Abstract
Na+/Ca2+ exchanger (NCX) proteins mediate Ca2+-fluxes across the cell membrane to maintain Ca2+ homeostasis in many cell types. Eukaryotic NCX contains Ca2+-binding regulatory domains, CBD1 and CBD2. Ca2+ binding to a primary sensor (Ca3-Ca4 sites) on CBD1 activates mammalian NCXs, whereas CALX, a Drosophila NCX ortholog, displays an inhibitory response to regulatory Ca2+. To further elucidate the underlying regulatory mechanisms, we determined the 2.7 Å crystal structure of mammalian CBD12-E454K, a two-domain construct that retains wild-type properties. In conjunction with stopped-flow kinetics and SAXS (small-angle X-ray scattering) analyses of CBD12 mutants, we show that Ca2+ binding to Ca3-Ca4 sites tethers the domains via a network of interdomain salt-bridges. This Ca2+-driven interdomain switch controls slow dissociation of “occluded” Ca2+ from the primary sensor and thus dictates Ca2+ sensing dynamics. In the Ca2+-bound conformation, the interdomain angle of CBD12 is very similar in NCX and CALX, meaning that the interdomain distances cannot account for regulatory diversity in NCX and CALX. Since the two-domain interface is nearly identical among eukaryotic NCXs, including CALX, we suggest that the Ca2+-driven interdomain switch described here represents a general mechanism for initial conduction of regulatory signals in NCX variants.
Introduction
The mammalian Na+/Ca2+ exchanger isoforms (NCX1–3) and their splice variants are expressed in a tissue-specific manner and catalyze electrogenic ion-exchange (3Na+:Ca2+) across the plasma membrane [1], [2], [3]. The recently determined crystal structure of an archaeal NCX from Methanococcus jannaschii (NCX_Mj) revealed the membrane domain architecture for this family, comprising ten transmembrane helices with a pseudo molecular dyad. Buried in the molecular center is a four-ion-binding site cluster with one site for Ca2+ and three for Na+. Two apparent passageways allow separate access for Na+ and Ca2+ ions [4]. The minimal structure supports Na+/Ca2+ exchange [4]. In contrast, the eukaryotic NCX has evolved an allosteric Ca2+ regulatory element encoded by a large cytosolic domain (∼500 amino acids) between transmembrane helices 5 and 6 i.e. the repeated membrane motif, designated as f-loop [5], [6], [7]. This large loop includes two Ca2+-binding regulatory domains (CBD1 and CBD2), connected by a short linker [6]. High-resolution NMR and crystal structures of isolated CBD1 and CBD2 domains of eukaryotic NCX revealed four Ca2+ sites (Ca1-Ca4) on CBD1 [8] and two Ca2+ sites (CaI-CaII) on CBD2 [6], [9].
Eukaryotic NCX variants differ in their response to regulatory Ca2+ [7], [10], [11], [12]. For example, Ca2+ interaction with CBD1 activates the brain (NCX1.4) and cardiac (NCX1.1) variants, whereas Ca2+ has no sustained effect on the kidney variant (NCX1.3) [7], [10]. In contrast, Ca2+ interaction with CBD1 of CALX1.1 (a Drosophila exchanger) inhibits the exchanger activity, whereas CALX1.2 is insensitive to regulatory Ca2+ [11], [12]. The Ca3-Ca4 sites of CBD1 govern Ca2+-dependent regulation either in NCX [13], [14] or CALX [15], [16]. Different lines of evidence suggest that CBD2 interacts with CBD1 to modify the equilibrium and kinetic properties of the primary Ca2+ sensor (Ca3-Ca4 sites), either in isolated CBD12 [17], [18], [19], [20] or in intact NCX [13], [14], [21]. These interdomain interactions are most prominently manifested as slow dissociation of “occluded” Ca2+ from Ca3-Ca4 sites of CBD12 [18], [20], which may represent the slow inactivation (I2 state) of intact NCX observed in electrophysiological experiments [10], [22].
Distinct techniques have been used to elucidate the structural effects of Ca2+ binding to isolated CBD12 [19], [21], [23]. SAXS analysis revealed that Ca2+ binding results in more compact conformation of CBD12 with decreased Dmax, suggesting that Ca2+ interaction with CBD1 changes the electrostatic potential to drive the conformational transition [19]. On the other hand, FRET studies on CBD12 revealed a small decrease in FRET upon Ca2+ binding to CBD1 and thus, suggest a more extended conformation of CBD12 [21]. Finally, NMR analysis of CBD12 suggested that Ca2+ binding to CBD1 results in rigidification of CBD12, by restricting the interdomain linker’s flexibility rather than inducing global changes in their relative reorientation [23].
Recently, high-resolution X-ray structures of CBD12 from CALX1.1 and CALX1.2 were obtained [24]. In the Ca2+-bound state, these structures display small differences in interdomain angles, which were suggested as the structural basis for their differential regulatory responses. According to this rationale, the interdomain angles of CBD12 might significantly differ in CALX1.1 and NCX1.4, since they exhibit diverse regulatory response to Ca2+. In light of these results, two critical mechanistic questions emerge. First, how precisely does Ca2+ binding/occlusion at the primary sensor (Ca3-Ca4 sites) couple conformational transitions in CBDs, and second, how can this coupling account for the regulatory diversity of NCX and CALX.
Here, we posit that the two-domain interface initiates conduction of regulatory signals upon Ca2+ binding to the primary sensor. To test this working hypothesis, we crystallized and determined the crystal structure of a two-domain CBD12 tandem from the brain splice variant (NCX1.4). In conjunction with stopped-flow kinetics and SAXS analyses of relevant mutants, we demonstrate that the two-domain interface controls Ca2+-induced tethering of CBDs and slow dissociation of occluded Ca2+. In Ca2+-bound conformations, the interdomain angle of CBD12 is very similar for NCX1.4 and CALX1.1, meaning that the interdomain angle and/or distance between the two domains cannot account for the regulatory diversity in NCX and CALX. Therefore, Ca2+-binding to a high-affinity sensor (Ca3-Ca4 sites) induces a disorder-to-order transition, producing a more rigid conformation of CBD12. This interdomain switch appears to be a common module for regulation even for NCX variants exhibiting opposite responses to regulatory Ca2+.
Results
Structure Overview
In order to crystallize the tandem, we employed a previously characterized mutant, E454K [25]. Our rationale was three-fold. (i) The E454K substitution replaces the Ca2+ ion at Ca1 site of CBD1, thus, stabilizing the protein’s structure through a charge compensation mechanism [25]. (ii) The full-length NCX1-E454K mutant exhibits WT [Ca2+]-dependent regulation in a cellular system [25]. (iii) The Ca3-Ca4 sites retain their affinity for Ca2+ binding in an isolated CBD1-E454K [20].
