Abstract
Studies of the familial Parkinson disease-related proteins PINK1 and Parkin have demonstrated that these factors promote the fragmentation and turnover of mitochondria following treatment of cultured cells with mitochondrial depolarizing agents. Whether PINK1 or Parkin influence mitochondrial quality control under normal physiological conditions in dopaminergic neurons, a principal cell type that degenerates in Parkinson disease, remains unclear. To address this matter, we developed a method to purify and characterize neural subtypes of interest from the adult Drosophila brain. Using this method, we find that dopaminergic neurons from Drosophila parkin mutants accumulate enlarged, depolarized mitochondria, and that genetic perturbations that promote mitochondrial fragmentation and turnover rescue the mitochondrial depolarization and neurodegenerative phenotypes of parkin mutants. In contrast, cholinergic neurons from parkin mutants accumulate enlarged depolarized mitochondria to a lesser extent than dopaminergic neurons, suggesting that a higher rate of mitochondrial damage, or a deficiency in alternative mechanisms to repair or eliminate damaged mitochondria explains the selective vulnerability of dopaminergic neurons in Parkinson disease. Our study validates key tenets of the model that PINK1 and Parkin promote the fragmentation and turnover of depolarized mitochondria in dopaminergic neurons. Moreover, our neural purification method provides a foundation to further explore the pathogenesis of Parkinson disease, and to address other neurobiological questions requiring the analysis of defined neural cell types.
Keywords: autophagy, fission, FACS, glia, flow cytometry
Parkinson disease (PD) is a common age-related movement disorder that exhibits selective degeneration of dopaminergic (DA) neurons in the substantia nigra. Our understanding of the molecular mechanisms underlying PD has been tremendously influenced by the discovery of genes involved in heritable forms of this disorder (1, 2). Studies of two of these genes, PINK1 and parkin, have strengthened the hypothesis that mitochondrial dysfunction may be a primary cause of PD (3) and have led to a model to explain how PINK1 and Parkin influence mitochondrial and neuronal integrity (4, 5). This model posits that PINK1, a mitochondrially targeted serine/threonine kinase, is selectively stabilized on the surface of depolarized mitochondria, where it recruits Parkin, a cytosolic ubiquitin ligase (6–8). Parkin then ubiquitinates the mitochondrial fusion-promoting factor Mitofusin (7, 9–13) and other mitochondrial proteins (14–16), leading to the fragmentation and autophagic turnover of the depolarized mitochondria (10, 17). Although there is substantial support for this model from studies involving nonneuronal cell lines and tissues, whether the PINK1/Parkin pathway plays this role in neurons under physiological conditions remains controversial (18, 19).
To test the hypothesis that PINK1 and Parkin promote the fragmentation and autophagic turnover of depolarized mitochondria in DA neurons, we developed a method to purify and characterize neural subtypes of interest from the adult Drosophila brain. Drosophila is an excellent model system for these studies because, like humans, PINK1 and parkin mutant flies exhibit selective degeneration of DA neurons (20, 21). Our neural purification method involves marking the neural cell types of interest with GFP using the UAS/GAL4 system (22), followed by tissue dissociation and FACS to analyze and purify the GFP+ cells. Using this method, we demonstrate that DA neurons purified from parkin mutants accumulate enlarged depolarized mitochondria, and that PINK1 overexpression increases the frequency of mitochondria with highly polarized membranes. We also find that the mitochondrial membrane potential (MMP) defect and neurodegenerative phenotype of parkin mutants are rescued by genetic manipulations that promote mitochondrial fragmentation and turnover. In contrast, we detect a less pronounced effect of parkin deficiency on mitochondrial morphology and MMP in cholinergic neurons, suggesting that mitochondria are damaged at higher rates in DA neurons, or that alternative mitochondrial repair or turnover pathways are more limited in this cell type. Taken together, our findings support the model that PINK1 and Parkin influence DA neuron integrity by inducing the fragmentation and turnover of depolarized mitochondria, and that mitochondria in DA neurons are particularly vulnerable to loss of the PINK1/Parkin pathway.
Results
Development of a Method to Purify and Characterize Specific Neural Cell Types from the Adult Drosophila Brain.
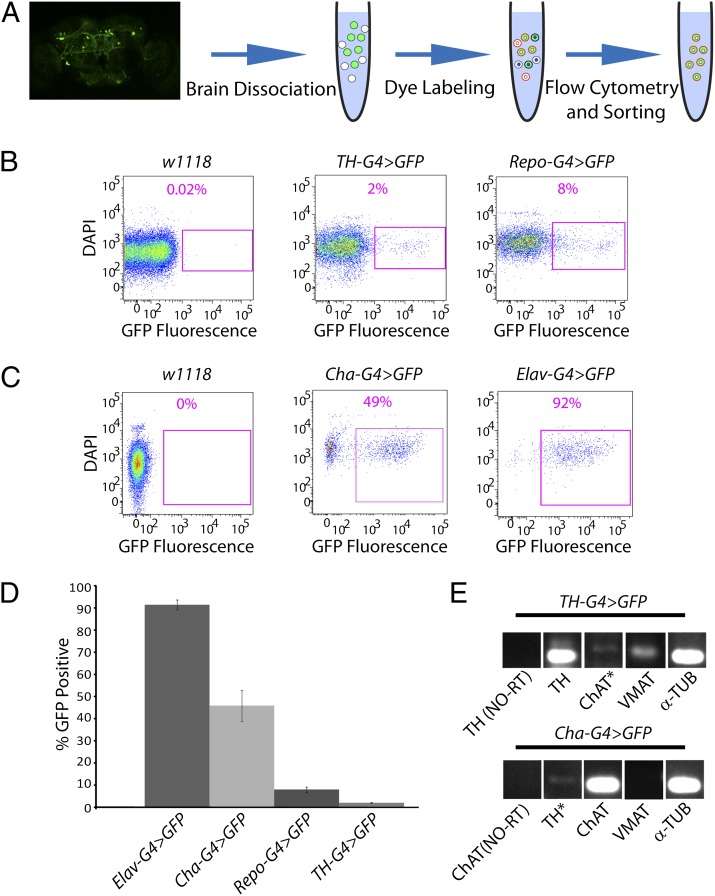
To test the model that DA neurons lacking Parkin degenerate because of a failure to fragment and degrade depolarized/damaged mitochondria, we developed a method to purify and characterize living DA neurons from the adult Drosophila brain (Fig. 1A). We dissected and dissociated the brains from flies selectively expressing GFP in DA neurons and treated the resulting cell suspension with the DNA-binding dye 4′,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI). Forward scatter selection of this cell suspension using flow cytometry revealed two distinct particle size distributions: a population of small particles, of which a high proportion exhibit bright DAPI fluorescence (DAPI+); and a population of larger particles, most of which stain weakly with DAPI (DAPI−) (Fig. S1A). The size range of the latter population is in accordance with the size of neuronal cell bodies in Drosophila (Fig. S1B) (23). Given that DAPI preferentially stains dead cells, our findings suggest that the small DAPI+ particles represent cell debris and dead cells, whereas the population of larger DAPI− particles represents living cells. All of our subsequent analyses involve only the larger DAPI− cell population.
