Abstract
Mutations in human leucine-rich-repeat kinase 2 (LRRK2) have been found to be the most frequent cause of late-onset Parkinson disease. Here we show that Dictyostelium discoideum Roco4 is a suitable model to study the structural and biochemical characteristics of the LRRK2 kinase and can be used for optimization of current and identification of new LRRK2 inhibitors. We have solved the structure of Roco4 kinase wild-type, Parkinson disease-related mutants G1179S and L1180T (G2019S and I2020T in LRRK2) and the structure of Roco4 kinase in complex with the LRRK2 inhibitor H1152. Taken together, our data give important insight in the LRRK2 activation mechanism and, most importantly, explain the G2019S-related increase in LRRK2 kinase activity.
Leucine-rich-repeat kinase 2 (LRRK2) belongs to the Roco family of proteins, which are characterized by the presence of leucine-rich repeats, a Ras-like G-domain (called “Roc”), a C terminal of ROC (COR) domain, and a kinase domain (1). Recently, missense mutations in LRRK2 have been linked to autosomal-dominant, late-onset Parkinson disease (PD) (2, 3). PD is a common neurodegenerative disorder characterized by a progressive loss of dopaminergic neurons of the substantia nigra, associated with the formation of fibrillar aggregates composed of α-synuclein and other proteins. PD is characterized clinically by tremor, bradykinesia, rigidity, and postural instability. The identification of missense mutations in LRRK2 has redefined the role of genetic variation in PD susceptibility. LRRK2 mutations initiate a penetrant phenotype with complete clinical and neurochemical overlap with idiopathic disease (4–6). The various mutations that have been identified in PD are concentrated in the central region of the protein; one residue mutated in the LRR region, one in the Roc domain (with multiple substitutions), one in the COR domain, and two in the kinase domain (7). The mutations are found in 5–6% of patients with familial PD and, importantly, also have been implicated in sporadic PD (8, 9). Although much progress has been made during the last few years, the exact pathogenic role and associated biochemical pathways responsible for LRRK2-linked disease are emerging only slowly (10). The multiple disease-linked mutations in LRRK2 represent a unique opportunity to explore the pathogenicity of LRRK2 biochemically and to identify therapeutic targets for this neurodegenerative disorder.
In the absence of suitable amounts of purified mammalian LRRK2 protein, and because recombinantly expressed full-length protein or any fragment thereof turned out to be unstable, insoluble, or permanently bound to chaperones, structural understanding of LRRK2 is very limited (11). Therefore, we used related proteins to investigate the complex structural regulatory mechanism of LRRK2. Previously we elucidated the structure of the RocCOR tandem of Chlorobium tepidum, which shows that the Roc domain is a Ras-like G domain tightly coupled to the COR domain as a dimerization device. Mutations analogous to Parkinson mutations were shown to be located in the Roc–COR interface. RocCOR proteins thus seem to belong to the G proteins activated by the nucleotide-dependent dimerization (GAD) class of molecular switches. PD-analogous mutations in Roc and COR alter the Roc–COR interface and result in decreased GTPase activity (12, 13). The structure of the Roc domain of human LRRK2 showed a domain-swapped dimeric G domain whose significance for the native protein is unclear (11, 13).
LRRK2 kinase activity is linked critically to clinical effects, and several pathogenic mutations in LRRK2 result in enhanced kinase activity, suggesting a possible PD-related gain of abnormal or toxic function (14–16). However, because of the lack of sufficient recombinant protein and physiological substrate, the published data regarding kinase activity of the PD-related mutants are conflicting (except for G2019S, which is associated consistently with an increased kinase activity) (17, 18).
Here we use Dictyostelium discoideum Roco4 as model to study the structural and biochemical characteristics of the LRRK2 kinase domain. We have solved the structure of Roco4 kinase wild-type and PD-related mutants G1179S and L1180T (G2019S and I2020T in LRRK2). A comparison of wild-type and mutant structures revealed that the PD mutants have different effects and, most importantly, explains the G2019S-related increase in LRRK2 kinase activity. Identifying small-molecule inhibitors of the kinase activity that specifically counteract the effect in vivo will be an important step towards finding a treatment for PD. The structure of Roco4 kinase in complex with the LRRK2 inhibitor H1152 shows that Roco4 is a suitable model system to obtain insight into the binding mechanism and to optimize current and identify new LRRK2 inhibitors.