The overall structure of CBD12-E454K (Figure 1A, Table 1) shows two nearly identical CBD domains, each composed of an anti-parallel β-sandwich, with root mean square deviation (r.m.s.d for 109 Cα atoms) of 1.3 Å. The domains in the tandem are also very similar to the isolated domains. The r.m.s.d between CBD1 in the tandem and isolated CBD1 (PDB 2DPK) is 0.85 Å (114 Cα atoms) while the r.m.s.d between CBD2 in the tandem and isolated CBD2 (PDB 2QVM) is 0.74 Å (114 Cα atoms).
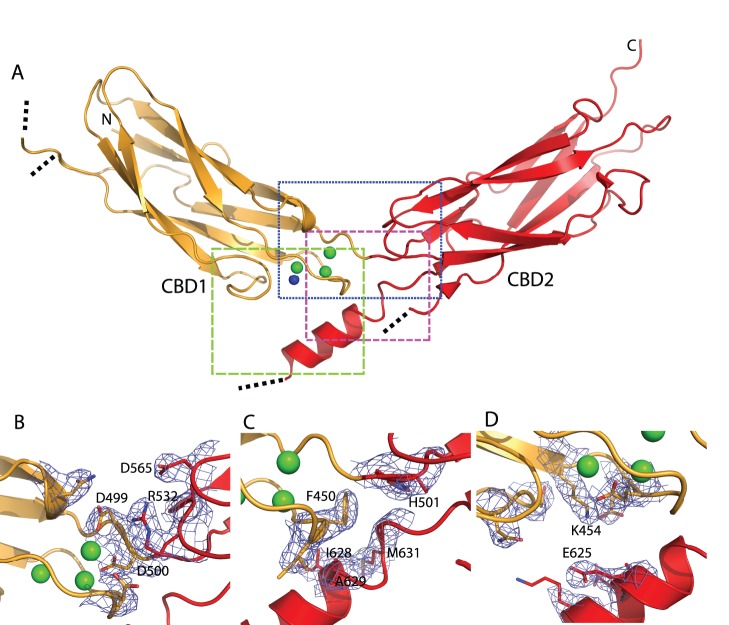
Figure 1. Structure of the CBD12 tandem.
(A) Crystal structure of CBD12-E454K in cartoon representation. CBD1 and CBD2 are colored orange and red, respectively. The rectangles frame a zoom perspective as depicted in panels B (blue), C (magenta) and D (green). Green and blue spheres depict Ca2+ ions and water molecules, respectively. Dotted black lines denote electron density chain breaks in the protein. (B–D) Residues with buried surfaces in the interface are depicted as sticks, with their electron density contoured at 1.5 σ (blue mesh).
Table 1. Crystallographic statistics.
| Data statistics | |
| Wavelength (Å) | 0.976 Å |
| Space group | C2221 |
| Unit cell parameters (Å) | a = 80.7, b = 152.5, c = 79.7 |
| Total reflections | 64902 |
| Unique reflections | 14002 |
| Completeness (%) | 90.0 (80.1)* |
| Rmerge (%) | 7.9 |
| I/σ | 6.1 |
| Resolution range (Å) | 50 - 2.6 |
| X-ray source | ESRF beamline ID29 |
| Refinement statistics | |
| No. of reflections (working/test) | 10459/898 |
| dmin (Å) | 2.68 |
| Rwork/Rfree (%) | 23.4/27.4 |
| RMS deviation from ideality: | |
| Bond lengths | 0.004 |
| Bond angles | 0.9 |
| B factors (Å2) (rmsd of bonded atoms-main/side chain) | 13.1/18.5 |
| Average B factor (Å2) | 84.3 |
| No. of protein atoms/solvent | 1890/15 |
after anisotropic scaling.
Similar to previously published crystal structures of isolated CBD domains, the FG loops of both domains (residues 469–481 and 600–618) cannot be fully visualized due to presumed high flexibility of these areas [9]. Additionally, the N-terminal His-tag, V371 and residues 653–657 are not observed in our structure. It has been shown by NMR that the CBD2 FG-loop contains an α-helix [6], that spans residues 625 through 630 [6], [19]. Our structure now reveals the presence of an α-helix in a similar position in CBD2, spanning residues 620–629, which has side chains that contribute to the interdomain interface, described in detail below.
Interdomain Interface
The interface has a buried surface area of ∼350 Å2. This small interface surface area is in agreement with our previous observations that isolated CBDs do not interact in solution and that a short linker is obligatory for interdomain interactions [20]. The interface involves mainly interactions of the CBD1 Ca2+ binding loops with the interdomain linker, the CBD2 flexible FG loop and the strictly conserved CBD2 BC loop (Figure 1). Interacting residues have low relative B-factors and are highly, if not absolutely conserved (Figure 2, S1 and Table S1), a hallmark of having an important structural or functional role.
Figure 2. Conservation analysis of CBD domains.
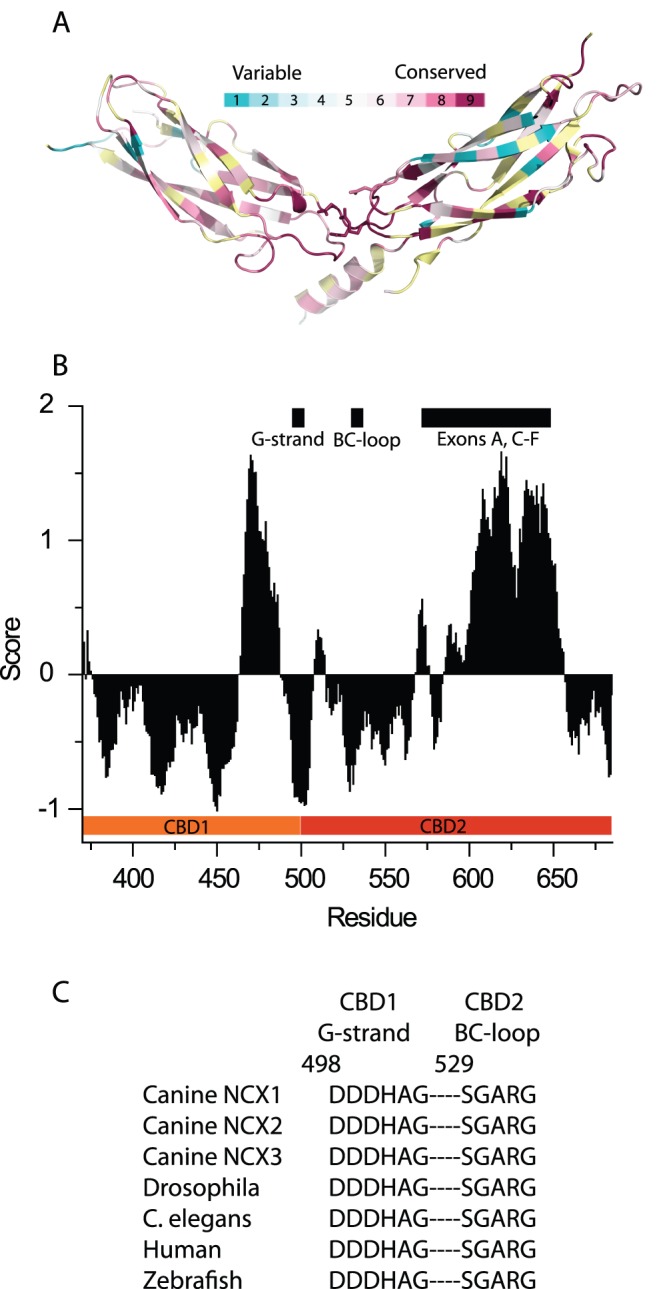
(A) CBD12-E454K structure colored according to the conservation score of each residue. (B) Conservation score for each residue. Negative values indicate conservation while positive values indicate variability. The sequence refers to the cardiac splice-variant, which is 35 residues longer than the brain spliced variant used in our study. (C) Conservation of interface residues located on the G-strand of CBD1 and BC-loop of CBD2.