Fig. 1.
A method for purification and analysis of neural subsets from the adult Drosophila brain. (A) Adult fly brains expressing GFP (green) in a cell type of interest are dissociated and labeled with the cell viability dye DAPI, as well as a MMP-dependent dye. The resulting cell suspension contains dead cells staining brightly for DAPI (white and green cells with blue nuclei), but lacking MMP-dependent staining, and living cells staining less brightly for DAPI (white and green cells with white nuclei) that exhibit MMP-dependent staining (red outlines). (Magnification: 20×.) (B) Neural preparations from nontransgenic flies (w1118) and from flies expressing GFP in DA neurons (TH-G4 > GFP) or glia (Repo-G4 > GFP) were analyzed by flow cytometry. Representative dot plots depict the DAPI−/GFP+ cell populations (purple boxed regions) and their frequency of abundance. (C) Neural preparations from nontransgenic flies (w1118) and from flies expressing GFP in CH neurons (Cha-G4 > GFP) or pan-neuronally (Elav-G4 > GFP) were analyzed by flow cytometry. Representative dot plots are as described in B. (D) The mean abundance of the indicated cell types is depicted. The number of biological replicates (n) and total number of cells analyzed (N) were as follows: Elav-G4 > GFP (n = 4; N = 12,563); CH-G4 > GFP (n = 5; N = 2,432); Repo-G4 > GFP (n = 4; N = 2,286); TH-G4 > GFP (n = 5; N = 842). (E) DA and CH neurons isolated by FACS were subjected to RT-PCR to detect transcripts corresponding to tyrosine hydroxylase (TH), choline acetyltransferase (ChAT), vesicular monoamine transporter (VMAT), and α-tubulin (α-TUB). RT-PCR reactions lacking reverse transcriptase (NO-RT) represent negative controls. Asterisks denote lanes with nonspecific bands of the incorrect size. The specific genotypes of the animals used in this and all other figures are fully defined in SI Materials and Methods.
To distinguish DA neurons from autofluorescent cells, we compared the GFP fluorescence distribution between suspensions prepared from animals expressing GFP selectively in DA neurons, and those prepared from nontransgenic control flies lacking GFP expression. This comparison enabled us to define gating parameters that largely distinguish GFP+ cells from autofluorescent cells, without overly compromising GFP detection efficiency. Applying these gating parameters to cells isolated from flies expressing GFP in DA neurons revealed that 2.0 ± 0.3% of the DAPI− population was GFP+ (Fig. 1 B and D). Similar results were obtained using independent GFP reporters, including cytosolic, plasma membrane bound, and mitochondrially localized forms of GFP (Fig. S1C). In contrast, only 0.04 ± 0.03% of the DAPI− cells derived from a nontransgenic control strain met the criteria of being GFP+ using identical laser and gating parameters (Fig. 1B). Thus, our gating parameters allow for the isolation of DA neurons with only 2% contamination by autofluorescent cells.
To explore the versatility of our neural purification method, we prepared and analyzed cell suspensions from flies expressing GFP under the control of glial, cholinergic (CH) neuron, and pan-neuronal specific GAL4 drivers. Our analyses of cell populations from flies bearing the glial GAL4 driver used gating parameters defined in experiments involving DA neurons to distinguish GFP+ from autofluorescent cells (Fig. 1B). However, in our analyses of cell populations from flies bearing the CH neuron and pan-neuronal drivers, we were able to define more stringent gating parameters by lowering laser voltage settings because of the relatively brighter GFP fluorescence generated from these drivers (Fig. 1C). We found that 7.9 ± 1.2%, 45.8 ± 6.9%, and 91.4 ± 2.2% of the DAPI− cell populations from flies bearing the glial, CH neuron, and pan-neuronal-specific GAL4 drivers, respectively, were GFP+ (Fig. 1D). Comparing these values to those obtained from applying the same gating parameters to nontransgenic control-cell populations indicated that contamination by autofluorescent cells was less than 0.5% in the purified glial cell preparation, and undetectable in the purified CH neuron and pan-neuronal preparations. These results demonstrate that our neural purification method is applicable to a variety of neural cell types.
To verify that the DAPI−/GFP+ cells purified using our method were indeed the cell types of interest, we performed RT-PCR to detect transcripts specific to DA and CH neurons. Transcripts corresponding to tyrosine hydroxylase and the vesicular monoamine transporter could be detected in isolated DA neurons, but not in CH neurons (Fig. 1E). Conversely, transcripts corresponding to choline acetyltransferase could be detected in isolated CH, but not DA neurons (Fig. 1E).
