Abstract
Tumor invasion and metastasis are strongly regulated by biophysical interactions between tumor cells and the extracellular matrix (ECM). While the influence of ECM stiffness on cell migration, adhesion, and contractility has been extensively studied in 2D culture, extension of this concept to 3D cultures that more closely resemble tissue has proven challenging, because perturbations that change matrix stiffness often concurrently change cellular confinement. This coupling is particularly problematic given that matrix-imposed steric barriers can regulate invasion speed independent of mechanics. Here we introduce a matrix platform based on microfabrication of channels of defined wall stiffness and geometry that allows independent variation of ECM stiffness and channel width. For a given ECM stiffness, cells confined to narrow channels surprisingly migrate faster than cells in wide channels or on unconstrained 2D surfaces, which we attribute to increased polarization of cell-ECM traction forces. Confinement also enables cells to migrate increasingly rapidly as ECM stiffness rises, in contrast with the biphasic relationship observed on unconfined ECMs. Inhibition of nonmuscle myosin II dissipates this traction polarization and renders the relationship between migration speed and ECM stiffness comparatively insensitive to matrix confinement. We test these hypotheses in silico by devising a multiscale mathematical model that relates cellular force generation to ECM stiffness and geometry, which we show is capable of recapitulating key experimental trends. These studies represent a paradigm for investigating matrix regulation of invasion and demonstrate that matrix confinement alters the relationship between cell migration speed and ECM stiffness.
Keywords: microchannels, glioblastoma, cytoskeleton, mechanobiology, polyacrylamide
Tumor cells exploit a variety of migration strategies to invade tissue and metastasize to distant anatomical sites, which in turn requires specific biochemical and biophysical interactions between these cells and the extracellular matrix (ECM) (1–4). Cell migration through the ECM may be regarded as a cyclic process that includes leading-edge extension of protrusions driven by actin polymerization, formation of cell-ECM adhesions, and trailing-edge retraction due to actomyosin contractility. While ECM-based biophysical cues have been demonstrated to influence all of these steps (5), ECM stiffness has emerged as a particular parameter of interest given the observations that tumors are frequently more rigid than normal tissue and that exogenous tissue stiffening can facilitate tumorigenesis (6–13). Much of the field’s understanding of matrix stiffness derives from studies in 2D culture, where polyacrylamide (PA) hydrogels conjugated with full-length ECM proteins have proven a versatile and robust paradigm for the independent control of ECM stiffness and biochemical ligand density (7–10, 13–15). For example, ECM stiffness strongly affects the migration and proliferation rate of glioma cells (8), the expression of prognostic markers in neuroblastoma (7), and the self-renewal and differentiation of stem cells (9, 16). Extension of these studies to 3D microenvironments characteristic of most tissues has proven extremely challenging given that manipulations normally used to vary ECM stiffness (e.g., variation of matrix and crosslink density) often concurrently alter matrix pore size, which can independently and significantly affect cell migration (4, 6). For example, reduction of matrix pore size has been shown to induce transitions between amoeboid and mesenchymal cell motility (3, 17, 18), and below a critical limit can slow or arrest cell invasion due to steric hindrance (6, 19). This convolution of parameters has motivated an interest in the development of 3D ECM scaffolds that permit independent variation of ECM stiffness and pore size, which could significantly improve the field’s understanding of the biophysical regulation of tumor cell invasion in tissue-like ECMs.
Several experimental strategies have been reported to study the regulation of tumor cell migration by ECM stiffness, porosity, or both. For example, polydimethylsiloxane (PDMS) microchannels of defined geometry have been used to simulate 3D matrix pores to study the dependence of cell migration on channel width (20–22). An important limitation of PDMS is that the stiffness of this material exceeds that of most soft tissues, strongly limiting its utility as a model of these tissues. In collagen matrices that span a more physiologically relevant range of ECM stiffness, pore size has been modified by manipulating collagen concentration, pH, and gelation temperature (23), and by adding agarose (6). Although these collagen-based scaffolds do permit indirect control of matrix porosity, their use is complicated by the fact that these manipulations also alter microscale and/or macroscale scaffold structure and mechanics. Moreover, most of these systems require retrospective determination of pore size based on empirical correlations and do not, in general, permit prospective imposition of some pore size of interest. As a way of addressing these limitations, “bead templating” approaches have recently been developed to yield hydrogels of defined stiffness containing a communicating network of pores whose characteristic size is dictated by the particles around which the scaffold is formed (24). In practice, the conduits between pores are so short that cells are observed to “jump” discontinuously between void chambers, offering limited opportunity to observe and characterize cells in confined geometries. Thus, the field could significantly benefit from a matrix platform that allows independent manipulation of ECM stiffness and pore size and also enables the monitoring of sustained cell migration in a confined microenvironment.