Although more than 20 residues are buried in the interface, only a few prominent interactions between residues from both domains are observed. The interface, whose electron density is of high quality, may be subdivided into three regions, two hydrophilic (Figure 1B, D) and one hydrophobic (Figure 1C). The first hydrophilic region (Figure 1B) includes a pivotal interdomain electrostatic network centered at R532 in CBD2. This arginine takes a conformation that forms bifurcated hydrogen-bonded and non-hydrogen-bonded salt-bridges with D499, D500 in CBD1 and D565 in CBD2. D499 and D500 also participate in the coordination of the Ca3-Ca4 sites, thereby playing a direct role at the primary Ca2+ sensor while concomitantly stabilizing the tandem domain interface. This interfacial region is the most highly conserved among NCX isoforms and orthologs (Figure 2). The network clearly acts as the principal linchpin holding the two CBDs together.
The hydrophobic interfacial region (Figure 1C) comprises residues from the Ca2+-binding EF loop of CBD1, the linker and the FG loop of CBD2. F450 serves as a core residue, forming van-der Waals interactions with H501, I628, A629, M631 and G632. It was shown that mutating residues participating in this region affect Ca2+ binding in CALX [24], even though they do not participate in Ca2+ binding directly. This finding suggests that Ca2+ binding affects the hydrophobic interfacial region and vice versa. Thus, this region may directly limit linker flexibility through the interaction of F450 with H501 in a Ca2+ dependent manner, as previously suggested by NMR data [23].
The second hydrophilic interfacial region (Figure 1D) is formed between the CBD1 CD and EF loops and the CBD2 FG loop helix. This interfacial region contains few specific interactions, although its residues are largely not surface accessible. Notably, K454 and E625 form a non-hydrogen-bonded salt-bridge (distance of 4.7 Å), a result of the mutant E454K protein used for crystallization. This salt bridge has a similar counterpart in CALX, formed between R673 and E521 [24].
Ca2+ Binding Sites
In agreement with the previously published crystal structure of CBD1-E454K [25], our structure reveals three Ca2+ ions in CBD1 that correspond to Ca2, Ca3 and Ca4 in CBD1-WT (Figure 3). Their low solvent accessibility indicates the structural role of Ca2+ binding for CBD1, which loses its structural integrity in the apo-form [6], [8].
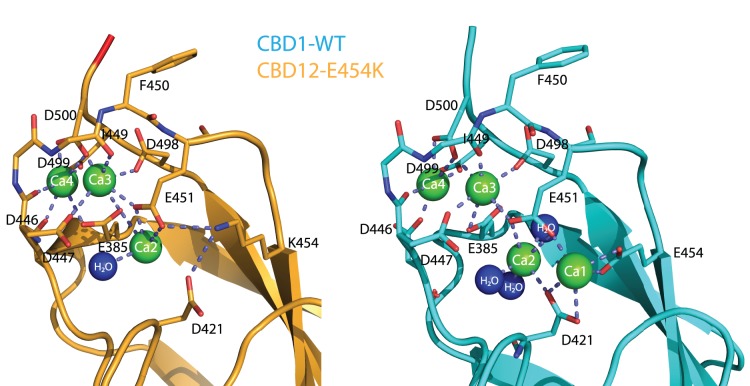
Figure 3. Ca2+ binding sites.
Ca2+ coordination in the CBD12-E454K crystal structure (orange) and in the CBD1-WT crystal structure (cyan, PDB 2DPK). Residues coordinating Ca2+ are depicted as sticks.
Ca2 is coordinated by E385, E451 and a water molecule, consistent with low affinity binding at this site. Residues E385, D447, I449, E451, D498 and D500 coordinate Ca3, whereas E385, D446, D447, D499 and D500 coordinate Ca4. E454K forms salt-bridges with D421 (non-hydrogen-bonded) and E451 (hydrogen-bonded), replacing Ca1 of CBD1-WT. The overall coordination is similar to that reported for isolated CBD1 [8] (Figure 3), with a few exceptions. E385 coordinates only Ca3 in isolated CBD1, while it coordinates Ca2, Ca3 and Ca4 in our structure. D421 coordinates Ca2 in CBD1, but here it only forms salt-bridge with K454, in agreement with the structure of isolated CBD1-E454K [25]. Notably, there is no indication for bound Ca2+ ions in CBD2 of our structure. We ascribe the lack of bound Ca2+ in CBD2 to crystal packing between symmetry mates. This intermolecular interaction utilizes residues that coordinate Ca2+ in the context of the isolated CBD2 [9] (Figure S2).
Ca2+ Dissociation kinetics of CBD12 Mutants
We have previously shown that in CBD12, bi-phasic dissociation kinetics of Ca2+ from the Ca3-Ca4 sites are observed, with slow dissociation of one Ca2+ ion (k s = 0.5 s−1). This sequential dissociation of Ca2+ is not observed either in isolated CBD1, mixture of isolated CBD1 and CBD2 or in CBD12-7A (containing additional seven alanine residues in the linker), and thus, serves as a functional hallmark of domains coupling [20] (Figure 4A). To test whether CBD12-E454K retains sequential dissociation kinetics, we performed stopped-flow assays. In agreement with the intact allosteric regulation of NCX1-E454K, sequential dissociation of Ca2+ is observed, although with ∼ 10 fold increase in k f (fast phase) representing the dissociation of the first Ca2+ ion from Ca3-Ca4 sites of CBD12 (Figure 4B,C). To test the role of the observed electrostatic network in our structure (Figure 1B), we also tested the mutant CBD12-R532A. Strikingly, monophasic dissociation is observed, with a rate constant similar to that of isolated CBD1 [17], [20] (Figure 4B,C) despite the fact that R532 does not directly coordinate Ca2+ at the Ca3-Ca4 sites. Thus, the electrostatic interdomain network has a role in functional coupling of CBDs.
