these Hox genes was eliminated, Drosophila leg motor neurons underwent apoptosis and axons showed arborization defects. Levels of Hox and Hox cofactor expression vary between individual Lin A motor neurons, and altering levels of Antp expression in Lin A cells results in axon targeting errors. By removing expression of the thoracic Hox genes (Scr, Antp, and Ubx) or hth function, the number of Lin A motor neurons in all three thoracic segments is reduced. For Lin B, Antp is also required for motor neuron survival, and hth is required for motor circuit development (33). In thinking about the manner in which Hox genes specialize regions of the Drosophila CNS, it is important to remember that Drosophila (as well as all other six-legged insects) appears to have evolved from an arthropod ancestor in which there were once legs on every segment; thus many aspects of motoneuron specialization in the Drosophila abdomen involve “sculpting” back from a thoracic-type pattern during development.
Fig. 4.
Summary of Hox gene expression patterns during the larval and pupal stages of Drosophila. The Hox genes Antp, Ubx, and Pb are expressed in the leg motor neuron containing thoracic segments (T1–T3; position of leg motor neurons indicated by circles). (Adapted from ref. 33.)
The influence of Hox in the specification of region-specific motor neurons is not limited to arthropods. In vertebrates, neurons are organized into distinct columns. Along the spinal cord, motor neurons acquire distinct columnar identities relative to their position along the rostrocaudal (anteroposterior) axis, and each columnar subtype innervates distinct muscle targets (reviewed by ref. 34). Interestingly, postmitotic motor neurons express Hox-c patterns relative to their rostrocaudal position (35), and these expression patterns appear to specify columnar fate. The misexpression of Hoxc6 (members of the Antp group of Hox genes) and Hoxc9 (members of the Abd-B group) elicits rostrocaudal shifts in thoracic- and limb-level identities, suggesting the role of Hox genes in the specification of motor neuron columnar subtypes (34). Furthermore, the rostrocaudal positioning of the lateral motor column (LMC) by Hox6 initiates subsequent axon projections along the dorsoventral axis of a limb (36), and the inactivation of Hoxa10 and Hoxd10 (members of the Abd-B group) causes defects in hind limb innervation (37).
Extension from Drosophila to Other Arthropods
As described previously, there are some notable differences between the process of neurogenesis in Drosophila and that in other arthropods, which have interesting implications for the notion that homologous structures need not share identical developmental pathways. Homology is an important concept in understanding evolution, and a deeper insight into morphogenesis from a developmental and molecular approach may serve to strengthen an abstract definition by referencing concrete operational mechanisms. In the case of insects and crustaceans, there are very clear homologies at the level of neuroblasts, differentiated neurons, and axonal projections (38). Further studies are still needed in myriapods and chelicerates to determine if one-to-one homologies can be extended to these arthropods. In either case, the segmental nature of the neuromeres is apparent for all arthropods. The early steps in the segmentation process vary significantly between arthropod groups, but there is significant conservation at the level of segment polarity gene expression. For example, the segment polarity gene gsb is expressed in the posterior portion of each ectodermal segment and in the underlying neuroblasts of rows 5 and 6. As shown in Fig. 5, the expression pattern of this gene is well conserved in insects, crustaceans, myriapods, and chelicerates. Thus, there are clear molecular similarities in the mechanisms that create the pattern of both body segments and neuromeres in all arthropods. The same also holds true for the genetic system that acts to make neuromeres and body segments different from one another. In this case, the conserved function of the Hox genes appears to control regionalization of both the external body segments (including appendages) and the nervous system in all arthropods studied so far.
