Abstract
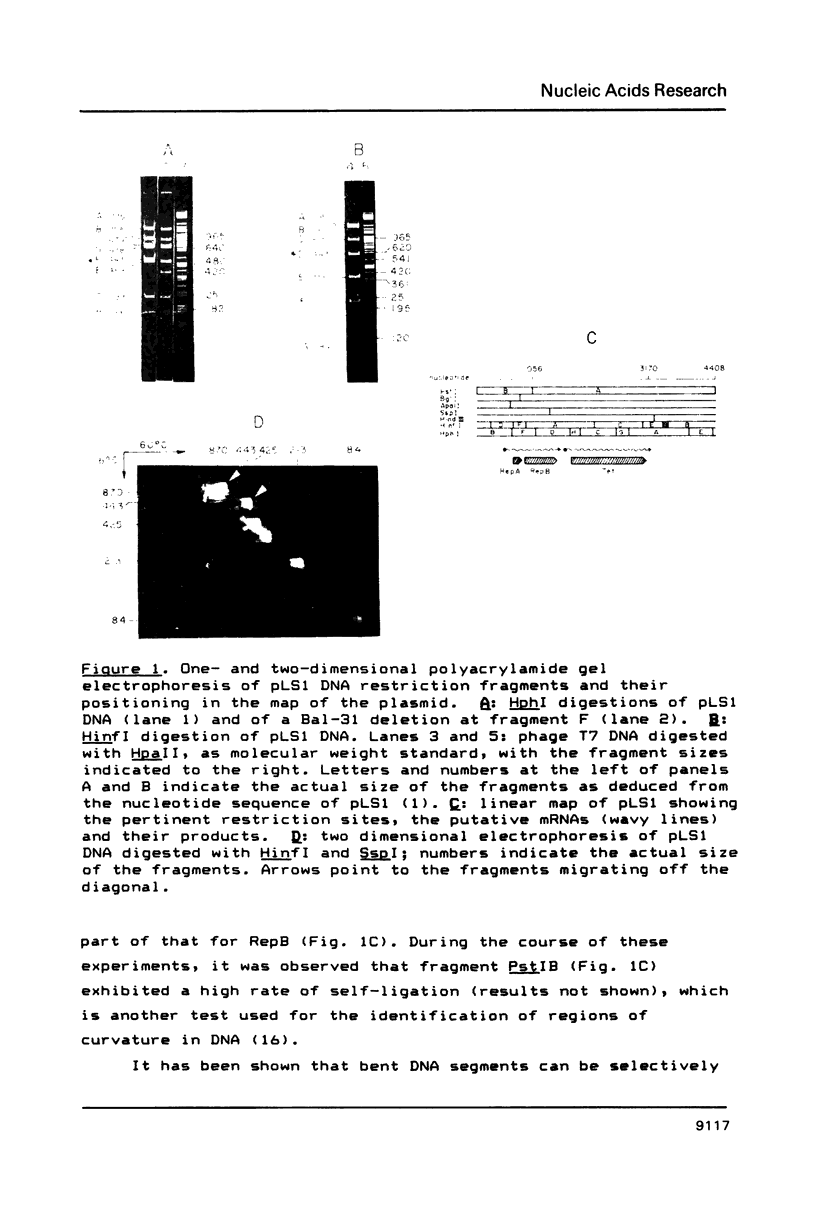
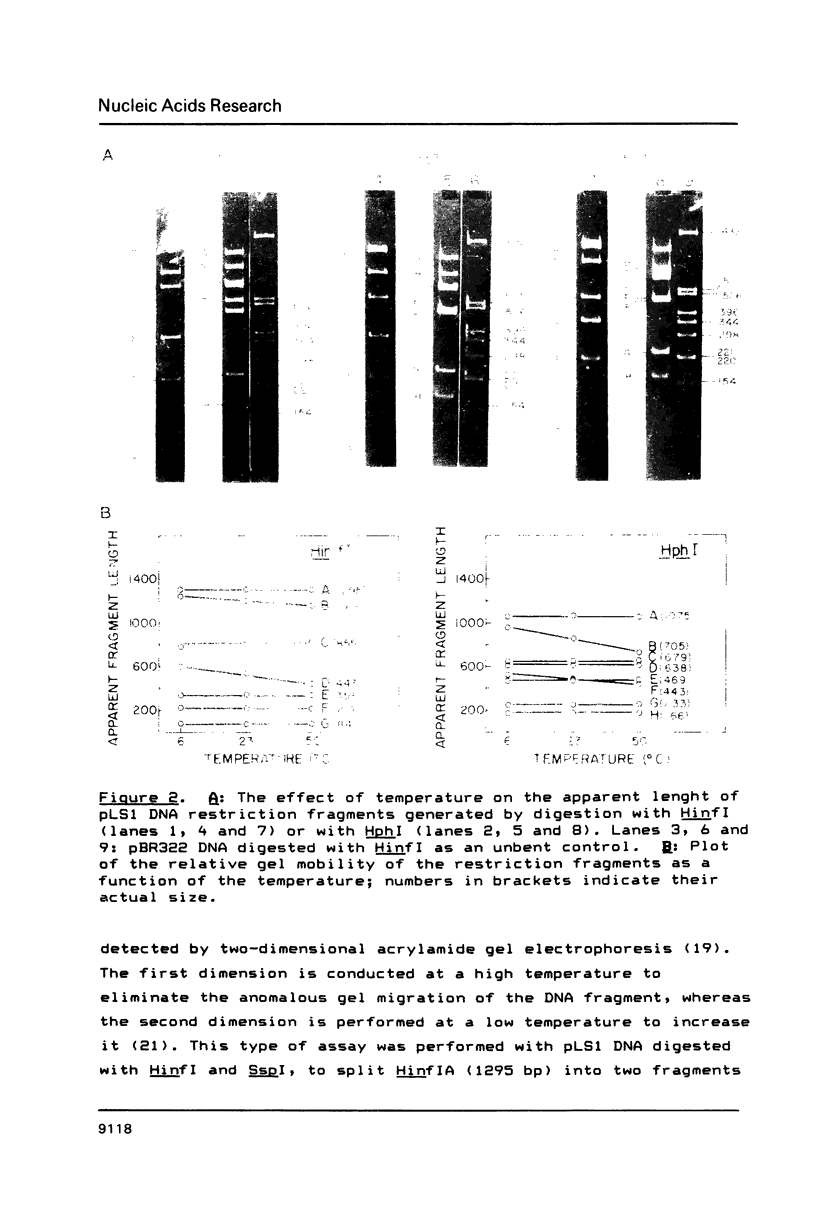
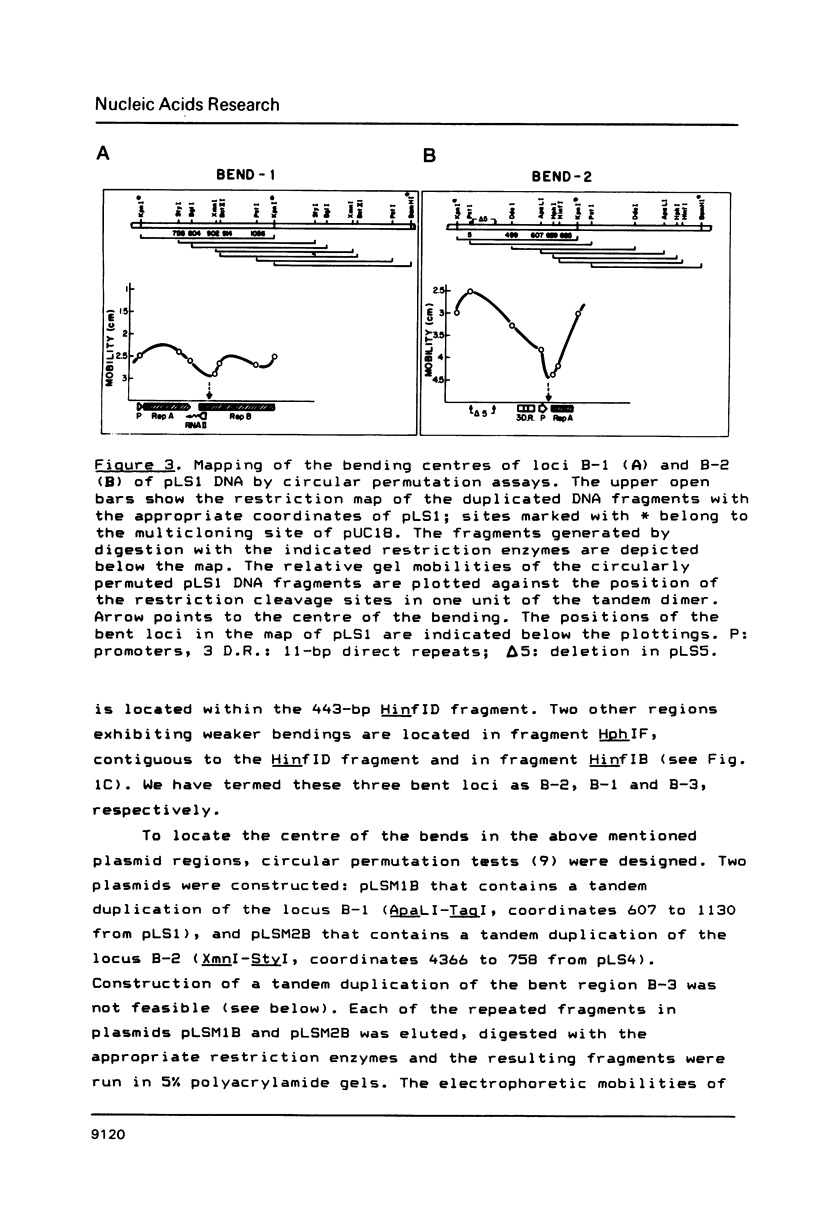
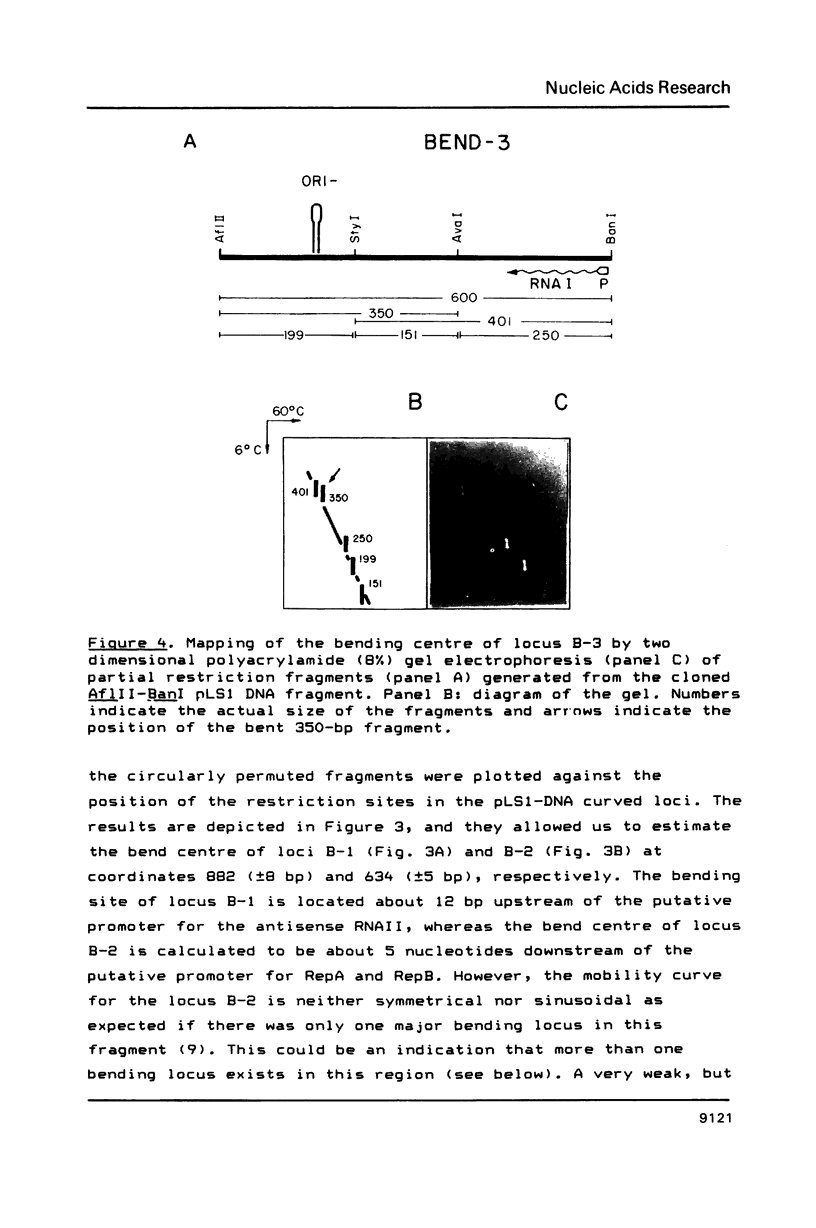
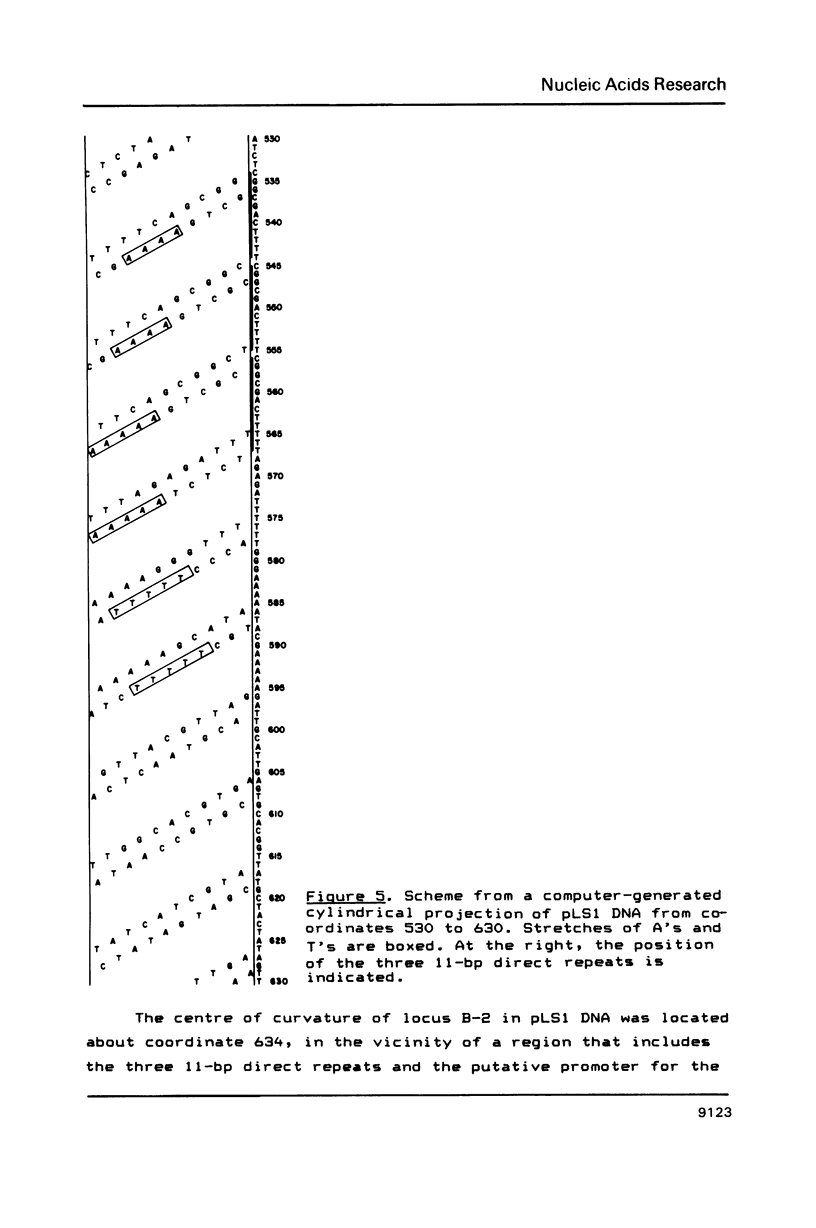
Three regions showing abnormal electrophoretic mobility, which is an indication of the existence of bends in DNA, have been observed in the DNA of plasmid pLS1. These loci have been characterized by assays designed to detect sequence-directed bending in DNA (temperature-dependence migration and two dimensional electrophoresis). The first region (locus B-1) was located within a fragment that contains a proposed inhibitor countertranscribed RNA (RNAII). The second locus (B-2) contains the plasmid plus origin of replication and the third region (locus B-3) was located in the vicinity of a putative antisense RNA (RNAI) of unknown function. The centres of the first two bent DNA regions were located by circular permutation assays at nucleotides 882 (locus B-1) and 634 (locus B-2). The bend centre of locus B-1 was found to be upstream of the promoter for the putative antisense RNAII. The centre of curvature in locus B-2 was located in the vicinity of the putative promoter of the replication proteins RepA and RepB and of a sequence that has