Abstract
We describe a directed genome-engineering approach that combines genome-wide methods for mapping genes to traits [Warner JR, Reeder PJ, Karimpour-Fard A, Woodruff LBA, Gill RT (2010) Nat Biotechnol 28:856–862] with strategies for rapidly creating combinatorial ribosomal binding site (RBS) mutation libraries containing billions of targeted modifications [Wang HH, et al. (2009) Nature 460:894–898]. This approach should prove broadly applicable to various efforts focused on improving production of fuels, chemicals, and pharmaceuticals, among other products. We used barcoded promoter mutation libraries to map the effect of increased or decreased expression of nearly every gene in Escherichia coli onto growth in several model environments (cellulosic hydrolysate, low pH, and high acetate). Based on these data, we created and evaluated RBS mutant libraries (containing greater than 100,000,000 targeted mutations), targeting the genes identified to most affect growth. On laboratory timescales, we successfully identified a broad range of mutations (>25 growth-enhancing mutations confirmed), which improved growth rate 10–200% for several different conditions. Although successful, our efforts to identify superior combinations of growth-enhancing genes emphasized the importance of epistatic interactions among the targeted genes (synergistic, antagonistic) for taking full advantage of this approach to directed genome engineering.
Keywords: recombineering, directed evolution, lignocellulosic hydrolysate
Directed evolution is fundamentally concerned with how to most efficiently search combinatorial mutation space on laboratory timescales (1–5). Many innovative approaches for improved searching using rational design, screening, and/or exhaustive residue mapping have been demonstrated at the level of individual proteins (3, 6–9). However, extensions to multiprotein complexes and/or pathways that are affected by a large number of possible mutations have remained a challenge. Advances in multiplex DNA synthesis along with several recently reported methods for rapidly modifying genomes have opened up the possibility of applying directed evolution search strategies at a scale far beyond what was previously possible (5, 10–23).
In particular, the trackable multiplex recombineering (TRMR) approach allows one to simultaneously map the effect of increased or decreased expression onto a trait of interest for every gene in the Escherichia coli genome (22). This allows one to perform high-throughput screens or growth selections, resulting in the identification of a subset of genes with direct relevance to a particular trait (i.e., acetate tolerance). As a complement, multiplex automated genome engineering (MAGE) allows one to generate billions of E. coli strains containing combinations of mutations targeting a subset of genes (up to dozens of genes) (5), thus allowing one to search for combinatorial mutants with superior performance relative to the wild-type (WT) strain or strains containing only a single mutation.
We hypothesized that these two methods could be combined into a rational approach for engineering complex traits in a manner conceptually similar to how such searches have been performed at the level of individual proteins. That is, we used the TRMR method to first perform a comprehensive mapping of the effect of changes in the expression (up or down) of individual genes on targeted traits, used these results to assign relevance to target genes, and then used MAGE-like recursive multiplex recombineering to create mutant libraries containing combinations of genes indicated by the TRMR studies (see Fig. 1). These libraries were then subjected to further growth selections to identify mutations, and combinations thereof, conferring further growth advantages. This approach, thus, mimics various combinatorial protein engineering strategies (4, 6, 9, 24) that were designed to address the same combinatorial search space challenges by first identifying relevant individual residue modifications and then constructing and searching combinations of such residue modifications. We expected that the demonstration of our genome-scale search strategy would be complicated by a range of factors, including the selective pressure used in the initial TRMR selections, the level of combinatorial diversity achieved using recursive multiplex recombineering, the epistatic effects of combined mutations (synergistic, additive, antagonistic), and the selective pressure used upon the combinatorial mutant libraries. With this in mind, we report here the demonstration of a combined genome search and combinatorial optimization approach through the engineering of several model traits (acetate tolerance, growth at pH 5, and cellulosic hydrolysate tolerance).
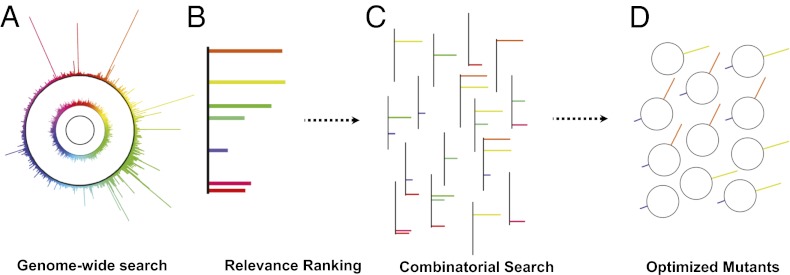
Fig. 1.