PINK1/Parkin Pathway Influences Mitochondrial Membrane Potential in DA Neurons.
Studies in cultured cell lines indicate that the PINK1/Parkin pathway promotes the selective turnover of depolarized mitochondria (17). A prediction of this model is that DA neurons from parkin mutants should accumulate depolarized mitochondria. To address this prediction, we first tested whether the mitochondria in our cell preparations maintained a membrane potential. We stained a cell suspension with the MMP-dependent dye tetramethylrhodamine ethyl ester (TMRE) before flow analysis and observed robust labeling in the DAPI− cells, but little to no staining in the DAPI+ cells (Fig. S2A). These findings suggest that mitochondria in the DAPI− cells maintain a membrane potential, whereas those in the DAPI+ cell population do not, confirming that the DAPI− and DAPI+ cells represent living and dead neurons, respectively. To control for the MMP dependency of dye labeling, we treated the cell suspensions with the mitochondrial uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) to dissipate the MMP before dye staining and flow analysis. Although the DAPI− DA neurons in the vehicle- (DMSO) treated preparations exhibited robust staining with both TMRE and another potentiometric dye, Mitotracker Deep Red (Mt-DR), those in the CCCP-treated cell suspensions displayed greatly reduced TMRE and Mt-DR fluorescence (Fig. S2B). Taken together, our findings demonstrate that TMRE and Mt-DR staining depends on MMP, and that the DAPI− DA neurons purified using our method bear mitochondria with an intact MMP.
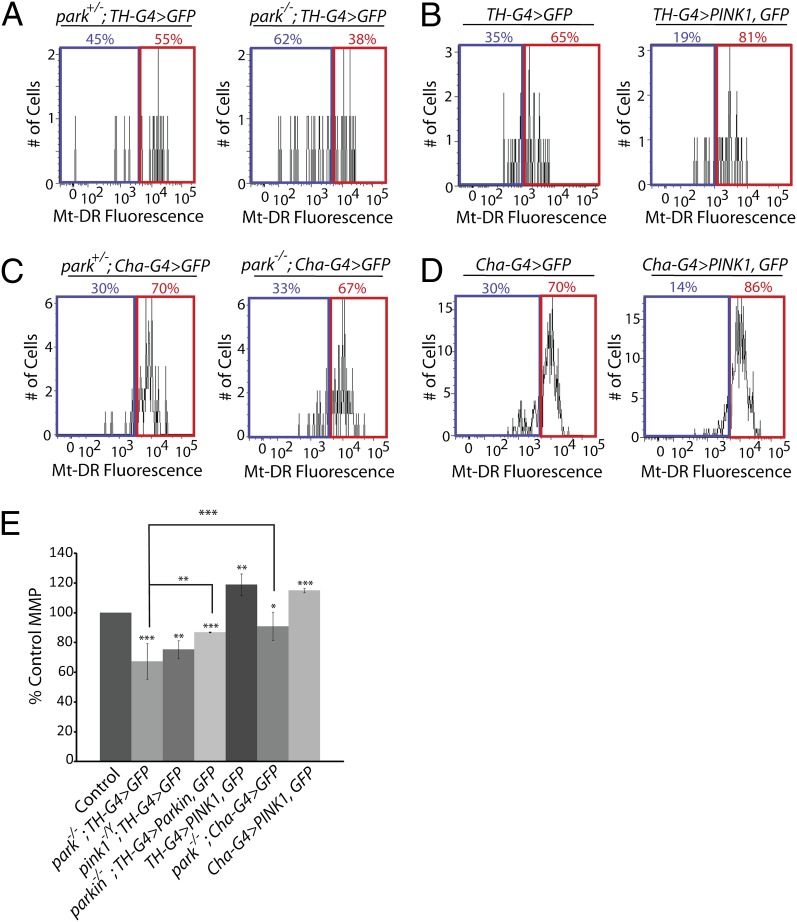
We next used our neural purification method to compare MMP in DA neurons obtained from parkin mutants and controls. DA neurons from parkin mutants exhibited a significant reduction in the mean Mt-DR fluorescence intensity relative to controls (Fig. 2 A and E), and this result was confirmed using TMRE (Fig. S3). The decreased Mt-DR fluorescence intensity of DA neurons from parkin mutants was rescued by transgenic expression of Parkin, demonstrating that this phenotype is a cell autonomous consequence of parkin deficiency. We also detected a similar decrease in Mt-DR fluorescence intensity in DA neurons from PINK1 mutants (Fig. 2E). Although these findings suggest that DA neurons from PINK1 and parkin mutants accumulate depolarized mitochondria, they are also consistent with a decrease in mitochondrial abundance. To distinguish between these possibilities we performed two experiments. First, we compared mitochondrial DNA (mtDNA) abundance in DA neurons from parkin mutants and controls using quantitative PCR. We found that DA neurons from parkin mutants contain more mtDNA than heterozygous controls (Fig. S4A), suggesting that mitochondrial abundance may actually be elevated in parkin mutants relative to controls. Second, we used the fluorescence intensity of mitochondrially-targeted GFP (mito-GFP) to compare mitochondrial abundance in DA neurons from parkin mutants and controls. Although we did not detect a correlation between Mt-DR fluorescence intensity and mitochondrial abundance in DA neurons (Fig. S4B), the mito-GFP fluorescence intensity of DA neurons from parkin mutants was modestly decreased relative to controls (Fig. S4C). However, the magnitude of the decrease in mito-GFP fluorescence intensity of DA neurons from parkin mutants (5%) was substantially smaller than the magnitude of the decrease in Mt-DR fluorescence intensity (33%) between parkin mutants and controls. Taken together, our findings indicate that an increase in the fraction of depolarized mitochondria is the main contributor to the decreased Mt-DR and TMRE fluorescence intensity of DA neurons from parkin mutants. The seemingly paradoxical findings of increased mtDNA abundance and decreased mito-GFP fluorescence intensity in DA neurons from parkin mutants may be explained by decreased import of mito-GFP into depolarized mitochondria.
