Abstract
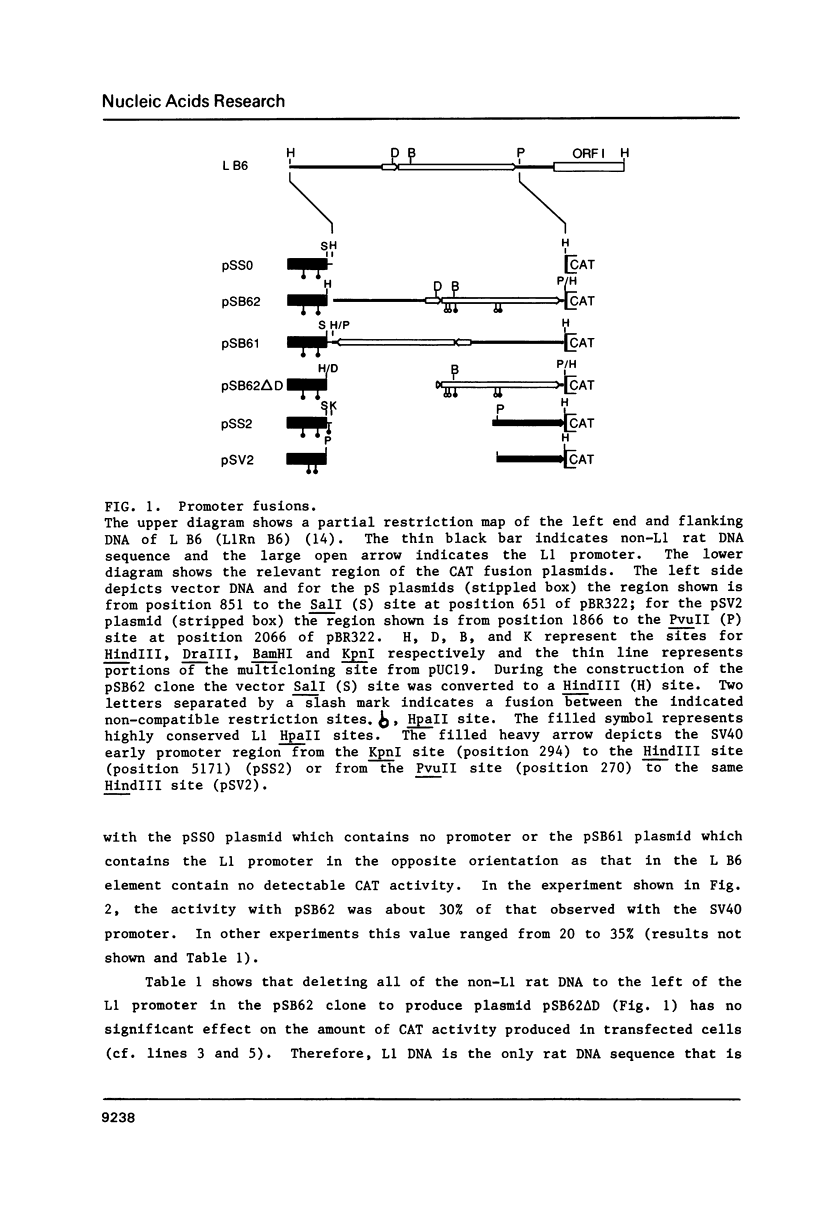
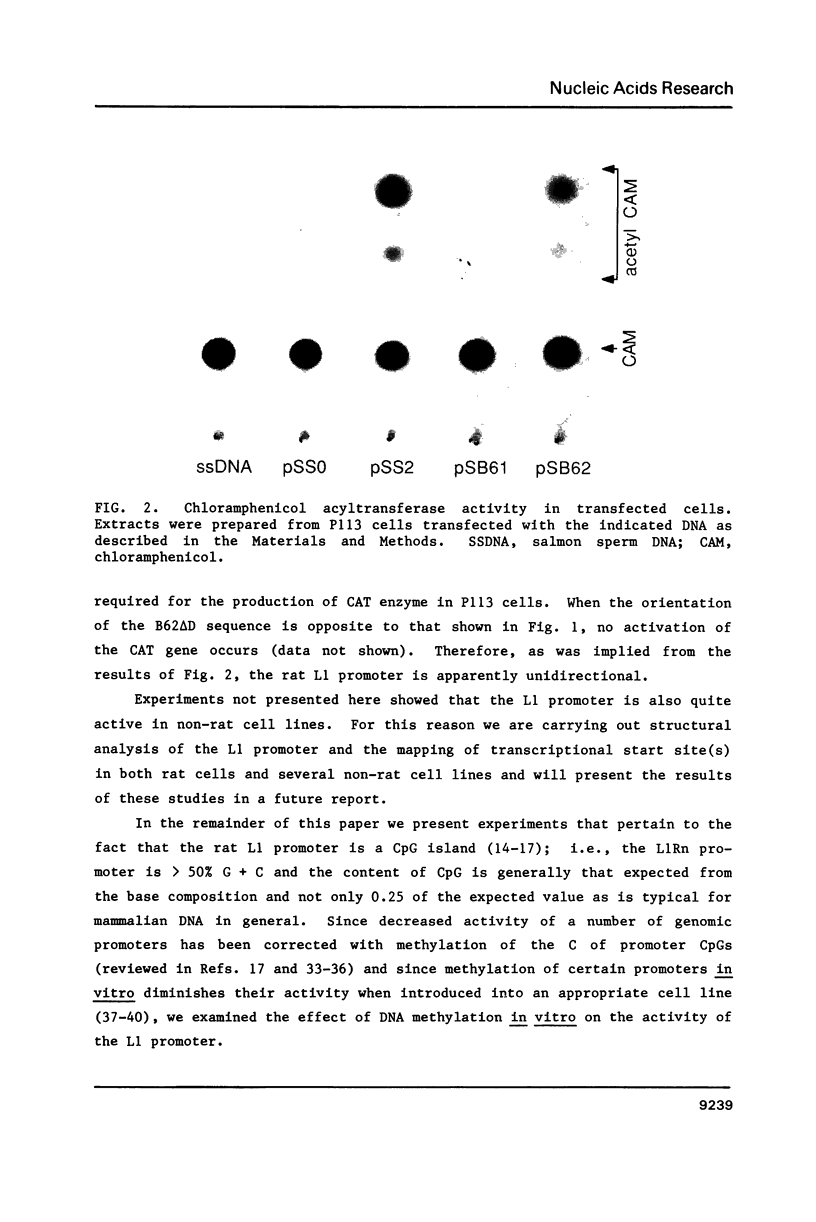
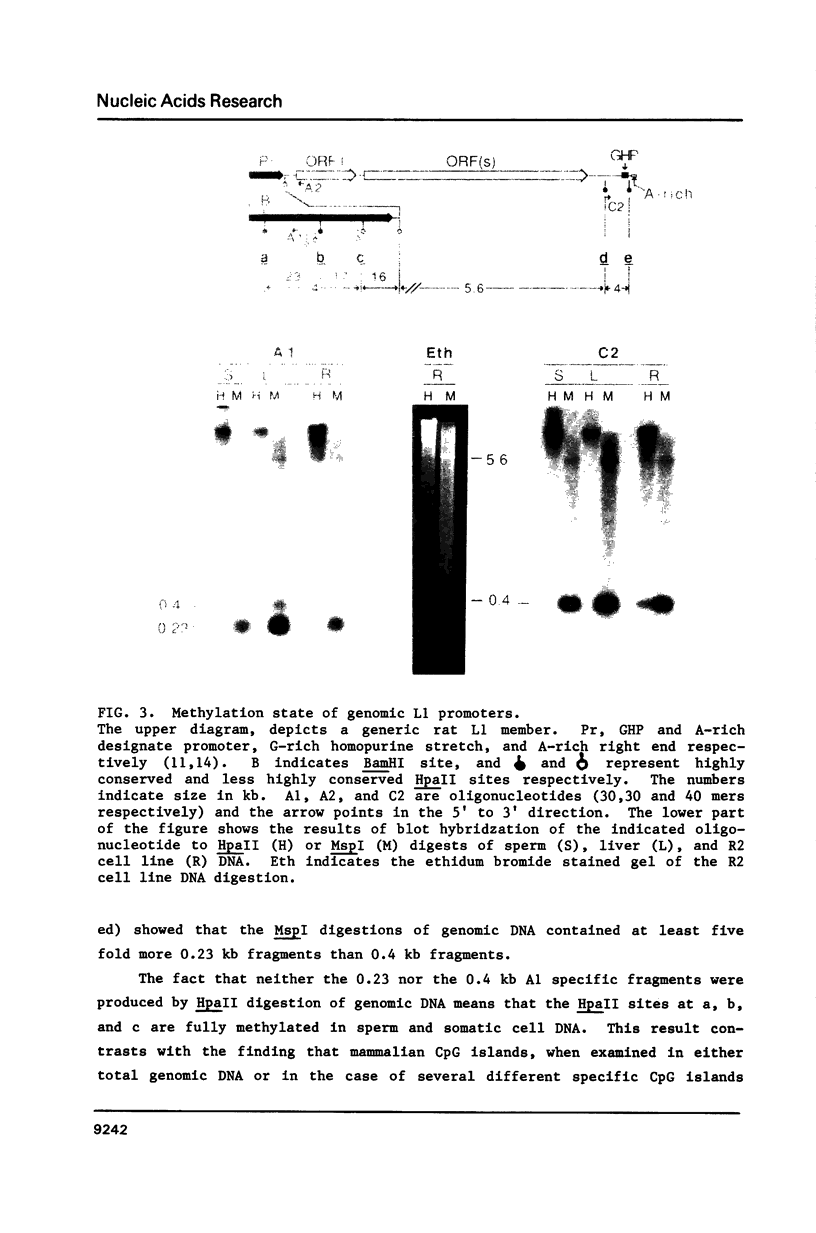
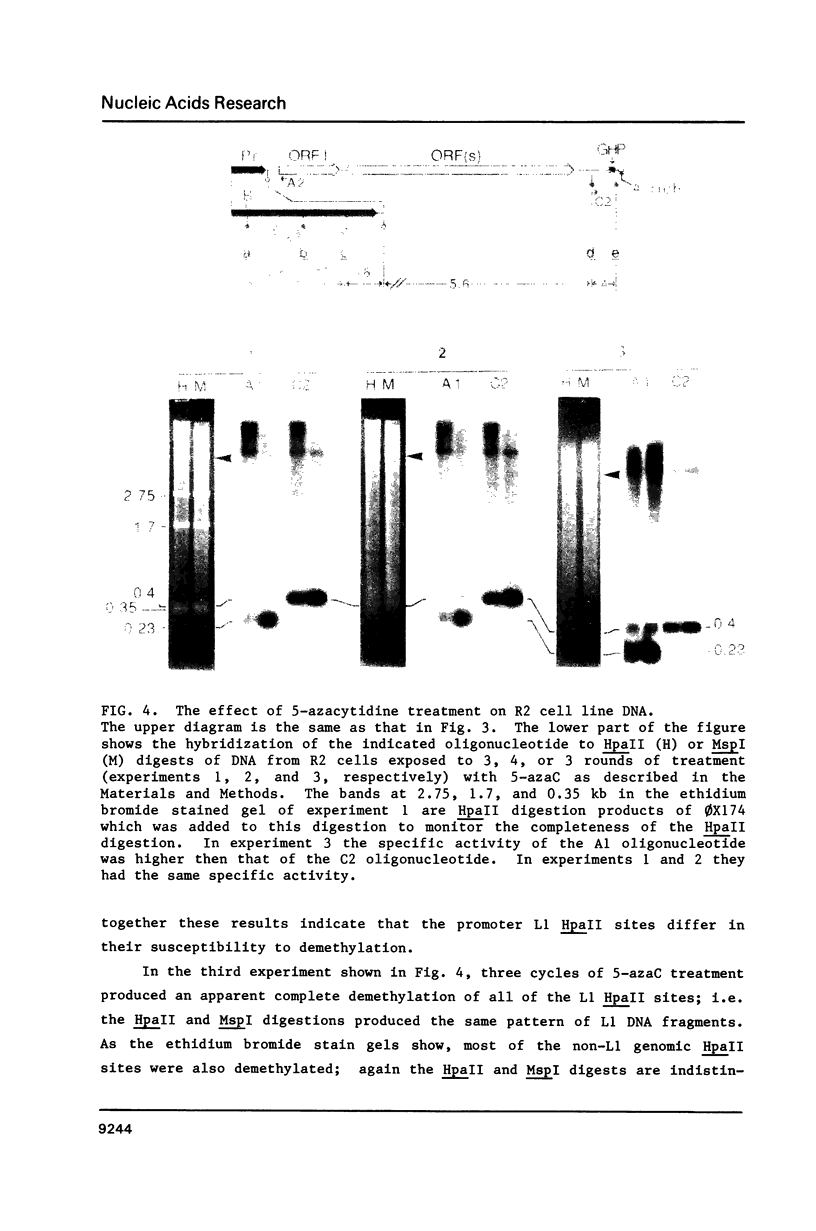
Here we report that the 600 bp promoter-like region at the left end of a newly isolated and characterized rat L1 DNA element can activate the prokaryotic chloramphenicol acyltransferase gene in a rat cell line. Activation only occurs when the promoter region is oriented to the transferase gene as it is to the L1 protein encoding sequences and is 75% inhibited by methylation of just 5 of the 22 CpGs present in the promoter. The G + C rich promoter contains enough CpGs to qualify it as a CpG island, but in contrast to other CpG islands, genomic L1 promoters are fully methylated in both somatic cell and sperm DNA as judged by restriction enzyme analysis. Partial demethylation of the genomic promoters by treatment with 5-azacytidine failed to produce discrete L1 transcripts. The relationship of methylation to the evolutionary history and fate of the rat L1 promoter is discussed.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird A. P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980 Apr 11;8(7):1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A., Taggart M., Frommer M., Miller O. J., Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985 Jan;40(1):91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Burton F. H., Loeb D. D., Chao S. F., Hutchison C. A., 3rd, Edgell M. H. Transposition of a long member of the L1 major interspersed DNA family into the mouse beta globin gene locus. Nucleic Acids Res. 1985 Jul 25;13(14):5071–5084. doi: 10.1093/nar/13.14.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton F. H., Loeb D. D., Voliva C. F., Martin S. L., Edgell M. H., Hutchison C. A., 3rd Conservation throughout mammalia and extensive protein-encoding capacity of the highly repeated DNA long interspersed sequence one. J Mol Biol. 1986 Jan 20;187(2):291–304. doi: 10.1016/0022-2836(86)90235-4. [DOI] [PubMed] [Google Scholar]
