Abstract
The IFN family of cytokines operates a frontline defense against pathogens and neoplastic cells in vivo by controlling the expression of several genes. The death-associated protein kinase 1 (DAPK1), an IFN-γ–induced enzyme, controls cell cycle, apoptosis, autophagy, and tumor metastasis, and its expression is frequently down-regulated in a number of human tumors. Although the biochemical action of DAPK1 is well understood, mechanisms that regulate its expression are unclear. Previously, we have shown that transcription factor C/EBP-β is required for the basal and IFN-γ–induced expression of DAPK1. Here, we show that ATF6, an ER stress-induced transcription factor, interacts with C/EBP-β in an IFN-stimulated manner and is obligatory for Dapk1 expression. IFN-stimulated proteolytic processing of ATF6 and ERK1/2-mediated phosphorylation of C/EBP-β are necessary for these interactions. More importantly, IFN-γ failed to activate autophagic response in cells lacking either ATF6 or C/EBP-β. Consistent with these observations, the Atf6−/− mice were highly susceptible to lethal bacterial infections compared with the wild-type mice. These studies not only unravel an IFN signaling pathway that controls cell growth and antibacterial defense, but also expand the role of ATF6 beyond ER stress.
Keywords: signal transduction, tumor suppression, gene expression, innate immunity
Steady-state IFN production suppresses neoplastic cell growth in vivo (1). IFN-stimulated genes (ISGs) regulate innate and acquired immune responses against pathogens and tumors (2). Although IFN-induced acute innate immune responses that control pathogens through JAK-STAT pathways are well defined (2), chronic IFN responses that regulate neoplastic cell growth, via direct effects on tumor cells, are poorly understood. IFN-γ regulates certain genes in a STAT1-independent manner through transcription factor C/EBP-β (3). Previously, we have shown that the expression of DAPK1, a cell death-inducing Ca+2/calmodulin-regulated serine/threonine kinase, critically depends on C/EBP-β (4). DAPK1, originally isolated as an IFN-regulated growth suppressor (5), promotes apoptosis and autophagy and suppresses cell division (6, 7). DAPK1 expression is lost in a number of advanced human cancers (6). Although promoter methylation appears to be one major mechanism, in several cases DAPK1 expression is silenced without apparent DNA methylation (6). Mutational inactivation of DAPK1, except the one that diminishes expression in a form of CLL (8), is almost rare in most cancers. Thus, DAPK1 regulation needs to be fully understood to define the mechanisms that cause a loss of its expression in tumors and to develop agents that can up-regulate it.
Although the basal and IFN-γ–induced Dapk1 expression critically depends on C/EBP-β (4), the IFN-induced regulatory elements that respond to it are not classical C/EBP-β–binding sites. Furthermore, depending on the cell type and signaling factors, C/EBP-β both negatively and positively regulates gene expression and cell growth (9, 10), which has been attributed to the ability of C/EBP-β to interact with various transcription factors outside its family (3) after certain modifications. Unlike that of STAT1, IFN-γ–dependent recruitment of C/EBP-β to the Dapk1 promoter occurs in a kinetically slower manner, suggesting the requirement/activation of other factor(s) (4). One such posttranslational modification is the phosphorylation of C/EBP-β by ERK1/2 (4), which allows its association with other proteins. Basing on this hypothesis, we searched for other factor(s) that cooperate with C/EBP-β to up-regulate Dapk1. In this investigation, we show that ATF6 (11), a key ER stress-activated transcription factor, associates with C/EBP-β in an IFN-γ–stimulated manner to promote Dapk1 expression.
In the steady state, ATF6 exists as an ER membrane-anchored precursor, with its C terminus in the ER lumen and the N-terminal half (containing the transcription factor activity) facing the cytosol (11). After ER stress, ATF6 dissociates from its inhibitor BiP (12) that unmasks the Golgi localization signals (GLS) of ATF6 and allows its translocation to Golgi apparatus (12). In Golgi apparatus, the transcriptionally active ATF6 is generated via proteolysis (11), permitting its nuclear translocation and stimulation of target genes (13, 14). Here, we report a signaling cascade wherein the IFN-γ–induced proteolytic processing of ATF6 and ERK1/2-dependent phosphorylation of C/EBP-β converge on and control the expression of Dapk1 and autophagy. We also show that the Atf6−/− mice are highly susceptible to lethal bacterial infections. This study establishes a connection between ER-stress regulators and cytokine signaling pathways.
Results
ATF6 Promotes IFN-γ–Stimulated Dapk1 Expression.
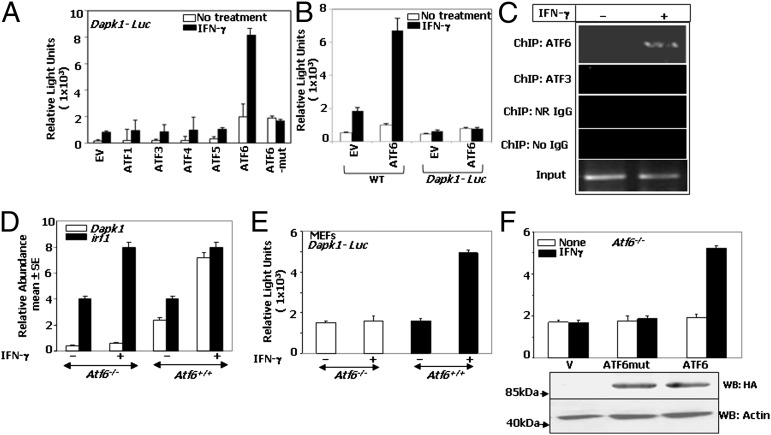
We first investigated whether ATF family members could induce the Dapk1 promoter. Reporter gene cotransfection assays with Dapk1-Luc and several ATF family members revealed that only ATF6 could robustly promote the IFN-γ–inducible expression of Dapk1 promoter (Fig. 1A). Furthermore, an ATF6 mutant (ATF6mut; lacking the critical S1 cleavage site, therefore, could not enter the nucleus) (15) failed to promote Luciferase expression, indicating the specificity (Fig. 1A). To determine whether the ATF6-dependent transcriptional response also required the CRE/ATF site that bound C/EBP-β, we compared the effect of ATF6 on wild type (WT) and a CRE/ATF site mutant of the Dapk1 promoters (Fig. 1B). ATF6 induced the luciferase expression driven by the WT but not the mutant promoter. ChIP assays using primers flanking the CRE/ATF site demonstrated that endogenous ATF6 bound to the Dapk1 promoter in response to IFN-γ (Fig. 1C). However, ChIPs assays with normal rabbit (NR) IgG and no-IgG controls and ATF3-specific antibodies did not yield any bands, demonstrating a selective recruitment of ATF6 to Dapk1 promoter.