Overview of strategy. (A) The TRMR library (middle circle) is generated by introducing mutations into WT E. coli (inner circle). A selection is performed yielding data on high-fitness mutants (outer circle). (B) A few mutants are chosen to be targets for further study. (C) A recursive multiplex recombineering library is constructed, generating a large diversity of clones with one or more mutations. (D) A selection is performed on the library to yield the most tolerant clones.
Results and Discussion
Multiplex DNA synthesis and recombineering advances have enabled the rapid construction of E. coli strains containing billions of specific mutations (5, 22, 25). Here, we used such technologies to develop an approach to genome engineering that starts with a broad-based search that maps individual genes to traits at the genome-scale and follows with an in-depth search of the combinatorial space comprising the subset of genes identified to have the largest effect on the trait of interest. We used three model traits to develop this search strategy: acetate tolerance, corn stover hydrolysate tolerance, and growth at low pH (pH 5). These traits were picked because of their broader relevance (i.e., sustainable fuels/chemicals) and differing levels of complexity in toxicity mechanisms, which we expected would aid in the generalization of our approach.
Broad-Based Searching to Map Genes to Traits at the Genome Scale.
The TRMR technology employs barcoded promoter replacement libraries to simultaneously map the effect of increased or decreased expression onto a selected trait for nearly every gene in the genome of E. coli (22). Here, we applied this technology to track library population dynamics in growth selections using three different selective environments (acetate, hydrolysate, pH 5). In so doing, we are able to identify promoter replacement alleles that are enriched or diluted at different time points in the growth selections, which enables the rapid identification of genes for which increased or decreased expression confers a growth advantage.
A critical aspect of our approach involves understanding selective pressure and, in particular, how to measure selective pressure in a way that aids the design of integrated broad-based (TRMR) and in-depth (MAGE) search strategies. The TRMR technology provides a control for direct measurement of selective pressure across any selection. Specifically, the E. coli strain JWKAN, which has a silent barcoded insertion near the attTn7 site (22), is mixed into the initial library population at a known concentration. This barcode insertion is silent in that no promoter or ribosomal binding site (RBS) is affected because of its location, rendering it useful only for tracking the allele frequency of a strain that grows similarly to the WT parental strain. Using this control strain, we can directly calculate fitness and quantitatively characterize selective pressure in a manner identical to standard competition experiments (26). The fitness of the control strain (WJWKAN), and all other strains in the library, is calculated by dividing the postselection barcoded allele frequency by the initial barcoded allele frequency. Because the choice of genes for combinatorial studies is largely based upon the TRMR based fitness measurements, and because the distribution of such fitness measurements is controlled by selective pressure, it is important to understand how strong or weak a particular growth selection was when interpreting fitness data for the ranking of genes for combinatorial studies.
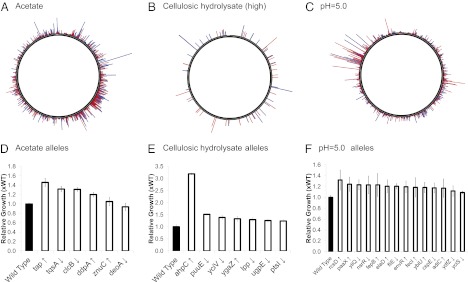
In the case of the acetate selections (performed for 69 h at 16 g/L acetate in 3-(N-morpholino)propanesulfonic acid (MOPS) minimal medium as previously described by Neidhardt et al. (28); Fig. 2A), the control strain fitness was calculated as WJWKAN = 0.024 (i.e., a 40-fold dilution of JWKAN), indicating a strong selective pressure. For hydrolysate selections (15–17% or 18–20% hydrolysate in MOPS minimal media; Fig. 2B), the averaged estimated control fitness for these selections were WJWKAN = 11.1 for 15–17% hydrolysate and WJWKAN = 0.079 for the 18–20% hydrolysate, indicating minimal selective pressure for the low concentrations and strong selective pressure for the high concentrations. For growth at pH 5 (15 h in minimal medium; Fig. 2C), the control fitness was calculated as WJWKAN = 0.25, indicating a moderate selective pressure. Each selection reduced the diversity of the population, yet the strength of selection differed in each case (see Fig. 2 A–C and Fig. S1). This resulted not only in the enrichment for a set of potential targets specific to each environment but also, more generally, in a different distribution of potential targets (see Fig. 2 A–C and Fig. S1).
Fig. 2.