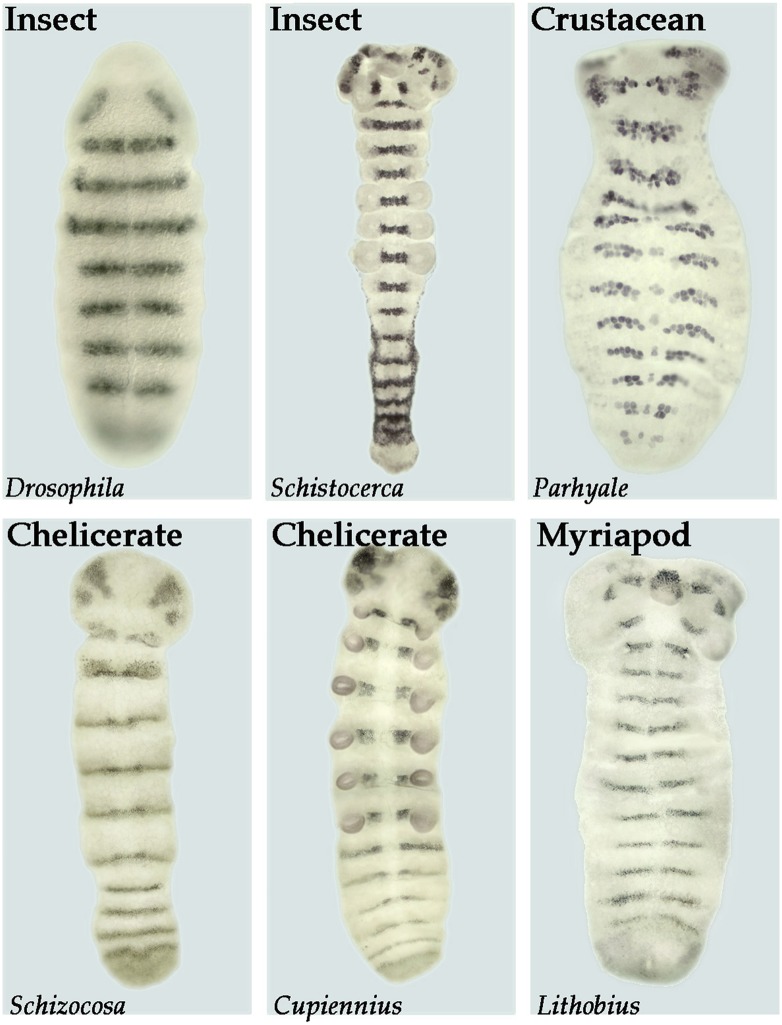
Fig. 5.
Similarity in segment polarity gene expression in the ectoderm and developing nervous system of various arthropods. The segment polarity genes function to maintain and refine segments within both the nervous system and the ectoderm of Drosophila. Shown here is the expression of the segment polarity gene gooseberry (gsb) in Drosophila. Similar patterns of striped expression of gsb homologs through both the ectoderm and the neurogenic region are seen in the grasshopper (Schistocerca), a crustacean (Parhyale), two species of spiders (Schizocosa and Cupiennius), and a centipede (Lithobius).
Summary
As we have described, the mechanisms of segmentation and body regionalization in arthropods function in a manner that allows developmental coordination between ectodermal structures, such as appendages, and the underlying nervous system. We suggest that subdivision of both the body into segments and the nervous system into neuromeres also provides evolutionary flexibility through modular design—any change in one will be mirrored by changes in the other. If segment number is varied by increasing the number of stripes of segment polarity gene expression, the number of neuromeres will also change so that there is still a one-to-one relationship between neural and ectodermal segments. Likewise, a homeotic shift that alters appendage morphology can simultaneously result in a shift in the pattern of neural regionalization. The next step will be to test these ideas in the context of arthropod evolution. Are homeotic-type shifts in appendage specialization during arthropod evolution accompanied by matching shifts in the nervous system that allow coordinated evolution of both appendage morphology and the neural mechanisms that control the locomotion of these appendages? For example, when a crustacean locomotory appendage is transformed to a feeding appendage, is the underlying neural pattern changed as well to ensure that the transformation is functional, not just morphological? Answering these questions will advance our understanding of how macroevolutionary changes in body plans might occur and ultimately help explain how complex nervous systems and behaviors evolve within animals.
Acknowledgments
We thank Greg Davis for providing the images shown in Fig. 5 and the other members of the N.H.P. laboratory for helpful comments and discussion.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution VI: Brain and Behavior,” held January 19–21, 2012, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at www.nasonline.org/evolution_vi.
This article is a PNAS Direct Submission.