Fig. 2.
The PINK1/Parkin pathway influences MMP in neurons. (A–D) Representative histograms show the percentage of neurons that exhibit Mt-DR fluorescence intensities above (red boxes) and below (blue boxes) the assigned cut-off intensity value determined by flow cytometry analysis of the following animals: (A) parkin-null heterozygote controls and sibling parkin-null homozygotes expressing GFP in DA neurons (park+/−; TH-G4 > GFP and park−/−; TH-G4 > GFP, respectively). (B) Control flies lacking PINK1 overexpression (TH-G4 > GFP) and siblings overexpressing PINK1 in DA neurons (TH-G4 > PINK1, GFP). (C) parkin-null heterozygote controls and sibling parkin-null homozygotes expressing GFP in CH neurons (park+/−; Cha-G4 > GFP and park−/−; Cha-G4 > GFP, respectively). (D) Control flies lacking PINK1 overexpression in CH neurons (Cha-G4 > GFP), and siblings overexpressing PINK1 (Cha-G4 > PINK1, GFP). (E) Mean MMP in DA or CH neurons from animals of the indicated genotypes relative to their respective sibling controls. The number of biological replicates (n) and total number of cells analyzed (N) were as follows: park−/−; TH-G4 > GFP (n = 4; N = 230); pink1−/Y; TH-G4 > GFP (n = 3; N = 353); park−/−; TH-G4>Parkin, GFP (n = 3; N = 156); park−/−; Cha-GAL4 > GFP (n = 4; N = 6,434); TH-G4 > PINK1, GFP (n = 4; N = 258); Cha-G4 > PINK1, GFP (n = 4; N = 4,857). Statistical tests used in this work are described in Materials and Methods. For all experiments and figures: *P < 0.05, **P < 0.01, ***P < 0.001.
Another prediction of the model that the PINK1/Parkin pathway promotes the selective turnover of depolarized mitochondria is that activating this pathway should preferentially eliminate those mitochondria with the lowest MMP, and thus shift the MMP distribution of DA neurons toward higher potentials. Because previous work has shown that PINK1 overexpression is sufficient to induce the recruitment of Parkin to mitochondria and their subsequent turnover (6, 8), we explored the effects of PINK1 overexpression on MMP in DA and CH neurons. We found that PINK1 overexpression significantly increased the mean MMP of both wild-type DA and CH neurons, without obviously changing maximal MMP values, suggesting that those mitochondria with the lowest MMP were selectively eliminated upon activation of the PINK1/Parkin pathway (Fig. 2 B, D, and E). Taken together, our findings are consistent with the model that the PINK1/Parkin pathway promotes the selective turnover of depolarized mitochondria in DA neurons.
The PINK1/Parkin Pathway Selectively Influences the Integrity of Mitochondria in DA Neurons.
Mutations in parkin selectively affect the survival of DA neurons in the human and fly brain (21, 24). This selective vulnerability may involve a specific effect of parkin deficiency on the integrity of mitochondria in DA neurons, or increased sensitivity of DA neurons to a mitochondrial insult that is similar in magnitude in all neurons. To distinguish these possibilities we compared MMP in CH neurons from parkin mutants and controls and observed that CH neurons also accumulate depolarized mitochondria relative to controls (Fig. 2C). However, the magnitude of the decrease in MMP in CH neurons from parkin mutants (91% of the value from control CH neurons) was significantly less than that of DA neurons from parkin mutants (67% of the value from control DA neurons) (Fig. 2E). We further confirmed the selective impact of parkin deficiency on the MMP of DA neurons using internally controlled experiments, where the MMP of all non–GFP-expressing cells was compared with GFP-expressing DA neurons within the same preparation (Fig. S5). These findings indicate that the PINK1/Parkin pathway functions in both CH and DA neurons, but that mitochondria in DA neurons are more sensitive to an absence of Parkin activity than those in CH neurons.
We next assessed mitochondrial morphology in DA and CH neurons. Previous work has shown that DA neurons from Drosophila parkin mutants accumulate fused mitochondria (20), and we independently confirmed this finding by comparing the proportion of fused mitochondria in DA neurons from parkin mutants and their controls using confocal microscopy. Our experiments revealed that an average Drosophila DA neuron cell body contains ∼one to three distinct mitochondria, and that DA neurons from parkin mutants are more likely to contain a single fused mitochondrion (Fig. S4D). We also detected a trend toward accumulation of fused mitochondria in CH neurons from parkin mutants, although this finding did not reach significance (Fig. S4D). We also measured mitochondrial length as an independent indicator of mitochondrial morphology in DA and CH neurons from parkin mutants and controls. We found that mitochondrial length was significantly increased in DA neurons isolated from parkin mutants relative to heterozygous controls. We also detected a trend toward increased mitochondrial length in CH neurons from parkin mutants, but this trend did not reach significance (Fig. S4E). Taken together, our findings suggest that the selective vulnerability of DA neurons to parkin deficiency derives from an increased rate of formation of damaged mitochondria in DA neurons relative to CH neurons, or an inability of DA neurons to efficiently eliminate or repair damaged mitochondria in the absence of Parkin.
Genetic Perturbations That Increase Mitochondrial Fragmentation and Turnover Rescue the Neuronal Phenotypes of parkin Mutants.