- Busslinger M., Hurst J., Flavell R. A. DNA methylation and the regulation of globin gene expression. Cell. 1983 Aug;34(1):197–206. doi: 10.1016/0092-8674(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cooper D. N., Taggart M. H., Bird A. P. Unmethylated domains in vertebrate DNA. Nucleic Acids Res. 1983 Feb 11;11(3):647–658. doi: 10.1093/nar/11.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio E., Waitzkin S. D., Witney F. R., Salemme A., Furano A. V. Structure of the highly repeated, long interspersed DNA family (LINE or L1Rn) of the rat. Mol Cell Biol. 1986 Feb;6(2):411–424. doi: 10.1128/mcb.6.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Economou-Pachnis A., Lohse M. A., Furano A. V., Tsichlis P. N. Insertion of long interspersed repeated elements at the Igh (immunoglobulin heavy chain) and Mlvi-2 (Moloney leukemia virus integration 2) loci of rats. Proc Natl Acad Sci U S A. 1985 May;82(9):2857–2861. doi: 10.1073/pnas.82.9.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein D. A., Witney F. R., Furano A. V. The spread of sequence variants in Rattus satellite DNAs. Nucleic Acids Res. 1984 Jan 25;12(2):973–988. doi: 10.1093/nar/12.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T., Singer M. The LINE-1 DNA sequences in four mammalian orders predict proteins that conserve homologies to retrovirus proteins. Nucleic Acids Res. 1987 Mar 11;15(5):2251–2260. doi: 10.1093/nar/15.5.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher P. B., Babiss L. E., Weinstein I. B., Ginsberg H. S. Analysis of type 5 adenovirus transformation with a cloned rat embryo cell line (CREF). Proc Natl Acad Sci U S A. 1982 Jun;79(11):3527–3531. doi: 10.1073/pnas.79.11.