References
- 1.Brusca RC, Brusca GJ. Invertebrates. 2nd Ed. Sunderland, MA: Sinauer; 2003. pp. 479–482. [Google Scholar]
- 2.Hughes CL, Kaufman TC. Hox genes and the evolution of the arthropod body plan. Evol Dev. 2002;4:459–499. doi: 10.1046/j.1525-142x.2002.02034.x. [DOI] [PubMed] [Google Scholar]
- 3.Averof M, Patel NH. Crustacean appendage evolution associated with changes in Hox gene expression. Nature. 1997;388:682–686. doi: 10.1038/41786. [DOI] [PubMed] [Google Scholar]
- 4.Liubicich DM, et al. Knockdown of Parhyale Ultrabithorax recapitulates evolutionary changes in crustacean appendage morphology. Proc Natl Acad Sci USA. 2009;106:13892–13896. doi: 10.1073/pnas.0903105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pavlopoulos A, et al. Probing the evolution of appendage specialization by Hox gene misexpression in an emerging model crustacean. Proc Natl Acad Sci USA. 2009;106:13897–13902. doi: 10.1073/pnas.0902804106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodman CS, Doe CQ. In: The Development of Drosophila melanogaster. Bate M, editor. Plainview, NY: Cold Spring Harbor Lab Press; 1993. pp. 1131–1206. [Google Scholar]
- 7.Campos-Ortega JA. Genetic mechanisms of early neurogenesis in Drosophila melanogaster. Mol Neurobiol. 1995;10:75–89. doi: 10.1007/BF02740668. [DOI] [PubMed] [Google Scholar]
- 8.Doe CQ, Goodman CS. Early events in insect neurogenesis. I. Development and segmental differences in the pattern of neuronal precursor cells. Dev Biol. 1985;111:193–205. doi: 10.1016/0012-1606(85)90445-2. [DOI] [PubMed] [Google Scholar]
- 9.Scholtz G, Dohle W. Cell lineage and cell fate in crustacean embryos—a comparative approach. Int J Dev Biol. 1996;40:211–220. [PubMed] [Google Scholar]
- 10.Kadner D, Stollewerk A. Neurogenesis in the chilopod Lithobius forficatus suggests more similarities to chelicerates than to insects. Dev Genes Evol. 2004;214:367–379. doi: 10.1007/s00427-004-0419-z. [DOI] [PubMed] [Google Scholar]
- 11.Dove H, Stollewerk A. Comparative analysis of neurogenesis in the myriapod Glomeris marginata (Diplopoda) suggests more similarities to chelicerates than to insects. Development. 2003;130:2161–2171. doi: 10.1242/dev.00442. [DOI] [PubMed] [Google Scholar]
- 12.Stollewerk A, Weller M, Tautz D. Neurogenesis in the spider Cupiennius salei. Development. 2001;128:2673–2688. doi: 10.1242/dev.128.14.2673. [DOI] [PubMed] [Google Scholar]
- 13.Weller M, Tautz D. Prospero and Snail expression during spider neurogenesis. Dev Genes Evol. 2003;213:554–566. doi: 10.1007/s00427-003-0362-4. [DOI] [PubMed] [Google Scholar]
- 14.Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- 15.Boyan GS, Ball EE. The grasshopper, Drosophila and neuronal homology (advantages of the insect nervous system for the neuroscientist) Prog Neurobiol. 1993;41:657–682. doi: 10.1016/0301-0082(93)90030-v. [DOI] [PubMed] [Google Scholar]
- 16.Chu-LaGraff Q, Doe CQ. Neuroblast specification and formation regulated by wingless in the Drosophila CNS. Science. 1993;261:1594–1597. doi: 10.1126/science.8372355. [DOI] [PubMed] [Google Scholar]
- 17.Duman-Scheel M, Li X, Orlov I, Noll M, Patel NH. Genetic separation of the neural and cuticular patterning functions of gooseberry. Development. 1997;124:2855–2865. doi: 10.1242/dev.124.15.2855. [DOI] [PubMed] [Google Scholar]
- 18.Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol. 2004;20:619–647. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- 19.Technau GM, Berger C, Urbach R. Generation of cell diversity and segmental pattern in the embryonic central nervous system of Drosophila. Dev Dyn. 2006;235:861–869. doi: 10.1002/dvdy.20566. [DOI] [PubMed] [Google Scholar]
- 20.Kannan R, Berger C, Myneni S, Technau GM, Shashidhara LS. Abdominal-A mediated repression of Cyclin E expression during cell-fate specification in the Drosophila central nervous system. Mech Dev. 2010;127:137–145. doi: 10.1016/j.mod.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Prokop A, Bray S, Harrison E, Technau GM. Homeotic regulation of segment-specific differences in neuroblast numbers and proliferation in the Drosophila central nervous system. Mech Dev. 1998;74:99–110. doi: 10.1016/s0925-4773(98)00068-9. [DOI] [PubMed] [Google Scholar]