Figure 4. Stopped flow analysis of CBD12-E454K and CBD12-R532A.
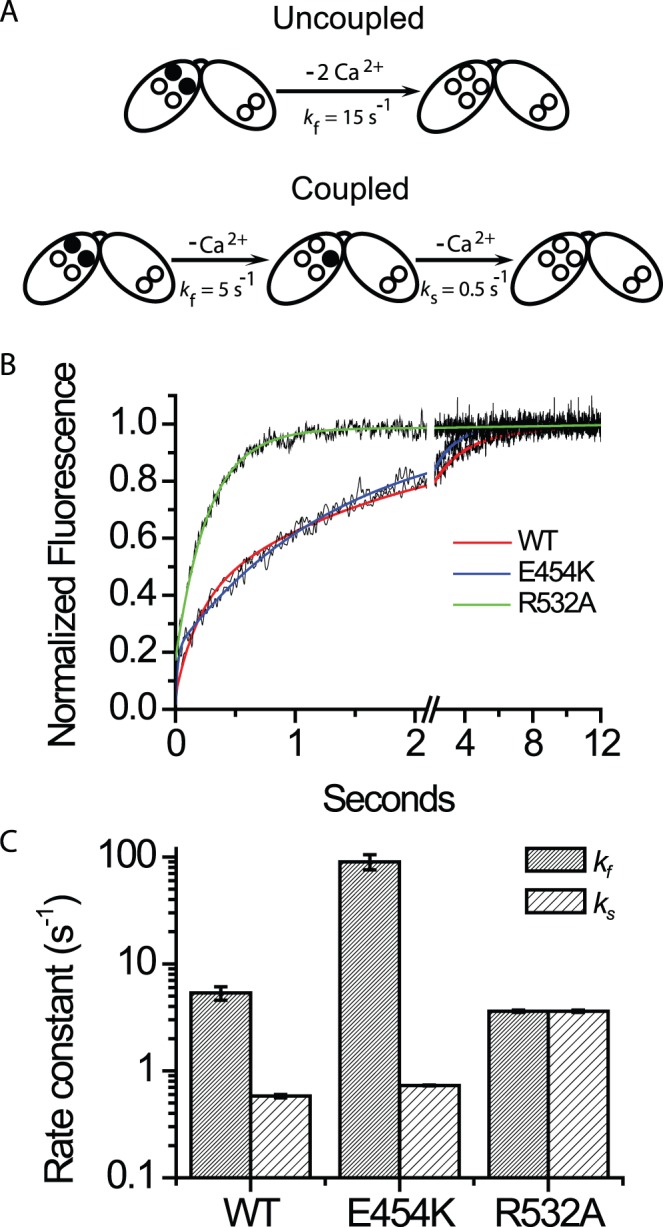
. (A) Monophasic (uncoupled CBDs) and biphasic (coupled CBDs) dissociation kinetics of two Ca2+ ions from the Ca3-Ca4 sites, measured by stopped-flow techniques. Occupied sites are denoted by filled circles, whereas open circles represent empty sites. (B) Representative traces of Ca2+ dissociation kinetics from CBD12-WT, CBD12-E454K and CBD12-R532A. Ca2+ dissociation kinetics of CBD12-WT were fit to a double exponential curve with kf = 5.3±0.04 s−1 and ks = 0.57±0.001 s−1. The trace of CBD12-E454K was fit to a double exponential curve with kf = 52.2±1.04 s−1 and ks = 0.73±0.001 s−1. The representative trace of CBD12-R532A was fit to a single exponential curve with kf = 3.6±0.01 s−1. (C) Bars represent the mean ± S.E values of the “fast” phase (kf) of Ca2+ dissociation (n = 6) and the mean ± S.E of slow off-rates (ks) (n = 6). For CBD12-R532A, in which monophasic dissociation is observed, k f and k s are identical.
SAXS Analysis of CBD12 Mutants
We utilized SAXS to detect Ca2+-dependent reorientation of the CBDs. The pair distance distribution function (PDDF) of CBD12-WT (Figure S3) reveals a conformational “switch” upon Ca2+ binding, with Dmax = 130±1 Å and Dmax = 107±1 Å for the Ca2+-free and Ca2+-bound forms, respectively (Figure 5A). These data are in a good agreement with previous SAXS measurements [19].
Figure 5. SAXS analysis of CBD12 proteins.
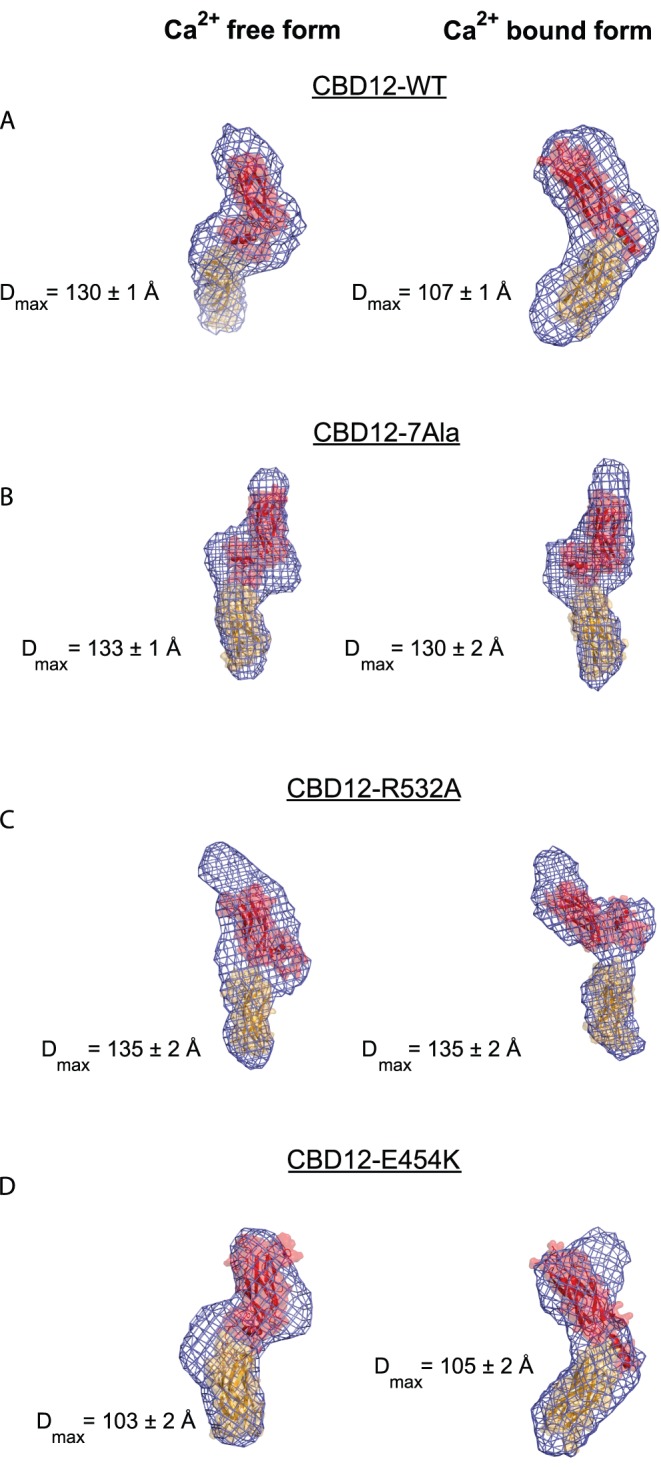
Bead model reconstruction was performed for each protein on the basis of experimental SAXS measurements. The experiments were done in the absence (left column) and presence (right column) of Ca2+, for the wild-type CBD12-WT (A), CBD12-7A (B) CBD12-R532A (C) and CBD12-E454K (D). The CBD1 domain (orange, PDB code: 2FWS) and the CBD2 domain (red, PDB code: 2FWU) are shown as cartoon and surface, and manually fit to the bead model shape (blue mesh), excepting panel A and D, right, where the cartoon is a depiction of the current crystal structure.