three 11-bp direct repeats. The DNA sequence at this region showed the existence of A.T tracts, with an internal repeat of 10 to 11 base pairs, for five helix turns. A complex curvature in the DNA of pLS1 at locus B-2 that may have a regulatory role in plasmid replication is postulated.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Calladine C. R., Drew H. R., McCall M. J. The intrinsic curvature of DNA in solution. J Mol Biol. 1988 May 5;201(1):127–137. doi: 10.1016/0022-2836(88)90444-5. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid. 1983 Nov;10(3):251–259. doi: 10.1016/0147-619x(83)90039-2. [DOI] [PubMed] [Google Scholar]
- Koepsel R. R., Khan S. A. Static and initiator protein-enhanced bending of DNA at a replication origin. Science. 1986 Sep 19;233(4770):1316–1318. doi: 10.1126/science.3749879. [DOI] [PubMed] [Google Scholar]
- Koepsel R. R., Murray R. W., Rosenblum W. D., Khan S. A. The replication initiator protein of plasmid pT181 has sequence-specific endonuclease and topoisomerase-like activities. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6845–6849. doi: 10.1073/pnas.82.20.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Crothers D. M. Calibration of DNA curvature and a unified description of sequence-directed bending. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1763–1767. doi: 10.1073/pnas.85.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Lacks S. A., Lopez P., Greenberg B., Espinosa M. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J Mol Biol. 1986 Dec 20;192(4):753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- Levene S. D., Crothers D. M. A computer graphics study of sequence-directed bending in DNA. J Biomol Struct Dyn. 1983 Oct;1(2):429–435. doi: 10.1080/07391102.1983.10507452. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T. Random cloning of bent DNA segments from Escherichia coli chromosome and primary characterization of their structures. Nucleic Acids Res. 1987 Sep 11;15(17):6827–6841. doi: 10.1093/nar/15.17.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Patel I., Bastia D. Conformational changes in a replication origin induced by an initiator protein. Cell. 1985 Nov;43(1):189–197. doi: 10.1016/0092-8674(85)90023-6. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Plasmid incompatibility. Microbiol Rev. 1987 Dec;51(4):381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaskon R. R., Wartell R. M. Sequence distributions associated with DNA curvature are found upstream of strong E. coli promoters. Nucleic Acids Res. 1987 Jan 26;15(2):785–796. doi: 10.1093/nar/15.