Here we present a culture platform for the study of tumor cell invasion in which matrix stiffness and confinement (pore size) may be varied independently of one another. Our approach is based on polymerization and gelation of defined-stiffness PA hydrogels around silicon-based scaffolds of defined microtopography, such that the surface of the PA gel adopts the inverse contours of the master, resulting in channels of defined geometry. In other words, the elasticity of these micro-PA channels (μPACs) is entirely determined by the choice of PA formulation, and the channel geometry is entirely dictated by the silicon master, with the two subject to independent control. Using this μPAC platform, we investigated the relationship between ECM stiffness, pore size, and cell migration speed, with a specific focus on understanding how the regulation of cell migration on ECM stiffness is affected by the degree of matrix confinement. Our results demonstrate that matrix confinement fundamentally alters the classical biphasic relationship between ECM stiffness and migration speed and that this phenomenology depends on nonmuscle myosin II (NMMII)-based contractility. We hypothesize that this effect originates from the fact that confinement forces polarization of traction forces along a single dimension, and we test this hypothesis using a multiscale computational model of a cell migrating through a pore of defined geometric and mechanical properties.
Results
Fabrication of μPAC Platform.
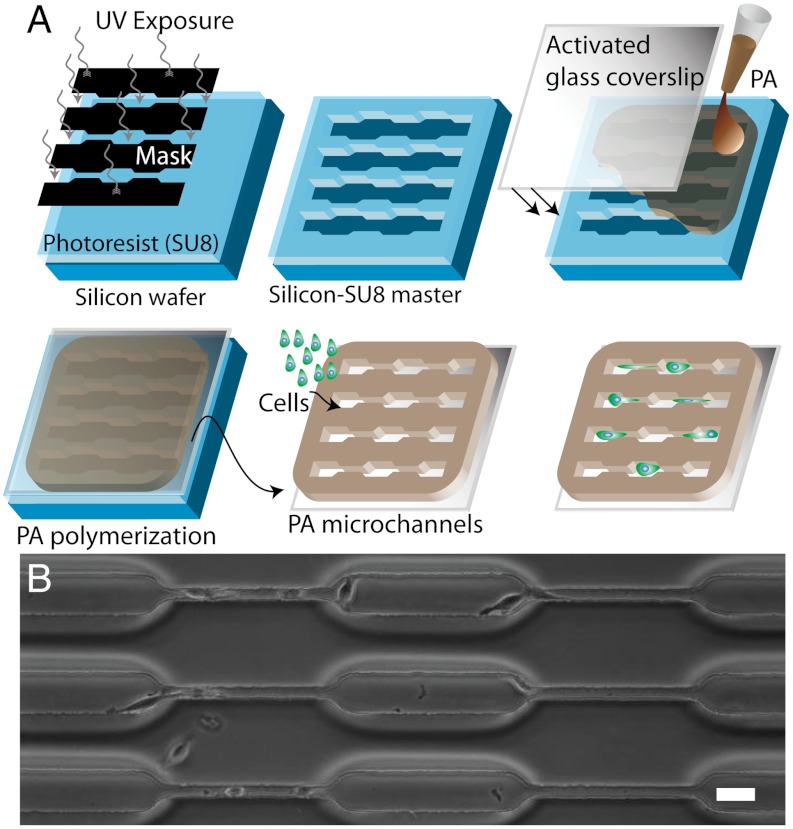
To study cell migration in a system in which ECM stiffness and confinement could be controlled independent of one another, we combined photolithography techniques and controlled polymerization of PA hydrogels to construct microchannels of varying width embedded in PA gels of specified stiffness (microfabricated polyacrylamide channels, or μPACs, Fig. 1A). In this approach, we allowed a PA precursor solution (acrylamide and bisacrylamide at a defined ratio plus initiators TEMED and ammonium persulfate) to polymerize and gel against a microfabricated silicon master with predefined topographical patterns of variable channel width (cw). This procedure resulted in PA hydrogels spanning a range of stiffness values whose surfaces were marked with channels of defined size that matched the contours of the silicon master, analogous to past uses of PA for micromolding applications (25, 26). We designed the device to have ≥75-μm-thick walls between all channels to minimize mechanical transmission across the channel wall (27). We confirmed this expectation by using atomic force microscopy (AFM) to measure ECM stiffness in the plateau regions between wide and narrow channels (Fig. S1), which indeed verified that ECM stiffness was uniform across the surface of the device. We subsequently functionalized these scaffolds with full-length ECM proteins to render them suitable for cell adhesion and migration. The subsequent spontaneous migration of cells within the channels enabled us to track cell motility in μPACs of varying stiffness and channel width over a period of several hours (Fig. 1B). Confocal microscopy revealed that cells in microchannels intimately associate with all three channel surfaces and form vinculin-positive adhesions on side walls, consistent with a 3D morphology (Fig. S2).
Fig. 1.
Design and creation of microfabricated polyacrylamide channels (μPACs). (A) Schematic of process, including fabrication of a silicon master with topographic patterns of defined size and shape, formation of a PA hydrogel of prescribed elasticity around the features, separation of the PA hydrogel from the master, and seeding of cells. (B) Phase contrast images of device fabricated from 120 kPa PA hydrogel and containing channels composed of pores of varying dimensions. Scale bar = 40 μm.
Enhanced Cell Migration in Narrow Channels.