Results and Discussion
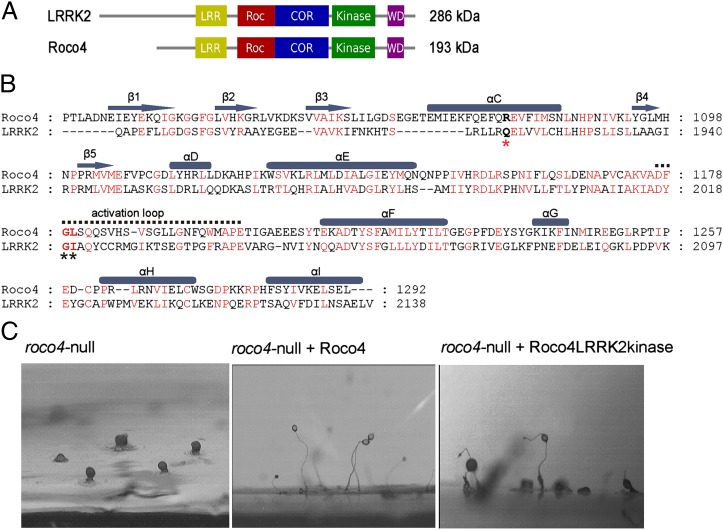
Vertebrates possess four Roco proteins, LRRK1, LRRK2, DAPK1, and MFHAS1. Remarkably, the social amoeba Dictyostelium contains 11 Roco family members that contain a large variety of domains and have been studied in detail (19–21). In this study we used Dictyostelium Roco4, which has the same domain topology as LRRK2 (Fig. 1A) but is biochemically more tractable. The kinase domain is well conserved in the Roco family of proteins, and the Roco4 kinase domain (amino acids 1018–1292) has a similarity of 47% to LRRK2. Unlike LRRK2, the Roco4 kinase domain could be expressed in Escherichia coli and isolated as a soluble and stable protein (Fig. 1B). Dictyostelium are single-celled amoeba that feed on bacteria. Upon starvation, cells enter a tightly regulated developmental process, resulting in multicellular fruiting bodies. Dictyostelium cells with a disruption in the roco4 gene cannot synthesize cellulose, resulting in instable stalks that are unable to lift the spore head (Fig. 1C) (20). This strong developmental defect of roco4-nulls cells can be rescued completely by the expression of wild-type Roco4 and also by a chimeric protein of Roco4 in which the kinase domain is replaced by that of LRRK2 (Fig. 1C). As a further demonstration of the similarity between the proteins, Roco4 kinase is able to phosphorylate LRRKtide, an artificial specific substrate of LRRK2. Also, Roco4 kinase activity is inhibited by various LRRK2 inhibitors (see below). Taken together, these data show that Roco4 can serve as model to study the complex regulatory mechanism of LRRK2.
Fig. 1.
Dictyostelium Roco4 as model for LRRK2. (A) Domain organization of LRRK2 and Dictyostelium Roco4. (B) Alignment of Roco4 kinase (amino acids 1018–1292) and LRRK2 kinase (amino acids 1879–2138) domains. Conserved residues are shown in red, PD-related mutations are indicated by black asterisks, the hydrogen bond-accepting residue specific for PD-related G2019S is indicated by a red asterisk. (C) Phenotype of roco4-null cells and rescue experiments. roco4-null cells and roco4-null cells expressing Roco4 or Roco4LRRK2 kinase, in which the kinase domain of Roco4 has been replaced with the kinase domain of LRRK2, were allowed to develop for 48 h on nutrient-free agar. Pieces of agar were excised and photographed from the side.
The purified Roco4 kinase domain (residues 1018–1292) was found to be autophosphorylated and was additionally incubated with ATP. It was dephosphorylated by incubation with alkaline phosphatase. As shown below, phosphorylated Roco4 appears to be the active form of the protein.