(A–C) Circle plots showing the result of TRMR analyses for acetate (16 g/L), hydrolysate (18–20%), and low pH. Clone fitness is mapped over the E. coli genome. Peak location represents location of clone in E. coli genome; peak size is relative to fitness. Colors denote the type of mutation in the clones: red spikes indicate an up mutation, blue spikes are down mutations. [B is adapted from Warner et al. (1).] (D–F) Growth studies of individual TRMR mutants. (D) Twenty-four-hour growth in 16 g/L acetate. (E) Fourteen-hour growth in 27.5% hydrolysate. (F) Twelve-hour growth in pH 5.0 M9 media. Error bars represent 1 SD.
The TRMR datasets were used to identify genes to target in combinatorial library design. Genes were selected primarily based on their measured fitness (see Fig. 2 A–C), as well as the level of improvement measured in individual growth studies (Fig. 2 D–F). The eight acetate targets (six “down” and two “up”) were chosen from the highest fitness mutants in the TRMR selection. The low pH targets (14 total: 6 down; 8 up) were selected by a combination of fitness and their relative abundance in the postselection population. The hydrolysate targets (27 total: 10 down; 17 up) were chosen by primarily by high fitness in the previously described TRMR selection (22). Two hydrolysate targets (talB and lpcA) were also included based on their close metabolic relationship to another target, talA, which was found with high fitness. All targets are listed in Table S1. As shown in Fig. 2, these TRMR studies resulted in the identification of a range of mutations that were confirmed to increase growth in several broadly relevant conditions, thus confirming our control calculations above that indicated medium to strong selective pressure.
Design Considerations for Combinatorial Searches.
Based on our TRMR search results and follow-up target identification efforts, we used recursive multiplex recombineering strategies identical to those described by Wang et al. (5) to generate several different combinatorial RBS libraries specific to acetate, hydrolysate, or low pH tolerance. In brief, ssDNA cassettes were designed to replace the RBS of relevant subsets of 8–27 target genes with a degenerate set of RBS sequences expected to span a range of increased or decreased expression levels relative to the WT RBS. Although the generation of combinatorial libraries was straightforward, both the efficiency of recombination and the extent of any epistatic relationships among targeted genes play key roles in dictating the effectiveness of our combinatorial search strategy.
The goal here is to perform growth selections upon combinatorial libraries to identify clones containing combinations of mutations that demonstrate superior growth relative to the WT or a strain containing individual promoter mutations. Enrichment for such improved clones depends upon the number of RBS mutations in a clone, the growth advantage conferred by individual RBS mutations, and the epistatic nature of the mutations within the clone. Although we can calculate the fraction of populations containing single, double, triple, etc. mutations (Fig. 3A), and we can measure the growth effect of individual mutations (as in Fig. 2 D–F), no current strategy exists for predicting the nature and extent of epistatic interactions among targeted genes. This challenges the design of growth selections that will identify superior combinations because it is not possible to then predict the selective pressure required to identify combinations present at a given sequencing depth.
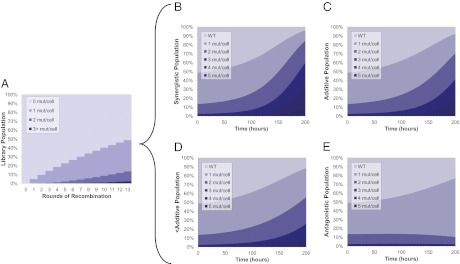
Fig. 3.
Model of recursive multiplex recombineering library growth. (A) Construction of the library. Recombination efficiencies of 2–10% were routinely achieved, as quantified using an oligo that restores operational sequence of the galK gene in the SIMD70 strain (oligo 478 in ref. 19). Shown is the theoretical population distribution of the library where recombination efficiency is 5.0%. After 13 rounds of recombination, single, double, and triple mutants represent 35%, 11%, and 2%, of the total library populations, respectively. (B–E) Four cases of varying epistasis in a growth selection. WT growth rate was set to 0.05 1/h (typical for 40% hydrolysate growth). Mutations were modeled to be either beneficial (35% increase in growth rate over control; 10% of mutations) or neutral (no change in growth rate; 90% of mutations). (B) Synergistic: combinations of mutations increase in growth 10% more than additive. (C) Additive: benefits of individual mutations are additive. (D) Less-than-additive: combinations 10% less than additive. (E) Antagonistic: combinations of mutations reduce growth rate by 15% compared with individual mutations.