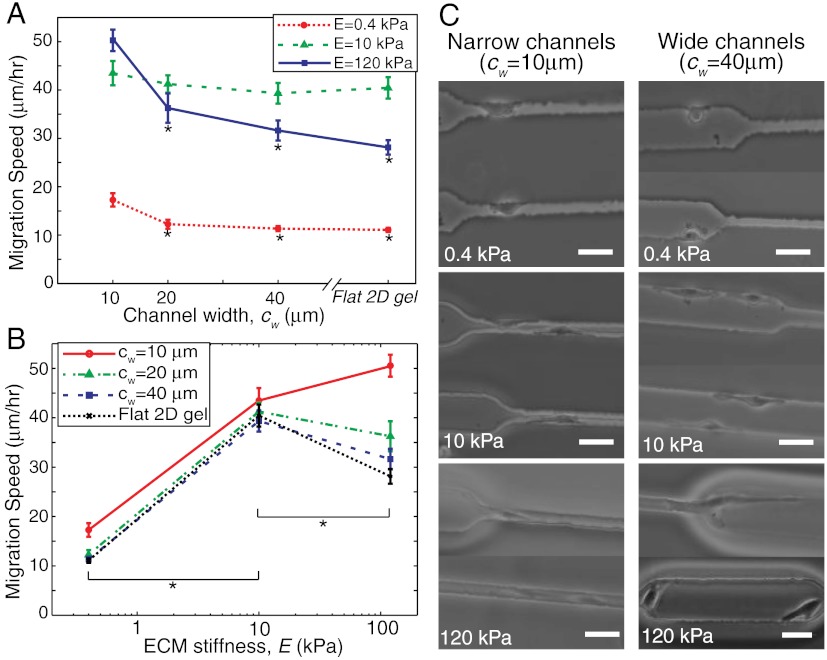
We began by investigating the dependence of migration speed of U373-MG human glioma cells on channel stiffness and width by creating a series of μPAC scaffolds in which stiffness was varied between 0.4 and 120 kPa and channel width was varied between 10 and 40 μm, with unconstrained (flat) gels included as a control (Fig. 2 A and B; these plots are the same dataset depicted two ways for clarity). We chose this stiffness range because it spans the regime of maximal mechanosensitivity for human glioblastoma cells (8, 28) and other mammalian cell types (10, 13, 14), and we chose these channel widths in order to sample values on the same length scale as the cell. Somewhat surprisingly, for a fixed stiffness, we observed that migration speed fell with increasing channel width (Fig. 2A), with the most pronounced confinement dependence being observed on the stiffest matrix (120 kPa). For a fixed pore diameter, we observed a diversity of relationships between migration speed and stiffness; specifically, migration speed varied biphasically (i.e., passed through a single maximum) with stiffness for the two largest channel widths (cw = 20 and 40 μm) and monotonically increased with stiffness for the smallest channel width (cw = 10 μm) (Fig. 2B). In narrow channels, migration speed increased but did not show biphasic behavior over the range of ECM stiffnesses examined; it is possible that a local maximum may theoretically exist on significantly stiffer ECMs (i.e., > 150–200 kPa), but these stiffness values exceed both peak values typically achieved with the PA system (8, 10, 12, 15) and the normal stiffness range of almost all soft tissues (29). Importantly, we also observed a biphasic migration pattern on unconfined ECMs, consistent with our own and others’ previous observations (14, 19, 28). In other words, confinement not only increases migration speed overall but also fundamentally alters the manner in which ECM stiffness governs migration speed. Phase contrast imaging provided additional insight into this observation and revealed that the degree to which a cell interacts with the channel depends on channel width, with wide channels permitting cells to preferentially adhere to one channel wall and narrow channels forcing cells to adhere to multiple walls and adopt the shape of the channel (Fig. 2C and Movies S1 and S2).
Fig. 2.
Migration speed versus ECM stiffness and channel width. (A) Migration speed versus channel width for soft, intermediate and stiff ECMs, corresponding to E = 0.4, 10, and 120 kPa. *p < 0.05 with respect to narrow (cw = 10 μm) channels. (B) Migration speed versus ECM stiffness for narrow, intermediate, and wide channels (cw = 10, 20, and 40 μm) and flat 2D gels. *p < 0.05 for pairwise comparison between two indicated stiffness values for all given channel widths, including 2D flat gel. n > 35 cells per condition over multiple channels and devices. (C) Phase contrast images of cells migrating inside channels of varying stiffness and width. Scale bar = 40 μm.
Relationship Between Matrix Stiffness, Cell Polarization, and Migration Speed.