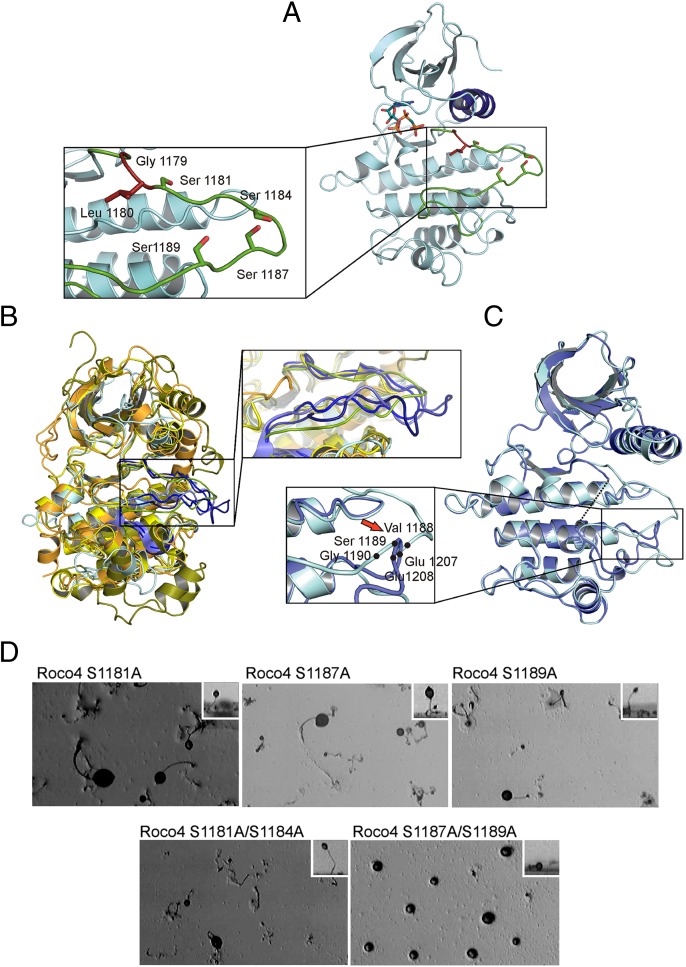
The active, phosphorylated form was crystallized in the presence of the ATP analog AppCH2p, the dephosphorylated protein without ATP. The crystals with space group P4(3)3(2)2 had similar unit cell parameters (42, 42, 340Å) (Table 1). Structures were solved by molecular replacement using MLK1 (3DTC) as the search model (Table S1). Roco4 kinase has a canonical kinase fold with a mostly β-sheet–containing N-terminal lobe and a highly α-helical C-terminal lobe. The nucleotide is located in the conventional nucleotide-binding site (Fig. 2A). In many kinases, the activation loop is a highly flexible element and contains the primary activity-related phosphorylation sites. In the unphosphorylated, inactive state, this loop often is disordered, but upon phosphorylation it reorients into an ordered, active conformation (22, 23). An overlay of phosphorylated Roco4 with ERK2, PKA, and DAPK1 in the active conformations highlights this conservation of the activation loop conformation (Fig. 2B). Because of its flexibility, the activation loop is not visible in the structure of dephosphorylated Roco4, and in the active form only the main chain can be traced. An overlay of the active and inactive Roco4 structures (shown in cyan and blue, respectively, in Fig. 2C) shows that the residues E1207/E1208 (inactive) and V1188, S1189, G1190 (active) would clash, indicating that the nonresolved loop must be in a different position in the inactive conformation than in the active conformation (Fig. 2C).
Table 1.
X-ray refinement statistics
| Crystal | Active | Inactive | G1179S | L1180T | H1152 |
| Resolution (Å) | 1.8 | 2.0 | 2.0 | 2.3 | 2.3 |
| Rwork/Rfree | 0.19/0.24 | 0.23/0.26 | 0.26/0.29 | 0.25/0.29 | 0.19/0.23 |
| Number of atoms | |||||
| Protein | 2,208 | 2,113 | 2,219 | 2,207 | 2,160 |
| Ligand/ion | 31 | — | 46 | 31 | 44 |
| Water | 211 | 62 | 96 | 165 | 54 |
| B-factors | |||||
| Protein | 29.9 | 43.6 | 26.1 | 32.8 | 45.6 |
| Ligand/ion | 55.3 | — | 24.6 | 70.8 | 70.3 |
| Water | 31.5 | 30.2 | 25.7 | 34.1 | 31.2 |
| R.m.s deviations | |||||
| Bond lengths (Å) | 0.027 | 0.007 | 0.006 | 0.006 | 0.024 |
| Bond angles (°) | 2.246 | 0.987 | 0.956 | 0.958 | 1.953 |
Fig. 2.
(A) Ribbon diagram of Roco4 kinase in the active state with AppCH2p (red ball and stick model) bound in the nucleotide-binding pocket. The activation loop and the regulatory αC-helix are shown in green and blue, respectively. Relevant side chains are indicated. In the enlarged view, residues homologous to relevant PD mutations (in blue/red) and putative phosphorylation sites within the activation loop (in red) are highlighted. (B) Overlay of active phosphorylated Roco4 (cyan) with several kinase structures in the active conformation: ERK2 (2ERK) (green), PKA (2CPK) (orange), and DAPK1 (3GU5) (yellow). The enlarged view highlights the activation loops. (C) Overlay of Roco4 in the active (cyan) and inactive (blue) forms. The enlarged view highlights the clash of the residues 1207/1208 (inactive) with 1188–1190 (active). (D) Analysis of Roco4 autophosphorylation sites in vivo. Roco4-null cells expressing the indicated serine mutants were analyzed for development as described in Fig. 1C. Pictures were taken from above. Insets show a side view of a fruiting body. The double-mutant S1187A/S1189A is not able to rescue the developmental defect of roco4-null cells.