Our strategy for addressing this limitation was to predict library population evolution using several different possibilities regarding epistasis. Specifically, we categorized epistatic interactions as either antagonistic, less-than-additive, additive, or synergistic (SI Selection Model with Varying Epistasis). Through a simple mathematical model, we show how these different scenarios alter population dynamics in a growth selection (Fig. 3 B–E) and how deep one must sequence to identify combinations containing 1, 2, 3, etc. genes. The model assumes the culture is in the exponential phase growth at the maximum specific growth rate (27), because our selections were serially transferred so that the cultures did not reach stationary phase. No interclonal interactions were assumed or taken into account here. In the synergistic model, after 200 h of selection, double and triple mutants quickly become a significant portion of the population and would be easily identified through the sequencing of a random collection of only a few clones (Fig. 3B). This enrichment, of course, happens more slowly in the additive and less-than-additive models (Fig. 3 C and D). In the antagonistic interaction model, the double and triple mutants are depleted in the population, and thus require more extensive sequencing for identification (Fig. 3E).
As a further complication, the number of targets chosen will have an effect on search results. Specifically, the number of possible double and triple combinations scales nearly exponentially with the number of targets. The relevance here is that although some double or triple mutants may have synergistic epistatic interactions, such mutants may represent an exceedingly small fraction of the population of possible double or triple mutants. As an example, 27 targets were chosen for recursive recombination in the hydrolysate library, allowing for 702 and 17,550 possible double and triple mutant combinations, respectively.
In the absence of a priori knowledge of epistatic interactions, it is difficult to design selections that will uniformly result in the identification of combinatorial mutants. However, as shown in Fig. 3, it is possible to make predictions that both provide a guide in selection design and a framework for rapidly interpreting selection results, which are critical to any future efforts to implement combinatorial genome-engineering as we describe next.
Construction and Searching of Combinatorial Mutant Libraries.
Hydrolysate, low pH, and acetate combinatorial libraries were constructed via recursive, multiplex recombineering and subjected to growth selections to identify further improved mutants. For the hydrolysate library, 13 rounds of multiplex recombination were performed with the mixed pool of oligonucleotides, as well as a galK+ marker to track recombination efficiency. Efficiency was measured at an average of 5.0% per round; thus, we estimate the combinatorial population to contain 35% single mutants, 11% double mutants, 2.1% triple mutants, and 0.3% quadruple mutants (similar to Fig. 2A). This library population was grown via 11 serial transfers for a total of 232 h in MOPS minimal medium containing 40% corn stover hydrolysate (as described in ref. 28). The growth rate of the final population doubled compared with the initial batch growth rate. After this time, samples were taken and culture was plated to obtain individual colonies.
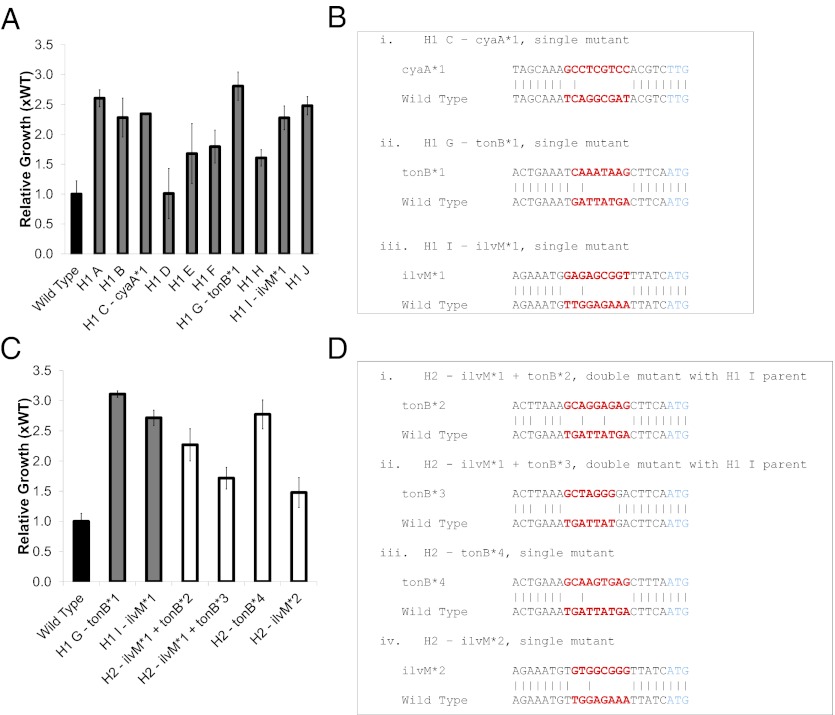
Based on the mathematics detailed above (and in Fig. 3), we picked 10 individual colonies for further examination (which would have contained 3–4 single mutants and 1 double mutant preselection as in Fig. 3A and as many as 2–5 single, double, triple, and quadruple mutations postselection as in Fig. 3 B–E). Six of the 10 picked colonies contained mutants that grew significantly better (as much as 200%) than the parent SIMD 70 strain (P value < 0.05; Fig. 4A), thus confirming that the same selection strategy we used successfully in the TRMR studies worked here to enrich for mutants with improved growth. The RBS regions corresponding to all 27 targeted regions were sequenced for each of these 6 improved mutants (H40 A, B, C, G, I, and J). The “H40 C” clone was modified in the target cyaA RBS, the “H40 G” clone was modified in the tonB RBS, and the “H40 I” clone in the ilvM