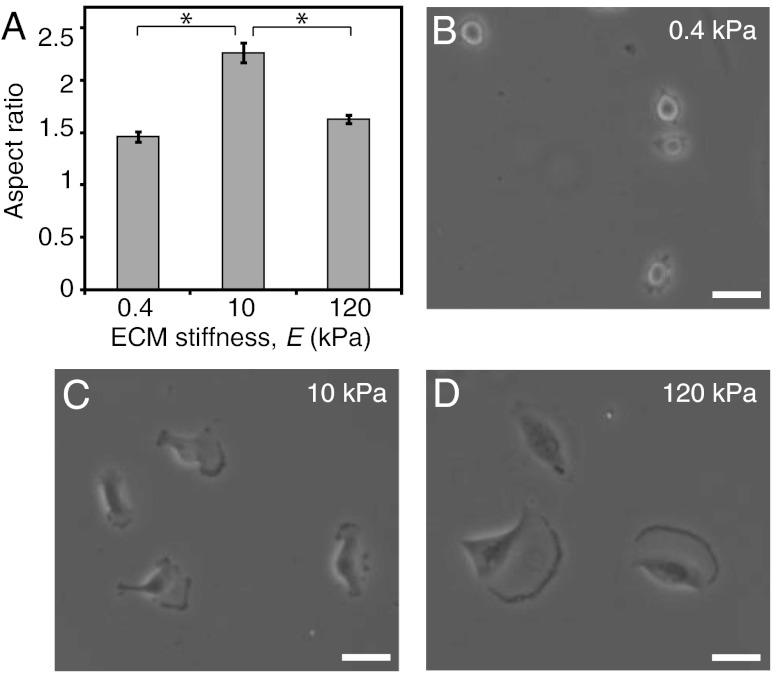
Given that very narrow channels promote cell polarization and also enhance migration speed, we next decided to investigate whether migration speed might correlate with cell polarization on matrices lacking geometric constraints on cell polarization: i.e., unconfined (flat) scaffolds (Figs. 3 and 4). As described earlier, migration speed depends biphasically on ECM stiffness (Fig. 2B). When we performed morphometric analysis on these cells, we confirmed that, as expected, projected cell area increased with increasing matrix stiffness, with cells failing to engage the most compliant ECMs and spreading extensively on the stiffest ECMs (Fig. 4D). Interestingly, however, we discovered that even though spreading area increased monotonically with ECM stiffness, cell polarization exhibited a biphasic dependence on ECM stiffness, similar to migration speed (Fig. 3A). Specifically, cells adopted isotropic morphologies on the softest and stiffest matrices (albeit with much different degrees of spreading in each case) and a highly polarized, spindle-like morphology on intermediate-stiffness matrices (Fig. 3 B–D). In other words, in the complete absence of geometric limitations on cell adhesion and spreading, morphological polarization predicts migration speed, with increases in one parameter correlated with increases in the other.
Fig. 3.
Morphological polarization of U373 cells on flat 2D gels. (A) Cell polarization measured as aspect ratio of cells cultured on flat unconfined (2D) gels. *p < 0.05 for indicated pairwise comparison; n > 35 cells per condition. (B, C, D) Phase contrast images of cells on 2D flat gels of varying stiffness. Scale bar = 40 μm.
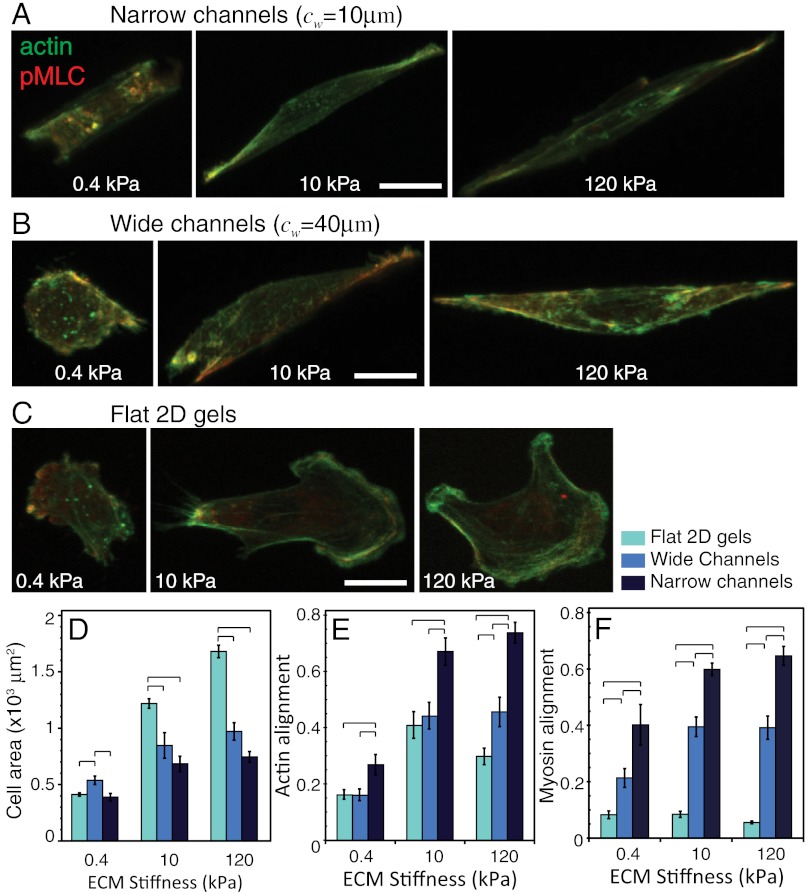
Fig. 4.