Fig. 1.
ATF6 is essential for IFN-γ–stimulated Dapk1 expression. (A) Indicated ATF expression vectors were cotransfected into MEFs along with the Dapk1-Luc and a β-actin–β-galactosidase reporter. n = 5 for each data point. Luciferase activity was normalized to that of cotransfected β-galactosidase. (B) Effect of ATF6 on WT and CRE/ATF site mutant Dapk1-Luc (n = 5 per data point). (C) ChIP assay with CRE/ATF-specific primers. IFN-γ treatment was performed for 8 h. For input control reactions, one of five of the soluble chromatin used for the ChIP analysis was used. Normal rabbit IgG (NR-IgG), ATF6-specific IgG, and ATF3-specific IgG were used at 10 μg each per reaction. (D) qPCR analysis of Dapk1 and Irf1 transcripts in MEFs. (E and F) MEFs were transfected with the Dapk1-Luc, and mean luciferase activity ± SE (n = 6) is presented. (F) Effect of an ATF6 mutant on Dapk1-Luc expression. Lower shows Western blot analysis of the same samples.
ATF6 Is Essential for IFN-γ–Stimulated Dapk1 Expression.
To establish a critical role for ATF6 in Dapk1 expression, we analyzed the expression of endogenous Dapk1 mRNA (Fig. 1D). The Dapk1 mRNA was not induced in the absence of ATF6 (Fig. 1D). Loss of Atf6 did not affect the IFN-induced expression of Irf1 (Fig. 1D). Consistent with these observations, the IFN-γ–induced expression of Dapk1-Luc occurred only in wild-type but not in Atf6−/− MEFs (Fig. 1E). More importantly, restoration of Atf6−/− MEFs with ATF6, but not ATF6mut, reinstated Dapk1-Luc expression (Fig. 1F). The distinct effects of ATF6 and ATF6mut on Dapk1-Luc levels were not due to their differential expression. The importance of ATF6 for IFN-induced expression of DAPK1 was also ascertained by using RNAi in human and mouse cells (Fig. S1).
IFN-γ–Induced Proteolytic Activation of ATF6 and Its Translocation to Nucleus via Golgi apparatus.
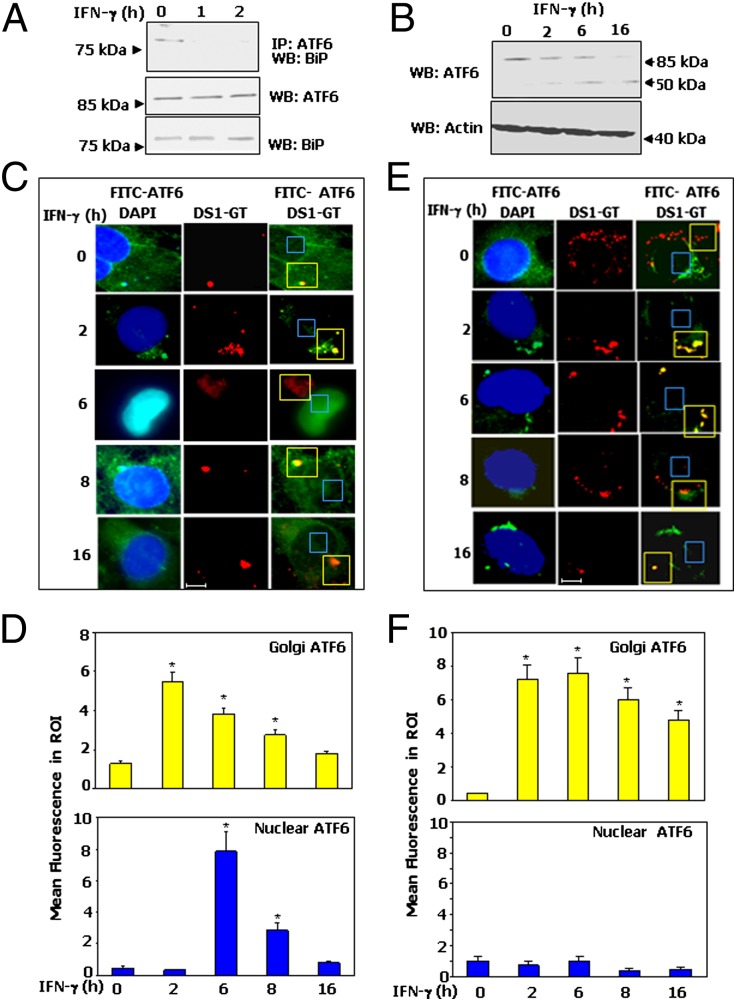
Because the activation of ATF6 requires dissociation from its inhibitor BiP in the ER (12), we next probed whether IFN-γ caused it. Immunoprecipitation analyses with endogenous ATF6 and BiP revealed that IFN-γ treatment for 1 h is sufficient to dissociate ATF6 from BiP (Fig. 2A). The bottom two images show comparable ATF6 and BiP levels, suggesting dissociation is not due to degradation of either protein.
Fig. 2.
IFN-γ–induced activation of ATF6. (A) MEFs were treated with IFN-γ, and lysates were subjected to immunoprecipitation and Western blot analyses with the indicated antibodies. (B) Effect of IFN-γ on ATF6 cleavage in MEFs. IFN-γ–induced translocation of ATF6 (C and D) and ATF6 mutant (E and F) into the nucleus. (C and E) Immunofluoresence images of DAPI, DS1-GT (Golgi marker), and FITC-ATF6. (Scale bar: 10 μm.) (D and F) Quantified fluorescence of FITC in regions of interest (ROI) after background correction. Bars are mean ± SE (n = 10 fields; *P < 0.001).