Phosphorylation of serine and threonine residues in kinase-activation loops is important for regulating kinase activity in many protein kinases (22–25). Because the Roco4 kinase becomes autophosphorylated during kinase assays, it is technically difficult to measure the difference in kinase activity in the phosphorylated and dephosphorylated protein. In autophosphorylation assays Roco4 incorporates maximally 2.04 ± 0.33 mol (n = 5) of phosphate per mole of protein. Roco4 kinase contains four putative phosphorylation sites in the activation loop: S1181, S1184, S1187, and S1189 (Figs. 1B and 2A). To characterize the Roco4 activation mechanism, we reexpressed Roco4 constructs with S-to-A mutations in roco4-null cells and analyzed them for development (Fig. 2D). By reexpressing single mutants and double mutants, we find that the double mutant S1181/1184, but not S1187/1189, rescues the developmental phenotype. Consistently, the purified S1187 and S1189 single mutants and the double mutant S1187/1189 have hardly any kinase activity, whereas the double mutant S1181/1184 shows wild-type activity (Fig. S1). The data show that Roco4 incorporates approximately two moles of phosphate and that serine 1187 and serine 1189 are essential for kinase activity. This finding supports the notion that autophosphorylation in the activation loop is required to induce the active conformation of the kinase.
The putative autophosphorylation sites are not conserved between Roco4 and LRRK2 (Fig. 1B). LRRK2 contains T2031/S2032/T2035 (Fig. 1B), three potential phosphorylation sites in the activation loop. Studies using phosphospecific antibodies have shown that all three sites are phosphorylated, but as in Roco4, only the latter two sites, S2032 and T2035, are important for LRRK2 activity in vivo (26, 27).
PD-linked mutations have been identified throughout the LRRK2 gene; one residue is mutated in the LRR region, one in the Roc domain (with multiple substitutions), one in the COR domain, and two in the kinase domain (7). The most prevalent PD mutation is G2019S in the kinase domain, which enhances kinase activity, whereas the PD-related mutation I2020T shows slightly decreased activity (15, 16, 28–30). The LRRK2 G2019 and I2020 residues are conserved in Roco4 and correspond to G1179 and L1180, respectively (Fig. 1B). We have created the corresponding mutations in Roco4 and determined their biochemical and structural properties.
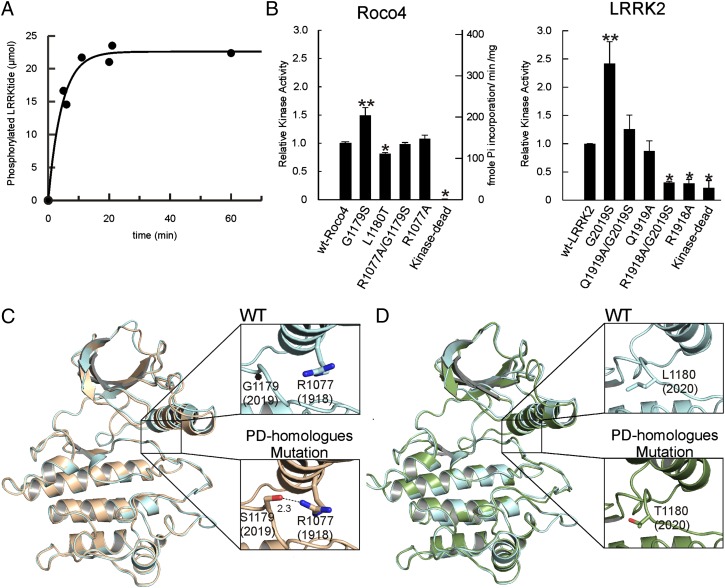
Active phosphorylated Roco4 kinase phosphorylates LRRKtide with a rate constant of 1.5⋅103 s−1 (± 427) (Fig. 3A). Like LRRK2, the Roco4 G1179S mutant showed a 1.5 ± 0.13-fold increased activity in autophosphorylation, relative to wild-type kinase, whereas the L1180T mutant shows a slightly but significantly decreased activity (0.8 ± 0.02) (Fig. 3B). The structures of the Roco mutants L1180T and G1179S were solved by molecular replacement to a resolution of 2.3 and 2.04 Å, respectively. Comparison of wild-type and mutant structures did not show large differences in the overall structure (rmsd 0.6) (Fig. 3 C and D). An overlay of wild-type and the most prevalent PD homolog mutation, G1179S, revealed an additional hydrogen bond between the mutated S1179 and an R1077 from the regulatory αC-helix, (Fig. 3C, Lower Right). We reasoned that this additional hydrogen bond stabilizes the active configuration involving the DFG motif, the activation loop, and the αC-helix so that kinase activity is increased. To test our hypothesis, we constructed a double mutant, G1179S/R1077A, in which the additional hydrogen bond cannot be formed, and measured kinase activity. Consistent with our model the G1179S/R1077A mutant has nearly wild-type activity (0.91 ± 0.02).
Fig. 3.