RBS (Fig. 4B). The cyaA gene codes for adenylate cyclase, which synthesizes cAMP, the signaling molecule. The membrane-bound protein TonB plays a role in the import of siderophore-bound iron and vitamin B12 (29, 30). The ilvM gene codes for a subunit of acetohydroxyacid synthase II (in the isoleucine synthesis pathway) (31). Interestingly, this enzyme should not be fully functional in K12 derivatives because of a frameshift mutation in the ilvG gene, which codes for the other subunits (31). The H40 A, B, and J clones were found with no mutations in the target regions, which may suggest spontaneous generation of tolerance over the 232 h selection. Because three unique single mutants were identified, it suggests that the end population was still genotypically diverse, even after a 10-d serial transfer selection. To check this, we PCR amplified and sequenced the RBS region from one of the targeted genes, lpp, from the mutant mixture remaining after selection. We confirmed that variation in the lpp RBS sequence remained after selection (see the chromatograph in Fig. S2). It is interesting to note the mutations found here are not the same as the best mutations found from the TRMR selection. Although these tools are similar, TRMR replaces both the promoter and the RBS, whereas MAGE replaces only the RBS; thus it is not surprising that the two methods do not yield identical results.
Fig. 4.
Recursive multiplex recombineering hydrolysate selection isolates. (A) Relative growth over 44 h of 10 isolated clones taken from sample plates after the initial selection. (B) Mutations in RBS of tolerant clones. (C) Relative growth over 20 h of four mutants isolated after the second selection compared with WT and H1 G and I (isolated from the first selection). (D) Mutations in RBS of the second combinatorial library clones isolated after the second selection. Error bars represent 1 SD.
Our results here suggested that the growth advantage conferred by the individual mutations was similar to the advantage conferred by the best combinations of such mutations (i.e., the scenario of Fig. 3 D or E). To study this possibility in greater depth, and to provide guidance for future applications of this approach, we again used recursive multiplex recombineering to generate combinatorial libraries but this time started with a mixture of the WT sensitive strain and two individual mutants we had confirmed to have a growth advantage. We also chose to target a smaller number of genes. Combined, these changes in design not only allowed us to decrease library diversity in a directed manner but also provided built in positive and negative controls that allowed us to discriminate between strength of selection effects and epistatic effects when analyzing our selection results.
This library started with an equal mix of the SIMD70 strain, the H40G clone (with the tonB RBS mutation), and the H40I clone (with the ilvM RBS mutation). We targeted the RBS region of 4 (lpcA, lpp, tonB, and ilvM) of the previous set of 27 hydrolysate tolerance targets. The lpcA target was chosen because it had previously been seen to also confer resistance to both acetate and furfural (32). The lpp target was chosen because its RBS region was diverse in the end population described above. Six rounds of recombination were performed with a measured efficiency of ∼7.0%. Because two-thirds of the initial population already had a mutation, we estimated that only 21.5% of the population had no mutations, 52.9% had a single mutation, 21.3% were double mutants, and 3.9% were triple mutants, with the remainder having four mutations per clone (Fig. S3). The population was subjected to three serial transfer selections in 40% hydrolysate over a total of 95 h (using the same conditions as above). After plating of selected samples, 17 colonies were picked, and their 4 target RBS regions were sequenced (i.e., completely genotyped). Eleven of the 17 were identical to the H40 I clone (i.e., each had the same ilvM RBS mutation), 2 clones had single mutations that were different compared with the H40 G (tonB) or H40 I (ilvM) starting clones but were in the tonB or ilvM RBS, and 4 of the picked clones were indeed double mutants. All double mutants had the H40 I clone ilvM mutation, presumably the parent of the two, and a unique tonB mutation as well (Fig. 4D).