After its dissociation from BiP, ATF6 migrates to the Golgi apparatus, where it undergoes cleavage by resident proteases to generate mature ATF6 (50 kDa) that enters the nucleus (11). Therefore, MEFs were stimulated with IFN-γ for 6 h or as indicated and analyzed by Western blot (Fig. 2B) with an ATF6 antibody that detects both full length (90 kDa) and mature (50 kDa) forms of ATF6. IFN-γ treatment caused a progressive decline in the ATF6 90kDa form and an emergence of the 50-kDa mature product between 2–16 h (Fig. 2B). IFN-γ–stimulated the cleavage of ATF6, comparable to that of Tunicamycin A (TMA), an ER stress inducer (Fig. S2C).
We next investigated whether IFN-γ promoted nuclear localization of ATF6 via the Golgi apparatus. We transfected FLAG-epitope–tagged ATF6 (Fig. 2C) and ATF6 mut (Fig. 2E) constructs into Atf6−/− cells (Fig. S2 A and B). Because of a lack of mutant specific antibody, immunofluoresence analyses were performed by using FLAG-antibody after IFN-γ treatment. To mark the Golgi apparatus, we transfected the DS1-GT expression vector into these cells (15). Steady-state ATF6 was detected in the perinuclear and cytoplasmic regions in a manner consistent with ER localization. IFN-γ treatment for 2 h caused an accumulation of ATF6 in the Golgi apparatus as indicated by the strong overlap of FITC (ATF6) and red (DS1-GT) fluorescence. Moreover, IFN-γ stimulation for 6 and 8 h caused higher nuclear ATF6 levels (Fig. 2C). The cleaved ATF6 (mature) becomes unstable overtime. At 16 h, ATF6 signals were found in cytoplasm, which could be due to a resynthesis of ATF6 from preexisting pools of mRNA. In contrast, the ATF6mut, although moved to the Golgi apparatus like the WT protein, failed to enter the nucleus even after 16 h of IFN-γ treatment (Fig. 2E). Fig. 2 D and F show the quantified fluorescence intensities of several fields (n = 10). These results suggest that IFN-γ treatment requires a Golgi-dependent proteolytic processing of ATF6 before its nuclear entry. A similar pattern of IFN-γ–induced translocation of endogenous ATF6 to nucleus via Golgi apparatus was observed in MEFs (Fig. S3 A and B).
IFN-γ−Stimulated Heteromerization of ATF6 and C/EBP-β.
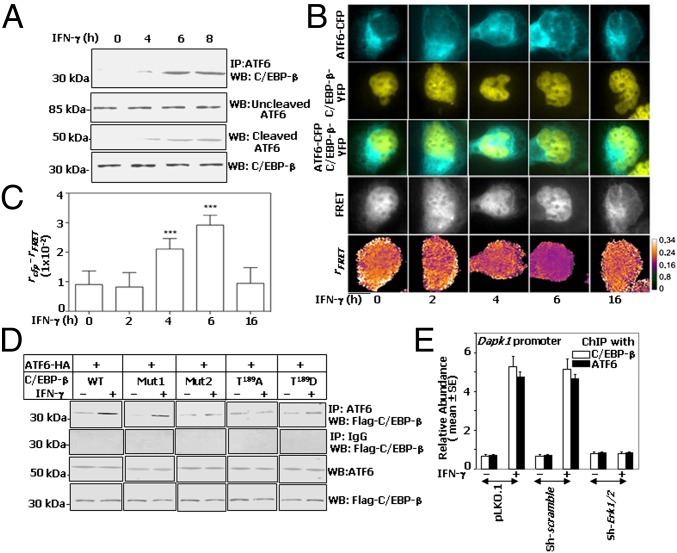
Because ATF6 and C/EBP-β required the same site on Dapk1 promoter for transcriptional activation, we asked whether ATF6 heteromerizes with C/EBP-β in response to IFN-γ. To test this hypothesis, MEFs were treated with IFN-γ and the cell lysates were subjected to immunoprecipitation and Western blot analysis with the indicated antibodies. IFN-γ enhanced the association of endogenous ATF6 with C/EBP-β (Fig. 3A). A similar association of C/EBP-β and ATF6 was observed in a variety of cells (Fig. S4A).
Fig. 3.
IFN-γ–induced heteromerization of ATF6 and C/EBP-β. (A) Immunoprecipitation and Western blot analyses of MEF lysates with the indicated antibodies. (B) ATF6-CFP and C/EBP-β-YFP were coexpressed in Cebpb−/− MEFs and examined for interaction by using the anisotropy (r) FRET method as discussed in SI Materials and Methods. CFP, YFP, and FRET (CFP excitation, YFP collection) images were taken under polarized light illumination, for cells treated with IFN-γ as indicated. Decreased rFRET is observed in the nucleus of INF-γ–treated cells indicating FRET and protein–protein interaction. The pseudocolor scale represents anisotropies. (Scale bar: 10 μm.) (C) FRET between ATF6-CFP and C/EBP-β–YFP is indicated by significant difference between rCFP and rFRET (***P < 0.001, n = 10, Student's t test vs. 0). (D) Immunoprecipitation and Western blot analyses of Cebpb−/− MEFs transfected with HA-ATF6 (373) and C/EBP-β mutants and then stimulated with IFN-γ. (E) ChIP assays with the indicated antibodies were performed, and the products were quantified by using qPCR (n = 9 per sample). Input normalized soluble chromatin was used for ChIP assays from each sample.
To demonstrate in situ interactions between C/EBP-β and ATF6, a FRET analysis was performed by using the ATF6-CFP and C/EBP-β–YFP constructs. After cotransfection and stimulation with IFN-γ, cells were examined for interaction(s) by using the anisotropy (r) FRET method, which showed an interaction between ATF6 and C/EBP-β in the nucleus starting from 4 h that reached a maximum plateau around 6 h and returned to basal level at 16 h of IFN-γ treatment (Fig. 3B). Quantification of FRET signals between ATF6-CFP and C/EBP-β–YFP indicated a significant difference between rCFP and rFRET at 4 and 6 h of IFN-γ treatment (Fig. 3C). These observations clearly show the in situ interactions of ATF6 and C/EBP-β in the presence of IFN-γ.
ERK1/2-Regulated Site of C/EBP-β Is Critical for IFN-γ–Stimulated Interaction with ATF6.