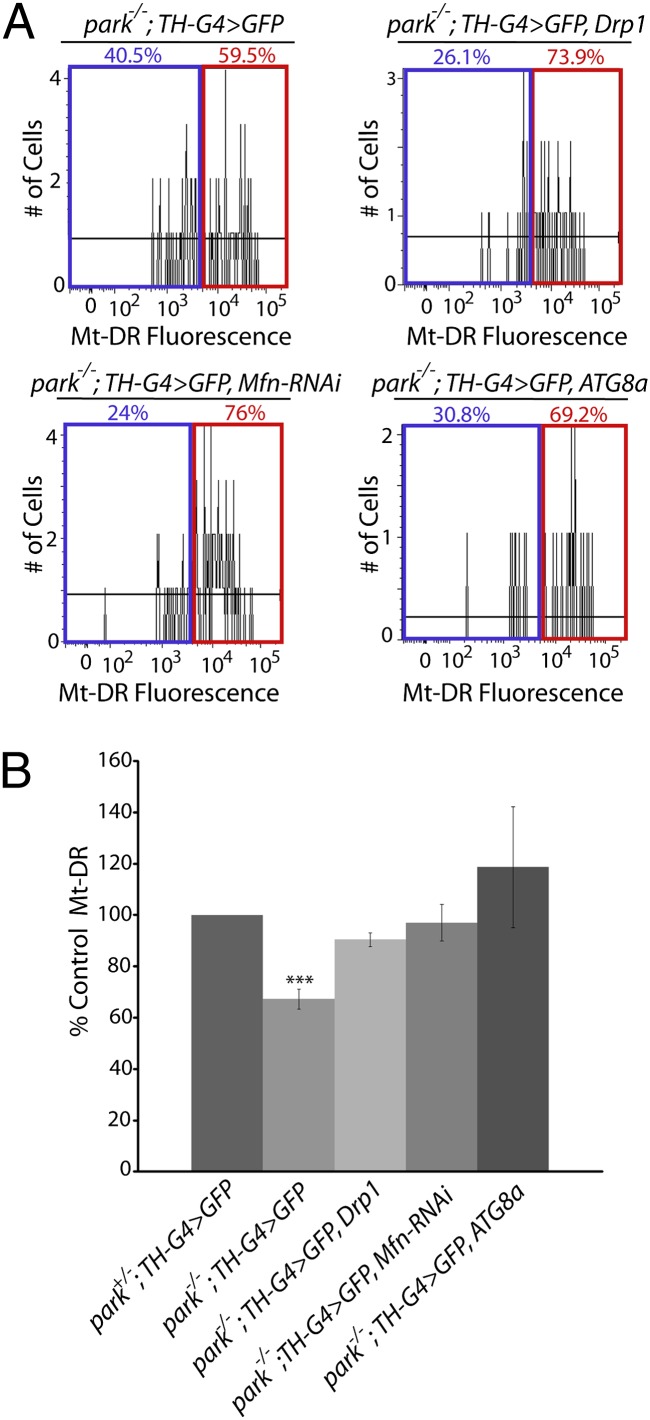
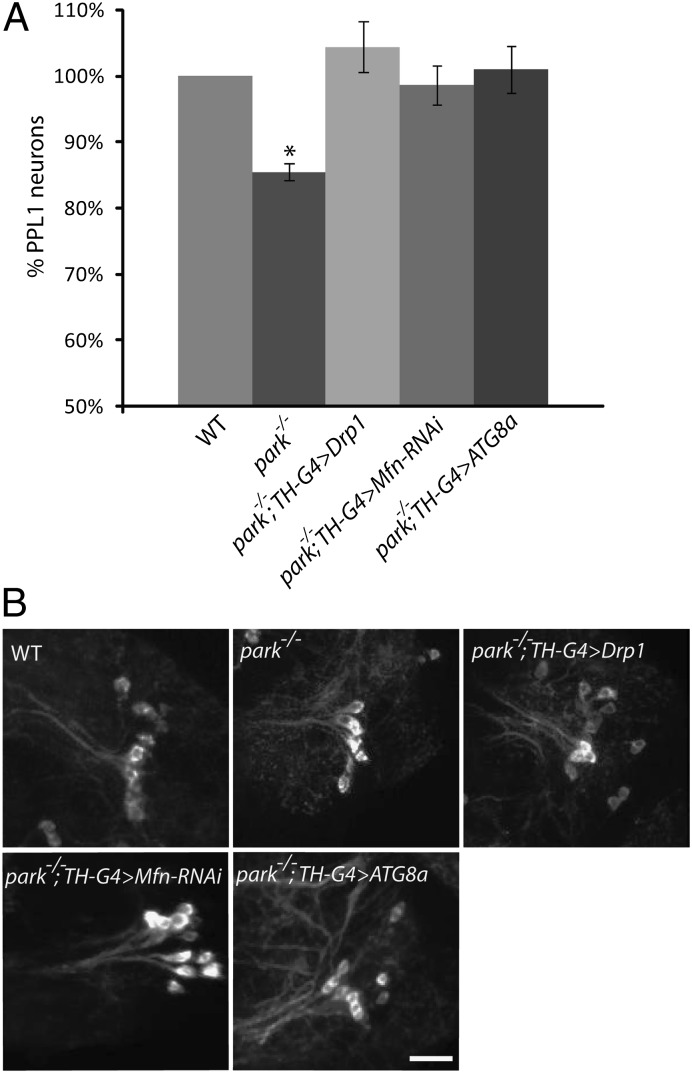
Studies in Drosophila have shown that Parkin promotes the ubiquitin-mediated turnover of the mitochondrial fusion-promoting factor Mitofusin (dMfn), and that genetic manipulations that induce mitochondrial fragmentation can suppress many of the parkin mutant phenotypes (7, 9, 25–28). Further experiments with vertebrate cell lines have shown that Parkin promotes mitochondrial fragmentation to segregate fusion-incompetent, depolarized mitochondria for turnover (10). However, previous work has not explored the influence of genetic perturbations that increase mitochondrial fragmentation or turnover on the integrity of mitochondria in DA neurons, or on the survival of this cell type in parkin mutants. We therefore examined the effects of genetic manipulations that increase mitochondrial fragmentation on the MMP defect of DA neurons from parkin mutants. These experiments were performed by overexpressing the mitochondrial fission-promoting factor dynamin related protein-1 (Drp1), and by inactivating dMfn using RNAi. Both of these perturbations fully corrected the MMP defect in DA neurons from parkin mutants (Fig. 3). We next tested whether overexpression of the autophagy-promoting factor ATG8a would influence the MMP defect of parkin mutants. This manipulation also fully rescued the MMP defect in DA neurons (Fig. 3). Finally, we tested whether these same genetic manipulations would also influence the survival of DA neurons. Our previous work has demonstrated that a subset of DA neurons in the protocerebral posterior lateral 1 (PPL1) cluster degenerate in Drosophila parkin mutants (21). Thus, we tested whether this neurodegenerative phenotype could be prevented by expressing the Mfn-RNAi construct in DA neurons or by overexpressing Drp1 or ATG8a in DA neurons. All three of these manipulations fully rescued the neurodegenerative phenotype of parkin mutants (Fig. 4).