Stress fiber alignment and spreading area of cells in defined confinement. Confocal images of F-actin and pMLC distribution for U373-MG cells in (A) narrow channels (cw = 10 μm), (B) wide channels (cw = 40 μm), and (C) unconfined flat 2D gels of varying ECM stiffness. Scale bar = 20 μm. (D) projected cell area, (E) F-actin alignment, and (F) coalignment of pMLC immunofluorescence with channel axis as a function of ECM stiffness for varying degrees of confinement. Statistically different pairs (p < 0.05) are indicated by horizontal square brackets. n > 12 cells per condition.
Regulation of Cytoskeletal Architecture by Matrix Stiffness and Confinement.
To gain additional insight into the relationship between channel width and cell polarization, we used confocal microscopy to visualize the intracellular distribution of phosphorylated myosin light chain (pMLC) and F-actin (Fig. 4 and Fig. S3). Cells in wide channels (cw = 40 μm) and on flat (2D) substrates adopted highly spread morphologies compared to cells in narrow channels (cw = 10 μm), with differences most apparent on the stiffest matrices (120 kPa) (Figs. 4 A–D and Movies S1 and S2). Moreover, automated image correlation analysis of F-actin and pMLC fluorescence revealed that cells on flat substrates and inside wide channels oriented their stress fibers in a much more radially uniform fashion than cells in narrow channels regardless of matrix stiffness (Fig. 4 E and F), with stress fibers in narrow channels strongly coaligned with the long axis of the channel. Thus, just as on unconfined matrices, stress fiber alignment strongly predicts migration speed, with the most highly aligned stress fibers—and fastest motility—observed in narrow, stiff channels. In other words, migration speed tracks with the degree to which actomyosin traction force is polarized along a single axis, which confinement in a narrow channel appears to promote.
Myosin II-based Traction Polarization is Essential for Confinement Sensitivity.
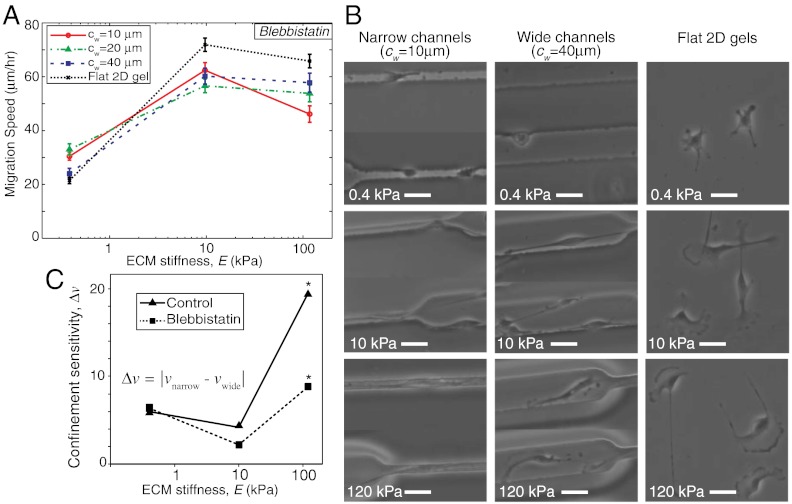
Based on the preceding observations, we hypothesized that the enhanced migration rates in the narrowest channels resulted directly from enhanced polarization of actomyosin traction forces in these scaffolds. If this hypothesis is correct, targeted disruption of this polarization would be expected to offset this enhanced migration. Inhibition of myosin II is known to disrupt cytoskeletal coherence (30, 31) and abrogate stress fibers, both of which are expected to be key to polarization of traction forces (Figs. 2 and 4). Thus, we repeated our studies in the presence of the NMMII ATPase inhibitor blebbistatin (32) (Fig. 5). Confocal imaging of F-actin distributions in blebbistatin-treated cells confirmed dissipation of stress fibers under all ECM conditions (Fig. S4). We then measured the migration speed of blebbistatin-treated cells for various ECM properties in our μPAC system (Fig. 5A), and found two important differences from the control case (Fig. 2). First, blebbistatin-treated cells migrated faster than control cells for most ECM conditions.(Figs. 2B, and 5A, and Fig. S5), similar to previous reports (8, 33, 34). This effect has been attributed to the ability of NMMII to periodically interrupt actin polymerization and hinder protrusions at the leading edge (35). Consistent with this role, we observed significantly longer protrusions in blebbistatin-treated cells (Fig. 5B and Fig. S4) than in untreated control cells (Figs. 2–4). Second, NMMII inhibition fundamentally altered the role of channel width in modulating the relationship between migration speed and ECM stiffness. In untreated controls (Fig. 2), we had observed a biphasic dependence of migration speed on ECM stiffness in unconfined 2D matrices and wide channels, whereas migration speed increased monotonically with ECM stiffness in narrow channels in the stiffness range examined. Conversely, in the setting of NMMII inhibition, all ECM conditions gave rise to a biphasic or saturating relationship, with the strongest biphasic relationship observed in the narrowest channels (Fig. 5A). This stark difference between blebbistatin-treated and untreated cells supports our hypothesis that traction polarization critically underlies the ability of narrow channels to increase migration speed and overcome motility limitations associated with stiff substrates. In other words, NMMII inhibition renders migration speed significantly less sensitive to confinement, a concept we quantify as “confinement sensitivity” and define as the difference between migration speed in wide and narrow channels for a given matrix stiffness (Fig. 5C).