Previously, we showed an important role for an ERK1/2 target site, within the regulatory domain 2 of C/EBP-β, in inducing Dapk1 expression in response to IFN (4). Therefore, we asked whether this site is necessary for interaction with ATF6. We transfected Cebpb−/− MEFs, with expression vectors coding for either wild-type or mutant C/EBP-β constructs along with HA-ATF6 (mature form). Lysates from these cells were subjected to immunoprecipitation with HA-ATF6, and the products were probed for C/EBP-β. Wild-type C/EBP-β and Mut1 (with an intact ERK1/2 phosphorylation site) weakly interacted with ATF6 at steady state (Fig. 3D), and IFN-γ treatment further enhanced it. In contrast, Mut2 (which entirely lacked the critical ERK1/2 phosphorylation motif, GTPS) interacted with ATF6 at steady state, but IFN-γ treatment failed to enhance it. Similarly, IFN-γ stimulation failed to promote interaction of mutant T189A, which lacked the critical threonine, with ATF6. In contrast, the phosphomimetic mutant T189D readily bound to ATF6, which increased further upon IFN-γ stimulation (Fig. 3D). This observation may indicate additional phosphorylation occurring either on C/EBP-β or ATF6, which is unclear at this stage. As expected, when these lysates were subjected to immunoprecipitation with a control IgG and the products were probed for C/EBP-β, no detectable bands appeared on the blot (Fig. 3D). The bottom two images in Fig. 3D show that such differential interactions are not due to expression differences of C/EBP-β or ATF6. Because ERK1/2 phosphorylated C/EBP-β at T189 (4), we examined whether IFN-γ–stimulated recruitment of ATF6 and C/EBP-β to the Dapk1 promoter and whether it required ERK1/2. A quantitative PCR (qPCR) analysis of ChIP products from MEFs stably expressing either vector alone or scrambled shRNA or Erk1/2-specific shRNA (4) revealed that IFN-γ stimulated the recruitment of ATF6 and C/EBP-β to the Dapk1 promoter, but not in those depleted of ERK 1/2 (Fig. 3E). Thus, IFN-γ–stimulated ATF6–C/EBP-β interaction with the Dapk1 promoter requires ERK1/2 and C/EBP-β-T189.
ATF6–C/EBP-β Complex Binds to CRE/ATF Elements in Dapk1 Promoter.
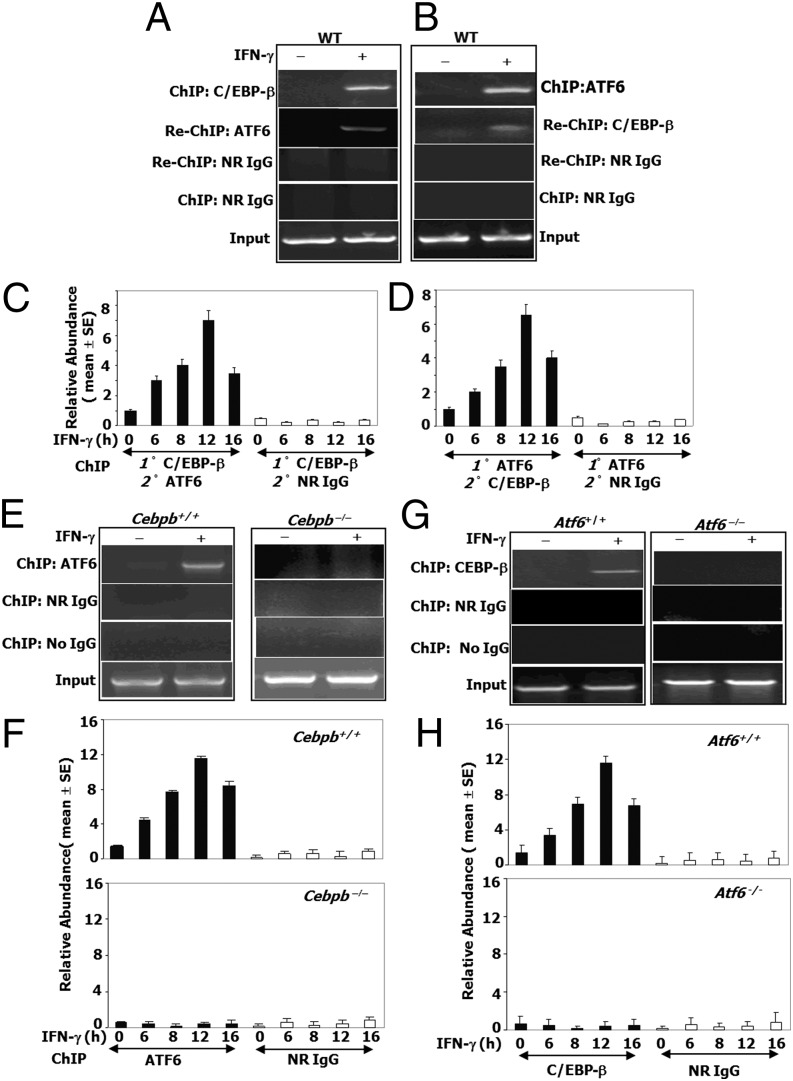
To address whether ATF6–C/EBP-β complex is recruited to Dapk1 promoter, we performed ChIP assays. We first immunoprecipitated the chromatin with a C/EBP-β antibody and then re-ChIPed the products with an ATF6 antibody and vice versa (Fig. 4 A and B). The reactions were Dapk1 promoter positive only upon IFN-γ treatment. As expected, PCR products were not detected in the control reactions where normal IgG (nonimmune) was used for re-ChIPing or for direct ChIP. Quantitative ChIP assays showed a time course of IFN-γ treatment (Fig. 4 C and D). Thus, both ATF6 and C/EBP-β are in the same promoter complex.
Fig. 4.
IFN-γ–induced ATF6–C/EBP-β complex binds to the Dapk1 promoter. ChIP assays with the indicated antibodies were performed as described under SI Materials and Methods using isogenic MEFs from WT (wild-type) and knockout mice lacking Atf6 or Cebpb. (A–D) Primary ChIP products (1°) were subjected to re-ChIP (2°) with the indicated antibodies. (A and B) Typical PCR patterns after ChIP and re-ChIP. IFN treatment was for 12 h. (C and D) qPCR analyses of the re-ChIP products with Dapk1 promoter-specific primers (n = 9 per sample). (E and G) ChIP assays. (F and H) A qPCR analysis of the ChIP products with Dapk1 promoter-specific primers (n = 9 per sample).
ATF6 and C/EBP-β Depend on Each Other for Binding to Dapk1 Promoter.