Fig. 3.
Genetic perturbations that increase mitochondrial fragmentation and turnover rescue the MMP defect of parkin mutants. (A) Neural cultures from parkin-null homozygotes expressing GFP (park−/−; TH-G4 > GFP), from parkin-null homozygotes overexpressing GFP and Drp1 (park−/−; TH-G4 > GFP, Drp1), from parkin-null homozygotes overexpressing GFP and an RNAi construct targeting Mfn (park−/−; TH-G4 > GFP, Mfn-RNAi), or from parkin-null homozygotes overexpressing GFP and ATG8a (park−/−; TH-G4 > GFP, ATG8a) in DA neurons were labeled with Mt-DR and analyzed by flow cytometry. Representative histograms show the percentage of neurons that exhibit Mt-DR fluorescence intensities above and below the cutoff intensity value. (B) Depiction of the mean MMP of DA neurons from animals of the indicated genotypes relative to parkin-null heterozygotes (park+/−; TH-G4 > GFP). The number of biological replicates (n) and total number of cells analyzed (N) for the following genotypes were as follows: park−/−; TH-G4 > GFP (n = 3; N = 299); park−/−; TH-G4 > GFP, Drp1 (n = 3; N = 189); park−/−; TH-G4 > GFP, Mfn-RNAi (n = 3; N = 218); park−/−; TH-G4 > GFP, ATG8a (n = 5; N = 154). ***P < 0.001.
Fig. 4.
Genetic perturbations that increase mitochondrial fragmentation and turnover rescue the neurodegenerative phenotype of parkin mutants. (A) The relative percentage of DA neurons in the PPL1 cluster from wild-type flies, parkin-null homozygotes (park−/−), parkin-null homozygotes overexpressing Drp1 in DA neurons (park−/−; TH-G4 > Drp1), parkin-null homozygotes overexpressing an RNAi construct targeting Mfn in DA neurons (park−/−; TH-G4 > Mfn-RNAi), or from parkin-null homozygotes overexpressing ATG8a in DA neurons (park−/−; TH-G4 > ATG8a) are shown. The number of biological replicates (n) and total number of cells analyzed (N) were as follows: WT (n = 3; N = 88); park−/− (n = 6; N = 122); park−/−; TH-G4 > Drp1 (n = 3; N = 69); park−/−; TH-G4 > Mfn-RNAi (n = 2; N = 34); park−/−; TH-G4 > ATG8a (n = 3; N = 40). Error bars represent SE. *P < 0.05 (B) Representative confocal images of PPL1 DA neurons from the genotypes described in A are shown in brains stained with an antiserum against tyrosine hydroxylase. (Scale bar, 10 μm.)
Taken together, our findings support the model that the PINK1/Parkin pathway influences DA neuron integrity by promoting the selective fragmentation and turnover of depolarized mitochondria in DA neurons, and that DA neurons have an increased dependency on the PINK1/Parkin mitochondrial quality control pathway for their survival relative to other neuronal subtypes.
Discussion
To address previous conflicts concerning the role of the PINK1/Parkin pathway in mitochondrial quality control (4, 18, 19, 29), we developed a neural purification method to study the effects of perturbations targeting this pathway in DA neurons using Drosophila. Although several methods have been described to isolate neural subsets from Drosophila, including manual dissection of marked neurons (30) and laser-capture microdissection (31), these methods have limitations that precluded their use in our studies. In particular, manual dissection has a low throughput, and laser-capture microdissection requires processing steps that render the isolated cells refractory to many cell biological analyses, including the measurement of MMP. Using flow cytometry-based neural purification, we find that DA neurons from both parkin and pink1-null mutants accumulate depolarized mitochondria, and that activation of the PINK1/Parkin pathway increases the fractional abundance of polarized mitochondria in wild-type animals. We also find that genetic perturbations that promote mitochondrial fragmentation and turnover rescue the mitochondrial depolarization and neurodegenerative phenotypes of parkin mutants. Thus, our findings support the model that the PINK/Parkin pathway influences DA neuron survival by promoting mitochondrial quality control.
An important question that arises from our findings is why other recent studies failed to detect a role for PINK1 and Parkin in mitochondrial quality control in neurons. One such study that examined Parkin-mediated mitophagy in CCCP-treated neurons concluded that the reliance of neurons on oxidative respiration for energy production prevents Parkin-mediated mitophagy following CCCP treatment because mitophagy is energy-dependent (19). However, it seems unlikely that the normal physiological processes that damage mitochondria in the intact brain would result in a complete collapse of MMP akin to CCCP treatment. Another study that failed to detect Parkin-mediated mitophagy in neurons showed that the mitochondrial pathology resulting from a DA-specific knockout of the mitochondrial transcription factor A (TFAM) gene in mice was unaffected by simultaneously inactivating parkin (18). However, because parkin knockout mice do not exhibit neurodegeneration (32), it is unclear whether these findings from mice can be extrapolated to humans. In contrast to these studies, our work explored the effects of perturbations targeting the PINK1/Parkin pathway under physiological conditions, in an animal model that exhibits robust parkin loss-of-function phenotypes, including DA neuron degeneration (21, 33).