Structure and properties of PD-related mutants. (A) Roco4-catalyzed phosphorylation of LRRKtide was measured by incubating 3 μM Roco with 150 μM LRRKtide and 25 μM radioactive ATP. Incorporation of radioactive phosphate was measured as described in Material and Methods. The data were fitted to the equation At = A∞(1-e−kt). (B) Kinase activity measured by autophosphorylation of wild-type and mutant Roco4 (Left) and wild-type and mutant LRRK2 (Right) as indicated. For purified Roco4 kinase, absolute rates are presented. For partially purified flag-LRRK2, the activity of the mutants is presented relative to the activity of the wild type. Data shown are the means and SD of at least three independent experiments. *P < 0.001; **P < 0.01 significantly different from wild-type control using ANOVA. (C) Overlay of the ribbon diagrams of the wild-type (cyan) and G1179S mutant Roco4 kinase equivalent to the LRRK2 PD-mutant G2019S (tan). The enlarged views of wild type (Upper) and G1179S mutant (Lower) reveal that the 1179S mutant has an additional hydrogen bond (red) with an arginine (blue) of the αC-helix that stabilizes the activation loop. The numbers in brackets give the corresponding numbers in LRRK2. (D) Overlay of the ribbon diagrams of the wild-type Roco4 kinase (cyan) and the PD-related mutant L1180T (LRRK2 I2020T) (green). Panels on the right represent enlarged views of the mutational site and show that L1180 (cyan) and T1180 (red) point into the solvent.
We then tested whether the above conclusion holds true for LRRK2. Roco4 R1077 corresponds to LRRK2 Q1919, but, because the adjacent R1918 also can form a potential hydrogen bond with S2019, both were mutated to alanine in wild-type and PD-mutant LRRK2 (Fig. 1B). Activity of immunoprecipitated LRRK2 was determined by autophosphorylation (Fig. 3B Right and Fig. S2). Although the R1918A mutation does reduce the activity of G2019S, R1918A itself has very low activity (Fig. 3B). The Roco4 analog, Q1076, points into the solvent; therefore R1918 is unlikely to interfere directly with active site configuration (Fig. 3C and Fig. S3). Dimerization is a common mechanism to regulate kinase activity (23). LRRK2, and most likely all Roco proteins, are dimer in solutions using the COR domain as dimerization interface (12, 13, 27). Autophosphorylation is essential for the function of LRRK2; however, data for autophosphorylation both in cis and in trans have been reported (15, 27). The recent structure of Aurora2 shows that the αC-helix is part of the kinase dimerization interface (31); because LRRK2 autophosphorylation can occur in trans (15), the R1918A mutation may disturb the putative dimer interface and subsequently reduce kinase activity.
As expected, the Q1919A mutation does indeed reduce the activity of the PD mutant almost to the wild-type level (1.26 ± 0.25) (Fig. 3B) and similar to the level of the Q1919A single mutant (0.87 ± 0.18), indicating that the PD-related increase in G2019S activity is indeed the result of an additional hydrogen bond with Q1919, which stabilizes the active conformation.
The structure of the Roco4 PD-homologous mutant L1180T shows that the T1180 side-chain points into the solvent and most likely is not involved directly in regulating kinase activity (Fig. 3D). For LRRK2, it has been postulated that the higher neurotoxicity of this mutant might be caused by the mutant’s greater susceptibility to intracellular degradation (28, 32). This notion seems unlikely, given that the disease phenotype is autosomal dominant and caused by a gain of function. Although the Roco4 structure does not reveal the exact mechanism, we speculate that, in analogy to the lower activity of B-Raf mutations, which are complemented by interaction with c-Raf, the kinase domains in LRRK2 work in tandem so that the interaction between wild-type and LRRK2-T1180 increases kinase activity (33). More importantly, the data do show that the PD-related effect of LRRK2 mutations results from different defects in the LRRK2 activation mechanism and suggest that the different LRRK2 mutations, such as S2019 and I2020, might require different methods of inhibition for the purpose of drug development.