We next determined whether ATF6 and C/EBP-β are recruited to the Dapk1 promoter independently of each other after IFN-γ treatment with ChIP assays for ATF6 by using Cebpb+/+ and Cebpb−/− MEFs. IFN-γ stimulated ATF6 recruitment to the Dapk1 promoter in Cebpb+/+, but not in Cebpb−/− MEFs (Fig. 4E). Fig. 4F shows quantified ChIP data after a time course of IFN-γ treatment. Similarly, we investigated whether C/EBP-β recruitment to Dapk1 promoter depended on ATF6. ChIP assays revealed that C/EBP-β was recruited to Dapk1 promoter only in Atf6+/+, but not in Atf6−/− MEFs upon IFN-γ treatment (Fig. 4G). Fig. 4H shows kinetics of C/EBP-β recruitment as shown by a quantitative ChIP. Thus, the IFN-γ–stimulated recruitment of ATF6 and C/EBP-β to the Dapk1 promoter appears to be mutually dependent. Further, IFN-β also induced Dapk1 expression and the recruitment of ATF6 and C/EBP-β to the Dapk1 promoter (Fig. S4 D and E).
Functional ATF6 and C/EBP-β Are Critical for IFN-γ–Induced Autophagy.
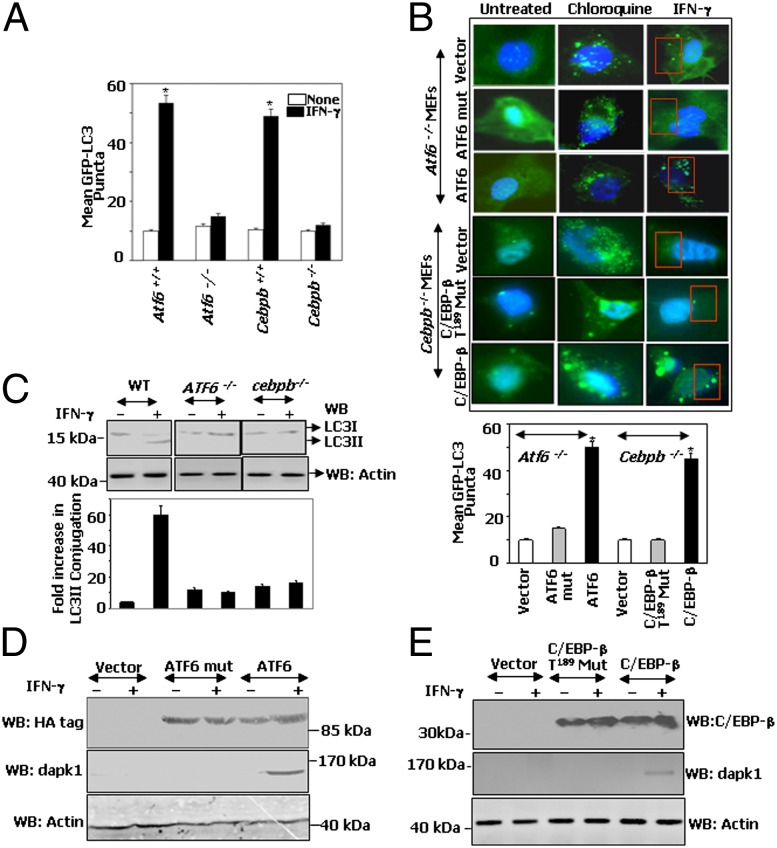
Because Atf6−/−MEFs showed very low levels of DAPK1 protein (Fig. S6A) and DAPK1 participates in autophagy (6, 7, 16), we next examined whether IFN-induced autophagy occurred in the absence of ATF6. Autophagy is characterized by redistribution of the microtubule-associated light chain 3 (LC3) protein into cytoplasmic puncta. To detect LC3 puncta, Atf6+/+, Atf6−/−, Cebpb+/+, or Cebpb−/− MEFs stably transfected with a GFP-LC3 expression vector (Fig. S6B) were treated with either IFN-γ or chloroquine (a positive control). Higher numbers of LC3 puncta were found in Atf6+/+ and Cebpb+/+ MEFs upon IFN-γ treatment compared with Atf6−/− or Cebpb−/− MEFs (Fig. 5A and Fig. S6B). We next performed a rescue experiment by transfecting these cells with corresponding wild-type or mutant versions of the missing gene products. IFN-γ failed to induce the puncta formation when the empty vector or ATF6mut was expressed into Atf6−/− cells (Fig. 5B). However, ATF6 restoration caused a significant rise in LC3 puncta in response to IFN-γ. Similarly, when Cebpb−/− cells were transfected with either an empty vector or C/EBP-β T189A mutant (which failed to interact with ATF6 upon IFN-γ treatment; see Fig. 3D), fewer LC3 puncta were observed (Fig. 5B). Restoration of wild-type C/EBP-β alone established IFN-γ−induced autophagy as observed by a significant increase in the number of LC3 puncta (Fig. 5B). Chloroquine (which blocks the fusion of autophagosomes with lysosomes) promoted LC3 puncta formation irrespective of ATF6 or C/EBP-β status, showing that there is no global deficiency of autophagy in these cells (Fig. 5B). Mean GFP-LC3 puncta formed upon IFN-γ treatment in each case (Fig. 5B, Bottom) showed significant differences. Alternatively, formation of LC3 II (generated by proteolysis and lipidation of LC3 I) upon IFN-γ treatment in WT, Atf6−/−, and Cebpb−/− MEFs was analyzed by Western blotting (Fig. 5C). The fold increase in LC3II levels (Fig. 5C, Lower), correlated well with puncta formation. Formation of LC3II in response to IFN-γ was significantly lost in the absence of either C/EBP-β or ATF6. Furthermore, formation of IFN-induced auotphagic puncta by endogenous LC3 was inhibited by 3-methyl adenine (Fig. S5A). Importantly, up-regulation of IFN-γ–induced autophagic response correlated with induction of DAPK1 levels in the presence of functional ATF6 (Fig. 5D) or C/EBP-β (Fig. 5E), but not by ATF6mut or C/EBP-β T189A mutant. Thus, functional ATF6 and C/EBP-β are necessary for IFN-γ–stimulated Dapk1 expression and induction of autophagy.
Fig. 5.