Because Parkin promotes mitochondrial fragmentation (10, 25–28, 34), but is also required for selective mitochondrial turnover downstream from fragmentation (10), another question raised by our findings concerns the mechanisms by which genetic perturbations that increase mitochondrial fragmentation rescue the parkin mutant phenotypes. One explanation for our findings is that smaller mitochondria may represent more efficient substrates for nonselective autophagy, and that genetic perturbations that increase mitochondrial fragmentation simply increase the likelihood that damaged mitochondria will be degraded through nonselective autophagy. Consistent with our interpretation, recent studies demonstrate that mitochondrial fusion represents a cellular strategy to prevent mitochondrial turnover during starvation-induced autophagy (35, 36), and that mitochondrial fragmentation promotes the segregation of dysfunctional mitochondria for turnover (10, 37, 38). Similarly, increasing the expression of ATG8a may result in an increased rate of nonselective autophagy, which would also presumably increase the rate at which damaged mitochondria are randomly selected for turnover, resulting in rescue of MMP in the face of ongoing biogenesis of healthy polarized mitochondria.
The mechanisms underlying cell type-specific vulnerability in PD and many other neurodegenerative disorders are poorly understood. Given that PINK1 and parkin are broadly expressed genes, the selective vulnerability of DA neurons to the loss of PINK1 or Parkin activity cannot readily be explained by the expression patterns of these genes. Our findings suggest that the enhanced sensitivity of DA neurons to the loss of the PINK1/Parkin pathway may derive from a lack of redundant mitochondrial quality control systems in DA neurons or an increased rate of mitochondrial damage in DA neurons. Although data are lacking to distinguish these models, previous work has shown that highly damaging species are formed as a byproduct of dopamine metabolism, potentially providing support for an increased rate of mitochondrial damage in DA neurons (39). Our neural purification and characterization procedure, coupled with the many powerful genetic tools available in Drosophila, should enable a test of these models in future work.
Materials and Methods
Drosophila Strains and Culture.
All Drosophila strains were maintained on standard cornmeal/molasses medium at 25 °C with a 12-h light-dark cycle. For brevity, the homozygous park25-null allele is designated as park−/− throughout the article, and the hemizygous PINK1B9 null allele as PINK1−/Y. Experiments involving PINK1 overexpression involved animals aged to at least 7 d before neural culture preparation. Experiments using parkin and pink1-null mutant animals used animals aged to at least 15 d.
Drosophila Brain Dissociation and Dye Labeling.
At least four dissected fly brains were placed into 1 mL of DME/Ham’s F-12 High Glucose media (Irvine Scientific) supplemented with 20 mM hepes (Sigma), 2.5 mM glutamine (Sigma), and 0.5% trypsin (Invitrogen), and mechanically dissociated by passage 30 times through a p200 pipette tip at room temperature. The cell suspension was then incubated at 37 °C for 45 min, with three additional pipetting steps executed at 15 min intervals. The cell suspension was then triturated ten times through a 15–45% (vol/vol) SurfaSil (Fisher) coated glass-flamed Pasteur pipette, and passed through a 70-μm mesh cell strainer (Fisher). The cell suspension was then treated with 10 nM Mt-DR (Invitrogen), or 150 nM TMRE (Enzo Life Sciences) for 10 or 20 min, respectively. Cells were spun at 700 × g for 2 min, and washed twice in supplemented media. Media was then aspirated, and the cells were resuspended in 250 μL of supplemented media containing 50 μg/mL insulin (VWR), 100 μM putrescine (Sigma), 20 ng/mL progesterone (Sigma), 100 μg/mL transferrin (Sigma), 2 μg/mL DAPI (Fisher), and 20 nM TMRE (TMRE-treated cultures only). For CCCP pretreatment, 40 μM or 100 μM of CCCP was added to culture media for 10 or 20 min before TMRE or Mt-DR addition, respectively. Cells were maintained at room temperature until flow analysis and sorting, which was always performed within 30 min upon completion of the protocol.
Flow Analysis.
Flow cytometry was performed using a BD FACSCanto, a BD LSRII, or a BD FACS Aria II cell sorter equipped with a 407-nm, a 488-nm, and a 635-nm laser for detection of DAPI, GFP/TMRE, and Mt-DR fluorescence, respectively. Flow cytometry measurements are expressed in arbitrary units throughout the paper. Dot plots and bar graphs were prepared using FloJo software (Treestar), with identical gating parameters used within each experiment.
RT-PCR.
For RT-PCR, 3,000–6,000 neurons were sorted into Cells Direct (Invitrogen) resuspension buffer supplemented with lysis enhancer buffer, and cDNA synthesis was performed according to the manufacturer’s instructions. cDNA was then preamplified using a fourfold dilution of a mixture of all of the Taqman qPCR assays (ABI) of interest in qPCR Master Mix (Roche) on a Roche LightCycler. Following a 17-cycle preamplification, aliquots were removed, diluted five fold, and subjected to standard qPCR on a Roche LightCycler using individual Taqman assays. PCR products were analyzed by gel electrophoresis to confirm that the predicted product sizes were obtained. Taqman assays used were Drosophila: ple/TH (Dm01842066_m1), VMAT (Dm01793487_g1), ChAT (Dm02134802_m1), α-tubulin (Dm02361072_s1), DDC (Dm01811001_m1), and parkin (Dm01798015_g1).