As based on the small interface surface area observed in our crystal structure and the absence of sequential dissociation kinetics [20], the insertion in CBD12-7A should abolish interaction between the two domains. Indeed, CBD12-7A lacks Ca2+ dependent reorientation of the CBDs (Figure 5B and Figure S3). Like CBD12-7A, CBD12-R532A does not show Ca2+ dependent reorientation of the CBDs (Figure 5C and Figure S3). Since CBD12-7A likely abolishes the interactions between the two domains, the similar phenotype exhibited by CBD12-R532A supports a key role for R532 in the interdomain network. Both of these mutants adopt only the extended conformations, observed for Ca2+-free CBD12-WT, correlating with their lack of Ca2+ occlusion. These results agree well with our crystallographic and biochemical data. Thus, the SAXS analysis appears to corroborate the structure-function relationship of the interdomain interface.
NCX1-E454K is regulated by Ca2+ as with NCX1-WT as assessed by electrophysiological studies [25]. In our SAXS analysis, however, CBD12-E454K shows no Ca2+ dependency for orientation of the CBDs (Dmax = 103±2 Å and Dmax = 105±2 Å for the Ca2+-free and Ca2+-bound forms, respectively) and maintains only the Ca2+-bound Dmax of CBD12-WT according to our SAXS analysis (Figure 5D and Figure S3). Again, this result correlates well with CBD12-E454’s Ca2+ occlusion behavior, supporting the hypothesis that interactions relevant for Ca2+ occlusion exist in the Ca2+ bound conformation. Table 2 summarizes Dmax and k s values of mutants analyzed by SAXS. Overall, SAXS parameters for CBD12 and its mutants are summarized in Table S2.
Table 2. Dmax and k s values of CBD12 mutants.
| Dmax (Å) | k s (s−1) | ||
| Ca2+-free | Ca2+-bound | ||
| WT | 130±1 | 107±1 | 0.58±0.02 |
| E454K | 103±2 | 105±2 | 0.73±0.007 |
| 7A | 133±1 | 130±2 | ∼17 (ref 18) |
| R532A | 135±2 | 135±2 | 3.61±0.09 |
Ca2+ -bound and -free refers to SAXS measurements performed in the presence of 10 mM CaCl2 or EDTA, respectively. k s values are the mean ± SEM from 6 independent measurements derived from stopped-flow experiments and represent the slowest rate constant measured.
Discussion
The Crystal Structure of Mammalian CBD12 Reveals a Ca2+-driven Interdomain Switch
The most important structural finding is that the two domains are tethered in a Ca2+-dependent manner, involving amino acids from both CBD1 and CBD2 (Figure 1). The buried R532, located in CBD2, is a central residue in the interface, tethering D565 in CBD2 and D499 and D500 in CBD1. Arginine, as opposed to lysine, appears to be required at this position due to the precise stereochemistry of the surrounding aspartates. Most importantly, the bifurcated salt-bridges between R532 and D499 and D500 support Ca2+ coordination at the Ca3-Ca4 sites. Therefore, Ca2+ binding to the Ca3-Ca4 sites couples directly to the interdomain interface to restrict the interdomain flexibility of the CBDs, as suggested by NMR (29). Consistent with this interpretation, Ca2+ binding to Ca3-Ca4 sites seems to be obligatory for robust interdomain interactions, since D499 and D500 are disordered in the apo-form [6], [16]. A recent NMR study revealed that Ca2+ binding to CBD1 restricts the flexibility of the CBD1-CBD2 linker to rigidify interdomain movements without global changes in CBDs reorientation [23]. The Ca2+-induced tethering of CBDs, described here, represents a new mode for ligand-induced rigidification of CBDs. Most probably, both mechanisms contribute in a cooperative way to the decoding of the regulatory message.
The CBD12-E454K mutant does not undergo global conformational changes in response to Ca2+ binding (Table 2), even though physiology shows NCX-E454K to be Ca2+ regulated [25]. This is not surprising, in light of the mutation’s stabilizing effect on the local order of CBD1 and possibly due to the salt-bridge formation with CBD2 (Figure 1D). It is possible that in the context of intact NCX, structural constraints would result in smaller or no change in Dmax. This possibility emphasizes the role of Ca2+-induced rigidification in allosteric regulation rather than large conformational changes. The reduced rigidity of CBD12-E454K in the absence of Ca2+ can be qualitatively assessed from the less-featured PDDF of its Ca2+-free form (Figure S3). Thus, our and NMR data point to a sequential scheme whereby Ca2+ occupation of Ca3-Ca4 sites orients side-chains D499 and D500 pairing with R532, and in turn restricts linker flexibility and interdomain motions in regulating NCX activity.
The Interface Governs Dynamic Aspects of the Primary Ca2+ Sensor in CBD1
Our findings clearly indicate that the interface participates in slow dissociation of occluded Ca2+ from CBD12. Our structure shows a proximity between two charged residues in CBD2 (R532 and D565) and two Ca2+ coordinating residues (D499 and D500) at Ca3-Ca4 sites of CBD1. The R532 mutant of CBD12 lacks the slow Ca2+ dissociation (Figure 3B,C) and Ca2+-dependent conformational transition observed by SAXS (Fig 4C). We speculate that after dissociation of the first Ca2+ ion, the interdomain salt-bridges prevent complete unfolding of Ca3-Ca4 sites in CBD1 by electrostatic compensation, thereby enabling occlusion of the remaining ion. Following dissociation of the second Ca2+ ion, CBD1 binding sites undergo further unfolding, resulting in the apo-form orientation (Dmax ∼ 130 Å) and no interaction of R532 with CBD1. Therefore, the R532 mutation results in “intrinsic uncoupling” of interdomain interactions, exhibiting a phenotype of non-interacting CBDs. Thus, CBD decoupling may impart decreased affinity for [Ca2+]-dependent activation in a NCX1-R532C mutant due to the low affinity binding of Ca2+ to CBD2 [14].