Functional ATF6 and C/EBP-β are critical for IFN-γ–induced autophagy. (A–C) MEFs stably transfected with GFP-LC3 were treated as indicated. (A) LC3 puncta were quantified and expressed as mean intensities (*P < 0.001, n = 10) (B) LC3 puncta formation in the presence of specific restoration vectors. (B Lower) LC3 puncta quantification (*P < 0.001, n = 10). (C) Indicated cell lines were treated with IFN-γ, and proteins lysates were subjected to Western blot analyses with specific antibodies. The fold LC3II formation was quantified (*P < 0.001). (D and E) Western blot analyses of lysates from Atf6−/− and Cebpb−/− MEFs, respectively, transfected with the indicated constructs.
Increased Mortality of Atf6−/− Mice upon Challenge with Bacillus anthracis (BA).
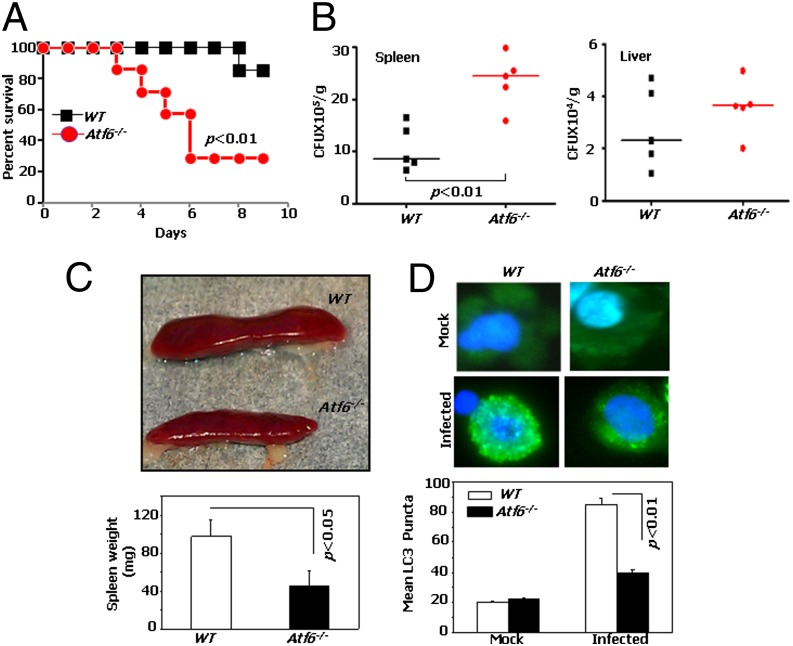
Because IFN-γ is a critical component of host defense against BA (17), we next investigated whether mice lacking ATF6 were sensitive to BA infection. Age-matched WT and Atf6−/− mice were infected i.p. with Gram-positive BA Sterne 34F2 spores (1 × 106 spores per mouse, a sublethal dose to WT mice) and monitored for signs of infection and survival. A significant percentage of Atf6−/− mice succumbed (71.5% mortality; P < 0.01) to infection, compared with the WT mice, which exhibited only 14.3% mortality after infection by the end of 9 d (Fig. 6A). Notably, mice started to die as early as the third day of infection in the Atf6−/− group, compared with day 8 in the WT group. To identify the bases for differences between Atf6−/− and WT mice in their responses to BA infection, bacterial loads were measured 4 d after infection. A significantly higher and a moderate increase in bacterial burden was observed in the spleens and livers, respectively, in Atf6−/− mice compared with the WT mice (Fig. 6B). The increased bacterial load in spleen corresponded to a dramatically diminished spleen size in Atf6−/− mice (Fig. 6C). Further, peritoneal macrophages collected from Atf6−/− mice showed significantly fewer numbers of LC3 puncta (P < 0.01) than those from WT mice (Fig. 6D). Among other bacteria, Salmonella typhimurium is also controlled by autophagy in the infected cells (18). We verified this observation by infecting WT and Atf6−/− MEFs with S. typhimurium. Unlike the WT MEFs, Atf6−/− MEFs formed significantly lower numbers of LC3 puncta upon infection with Salmonella in cell culture (Fig. S6C). Taken together, our findings suggest that ATF6 may function in the defense against microbial pathogens via induction of autophagy.
Fig. 6.
Atf6−/− mice are highly susceptible to lethal infection by B. anthracis. (A) Atf6−/− and WT mice (n = 7 mice per group) were challenged with BA Sterne Spores, were performed, and percent survival (Kaplan–Meier analyses) was plotted. (B) Mice were infected with 1 × 106 BA Sterne 34F2 spores i.p., and bacterial titers in organs (cfu/g of tissue) were determined at 96 hpi. Lines indicate median values. (C) Spleens of Atf6−/− and WT mice were shown at 96 hpi. C Lower shows quantification of the same (n = 5 per group); error bars represent SD. (D) Endogenous LC3 puncta formation in peritoneal macrophages of Atf6−/− and WT mice at 96 hpi. (D Lower) LC3 puncta quantification (n = 10 fields) from three independent mice per group. *P < 0.01.
Discussion
DAPK1 suppresses tumor growth and metastasis via induction of apoptosis and autophagy (5, 7, 16). DAPK1 is also a transcriptional target of p53 and p19ARF (6). In certain nonsmall cell lung cancer and neuroblastoma cell lines, DAPK1 is down-regulated by RASSF1A (19). Earlier, we have identified that the expression of Dapk1 critically depends on transcription factor C/EBP-β through a CRE/ATF element (4).
C/EBP-β is a major regulator of metabolism (3), cell differentiation, growth, immune responses, neoplastic growth, development of the reproductive system (9), and pro- and antigrowth pathways (20). Given these diverse activities, it is conceivable that C/EBP-β association with different cellular factors in a gene context and signal-specific manner may regulate it. Indeed, C/EBP-β can interact with transcription factors outside its family (3). Here, we have provided evidence for a unique pathway in which IFN-γ–stimulated proteolytic processing of ATF6 and ERK1/2-induced phosphorylation of C/EBP-β converge on and up-regulate Dapk1. ATF6 induces the XBP1 mRNA (coding for a transcription factor) in response to ER stress. Interestingly, in Xbp1−/− cells, expression of C/EBP-β is suppressed (21). An XBP1-binding site in the 3′ UTR of Cebpb is necessary for inducing transcription (21).