Brain Whole-Mount Immunostaining.
DA neurons were quantified in situ using anti-TH antiserum (Immunostar) and confocal microscopy, as previously described (40).
Statistical Tests.
All statistical tests involve Student's t test, and error bars represent SDs, unless stated otherwise. For all experiments *P ≤ 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Dr. T. Neufeld (University of Minnesota), Dr. M. Guo (University of California at Los Angeles), and Dr. J. K. Chung (Korea Advanced Institute of Science and Technology) for providing fly stocks; David Bell, Thane Mittlesteadt, and Dr. Michele Black (University of Washington) for flow cytometry support; Tarannum Dhillon (University of Washington), Dr. Adrian Ozinsky and Dr. Anne Grosse-Wilde (Institute for Systems Biology), and Dr. Kien Trinh and Dr. Jay Parrish (University of Washington) for technical support; and Evelyn S. Vincow and Dr. Ruth E. Thomas (University of Washington) for critical reading of the manuscript. This work was supported by a Parkinson Society Canada Research Fellowship (to J.L.B.); a Canadian Institutes of Health Research Fellowship (to J.L.B.); National Institutes of Health Grant 1R01GM086394-01 (to L.P.); and Muscular Dystrophy Association Grant 172828 (to L.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120688109/-/DCSupplemental.
References
- 1.Farrer MJ. Genetics of Parkinson disease: Paradigm shifts and future prospects. Nat Rev Genet. 2006;7:306–318. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- 2.Shulman JM, De Jager PL, Feany MB. Parkinson’s disease: Genetics and pathogenesis. Annu Rev Pathol. 2011;6:193–222. doi: 10.1146/annurev-pathol-011110-130242. [DOI] [PubMed] [Google Scholar]
- 3.Zhu J, Chu CT. Mitochondrial dysfunction in Parkinson’s disease. J Alzheimers Dis. 2010;20(Suppl 2):S325–S334. doi: 10.3233/JAD-2010-100363. [DOI] [PubMed] [Google Scholar]
- 4.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vives-Bauza C, Przedborski S. Mitophagy: The latest problem for Parkinson’s disease. Trends Mol Med. 2011;17:158–165. doi: 10.1016/j.molmed.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci USA. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS ONE. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka A, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glauser L, Sonnay S, Stafa K, Moore DJ. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J Neurochem. 2011;118:636–645. doi: 10.1111/j.1471-4159.2011.07318.x. [DOI] [PubMed] [Google Scholar]
- 12.Rakovic A, et al. Mutations in PINK1 and Parkin impair ubiquitination of Mitofusins in human fibroblasts. PLoS ONE. 2011;6:e16746. doi: 10.1371/journal.pone.0016746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gegg ME, et al. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan NC, et al. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geisler S, et al. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy. 2010;6:871–878. doi: 10.4161/auto.6.7.13286. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sterky FH, Lee S, Wibom R, Olson L, Larsson NG. Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proc Natl Acad Sci USA. 2011;108:12937–12942. doi: 10.1073/pnas.1103295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Laar VS, et al. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum Mol Genet. 2011;20:927–940. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 21.Whitworth AJ, et al. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson’s disease. Proc Natl Acad Sci USA. 2005;102:8024–8029. doi: 10.1073/pnas.0501078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 23.Sang TK, et al. A Drosophila model of mutant human parkin-induced toxicity demonstrates selective loss of dopaminergic neurons and dependence on cellular dopamine. J Neurosci. 2007;27:981–992. doi: 10.1523/JNEUROSCI.4810-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 25.Poole AC, et al. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci USA. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng H, Dodson MW, Huang H, Guo M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci USA. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y, et al. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci USA. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J, Lee G, Chung J. The PINK1-Parkin pathway is involved in the regulation of mitochondrial remodeling process. Biochem Biophys Res Commun. 2009;378:518–523. doi: 10.1016/j.bbrc.2008.11.086. [DOI] [PubMed] [Google Scholar]
- 29.Gusdon AM, Chu CT. To eat or not to eat: Neuronal metabolism, mitophagy, and Parkinson’s disease. Antioxid Redox Signal. 2011;14:1979–1987. doi: 10.1089/ars.2010.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kula-Eversole E, et al. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci USA. 2010;107:13497–13502. doi: 10.1073/pnas.1002081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vicidomini R, Tortoriello G, Furia M, Polese G. Laser microdissection applied to gene expression profiling of subset of cells from the Drosophila wing disc. J Vis Exp. 2010;(38) doi: 10.3791/1895. 1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez FA, Palmiter RD. Parkin-deficient mice are not a robust model of parkinsonism. Proc Natl Acad Sci USA. 2005;102:2174–2179. doi: 10.1073/pnas.0409598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greene JC, et al. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci USA. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu W, Sun Y, Guo S, Lu B. The PINK1/Parkin pathway regulates mitochondrial dynamics and function in mammalian hippocampal and dopaminergic neurons. Hum Mol Genet. 2011;20:3227–3240. doi: 10.1093/hmg/ddr235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci USA. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomes LC, Scorrano L. High levels of Fis1, a pro-fission mitochondrial protein, trigger autophagy. Biochim Biophys Acta. 2008;1777:860–866. doi: 10.1016/j.bbabio.2008.05.442. [DOI] [PubMed] [Google Scholar]
- 39.Barzilai A, Melamed E. Molecular mechanisms of selective dopaminergic neuronal death in Parkinson’s disease. Trends Mol Med. 2003;9:126–132. doi: 10.1016/s1471-4914(03)00020-0. [DOI] [PubMed] [Google Scholar]
- 40.Trinh K, et al. Induction of the phase II detoxification pathway suppresses neuron loss in Drosophila models of Parkinson’s disease. J Neurosci. 2008;28:465–472. doi: 10.1523/JNEUROSCI.4778-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.