Fig. 5.
Effect of nonmuscle myosin II inhibition on cell motility. (A) Migration speed versus ECM stiffness for varying channel width. n > 30 cells per condition. (B) Phase contrast images of cells inside narrow and wide channels and flat 2D gels of varying stiffness. Scale bar = 40 μm. (C) Confinement sensitivity, Δν, calculated as |νnarrow - νwide|, where νnarrow and νwide are average respective migration speeds for narrow (cw = 10 μm) and wide (cw = 40 μm) channels versus ECM stiffness for control and blebbistatin-treated cells. Statistical significance (*p < 0.05) determined by Student’s t test (unpaired, two-tailed, 95% confidence interval).
Computational Model of Migration in Channels of Defined Width and Stiffness.
Our experimental data are strongly consistent with a framework in which NMMII-based polarization of actomyosin traction forces is key to promoting rapid cell migration in narrow channels. To integrate all of these parameters into a coherent, quantitative paradigm, we developed a predictive computational model of a cell migrating in an ECM channel of defined width and stiffness (SI Text, Fig. S6). Using this model, we calculated migration speed (v) as a function of myosin activation and compared the model predictions with our experimental measurements on ECMs of defined stiffness and channel width. We also simulated the effects of high and low myosin activity for the control and the myosin knockdown cases, respectively.
In the control cases (i.e., no NMMII inhibition), our model predicted a biphasic dependence of migration speed on ECM stiffness on flat 2D gels and wide channels (Fig. 6A), which is consistent with our experiments (Fig. 2). The drag force (Fd) increased proportionally with ECM stiffness and cell size (36), which in turn reduced the migration speed on very stiff ECMs and caused the predicted biphasicity for wide channels and 2D gels (see cw≫10 μm in Fig. 6A). We could track concurrent changes in cell polarization by defining a polarization factor (ψ, see SI Methods), which depends on cell size and ECM confinement. In 2D-like settings (flat matrix or cw≫10 μm), ψ falls, which in turn reduces the polarized traction force (ψFc) and thus reduces migration speed. Conversely, narrow channels (cw ≈ 10 μm) impose constraints on cell spreading that increase the polarization factor (ψ) and thus also increase the polarized traction. This enhanced traction polarization inside narrow channels yielded faster migration than in wider channels (Fig. 6A) and abrogated the biphasic dependence of migration speed on ECM stiffness.
Fig. 6.
Model predictions and comparison with experiments. Migration speed (v) versus ECM stiffness under (A) control and (B) blebbistatin-treated conditions, where model predictions are plotted as solid lines color-coded for varying degrees of matrix confinement with overlaid symbols and dotted lines corresponding to experimental measurements presented in Figs. 2B and 5A.
Upon NMMII inhibition, the net contractile force (Fc) fell dramatically and reduced the impact of the polarized traction term (ψFc) on migration speed in SI Text, Eq. S5. Stated another way, the reduction in contractile force and traction polarization associated with NMMII inhibition offset any gains in traction polarization associated with a narrow channel. As a result, migration speed was primarily dictated by ECM stiffness and thus varied biphasically with ECM stiffness regardless of channel width (Fig. 6B). The model also predicted faster migration following NMMII inhibition under all ECM conditions (Fig. 6), which was caused by a drastic rise in protrusive force (Fp) due to the release of myosin-driven pauses in protrusions (35, 37). Additionally, we assumed that the number of protrusions at the leading edge of the cell would depend on cell area, which has been shown to increase with channel width (Fig. 4D). We captured this behavior in the model by prescribing a simple proportionality between the protrusive force and channel width, which in turn yielded higher migration speed in wider channels (Fig. 6B) in NMMII-inhibited cells where a protrusive mechanism was the most dominant regulator of motility.
Discussion
The invasion of a wide variety of tumors is strongly regulated by the biophysical properties of the ECM, including ECM elasticity and microstructure. However, the field’s ability to dissect the distinct contributions of these two parameters to tumor invasion and gain insight into the underlying molecular mechanisms has been severely limited by the absence of ECM scaffolds that enable their independent variation. The problem is further compounded by the lack of computational models of cell motility that integrate cellular adhesive and contractile mechanisms with ECM mechanics and geometry. To address these unmet needs in a systematic manner, we developed a unique microscale culture platform (μPACs) that enabled independent investigation of the effects of ECM stiffness and confinement on cell migration. As a result of these studies, we were able to show that matrix confinement profoundly alters the dependence of cell migration speed on ECM stiffness. We attribute this result to the effect of ECM stiffness and channel width on polarization of traction force; while overall cellular actomyosin contractility increases with ECM stiffness, isotropic spreading of the cell in 2D-likesettings (wide channels) spatially defocuses traction forces and reduces the net propulsive force in the direction of cell migration. Physical constraint of cells to a narrow channel forces polarization and alignment of these forces along the direction of the channel and promotes faster cell migration for a given ECM stiffness. Thus, narrow channels permit cells to exploit the high traction forces supported by stiff substrates without the loss of traction polarization associated with isotropic spreading observed on unconfined substrates. As a result, the fastest motility is observed in stiff, narrow channels.