Structural Comparison of the CBD12 Interface with other Proteins
Interface conservation and composition points to a general mechanism for the NCX family. Importantly, the architecture of this interface differs from the tandem C2 domains of synaptotagmin and PKC [26], implying a different mode of action. Rather, motif searches [27] of the PDB reveal a striking similarity with the cadherin extracellular domain, which bears multiple β-sandwich domains bridged by small interfaces, and which contains three Ca2+ sites. Cadherin studies demonstrated that Ca2+ rigidifies the protein [28], enabling cell-cell interactions. Furthermore, in vivo studies suggest that extracellular Ca2+ fluctuations may physiologically regulate cadherin activity [29], suggesting the relevance of Ca2+-dependent rigidification. In addition, the tandem architecture is reminiscent of the arrestin family, where tandem β sandwiches are apposed by a polar core of buried charged residues. Disruption of this polar core activates arrestin for high-affinity binding to its GPCR target [30]. These parallel actions share a common denominator of interfaces with charge interactions wherein charged ligands impart structural transitions.
A Common Mechanism Underlies Ca2+-driven Interdomain Switch in the NCX Family
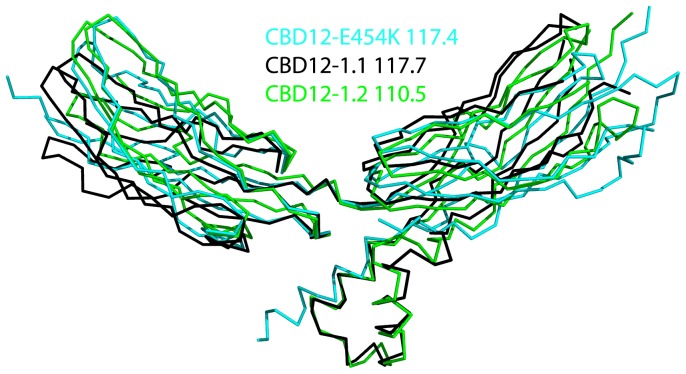
Recently, the crystal structures of CBD12 tandems from the two CALX splice variants (1.1 and 1.2) were determined [24]. CALX is a Na+/Ca2+ exchanger from Drosophila in which the binding of Ca2+ to CBDs inactivates the exchanger (1.1 variant) or has no effect (1.2 variant) [12]. Zheng and coworkers proposed that the stark regulatory differences between the two CALX splice variants arise from different hinge angles between the domains, thereby altering signal transmission to the membrane domain [24]. In agreement with the high degree of structural similarity between isolated CBD domains from NCX and CALX [8], [9], [15], [16], the structures of the tandem CBD12-E454K and CALX CBD12-1.1 molecules are similar (r.m.s.d of 1.86 Å for 214 Cα atoms) (Figure 6). Since the hinge angle is similar for CBD12-E454K, CALX CBD12-1.1 and CALX CBD12-1.2 (117.4°, 117.7°, and 110.5°, respectively) (Figure 6), and taking into account the existence of interdomain movements, the conformation of these domains is probably very similar in solution. The structural similarity between CBD12 from NCX and CALX (Figure 6) implies that the different responses to regulatory Ca2+ cannot be attributed solely to the orientation of CBDs in the CBD12 tandem. It is also possible that the conformational dynamic responses to Ca2+ binding differ in CBD12 of NCX and CALX, despite the similar orientation of CBDs present in these crystal structures. We hypothesize that additional structural elements in the regulatory f-loop and/or membrane domain are probably involved in specifying their differing regulatory effects. Identification of these structural elements is a crucial task for further defining the mechanism underlying Ca2+ regulation.
Figure 6. Superposition of CBD12 from NCX and CALX.
The structures of CBD12-E454K from NCX1 and of CBD12-1.1 and CBD12-1.2 from CALX (PDB codes 3RB5 and 3RB7, respectively) are colored cyan, black and green, respectively. The indicated values represent the hinge angle between CBD1 and 2 as defined by Cα atoms K373, H501 and E647 (NCX CBD12-E454K) and R443, H553 and I692 (CALX CBD12-1.1 and CBD12-1.2).
In summary, we solved the crystal structure of a two-domain tandem mutant from NCX1’s intracellular regulatory loop and found that the CBDs communicate via a complex network of electrostatic interactions at their interface. We found a correlation between the dynamic properties of CBD12 and structural data revealing Ca2+-driven communication between CBDs. Most importantly, occupation of Ca3-Ca4 sites by Ca2+ induces a disorder-to-order transition (40) due to charge neutralization and coordination, thereby constraining CBD conformational freedom, rigidifying the NCX1 f-loop, and triggering regulatory signal transmission to the membrane domain. Undoubtedly, additional structural motifs are involved in transmitting the regulatory signal in NCX variants.
Materials and Methods
Overexpression, Mutagenesis, and Purification of CBD12 Proteins
The DNA constructs of CBD12 (encoding residues 371–657) of canine NCX1 (accession code P23685; AD-splice variant) were cloned into pET23b vector and expressed in E. coli Rosetta2 (DE3) competent cells (Novagen), as described [17], [18], [20]. Mutations were introduced by QuickChange mutagenesis (Stratagene) and confirmed by sequencing. Overexpressed proteins were purified on Ni-beads followed by size exclusion chromatography (>95% purity, judged by SDS-PAGE).
Crystallization, Data Collection and Structure Determination
Purified CBD12-E454K at a concentration of ∼30 mg/mL was prepared in buffer composed of 100 mM KCl, 10 mM CaCl2, 10 mM β-mercaptoethanol and 10 mM Tris-HCl at pH 7.2. Initial crystallization screens were performed at 19°C with Macrosol MD1-22 screen (Molecular Dimensions, Inc.) using the sitting drop vapor diffusion method. Initial crystals were obtained with 12% PEG-8K and 200 mM NH4SO4. Crystallization was optimized by adding 10% sucrose and lowering the PEG-8K concentration to 7%. Crystals were slowly dehydrated by raising the PEG-8K concentration in the reservoir solution by 2%/day. When the PEG-8K concentration reached 30%–32%, crystals were flash-frozen in cryo-loops. Data were collected from the flash-frozen crystals cryocooled to 110° K at beamline ID29 of the European Synchrotron Radiation Facility (Grenoble, France).