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatau E., Gonzales F. A., Michalowsky L. A., Jones P. A. DNA methylation in 5-aza-2'-deoxycytidine-resistant variants of C3H 10T1/2 C18 cells. Mol Cell Biol. 1984 Oct;4(10):2098–2102. doi: 10.1128/mcb.4.10.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita A., Hattori M., Takenaka O., Sakaki Y. The L1 family (KpnI family) sequence near the 3' end of human beta-globin gene may have been derived from an active L1 sequence. Nucleic Acids Res. 1987 May 26;15(10):4007–4020. doi: 10.1093/nar/15.10.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hattori M., Hidaka S., Sakaki Y. Sequence analysis of a KpnI family member near the 3' end of human beta-globin gene. Nucleic Acids Res. 1985 Nov 11;13(21):7813–7827. doi: 10.1093/nar/13.21.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. The inheritance of epigenetic defects. Science. 1987 Oct 9;238(4824):163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- Katzir N., Rechavi G., Cohen J. B., Unger T., Simoni F., Segal S., Cohen D., Givol D. "Retroposon" insertion into the cellular oncogene c-myc in canine transmissible venereal tumor. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1054–1058. doi: 10.1073/pnas.82.4.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruczek I., Doerfler W. Expression of the chloramphenicol acetyltransferase gene in mammalian cells under the control of adenovirus type 12 promoters: effect of promoter methylation on gene expression. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7586–7590. doi: 10.1073/pnas.80.24.7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmikumaran M. S., D'Ambrosio E., Laimins L. A., Lin D. T., Furano A. V. Long interspersed repeated DNA (LINE) causes polymorphism at the rat insulin 1 locus. Mol Cell Biol. 1985 Sep;5(9):2197–2203. doi: 10.1128/mcb.5.9.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner K. D., Weyer U., Doerfler W. Trans effect of the E1 region of adenoviruses on the expression of a prokaryotic gene in mammalian cells: resistance to 5' -CCGG- 3' methylation. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1598–1602. doi: 10.1073/pnas.83.6.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay S., Bird A. P. Use of restriction enzymes to detect potential gene sequences in mammalian DNA. 1987 May 28-Jun 3Nature. 327(6120):336–338. doi: 10.1038/327336a0. [DOI] [PubMed] [Google Scholar]
- Loeb D. D., Padgett R. W., Hardies S. C., Shehee W. R., Comer M. B., Edgell M. H., Hutchison C. A., 3rd The sequence of a large L1Md element reveals a tandemly repeated 5' end and several features found in retrotransposons. Mol Cell Biol. 1986 Jan;6(1):168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthman H., Magnusson G. High efficiency polyoma DNA transfection of chloroquine treated cells. Nucleic Acids Res. 1983 Mar 11;11(5):1295–1308. doi: 10.1093/nar/11.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech M., Igo-Kemenes T., Zachau H. G. Nucleotide sequence of a highly repetitive component of rat DNA. Nucleic Acids Res. 1979 Sep 25;7(2):417–432. doi: 10.1093/nar/7.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Cedar H. DNA methylation in eukaryotic cells. Int Rev Cytol. 1984;92:159–185. doi: 10.1016/s0074-7696(08)61327-3. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Jones P. A. 5-methylcytosine, gene regulation, and cancer. Adv Cancer Res. 1983;40:1–30. doi: 10.1016/s0065-230x(08)60678-8. [DOI] [PubMed] [Google Scholar]