Collectively, these results confirmed the results from the original library studies above, suggesting that epistatic interactions indeed followed the less than additive or antagonistic models laid out in Fig. 3. This conclusion was further supported by substantial decrease in WT strains remaining in the population (0 of 17 identified, or <5–6% of population) after selection even though such strains made up ∼21.5% of the preselection population (see Fig. S3). However, to further test this suggestion, we measured the growth of a representative number of clones isolated from this second selection. As can be seen in Fig. 4C, the double mutants tested did not grow better than the single mutants from which they derive.
Although these results emphasize the importance of epistasis, it is possible that these results were specific to hydrolysate based toxicity, which involves a complex and incompletely understood model of inhibition involving the broad range of inhibitory compounds within the hydrolysate mixture. To investigate this possibility, we performed similar recursive multiplex recombineering and selection studies using the acetate and low pH TRMR results. Fourteen targets were chosen for the low pH combinatorial library (Table S1). Although slightly different strategies were used for library generation and selection (SI Materials and Methods), the same results were observed where sequencing results resulted in the identification of only a single RBS mutant: ybiU up. Similar to the hydrolysate studies, we performed further recombineering using the ybiU up strain as the basis for a second low pH library. After recombination and selection by serial cultures at pH 5, we identified a double mutant strain (ybiU, up; and ydfZ, down) that grows significantly better than WT at pH 5. The ybiU gene encodes an uncharacterized predicted protein, whereas the ydfZ gene encodes for an uncharacterized conserved protein; neither has been implicated previously in low pH tolerance. In contrast to the lack of positive epistasis observed in our hydrolysate studies, the double mutant demonstrated slightly positive epistasis here with a ca. 65% increased growth relative to the WT, whereas the best single mutant grew ca. 50% better (Fig. S4). For the acetate library, eight targets were chosen from the highest fitness TRMR alleles. Although this library underwent serial transfer selection in 10 g/L acetate for 110 h, none of the 23 colonies picked exhibited a further increase in growth rate. Four colonies were selected and each of their 8 target RBS regions sequenced (32 total sequencing reactions), but no mutations were found. This result reinforces our hydrolysate findings and further underlines the importance of developing a better ability to predict epistatic interactions.
Conclusions
We have presented data demonstrating an approach for directed genome-engineering that can be performed on laboratory timescales (weeks) by a few individuals (one scientist can perform several rounds of this approach in a month). Specifically, we quantified the effect of increased or decreased expression of nearly every gene in the E. coli genome (8,155 mutants) under three different conditions. Many of such growth selections can be performed in parallel, thus allowing one to rapidly map promoter mutations onto various phenotypes of interest. Analysis of these growth data led to the constructing of combinatorial libraries based on the best performers (more than 100,000,000 unique mutants were constructed). These mutants were constructed using readily available multiplex recombineering methods that can be performed recursively by hand or in an automated fashion (5) to generate billions of mutations in a few days. Although we were able to identify a broad range of previously unknown mutations affecting growth, our results suggested that the ability to take full advantage of this approach depends on the nature of interactions between the different mutations (synergistic, antagonistic, etc.). Such epistatic interactions are currently difficult or impossible to predict, which, thus, emphasizes the need for improved methods of assigning relevance to fitness of the individual mutants in relation to other high-ranking mutants.
In our studies, we explored several strategies for assigning relevance to individual genes that went beyond simply using the fitness data provided by our TRMR selections. These approaches included the use of online databases of gene classifications (Gene Ontology Enrichment Analysis Software Toolkit; Ecocyc) to attempt to provide additional information for selecting genes for combinatorial studies. Unfortunately, our efforts here did not successfully identify any core set of enriched genes that were disproportionately represented in certain functional, metabolic, or orthologous classifications. [In contrast to prior studies, we have reported linking-enriched genetic loci by metabolic pathways (32).] Given the clear epistatic complications that arise, and the strong effect these have on the success of this approach, methods for interpreting fitness data in a way that leads to improved target identification for combinatorial studies are critical. These could involve better understanding of biological networks at various levels of interaction (regulatory, protein–protein, metabolic, gene ontology, energy balancing, etc.). The ability to predict these interactions will be enhanced by the methods we described herein, where it is now possible to test a broad range of predictions in a short amount of time (i.e., by recursive multiplex recombineering). Taken together, the approach we describe herein, along with continued development in predicting target relevance, will not only allow for improved directed genome engineering but also for enabling a broad range of fundamental studies seeking to better understand how to construct complex phenotypes in general.