- 22.Rogulja-Ortmann A, Renner S, Technau GM. Antagonistic roles for Ultrabithorax and Antennapedia in regulating segment-specific apoptosis of differentiated motoneurons in the Drosophila embryonic central nervous system. Development. 2008;135:3435–3445. doi: 10.1242/dev.023986. [DOI] [PubMed] [Google Scholar]
- 23.Udolph G, Prokop A, Bossing T, Technau GM. A common precursor for glia and neurons in the embryonic CNS of Drosophila gives rise to segment-specific lineage variants. Development. 1993;118:765–775. doi: 10.1242/dev.118.3.765. [DOI] [PubMed] [Google Scholar]
- 24.Prokop A, Technau GM. Early tagma-specific commitment of Drosophila CNS progenitor NB1-1. Development. 1994;120:2567–2578. doi: 10.1242/dev.120.9.2567. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt H, et al. The embryonic central nervous system lineages of Drosophila melanogaster II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- 26.Rogulja-Ortmann A, Lüer K, Seibert J, Rickert C, Technau GM. Programmed cell death in the embryonic central nervous system of Drosophila melanogaster. Development. 2007;134:105–116. doi: 10.1242/dev.02707. [DOI] [PubMed] [Google Scholar]
- 27.Peterson C, Carney GE, Taylor BJ, White K. Reaper is required for neuroblast apoptosis during Drosophila development. Development. 2002;129:1467–1476. doi: 10.1242/dev.129.6.1467. [DOI] [PubMed] [Google Scholar]
- 28.Prokop A, Technau GM. The origin of postembryonic neuroblasts in the ventral nerve cord of Drosophila melanogaster. Development. 1991;111:79–88. doi: 10.1242/dev.111.1.79. [DOI] [PubMed] [Google Scholar]
- 29.Bello BC, Hirth F, Gould AP. A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron. 2003;37:209–219. doi: 10.1016/s0896-6273(02)01181-9. [DOI] [PubMed] [Google Scholar]
- 30.Miguel-Aliaga I, Thor S. Segment-specific prevention of pioneer neuron apoptosis by cell-autonomous, postmitotic Hox gene activity. Development. 2004;131:6093–6105. doi: 10.1242/dev.01521. [DOI] [PubMed] [Google Scholar]
- 31.Rogulja-Ortmann A, Technau GM. Multiple roles for Hox genes in segment-specific shaping of CNS lineages. Fly (Austin) 2008;2:316–319. doi: 10.4161/fly.7464. [DOI] [PubMed] [Google Scholar]
- 32.Landgraf M, Thor S. Development of Drosophila motoneurons: specification and morphology. Semin Cell Dev Biol. 2006;17:3–11. doi: 10.1016/j.semcdb.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Baek M. Development of leg motor neurons in Drosophila melanogaster. 2011 PhD dissertation (Columbia University, New York) [Google Scholar]
- 34.Dasen JS, Liu J-P, Jessell TM. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature. 2003;425:926–933. doi: 10.1038/nature02051. [DOI] [PubMed] [Google Scholar]
- 35.Liu JP, Laufer E, Jessell TM. Assigning the positional identity of spinal motor neurons: Rostrocaudal patterning of Hox-c expression by FGFs, Gdf11, and retinoids. Neuron. 2001;32:997–1012. doi: 10.1016/s0896-6273(01)00544-x. [DOI] [PubMed] [Google Scholar]
- 36.Kania A, Jessell TM. Topographic motor projections in the limb imposed by LIM homeodomain protein regulation of ephrin-A:EphA interactions. Neuron. 2003;38:581–596. doi: 10.1016/s0896-6273(03)00292-7. [DOI] [PubMed] [Google Scholar]
- 37.Wahba GM, Hostikka SL, Carpenter EM. The paralogous Hox genes Hoxa10 and Hoxd10 interact to pattern the mouse hindlimb peripheral nervous system and skeleton. Dev Biol. 2001;231:87–102. doi: 10.1006/dbio.2000.0130. [DOI] [PubMed] [Google Scholar]
- 38.Duman-Scheel M, Patel NH. Analysis of molecular marker expression reveals neuronal homology in distantly related arthropods. Development. 1999;126:2327–2334. doi: 10.1242/dev.126.11.2327. [DOI] [PubMed] [Google Scholar]
- 39.Stollewerk A, Chipman AD. Neurogenesis in myriapods and chelicerates and its importance for understanding arthropod relationships. Integr Comp Biol. 2006;46:195–206. doi: 10.1093/icb/icj020. [DOI] [PubMed] [Google Scholar]