Our work adds to a growing literature on the regulation of migration speed by ECM stiffness (8, 14, 28). A biphasic dependence of cell migration speed on ECM stiffness was first reported for vascular smooth muscle cells (SMCs) (14), and we have reported (and confirmed here) a similar relationship for human glioblastoma cells (28). Analogous to the biphasic dependence of migration speed on ECM ligand density (5, 36) this relationship is typically explained in terms of the balance between traction force generation and adhesion stability: Soft matrices do not support the traction forces needed for effective spreading and migration, resulting in unstable punctate focal adhesions, whereas stiff matrices support such high traction forces that adhesions are hyperstabilized to the point where they restrict motility. Consequently, an optimum migration exists at some intermediate ECM stiffness. Our data add to this understanding by elucidating the role of traction polarization, which is strongly affected both by stiffness and matrix confinement. Specifically, stiff ECMs reduce traction polarization by permitting isotropic spreading, but this can be overcome (rescued) by confining cells in narrow channels that force these traction forces to be highly aligned. Disruption of this polarization via myosin inhibition speeds motility and causes recovery of biphasic behavior in narrow channels. These findings also build upon recent studies demonstrating that restriction of cells to narrow ECM strips significantly increases migration speed, which may underlie the high, persistent motility rates observed in fibrillar ECMs (38, 39).
As more is understood about biophysical regulation of cell motility in 3D ECMs, it is becoming clear that ECM mechanics and architecture vary widely in both space and time, and that cell migration is closely attuned to these variations (17, 40). Given the diverse biophysical properties of the matrix present in vivo, it is critical to develop culture models that facilitate systematic in vitro deconstruction of how these inputs drive cell migration. We have now advanced these efforts by developing a paradigm for separating the roles of ECM stiffness and matrix confinement, using this system to discover a way to alter a previously established relationship between migration speed and ECM stiffness solely by varying matrix confinement, and identifying actomyosin contractility as an intracellular signaling pathway that enables confinement-mediated motility enhancement. We anticipate that the μPAC platform will facilitate the study of even more complex variations in ECM confinement and stiffness. Because of the high throughput and parallelizability of this microfluidic platform and its compatibility with most standard microscopy methods, our system also retains some of the key advantages of traditional 2D culture that may eventually play a role in high-throughput screening of intracellular and extracellular factors that modulate 3D invasion.
Materials and Methods
See SI Text for detailed descriptions of following methods: (a) Cell culture; (b) Live cell imaging and data analysis; (c) Immunofluorescence, confocal microscopy, and image analysis; (d) Statistical analysis; (e) Atomic force microscopy.
Fabrication of PA Microchannels.
Silicon masters were fabricated using standard photolithographic techniques (Fig. 1). A volume of PA precursor solution sufficient to achieve a gel of approximately 100 μm thickness was placed between a reactive glass surface and the silicon mold and allowed to polymerize. The PA precursor solutions were made as previously described (8) acrylamide/bisacrylamide (A/B) percentages of 4% A/0.2% B, 10% A/0.3% B, and 15% A/1.2% B corresponding to PA gels of elastic moduli of 0.4, 10, and 120 kPa (8, 15). Channel widths, cw, are reported with respect to the silicon master, not the final PA channels. See SI Text for a more detailed description.
Mathematical Model.
Our model assumes that cell migration is a result of several subcellular mechanisms working in tandem: stabilization of adhesions, extension of protrusions, and generation of actomyosin contractility to overcome drag forces (5, 28), as illustrated in Fig. S6 and described in more detail in SI Text.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by grants to SK from the National Institutes of Health (Director’s New Innovator Award 1DP2OD004213 and Physical Sciences-Oncology Center Award 1U54CA143836) and the National Science Foundation (CMMI 1105539). Confocal images were obtained at the CIRM/QB3 Stem Cell Shared Facility.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.E.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118073109/-/DCSupplemental.