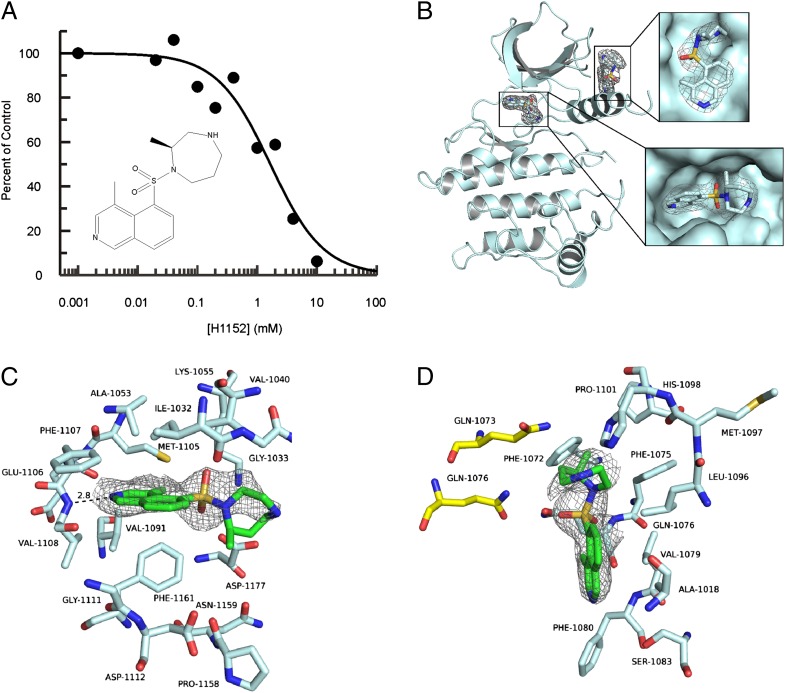
To date, several relatively nonspecific kinase inhibitors, such as H1152, staurosporine, sunitinib, and GW5074, and more specific LRRK2 inhibitors, such as LRRK2-IN-1, have been identified (34, 35). It is speculated that the ATP-binding site is the direct target for many of the inhibitors, but the exact binding mechanism is unknown. We were able to cocrystallize the Roco4 kinase domain with H1152, which originally was identified as a rho-associated protein kinase (ROCK) inhibitor but recently was reported to have nearly the same inhibitory effect on LRRK2 (35). H1152 also was found to inhibit Roco4 kinase activity, and binding is ATP competitive (Fig. 4A). The structure of Roco4 in complex with H1152 was solved by molecular replacement to a resolution of 2.3 Å and revealed two inhibitor binding sites (Fig. 4B). The first H1152 binding site, is in the nucleotide-binding pocket, as expected from its inhibitory mechanism, and is similar to the mode in which H1152 binds to ROCK1 (36). The binding site is formed by 17 residues, and the buried surface area is 280.6 Å2 (Fig. 4C). In the complex with ROCK1, the H1152 isoquinoline nitrogen accepts a main-chain hydrogen bond from M156; in Roco4 the same interaction takes place with V1108 (Fig. 4C). Hydrophobic interaction of the two H1152 C-4 methyl groups with both Roco4 and ROCK1 helps restrict the conformational freedom of the inhibitor. The second H1152 binding site is close to the αC-Helix and is formed by 14 residues in total; 12 are from one kinase molecule, and two are from a symmetry-related molecule within the crystal structure (Fig. 4D). The relevance of this second binding site for inhibition of kinase activity in solution is not clear but also was observed recently in the H1152 structure in complex with PKA (37). Roco4 kinase activity is inhibited by H1152 not only in vitro but also in vivo: Dictyostelium cells in the presence of 0.1 mM and 0.5 mM H1152 have a partial or complete roco4-null phenotype, respectively (Fig. S4). These results show that Roco4 can be used to characterize LRRK2 inhibitor binding in detail, biochemically and structurally. Furthermore, Roco4 structures will allow the construction of a reliable model of LRRK2 for computer-aided drug development. The biochemical tractability of Roco4 allows in vitro screening of inhibitor libraries, whereas the unique phenotype of roco4-null mutants and its rescue by the Roco4-LRRK2-kinase chimera (Fig. 1C) can be used for in vivo testing and screening.
Fig. 4.
Binding of the LRRK2-inhibitor H1152 to Roco4. (A) Kinase activity of Roco4 was measured in the presence or absence of the indicated concentration of H1152 (chemical structure is shown below the plot) in the presence of 25 μM ATP. The results are presented as percentage of kinase activity relative to the control. (B) Ribbon diagram of Roco4 in complex with H1152. Enlarged views of the two binding sites are shown in the right panels. (C and D) Detailed view of H1152 binding in the Roco4 nucleotide-binding pocket (C) and in the interface of two molecules in the crystal (D).
Take together our data give important insight into the mechanism of LRRK2 activation and into how a mutation in the kinase domain increases kinase activity. Although mutants spread over all parts of the multidomain protein LRRK2 produce a similar pathogenic output signal, i.e., PD, our structures show that different mutations have different defects in the activation mechanism. For a further understanding of other mutations, it will be important to characterize fully the intramolecular regulation of LRRK2 and show how the Roc domain might regulate kinase activity, the role that COR plays in this process, and how PD-linked missense mutations alter the interactions between the different domains. Our work shows that structures of the more tractable Roco4 and possibly other Roco proteins could be important in this enterprise.
Materials and Methods
Protein Purification and Radiometric Assays.