A direct role for ATF6 in IFN-induced Dapk1 expression was supported by (i) binding of ATF6 to CRE/ATF in Dapk1 promoter (Figs. 1 and 4); (ii) loss of DAPK1 induction in the presence of ATF6-specific shRNAs (Fig. 1); and (iii) restoration of DAPK1 levels and autophagy after expression of ATF6 in Atf6−/− cells (Fig. 5). Because the IFN-induced expression of Irf1 (22) was unaffected by the loss of either ATF6 (Fig. 1) or C/EBP-β (4), we believe there was no global loss of IFN-γ response in these cells. A number of stress conditions disrupt ER homeostasis and cause the activation of ATF6 (23). Indeed, IFN-γ has been reported to induce ER stress in oligodendrocytes, although the mechanisms are unclear (24). Thyroid cells undergoing ER stress stimulate IFN-γ production by NK cells (25). Interestingly, like the ER stress inducers, IFN-γ caused a dissociation of ATF6 from BiP and migration to the nucleus (Fig. 2). However, unlike the classical ER stress response, ATF6 required C/EBP-β to drive Dapk1 expression (Fig. 4). Although ATF6 exhibits some DNA-binding activity, it is known to partner with other DNA-binding proteins, such as NF-Y, YY1, and SRF, to modulate gene expression in other pathways (26, 27). ATF6/NF-Y and ESRE interactions are critical for transcriptional induction of not only ER chaperones but also of CHOP and XBP-1 (13). Whether IFN-γ crosses into these pathways remains unknown. The IFN-γ–induced activation of ATF6 is a mechanism of proteolytic processing of a transcription factor for this cytokine. Previously, an ER stress pathway that up-regulates CHOP (a target of ATF6), which plays a key role in nitric oxide-mediated apoptosis in murine macrophages, was reported (28). These studies, however, did not identify the mechanism(s) involved.
We have reported the critical dependence of an ERK1/2 phosphorylation site of C/EBP-β for transcriptional induction of Dapk1 and other genes earlier (4). The disruption of this site abolished IFN-γ–stimulated C/EBP-β interaction with ATF6 (Fig. 3). Consistent with this result, IFN-γ–stimulated recruitment of both ATF6 and C/EBP-β to Dapk1 promoter is severely ablated in ERK1/2-depleted cells (Fig. 3). Interestingly, IFN-γ stimulated ATF6–C/EBP-β interactions occurred in cells lacking either STAT1 or JAK1 (Fig. S4 B and C), indicating their independence from these classic IFN-regulated signaling factors. These interactions were slightly weaker (1.5-fold less) in the absence of JAK1 (U4A), which did not recover even after the rescue of JAK1 (U4A/R).
Our studies demonstrated C/EBP-β is critical for IFN-induced growth-suppressive pathway via DAPK1 (antigrowth pathway), whereas other studies showed Ras-mediated activation of C/EBP-β as important for keratinocyte survival (progrowth pathway) and skin tumorigenesis (20). These diverse effects of C/EBP-β on cell growth can now be explained in part by our current studies. ATF6-C/EBP-β promotes a growth-suppressive pathway, whereas C/EBP-β homodimers may promote growth response (29) in a signal and cell type-dependent manner. Importantly, ATF6 and C/EBP-β recruitment to the Dapk1 promoter depends on the presence of each other (Fig. 4). Such a mechanism is analogous to STAT1/STAT2 interaction with the ISREs in response to type I IFNs (2).
DAPK1 is implicated in early steps of autophagy (6, 7, 16). Indeed, DAPK1 constitutes a critical integration point in ER stress signaling and cell death that transmits these signals into two distinct directions, caspase activation and autophagy (16). DAPK1 requires Ca2+/Calmodulin for its activity and associates with cytoskeleton during cell death induction (6). It is likely that although ER membranes support the formation of autophagophores, Ca2+ derived from ER stores could activate DAPK1 and promote maturation of autophagosomes. Indeed, a recent study showed that DAPK1 binds to another regulator of autophagy, microtubule-associated protein 1B (MAP1B), and depletion of MAP1B attenuated DAPK-1–stimulated autophagy (30). MAP1B further recruits LC3 II (also known as ATG8), which permits the maturation of autophagic vesicles. It is clear that a number of steps in this process are signal-dependent. For example, Beclin1 is phosphorylated by DAPK1 to drive autophagy (31). Consistent with these observations, we found defective autophagy in cells lacking either ATF6 or C/EBP-β (Fig. 5 and Figs. S5 and S6). This notion is further bolstered by our observations that the Atf6−/− mice are relatively hypersensitive to bacterial infection-induced death (Fig. 6), even at sublethal doses. Such defective protection against infection appears in part due to a significantly poorer execution of autophagy by the Atf6−/− macrophages than the WT cells. Infection and comparison of WT and Atf6−/− MEFs in vitro with Salmonella also yielded very similar observations. As our paper was being revised, an independent group reported proteolytic activation of ATF6 and ER stress after infection with Listeria monocytogenes (32). Cells infected with Listeria also mount autophagy as a defense mechanism (33). Thus, ATF6 appears to be intimately associated with the autophagy. Interestingly, one of the major phenotypes of the Cebpb−/− mice is their inability to inhibit the multiplication of intracellular bacteria and kill heterologous tumors (10). If one closely observes the electron micrographs reported by Tanaka et al. (10), they seem more like defects in autophagy rather than phagosomes as suggested by the authors. Our data showing C/EBP-β-T189 and ERK1/2 requirement for IFN induced autophagosome formation (Fig. 5) consistent with the report of Tanaka et al. Importantly, although a connection between ER stress and autophagy has been suggested (34), how ER stress is coupled to autophagy is unknown. The participation of ATF6 in autophagy (this report) establishes this critical missing link.
Autophagy is crucial for a number of diverse physiological responses, including elimination of intracellular bacteria, viruses and parasitic pathogens, damaged organelles, antigen presentation, tumor suppression, and macrophage-dependent clearance of apoptotic cells (35, 36). It is interesting to note that IFNs play a major role in all these processes. The, IFN-γ–ATF6-C/EBP-β–DAPK1 axis may explain a route of control for regulating tumor and bacterial growth via autophagy.
Materials and Methods
All gene expression, Western blot, RNAi, transfection, ChIP assays, and quantitiative PCR analyses were performed as described in our earlier reports (4). ATF6-kncokout mice in the C57BL/6 background were described (37). All reagents and detailed methods and results are described in SI Materials and Methods and SI Results.
Supplementary Material
Acknowledgments
We thank Dr. Shreeram Nallar for a critical reading of this manuscript and Sharon Tennant for providing Salmonella enteria strain. These studies are supported by National Institutes of Health Grants CA78282 (to D.V.K.), DK077140 (to M.A.R.), and NIH2454AI057168-06 (to A.S.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119273109/-/DCSupplemental.