Data were processed using HKL2000 [31]. Due to anisotropic diffraction, ellipsoid truncation of the data was performed using the UCLA MBI Diffraction Anisotropy Server [32]. Resolution limits were 3.1, 2.9 and 2.68 Å along a*, b* and c* axes, respectively. This anisotropically scaled data were used for refinement of an initial model. The initial model was obtained using molecular replacement with the program PHASER [33] in the PHENIX suite [34] using a data set that diffracted to ∼3 Å. The coordinates of CBD1-E454K (PDB code 3GIN) and CBD2 (PDB code 2QVM) were used as search models. The model was refined by PHENIX [34] with multiple rounds of manual model building, performed using COOT [35]. Data and refinement statistics of the improved data set are summarized in table 1. The atomic coordinates and structure factors (PDB code 3US9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Stopped-flow Experiments
The stopped-flow assays were performed with a three-syringe/two mixer SFM-3 instrument (BioLogic, France) as described [17], [18], [20]. The data were analyzed with Bio-Kine 32 V4.45 (Bio-Logic, France). In the stopped-flow experiments, the Ca2+ dissociation from proteins was monitored with Quin-2 as described [17], [18], [20]. Briefly, 150 µl of 10 µM protein (pre-equilibrated with [Ca]free = 5–10 µM) was mixed with 150 µl of 200 µM Quin-2 in TK-buffer.
SAXS Data Collection, Data Analysis and Bead Model Reconstruction
SAXS data were measured at beamlines 12-ID-B and 12-ID-C of the Advanced Photon Source, Argonne National Laboratory, USA and at ID14-3 of the European Synchrotron Radiation Facility (Grenoble, France). The following samples were used for the experiments: the wild-type protein CBD12 and its mutants at three concentrations (2, 5 and 10 mg/ml) in buffers containing 20 mM Tris-HCl, pH 7.5, 100 mM KCl, 20 mM β-mercaptoethanol with10 mM EDTA or 10 mM Ca2+. A FoXS web-server was used for computation and fitting of SAXS profiles [36]. Further description of data collection, analysis and bead model reconstitution is described in the Information S1.
Conservation Analysis of CBD12
The Ensembl server [37] was used to search for canine cardiac NCX orthologs, which is the longest splice variant containing an additional 35 residues in CBD2. These sequences were used to generate a multiple sequence alignment using Clustalw [38]. The resulting alignment was used as input for Consurf [39], [40], [41], which outputs a conservation score for each residue.
Supporting Information
Multiple Sequence Alignment of CBD12 orthologs. Numbering according to canine NCX1 cardiac splice variant.
(PDF)
Crystal packing involves CBD2 Ca2+ binding sites. (A) Interaction between symmetry mates in the CBD12-E454K crystal. (B) Salt-bridges are formed between symmetry mates, involving residues that participate in coordination of Ca2+ in isolated CBD2 (E516, K585).
(EPS)
SAXS analysis. Comparison of SAXS scattering curves (left column) and normalized pair distance distribution functions (PDDF) (right column) for the wild type CBD12 (A, B), CBD12-7Ala (C, D) CBD12-R532A (E, F) and CBD12-E454K (G, H), respectively, in the absence (black) or presence of Ca2+ (red). The scattering profiles were normalized and superimposed against the first data points.
(EPS)
CBD12 Interface Conservation.
(DOCX)
Overall SAXS parameters for CBD12 and its mutants. R g, D max, and NSD are, respectively, the radius of gyration derived from Guinier plotting, maximum inter-atomic dimension, normalized shape discrepancy for DAMMIN calculation. Note that NSD scores for all bead models are less than 0.8, indicating high convergence of the bead model calculations.
(DOCX)
Experimental procedures.
(DOCX)
Acknowledgments
We are grateful to APS scientists, Soenke Seifert, Xiaobing Zuo, and Randall Winans for their kind help in the SAXS experiments. We thank the staffs of ID29 and ID14-3 at the ESRF for assistance with diffraction and SAXS experimentation, respectively.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was partially funded by the Israeli Ministry of Health Grant # 2010-3-6266, the USA-Israel Binational Foundation Research Grant # 2009-334, the Israel Science Foundation Grant # 23/10 to DK and a DIP-DFG grant to JAH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Philipson KD, Nicoll DA. Sodium-calcium exchange: a molecular perspective. Annu Rev Physiol. 2000;62:111–133. doi: 10.1146/annurev.physiol.62.1.111. [DOI] [PubMed] [Google Scholar]
- 2.Blaustein MP, Lederer WJ. Sodium/calcium exchange: its physiological implications. Physiol Rev. 1999;79:763–854. doi: 10.1152/physrev.1999.79.3.763. [DOI] [PubMed] [Google Scholar]
- 3.Lytton J. Na/Ca2+ exchangers: three mammalian gene families control Ca2+ transport. Biochem J. 2007;406:365–382. doi: 10.1042/BJ20070619. [DOI] [PubMed] [Google Scholar]
- 4.Liao J, Li H, Zeng W, Sauer DB, Belmares R, et al. Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger. Science. 2012;335:686–690. doi: 10.1126/science.1215759. [DOI] [PubMed] [Google Scholar]
- 5.Levitsky DO, Nicoll DA, Philipson KD. Identification of the high affinity Ca2+-binding domain of the cardiac Na+-Ca2+ exchanger. J Biol Chem. 1994;269:22847–22852. [PubMed] [Google Scholar]
- 6.Hilge M, Aelen J, Vuister GW. Ca2+ regulation in the Na+/Ca2+ exchanger involves two markedly different Ca2+ sensors. Mol Cell. 2006;22:15–25. doi: 10.1016/j.molcel.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Matsuoka S, Nicoll DA, Reilly RF, Hilgemann DW, Philipson KD. Initial localization of regulatory regions of the cardiac sarcolemmal Na+-Ca2+ exchanger. Proc Natl Acad Sci U S A. 1993;90:3870–3874. doi: 10.1073/pnas.90.9.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicoll DA, Sawaya MR, Kwon S, Cascio D, Philipson KD, et al. The crystal structure of the primary Ca2+ sensor of the Na+/Ca2+ exchanger reveals a novel Ca2+ binding motif. J Biol Chem. 2006;281:21577–21581. doi: 10.1074/jbc.C600117200. [DOI] [PubMed] [Google Scholar]