Roco4 kinase (amino acids 1018–1292) was cloned into a Gateway-compatible pGEX4T1 plasmid containing an N-terminal TEV cleavage side. Proteins were purified in the presence of 1 mM ATP by GSH affinity, cleavage, and size-exclusion chromatography. Dephosphorylated Roco4 kinase was obtained by incubating 1 mg isolated protein with 100 U alkaline phosphate for 1 h at 4 °C. Roco4 kinase activity was determined at 30 °C in kinase buffer consisting of 20 mM Tris (pH 7.5), 10 mM MgCl2, 1 mM EGTA, 1 mM sodium orthovanadate, 1 mM NaF, 5 mM β-glycerolphosphate, 0.02% Triton X-100, and 2 mM DTT. Autophosphorylation was measured with 1 mg/mL purified protein. The reaction was started by adding 50 μM (2 Ci/mmol) ATP-γ-32P (Perkin-Elmer) and was stopped by adding 100 mM ice-cold EDTA. Samples were spotted on nitrocellulose filters, washed with 50 mM phosphoric acid, and dried before scintillation counting (Perkin-Elmer). For LRRKtide, 32P incorporation assays similar to those described above were performed with 0.05 mg/mL kinase, 25 μM (2 Ci/mmol) ATP-γ-32P, and 150 μM LRRKtide. Kinase inhibition was determined by varying the concentration of H1152 (Tocris Bioscience). Human Flag-LRRK2 was expressed and isolated from HEK293T cells by immunoprecipitation using anti-Flag (M5) antibody (Sigma), and kinase activity was measured as previously described (30).
Crystallography.
Roco4 crystals were obtained in 100 mM 1,3-bis(tris(hydroxymethyl)methylamino)propane (pH 8,5), 200 mM Na/K tartrate, and 11% (wt/vol) PEG 3350 using the hanging drop/vapor diffusion method. For data collection, crystals were cryoprotected in reservoir solution containing 35% (wt/vol) PEG3350 as cryoprotectant. Datasets were collected on beam line ×10SA at the Swiss Light Source (Paul Scherrer Institut, Villigen, Switzerland) and were indexed, integrated, and scaled with the XDS package (38). The model was built in COOT (39) and refined with REFMAC5 using TLS-refinement (CCP4 suite) (40). Figures were generated using PYMOL (DeLano Scientific LLC).
Dictyostelium Aggregation Assays.
Development of Dictyostelium mutants was tested as previously described (20). Briefly, cells were harvested in phosphate buffer (11 mM KH2PO4, 2.8 mM Na2HPO4), plated at a density of 1 × 106 cells/cm2, and allowed to develop for 48 h on nutrient-free plates containing phosphate buffer supplemented with 15 g/L agar. Pieces of agar were excised and photographed from the top and side.
Supplementary Material
Acknowledgments
We thank Matthieu Bosman for his input in the project, Patricia Stege and Ineke Keizer-Gunnink for technical assistance, the X-ray communities of Max Planck Institutes Dortmund and Heidelberg, Eckhard Hofmann (University Bochum) and the beamline staff of X10SA (Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland) for support and data collection, and Maarten Linskens for reading the manuscript. Funding was provided by the Michael J. Fox Foundation for Parkinson’s Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structures reported in this article have been deposited in the Protein Data Bank, www.pdb.org [PDB code 4F0F (wild type active), 4F0G (wild type inactive), 4F1M (G1179S), 4F1O (L1180T), and 4F1T (wild type in complex with H1152)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203223109/-/DCSupplemental.
References
- 1.Bosgraaf L, Van Haastert PJ. Roc, a Ras/GTPase domain in complex proteins. Biochim Biophys Acta. 2003;1643:5–10. doi: 10.1016/j.bbamcr.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Paisán-Ruíz C, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 3.Zimprich A, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Aasly JO, et al. Clinical features of LRRK2-associated Parkinson’s disease in central Norway. Ann Neurol. 2005;57:762–765. doi: 10.1002/ana.20456. [DOI] [PubMed] [Google Scholar]
- 5.Hernandez DG, et al. Clinical and positron emission tomography of Parkinson’s disease caused by LRRK2. Ann Neurol. 2005;57:453–456. doi: 10.1002/ana.20401. [DOI] [PubMed] [Google Scholar]
- 6.Khan NL, et al. Mutations in the gene LRRK2 encoding dardarin (PARK8) cause familial Parkinson’s disease: Clinical, pathological, olfactory and functional imaging and genetic data. Brain. 2005;128:2786–2796. doi: 10.1093/brain/awh667. [DOI] [PubMed] [Google Scholar]
- 7.Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson’s disease. Nat Rev Neurosci. 2010;11:791–797. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilks WP, et al. A common LRRK2 mutation in idiopathic Parkinson’s disease. Lancet. 2005;365:415–416. doi: 10.1016/S0140-6736(05)17830-1. [DOI] [PubMed] [Google Scholar]