Materials and Methods
Bacteria, Plasmids, and Media.
E. coli K12 (ATCC no. 29425) was used for dsDNA TRMR clone reconstructions. Plasmid pSIM5 was provided by Donald Court (Gene Regulation and Chromosome Biology Laboratory, National Cancer Institute at Frederick, Frederick, MD) and used for the dsDNA recombination (20). Plasmids pKD13 and pKD46 were obtained from the Coli Genetic Stock Center (CGSC) (7633 and 7634) (14). E. coli strain SIMD 70, derived from strain SIMD 50, was graciously provided by Donald Court and was used for ssDNA recombination (33). Overnight cultures used Luria–Bertani (LB) medium. Samples on solid media were with LB media and agar unless the antibiotic blasticidin was used, in which case, a low-salt LB media with agar was used. MOPS minimal media was used as described (28). Pretreated corn stover cellulosic hydrolysate was provided by the National Renewable Energy Laboratory (Golden, CO). Blasticidin was used at a working concentration of 90 μg/mL. Chloramphenicol was used at a working concentration of 20 μg/mL.
TRMR Library and Selections.
The TRMR library was prepared in the laboratory of R.T.G. previous to this study (22). Preparation of the TRMR library for all selections and the sample populations for microarray analysis were done according to the instructions of the author (22). All TRMR selections were performed at 37 °C in a shaking incubator rotating at 225 rpm.
Selections in acetate were performed in 200 mL of MOPS minimal medium with 0.2% (mass/vol) glucose and 16 g/L acetate. Stock acetic acid solution was prepared by titrating HPLC-grade 50% (vol/vol) acetic acid solution (Fluka) on ice with 10 M KOH to neutral pH. Portions of the up and down libraries were mixed for an equal number of cells. The JWKAN strain was introduced into the library mix so that the JWKAN strain started with 20 times the number of the average TRMR mutant present in the library, or in a ratio of 1:400. This final mixture was introduced into the selection environment at a 2.5% inoculation. The acetate selection was performed for 69 h.
Selections in low pH were performed in 500 mL of M9 media (adjusted to pH 5 using 1 M H2SO4) in a 2-L flask at cell density of 2 × 106 cells/mL. Control strain JWKAN was mixed at the ratio of 1:8,000. Cells were grown for ∼8 h and transferred to fresh medium of pH 5 serially before cell growth enters stationary phase. Serial transfer was performed 10 times corresponding to about 30 generations.
Selections in hydrolysate were performed as described previously (22). Briefly, selections were performed in a decreasing concentration of hydrolysate over three batches.
TRMR Clone Reconstruction.
TRMR clones were reconstructed in E. coli K12 via recombination using the pSIM5 or pKD46 plasmid. TRMR up and down inserts were amplified from genomic DNA extracted from the library population. Fifty base pairs of homology were added to each end of the insert specific to the desired location of insertion. The homology regions chosen were identical to those in the original TRMR library. Recombination was performed at 30 °C as described previously (21). Recombination recovery cultures were plated onto low-salt LB with agar solid medium with blasticidin. To ensure the TRMR insert was located in the proper location and orientation in the genome, the surrounding region was amplified via PCR, purified, and sequenced. Growth study methods can be found in SI Materials and Methods.
Library Construction of Mutated RBS.
The mutated RBS libraries were constructed via λ-red recombination. SIMD 70 was used as the base strain for the acetate and hydrolysate libraries. ssDNA oligonucleotides were designed to replace the RBS of the previously selected gene target with either a partially or completely degenerate sequence according to the allele found in the TRMR selection. Design constraints of the oligos and quantification of efficiency can be found in the supplemental methods (SI Materials and Methods).
Multiple rounds of recombination were performed on SIMD 70 with the various mixed oligo pools. Recombination was performed at 30 °C as described previously (21). After each round of recombination, the recombined cultures were allowed to recover for 1–2 h in Terrific Broth. The recovery culture was then used to inoculate a Luria Broth culture for the next round of recombination. If the cells were not immediately recultured, the recovery culture was pelleted and chilled at 4 °C until further use. The galK+ oligo was spiked in at a rate of 1:20, so that the recombination frequency of the final library could be tested on MacConkey agar with 1% (mass/vol) galactose.