References
- 1.Gresser I, Belardelli F. Endogenous type I interferons as a defense against tumors. Cytokine Growth Factor Rev. 2002;13:111–118. doi: 10.1016/s1359-6101(01)00035-1. [DOI] [PubMed] [Google Scholar]
- 2.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Li H, Gade P, Xiao W, Kalvakolanu DV. The interferon signaling network and transcription factor C/EBP-beta. Cell Mol Immunol. 2007;4:407–418. [PMC free article] [PubMed] [Google Scholar]
- 4.Gade P, Roy SK, Li H, Nallar SC, Kalvakolanu DV. Critical role for transcription factor C/EBP-beta in regulating the expression of death-associated protein kinase 1. Mol Cell Biol. 2008;28:2528–2548. doi: 10.1128/MCB.00784-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deiss LP, Feinstein E, Berissi H, Cohen O, Kimchi A. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 1995;9:15–30. doi: 10.1101/gad.9.1.15. [DOI] [PubMed] [Google Scholar]
- 6.Bialik S, Kimchi A. The death-associated protein kinases: Structure, function, and beyond. Annu Rev Biochem. 2006;75:189–210. doi: 10.1146/annurev.biochem.75.103004.142615. [DOI] [PubMed] [Google Scholar]
- 7.Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol. 2002;157:455–468. doi: 10.1083/jcb.200109094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raval A, et al. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell. 2007;129:879–890. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sterneck E, Tessarollo L, Johnson PF. An essential role for C/EBPbeta in female reproduction. Genes Dev. 1997;11:2153–2162. doi: 10.1101/gad.11.17.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka T, et al. Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- 11.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida H, et al. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, et al. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- 15.Shen J, Prywes R. Dependence of site-2 protease cleavage of ATF6 on prior site-1 protease digestion is determined by the size of the luminal domain of ATF6. J Biol Chem. 2004;279:43046–43051. doi: 10.1074/jbc.M408466200. [DOI] [PubMed] [Google Scholar]
- 16.Gozuacik D, et al. DAP-kinase is a mediator of endoplasmic reticulum stress-induced caspase activation and autophagic cell death. Cell Death Differ. 2008;15:1875–1886. doi: 10.1038/cdd.2008.121. [DOI] [PubMed] [Google Scholar]
- 17.Kang TJ, et al. Murine macrophages kill the vegetative form of Bacillus anthracis. Infect Immun. 2005;73:7495–7501. doi: 10.1128/IAI.73.11.7495-7501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 19.Agathanggelou A, et al. Identification of novel gene expression targets for the Ras association domain family 1 (RASSF1A) tumor suppressor gene in non-small cell lung cancer and neuroblastoma. Cancer Res. 2003;63:5344–5351. [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu S, Yoon K, Sterneck E, Johnson PF, Smart RC. CCAAT/enhancer binding protein-beta is a mediator of keratinocyte survival and skin tumorigenesis involving oncogenic Ras signaling. Proc Natl Acad Sci USA. 2002;99:207–212. doi: 10.1073/pnas.012437299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C, Dudenhausen EE, Pan YX, Zhong C, Kilberg MS. Human CCAAT/enhancer-binding protein beta gene expression is activated by endoplasmic reticulum stress through an unfolded protein response element downstream of the protein coding sequence. J Biol Chem. 2004;279:27948–27956. doi: 10.1074/jbc.M313920200. [DOI] [PubMed] [Google Scholar]
- 22.Pine R, Canova A, Schindler C. Tyrosine phosphorylated p91 binds to a single element in the ISGF2/IRF-1 promoter to mediate induction by IFN alpha and IFN gamma, and is likely to autoregulate the p91 gene. EMBO J. 1994;13:158–167. doi: 10.1002/j.1460-2075.1994.tb06245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutkowski DT, Kaufman RJ. A trip to the ER: Coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 24.Lin W, et al. Interferon-gamma inhibits central nervous system remyelination through a process modulated by endoplasmic reticulum stress. Brain. 2006;129:1306–1318. doi: 10.1093/brain/awl044. [DOI] [PubMed] [Google Scholar]
- 25.Ulianich L, et al. ER stress impairs MHC Class I surface expression and increases susceptibility of thyroid cells to NK-mediated cytotoxicity. Biochim Biophys Acta. 2011;1812:431–438. doi: 10.1016/j.bbadis.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 26.Zhu C, Johansen FE, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, et al. ATF6 as a transcription activator of the endoplasmic reticulum stress element: Thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol Cell Biol. 2000;20:5096–5106. doi: 10.1128/mcb.20.14.5096-5106.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gotoh T, Oyadomari S, Mori K, Mori M. Nitric oxide-induced apoptosis in RAW 264.7 macrophages is mediated by endoplasmic reticulum stress pathway involving ATF6 and CHOP. J Biol Chem. 2002;277:12343–12350. doi: 10.1074/jbc.M107988200. [DOI] [PubMed] [Google Scholar]
- 29.Lee S, et al. RSK-mediated phosphorylation in the C/EBPbeta leucine zipper regulates DNA binding, dimerization, and growth arrest activity. Mol Cell Biol. 2010;30:2621–2635. doi: 10.1128/MCB.00782-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrison B, et al. DAPK-1 binding to a linear peptide motif in MAP1B stimulates autophagy and membrane blebbing. J Biol Chem. 2008;283:9999–10014. doi: 10.1074/jbc.M706040200. [DOI] [PubMed] [Google Scholar]
- 31.Zalckvar E, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pillich H, Loose M, Zimmer KP, Chakraborty T. Activation of the unfolded protein response by Listeria monocytogenes. Cell Microbiol. 2012;14:949–964. doi: 10.1111/j.1462-5822.2012.01769.x. [DOI] [PubMed] [Google Scholar]
- 33.Rich KA, Burkett C, Webster P. Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol. 2003;5:455–468. doi: 10.1046/j.1462-5822.2003.00292.x. [DOI] [PubMed] [Google Scholar]
- 34.Yorimitsu T, Klionsky DJ. Endoplasmic reticulum stress: A new pathway to induce autophagy. Autophagy. 2007;3:160–162. doi: 10.4161/auto.3653. [DOI] [PubMed] [Google Scholar]
- 35.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 36.Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto K, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.