- 9.Nalls MA, et al. International Parkinson Disease Genomics Consortium Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cookson MR, Bandmann O. Parkinson’s disease: Insights from pathways. Hum Mol Genet. 2010;19(R1):R21–R27. doi: 10.1093/hmg/ddq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng J, et al. Structure of the ROC domain from the Parkinson’s disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proc Natl Acad Sci USA. 2008;105:1499–1504. doi: 10.1073/pnas.0709098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A. It takes two to tango: Regulation of G proteins by dimerization. Nat Rev Mol Cell Biol. 2009;10:423–429. doi: 10.1038/nrm2689. [DOI] [PubMed] [Google Scholar]
- 13.Gotthardt K, Weyand M, Kortholt A, Van Haastert PJ, Wittinghofer A. Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase. EMBO J. 2008;27:2239–2249. doi: 10.1038/emboj.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greggio E, et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Luzón-Toro B, Rubio de la Torre E, Delgado A, Pérez-Tur J, Hilfiker S. Mechanistic insight into the dominant mode of the Parkinson’s disease-associated G2019S LRRK2 mutation. Hum Mol Genet. 2007;16:2031–2039. doi: 10.1093/hmg/ddm151. [DOI] [PubMed] [Google Scholar]
- 16.West AB, et al. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci USA. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anand VS, Braithwaite SP. LRRK2 in Parkinson’s disease: Biochemical functions. FEBS J. 2009;276:6428–6435. doi: 10.1111/j.1742-4658.2009.07341.x. [DOI] [PubMed] [Google Scholar]
- 18.Greggio E, Cookson MR. Leucine-rich repeat kinase 2 mutations and Parkinson’s disease: Three questions. ASN Neuro. 2009;1:1. doi: 10.1042/AN20090007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Egmond WN, et al. Intramolecular activation mechanism of the Dictyostelium LRRK2 homolog Roco protein GbpC. J Biol Chem. 2008;283:30412–30420. doi: 10.1074/jbc.M804265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Egmond WN, van Haastert PJ. Characterization of the Roco protein family in Dictyostelium discoideum. Eukaryot Cell. 2010;9:751–761. doi: 10.1128/EC.00366-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kicka S, et al. The LRRK2-related Roco kinase Roco2 is regulated by Rab1A and controls the actin cytoskeleton. Mol Biol Cell. 2011;22:2198–2211. doi: 10.1091/mbc.E10-12-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 23.Taylor SS, Kornev AP. Protein kinases: Evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams JA. Activation loop phosphorylation and catalysis in protein kinases: Is there functional evidence for the autoinhibitor model? Biochemistry. 2003;42:601–607. doi: 10.1021/bi020617o. [DOI] [PubMed] [Google Scholar]
- 25.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci USA. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Moore DJ, Xiong Y, Dawson TM, Dawson VL. Reevaluation of phosphorylation sites in the Parkinson disease-associated leucine-rich repeat kinase 2. J Biol Chem. 2010;285:29569–29576. doi: 10.1074/jbc.M110.127639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greggio E, et al. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J Biol Chem. 2008;283:16906–16914. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaleel M, et al. LRRK2 phosphorylates moesin at threonine-558: Characterization of how Parkinson’s disease mutants affect kinase activity. Biochem J. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith WW, et al. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 30.Anand VS, et al. Investigation of leucine-rich repeat kinase 2 : Enzymological properties and novel assays. FEBS J. 2009;276:466–478. doi: 10.1111/j.1742-4658.2008.06789.x. [DOI] [PubMed] [Google Scholar]
- 31.Fancelli D, et al. Potent and selective Aurora inhibitors identified by the expansion of a novel scaffold for protein kinase inhibition. J Med Chem. 2005;48:3080–3084. doi: 10.1021/jm049076m. [DOI] [PubMed] [Google Scholar]
- 32.Ohta E, Kubo M, Obata F. Prevention of intracellular degradation of I2020T mutant LRRK2 restores its protectivity against apoptosis. Biochem Biophys Res Commun. 2010;391:242–247. doi: 10.1016/j.bbrc.2009.11.043. [DOI] [PubMed] [Google Scholar]
- 33.Wan PT, et al. Cancer Genome Project Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 34.Deng X, et al. Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2. Nat Chem Biol. 2011;7:203–205. doi: 10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nichols RJ, et al. Substrate specificity and inhibitors of LRRK2, a protein kinase mutated in Parkinson’s disease. Biochem J. 2009;424:47–60. doi: 10.1042/BJ20091035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobs M, et al. The structure of dimeric ROCK I reveals the mechanism for ligand selectivity. J Biol Chem. 2006;281:260–268. doi: 10.1074/jbc.M508847200. [DOI] [PubMed] [Google Scholar]
- 37.Breitenlechner C, et al. Protein kinase A in complex with Rho-kinase inhibitors Y-27632, Fasudil, and H-1152P: Structural basis of selectivity. Structure. 2003;11:1595–1607. doi: 10.1016/j.str.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.