Recombineering for the low pH library was performed using the strain MG1655LRM with the same procedure as above. Methods for recombination strain construction can be found in SI Materials and Methods. Selection was performed by serial cultures in M9 media (pH 5) after two or three rounds of recursive recombineering.
Supplementary Material
Acknowledgments
We thank Dr. Donald Court for providing plasmid pSIM5 and strain SIMD70.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206299109/-/DCSupplemental.
References
- 1.Stemmer WP. DNA shuffling by random fragmentation and reassembly: In vitro recombination for molecular evolution. Proc Natl Acad Sci USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright MC, Joyce GF. Continuous in vitro evolution of catalytic function. Science. 1997;276:614–617. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]
- 3.Voigt CA, Mayo SL, Arnold FH, Wang ZG. Computational method to reduce the search space for directed protein evolution. Proc Natl Acad Sci USA. 2001;98:3778–3783. doi: 10.1073/pnas.051614498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloom JD, Arnold FH. In the light of directed evolution: Pathways of adaptive protein evolution. Proc Natl Acad Sci USA. 2009;106(Suppl 1):9995–10000. doi: 10.1073/pnas.0901522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Estell DA, Graycar TP, Wells JA. Engineering an enzyme by site-directed mutagenesis to be resistant to chemical oxidation. J Biol Chem. 1985;260:6518–6521. [PubMed] [Google Scholar]
- 7.Estell DA, Wells JA. 1988. United States patent 4,760,025 (7/26/1988)
- 8.Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 9.Fox RJ, et al. Improving catalytic function by ProSAR-driven enzyme evolution. Nat Biotechnol. 2007;25:338–344. doi: 10.1038/nbt1286. [DOI] [PubMed] [Google Scholar]
- 10.LeProust EM, et al. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res. 2010;38:2522–2540. doi: 10.1093/nar/gkq163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy KC. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 13.Muyrers JP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Muyrers JP, Testa G, Stewart AF. DNA cloning by homologous recombination in Escherichia coli. Nat Biotechnol. 2000;18:1314–1317. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
- 16.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci USA. 2001;98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costantino N, Court DL. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc Natl Acad Sci USA. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li XT, et al. Identification of factors influencing strand bias in oligonucleotide-mediated recombination in Escherichia coli. Nucleic Acids Res. 2003;31:6674–6687. doi: 10.1093/nar/gkg844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cleary MA, et al. Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nat Methods. 2004;1:241–248. doi: 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- 20.Datta S, Costantino N, Court DL. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 21.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: A homologous recombination-based method of genetic engineering. Nat Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warner JR, Reeder PJ, Karimpour-Fard A, Woodruff LBA, Gill RT. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat Biotechnol. 2010;28:856–862. doi: 10.1038/nbt.1653. [DOI] [PubMed] [Google Scholar]
- 23.Sawitzke JA, et al. Probing cellular processes with oligo-mediated recombination and using the knowledge gained to optimize recombineering. J Mol Biol. 2011;407:45–59. doi: 10.1016/j.jmb.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells JA. Systematic mutational analyses of protein-protein interfaces. Methods Enzymol. 1991;202:390–411. doi: 10.1016/0076-6879(91)02020-a. [DOI] [PubMed] [Google Scholar]
- 25.Isaacs FJ, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper VS, Lenski RE. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature. 2000;407:736–739. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- 27.Zwietering MH, Jongenburger I, Rombouts FM, van ’t Riet K. Modeling of the bacterial growth curve. Appl Environ Microbiol. 1990;56:1875–1881. doi: 10.1128/aem.56.6.1875-1881.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letain TE, Postle K. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Escherichia coli. Mol Microbiol. 1997;24:271–283. doi: 10.1046/j.1365-2958.1997.3331703.x. [DOI] [PubMed] [Google Scholar]
- 30.Braun V, Braun M. Active transport of iron and siderophore antibiotics. Curr Opin Microbiol. 2002;5:194–201. doi: 10.1016/s1369-5274(02)00298-9. [DOI] [PubMed] [Google Scholar]
- 31.Lawther RP, et al. Molecular basis of valine resistance in Escherichia coli K-12. Proc Natl Acad Sci USA. 1981;78:922–925. doi: 10.1073/pnas.78.2.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandoval NR, Mills TY, Zhang M, Gill RT. Elucidating acetate tolerance in E. coli using a genome-wide approach. Metab Eng. 2011;13:214–224. doi: 10.1016/j.ymben.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Datta S, Costantino N, Zhou X, Court DL. Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc Natl Acad Sci USA. 2008;105:1626–1631. doi: 10.1073/pnas.0709089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.