References
- 1.Kumar S, Weaver V. Mechanics, malignancy, and metastasis: The force journey of a tumor cell. Cancer Metast Rev. 2009;28:113–127. doi: 10.1007/s10555-008-9173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: Tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2006;22:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedl P, Wolf K. Tumour-cell invasion and migration: Diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 4.Pathak A, Kumar S. Biophysical regulation of tumor cell invasion: Moving beyond matrix stiffness. Integr Biol. 2011;3:267–278. doi: 10.1039/c0ib00095g. [DOI] [PubMed] [Google Scholar]
- 5.Lauffenburger DA, Horwitz AF. Cell migration: A physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 6.Ulrich TA, Jain A, Tanner K, Mackay JL, Kumar S. Probing cellular mechanobiology in three-dimensional culture with collagen-agarose matrices. Biomaterials. 2010;31:1875–1884. doi: 10.1016/j.biomaterials.2009.10.047. [DOI] [PubMed] [Google Scholar]
- 7.Lam W, et al. Extracellular matrix rigidity modulates neuroblastoma cell differentiation and N-myc expression. Mol Cancer. 2010;9:35. doi: 10.1186/1476-4598-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulrich TA, de Juan Pardo EM, Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69:4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 10.Yeung T, et al. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 11.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Engler A, et al. Substrate compliance versus ligand density in cell on gel responses. Biophys J. 2004;86:617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelham RJ, Wang Y-l. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J Cell Physiol. 2005;204:198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- 15.Saha K, et al. Substrate modulus directs neural stem cell behavior. Biophys J. 2008;95:4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keung AJ, de Juan-Pardo EM, Schaffer DV, Kumar S. Rho GTPases mediate the mechanosensitive lineage commitment of neural stem cells. Stem Cells. 2011;29:1886–1897. doi: 10.1002/stem.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beadle C, et al. The role of myosin II in glioma invasion of the brain. Mol Biol Cell. 2008;19:3357–3368. doi: 10.1091/mbc.E08-03-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf K, et al. Compensation mechanism in tumor cell migration: Mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaman MH, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci USA. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irimia D, Toner M. Spontaneous migration of cancer cells under conditions of mechanical confinement. Integr Biol. 2009;1:506–512. doi: 10.1039/b908595e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rolli CG, Seufferlein T, Kemkemer R, Spatz JP. Impact of tumor cell cytoskeleton organization on invasiveness and migration: A microchannel-based approach. PLoS ONE. 2010;5:e8726. doi: 10.1371/journal.pone.0008726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heuzé ML, Collin O, Terriac E, Lennon-Duménil A-M, Piel M. Cell migration in confinement: A micro-channel-based assay. In: Wells CM, Parsons M, editors. Methods in Molecular Biology™. Vol. 769. Clifton, NJ: Humana Press; 2011. pp. 415–434. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y-l, Motte S, Kaufman LJ. Pore size variable type I collagen gels and their interaction with glioma cells. Biomaterials. 2010;31:5678–5688. doi: 10.1016/j.biomaterials.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 24.Peyton SR, et al. Marrow-derived stem cell motility in 3D synthetic scaffold is governed by geometry along with adhesivity and stiffness. Biotechnol Bioeng. 2011;108:1181–1193. doi: 10.1002/bit.23027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kandow CE, et al. Methods in Cell Biology. Vol. 83. New York: Academic Press; 2007. Polyacrylamide Hydrogels for Cell Mechanics: Steps Toward Optimization and Alternative Uses; pp. 29–46. [DOI] [PubMed] [Google Scholar]
- 26.Charest JM, Califano JP, Carey SP, Reinhart-King CA. Fabrication of substrates with defined mechanical properties and topographical features for the study of cell migration. Macromol Biosci. 2012;12:12–20. doi: 10.1002/mabi.201100264. [DOI] [PubMed] [Google Scholar]
- 27.Engler AJ, Richert L, Wong JY, Picart C, Discher DE. Surface probe measurements of the elasticity of sectioned tissue, thin gels and polyelectrolyte multilayer films: Correlations between substrate stiffness and cell adhesion. Surf Sci. 2004;570:142–154. [Google Scholar]
- 28.Pathak A, Kumar S. From molecular signal activation to locomotion: An integrated, multiscale analysis of cell motility on defined matrices. PLoS ONE. 2011;6:e18423. doi: 10.1371/journal.pone.0018423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levental I, Georges PC, Janmey PA. Soft biological materials and their impact on cell function. Soft Matter. 2007;3:299–306. doi: 10.1039/b610522j. [DOI] [PubMed] [Google Scholar]
- 30.Cai Y, et al. Cytoskeletal coherence requires myosin-IIA contractility. J Cell Sci. 2010;123:413–423. doi: 10.1242/jcs.058297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poincloux R, et al. Contractility of the cell rear drives invasion of breast tumor cells in 3D Matrigel. Proc Natl Acad Sci USA. 2011;108:1943–1948. doi: 10.1073/pnas.1010396108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Straight AF, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 33.Even-Ram S, et al. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat Cell Biol. 2007;9:299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- 34.Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J Cell Biol. 2010;188:877–890. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giannone G, et al. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 2004;116:431–443. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 36.DiMilla PA, Barbee K, Lauffenburger DA. Mathematical model for the effects of adhesion and mechanics on cell migration speed. Biophys J. 1991;60:15–37. doi: 10.1016/S0006-3495(91)82027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vicente-Manzanares M, Zareno J, Whitmore L, Choi CK, Horwitz AF. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J Cell Biol. 2007;176:573–580. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doyle AD, et al. Microenvironmental control of cell migration: Myosin IIA is required for efficient migration in fibrillar environments through control of cell adhesion dynamics. J Cell Sci. 2012 doi: 10.1242/jcs.098806. 10.1242/jcs.098806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ananthanarayanan B, Kim Y, Kumar S. Elucidating the mechanobiology of malignant brain tumors using a brain matrix-mimetic hyaluronic acid hydrogel platform. Biomaterials. 2011;32:7913–7923. doi: 10.1016/j.biomaterials.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.