- Seed B., Sheen J. Y. A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene. 1988 Jul 30;67(2):271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- Shyman S., Weaver S. Chromosomal rearrangements associated with LINE elements in the mouse genome. Nucleic Acids Res. 1985 Jul 25;13(14):5085–5093. doi: 10.1093/nar/13.14.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. E., Summers M. D. The bidirectional transfer of DNA and RNA to nitrocellulose or diazobenzyloxymethyl-paper. Anal Biochem. 1980 Nov 15;109(1):123–129. doi: 10.1016/0003-2697(80)90019-6. [DOI] [PubMed] [Google Scholar]
- Soares M. B., Schon E., Efstratiadis A. Rat LINE1: the origin and evolution of a family of long interspersed middle repetitive DNA elements. J Mol Evol. 1985;22(2):117–133. doi: 10.1007/BF02101690. [DOI] [PubMed] [Google Scholar]
- Soberon X., Covarrubias L., Bolivar F. Construction and characterization of new cloning vehicles. IV. Deletion derivatives of pBR322 and pBR325. Gene. 1980 May;9(3-4):287–305. doi: 10.1016/0378-1119(90)90328-o. [DOI] [PubMed] [Google Scholar]
- Stein R., Razin A., Cedar H. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3418–3422. doi: 10.1073/pnas.79.11.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. M., Jones P. A. Mechanism of action of eukaryotic DNA methyltransferase. Use of 5-azacytosine-containing DNA. J Mol Biol. 1982 Dec 15;162(3):679–692. doi: 10.1016/0022-2836(82)90395-3. [DOI] [PubMed] [Google Scholar]
- Topp W. C. Normal rat cell lines deficient in nuclear thymidine kinase. Virology. 1981 Aug;113(1):408–411. doi: 10.1016/0042-6822(81)90168-9. [DOI] [PubMed] [Google Scholar]
- Witney F. R., Furano A. V. Highly repeated DNA families in the rat. J Biol Chem. 1984 Aug 25;259(16):10481–10492. [PubMed] [Google Scholar]