- 9.Besserer GM, Ottolia M, Nicoll DA, Chaptal V, Cascio D, et al. The second Ca2+-binding domain of the Na+-Ca2+ exchanger is essential for regulation: crystal structures and mutational analysis. Proc Natl Acad Sci U S A. 2007;104:18467–18472. doi: 10.1073/pnas.0707417104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyck C, Omelchenko A, Elias CL, Quednau BD, Philipson KD, et al. Ionic regulatory properties of brain and kidney splice variants of the NCX1 Na+-Ca2+ exchanger. J Gen Physiol. 1999;114:701–711. doi: 10.1085/jgp.114.5.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hryshko LV, Matsuoka S, Nicoll DA, Weiss JN, Schwarz EM, et al. Anomalous regulation of the Drosophila Na+-Ca2+ exchanger by Ca2+. J Gen Physiol. 1996;108:67–74. doi: 10.1085/jgp.108.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Omelchenko A, Dyck C, Hnatowich M, Buchko J, Nicoll DA, et al. Functional differences in ionic regulation between alternatively spliced isoforms of the Na+-Ca2+ exchanger from Drosophila melanogaster. J Gen Physiol. 1998;111:691–702. doi: 10.1085/jgp.111.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ottolia M, Nicoll DA, Philipson KD. Roles of two Ca2+-binding domains in regulation of the cardiac Na+-Ca2+ exchanger. J Biol Chem. 2009;284:32735–32741. doi: 10.1074/jbc.M109.055434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ottolia M, Nicoll DA, John S, Philipson KD. Interactions between Ca2+ binding domains of the Na+-Ca2+ exchanger and secondary regulation. Channels (Austin) 2010;4:159–162. doi: 10.4161/chan.4.3.11386. [DOI] [PubMed] [Google Scholar]
- 15.Wu M, Wang M, Nix J, Hryshko LV, Zheng L. Crystal structure of CBD2 from the Drosophila Na+/Ca2+ exchanger: diversity of Ca2+ regulation and its alternative splicing modification. J Mol Biol. 2009;387:104–112. doi: 10.1016/j.jmb.2009.01.045. [DOI] [PubMed] [Google Scholar]
- 16.Wu M, Le HD, Wang M, Yurkov V, Omelchenko A, et al. Crystal structures of progressive Ca2+ binding states of the Ca2+ sensor Ca2+ binding domain 1 (CBD1) from the CALX Na+/Ca2+ exchanger reveal incremental conformational transitions. J Biol Chem. 2010;285:2554–2561. doi: 10.1074/jbc.M109.059162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyman L, Mikhasenko H, Hiller R, Khananshvili D. Kinetic and equilibrium properties of regulatory calcium sensors of NCX1 protein. J Biol Chem. 2009;284:6185–6193. doi: 10.1074/jbc.M809012200. [DOI] [PubMed] [Google Scholar]
- 18.Boyman L, Hagen BM, Giladi M, Hiller R, Lederer WJ, et al. Proton-sensing Ca2+ binding domains regulate the cardiac Na+/Ca2+ exchanger. J Biol Chem. 2011;286:28811–28820. doi: 10.1074/jbc.M110.214106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilge M, Aelen J, Foarce A, Perrakis A, Vuister GW. Ca2+ regulation in the Na+/Ca2+ exchanger features a dual electrostatic switch mechanism. Proc Natl Acad Sci U S A. 2009;106:14333–14338. doi: 10.1073/pnas.0902171106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giladi M, Boyman L, Mikhasenko H, Hiller R, Khananshvili D. Essential role of the CBD1-CBD2 linker in slow dissociation of Ca2+ from the regulatory two-domain tandem of NCX1. J Biol Chem. 2010;285:28117–28125. doi: 10.1074/jbc.M110.127001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.John SA, Ribalet B, Weiss JN, Philipson KD, Ottolia M. Ca2+-dependent structural rearrangements within Na+-Ca2+ exchanger dimers. Proc Natl Acad Sci U S A. 2011;108:1699–1704. doi: 10.1073/pnas.1016114108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuoka S, Nicoll DA, Hryshko LV, Levitsky DO, Weiss JN, et al. Regulation of the cardiac Na+-Ca2+ exchanger by Ca2+. Mutational analysis of the Ca2+-binding domain. J Gen Physiol. 1995;105:403–420. doi: 10.1085/jgp.105.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salinas RK, Bruschweiler-Li L, Johnson E, Bruschweiler R. Ca2+ binding alters the interdomain flexibility between the two cytoplasmic calcium-binding domains in the Na+/Ca2+ exchanger. J Biol Chem. 2011;286:32123–32131. doi: 10.1074/jbc.M111.249268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu M, Tong S, Gonzalez J, Jayaraman V, Spudich JL, et al. Structural Basis of the Ca2+ Inhibitory Mechanism of Drosophila Na+/Ca2+ Exchanger CALX and Its Modification by Alternative Splicing. Structure. 2011;19:1509–1517. doi: 10.1016/j.str.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaptal V, Ottolia M, Mercado-Besserer G, Nicoll DA, Philipson KD, et al. Structure and functional analysis of a Ca2+ sensor mutant of the Na+/Ca2+ exchanger. J Biol Chem. 2009;284:14688–14692. doi: 10.1074/jbc.C900037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stahelin RV, Wang J, Blatner NR, Rafter JD, Murray D, et al. The origin of C1A-C2 interdomain interactions in protein kinase Cα. J Biol Chem. 2005;280:36452–36463. doi: 10.1074/jbc.M506224200. [DOI] [PubMed] [Google Scholar]
- 27.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 28.Haussinger D, Ahrens T, Sass HJ, Pertz O, Engel J, et al. Calcium-dependent homoassociation of E-cadherin by NMR spectroscopy: changes in mobility, conformation and mapping of contact regions. J Mol Biol. 2002;324:823–839. doi: 10.1016/s0022-2836(02)01137-3. [DOI] [PubMed] [Google Scholar]
- 29.Baumgartner W, Golenhofen N, Grundhofer N, Wiegand J, Drenckhahn D. Ca2+ dependency of N-cadherin function probed by laser tweezer and atomic force microscopy. J Neurosci. 2003;23:11008–11014. doi: 10.1523/JNEUROSCI.23-35-11008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. The 2.8 Å crystal structure of visual arrestin: a model for arrestin’s regulation. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flicek P, Amode MR, Barrell D, Beal K, Brent S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2: Unit 2 3. 2002. [DOI] [PubMed]
- 39.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, et al. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple Sequence Alignment of CBD12 orthologs. Numbering according to canine NCX1 cardiac splice variant.
(PDF)
Crystal packing involves CBD2 Ca2+ binding sites. (A) Interaction between symmetry mates in the CBD12-E454K crystal. (B) Salt-bridges are formed between symmetry mates, involving residues that participate in coordination of Ca2+ in isolated CBD2 (E516, K585).
(EPS)
SAXS analysis. Comparison of SAXS scattering curves (left column) and normalized pair distance distribution functions (PDDF) (right column) for the wild type CBD12 (A, B), CBD12-7Ala (C, D) CBD12-R532A (E, F) and CBD12-E454K (G, H), respectively, in the absence (black) or presence of Ca2+ (red). The scattering profiles were normalized and superimposed against the first data points.
(EPS)
CBD12 Interface Conservation.
(DOCX)
Overall SAXS parameters for CBD12 and its mutants. R g, D max, and NSD are, respectively, the radius of gyration derived from Guinier plotting, maximum inter-atomic dimension, normalized shape discrepancy for DAMMIN calculation. Note that NSD scores for all bead models are less than 0.8, indicating high convergence of the bead model calculations.
(DOCX)
Experimental procedures.
(DOCX)