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poljak L. G., Gralla J. D. The SV40 termination region exhibits an altered helical DNA conformation. Nucleic Acids Res. 1987 Jul 10;15(13):5433–5442. doi: 10.1093/nar/15.13.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puyet A., del Solar G. H., Espinosa M. Identification of the origin and direction of replication of the broad-host-range plasmid pLS1. Nucleic Acids Res. 1988 Jan 11;16(1):115–133. doi: 10.1093/nar/16.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder K., Silver S., DeLucia A. L., Fanning E., Tegtmeyer P. An altered DNA conformation in origin region I is a determinant for the binding of SV40 large T antigen. Cell. 1986 Mar 14;44(5):719–725. doi: 10.1016/0092-8674(86)90838-x. [DOI] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Curved DNA. CRC Crit Rev Biochem. 1985;19(2):89–106. doi: 10.3109/10409238509082540. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L. E., Trifonov E. N. Estimation of wedge components in curved DNA. Nature. 1987 Apr 16;326(6114):720–722. doi: 10.1038/326720a0. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Binding and bending of the lambda replication origin by the phage O protein. EMBO J. 1985 Dec 16;4(13A):3605–3616. doi: 10.1002/j.1460-2075.1985.tb04124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Direct evidence for DNA bending at the lambda replication origin. Science. 1987 Apr 24;236(4800):416–422. doi: 10.1126/science.2951850. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Sequence-induced DNA curvature at the bacteriophage lambda origin of replication. Nature. 1985 Oct 3;317(6036):451–453. doi: 10.1038/317451a0. [DOI] [PubMed] [Google Scholar]
- del Solar G. H., Puyet A., Espinosa M. Initiation signals for the conversion of single stranded to double stranded DNA forms in the streptococcal plasmid pLS1. Nucleic Acids Res. 1987 Jul 24;15(14):5561–5580. doi: 10.1093/nar/15.14.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Solar G., Diaz R., Espinosa M. Replication of the streptococcal plasmid pMV158 and derivatives in cell-free extracts of Escherichia coli. Mol Gen Genet. 1987 Mar;206(3):428–435. doi: 10.1007/BF00428882. [DOI] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Are single-stranded circles intermediates in plasmid DNA replication? EMBO J. 1986 Mar;5(3):631–637. doi: 10.1002/j.1460-2075.1986.tb04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]






