Abstract
Iron/sulfur cluster (ISC)-containing proteins are essential components of cells. In most eukaryotes, Fe/S clusters are synthesized by the mitochondrial ISC machinery, the cytosolic iron/sulfur assembly system, and, in photosynthetic species, a plastid sulfur-mobilization (SUF) system. Here we show that the anaerobic human protozoan parasite Blastocystis, in addition to possessing ISC and iron/sulfur assembly systems, expresses a fused version of the SufC and SufB proteins of prokaryotes that it has acquired by lateral transfer from an archaeon related to the Methanomicrobiales, an important lineage represented in the human gastrointestinal tract microbiome. Although components of the Blastocystis ISC system function within its anaerobic mitochondrion-related organelles and can functionally replace homologues in Trypanosoma brucei, its SufCB protein has similar biochemical properties to its prokaryotic homologues, functions within the parasite’s cytosol, and is up-regulated under oxygen stress. Blastocystis is unique among eukaryotic pathogens in having adapted to its parasitic lifestyle by acquiring a SUF system from nonpathogenic Archaea to synthesize Fe/S clusters under oxygen stress.
Keywords: iron/sulfur cluster biosynthesis, lateral gene transfer, parasite evolution, sulfur-mobilization machinery, oxygen stress adaptation
The iron/sulfur cluster (ISC) biosynthetic machinery is a fundamental component of cells; more than 110 proteins in Escherichia coli require Fe/S clusters to function, and the number of Fe/S proteins in eukaryotes is even greater (1). Fe/S proteins are responsible for central functions in enzymatic catalysis, electron transport, photosynthesis, nitrogen fixation, and the regulation of gene expression (2, 3), and they are found in all compartments of the eukaryotic cell. Assembly of Fe/S clusters in vitro occurs spontaneously under favorable conditions when large amounts of free iron and sulfide are available. However, in vivo, these elements are toxic to cells (4); thus, their concentrations for the various cellular processes have to be tightly regulated. The assembly of Fe/S clusters is a complex process involving a host of different systems, each made up of numerous specific proteins that are widespread across the tree of life (5, 6). In eukaryotes, Fe/S proteins are assembled by several different systems, including the ISC assembly and the sulfur-mobilization (SUF) machineries, which are found only in mitochondria and plastids, respectively. Two exceptions to this rule are found in Entamoeba histolytica and Mastigamoeba balamuthi, which possess nitrogen fixation-type Fe/S systems, although the subcellular localizations of these systems remain controversial (7, 8). The maturation of cytosolic and nuclear Fe/S proteins in eukaryotes is typically dependent on the cytosolic iron/sulfur assembly (CIA) machinery, which is essential in yeast and is found in all eukaryotes investigated so far (9).
We have chosen to investigate the Fe/S cluster biogenesis systems of Blastocystis spp., a poorly studied unicellular anaerobic intestinal parasite of humans (10). Blastocystis has unusual mitochondrion-related organelles (MROs) that contain cristae and DNA, but appear to lack classical aerobic mitochondrial pathways and function in the complete absence of oxygen (10, 11). To clarify the functions of these organelles, the first broad transcriptomic (12) and genomic (13) studies of Blastocystis were recently completed; these studies have identified 115 and 360 genes, respectively, encoding putative mitochondrial and hydrogenosomal proteins. Among these were mitochondrial ISC system proteins (12–15), suggesting that Fe/S cluster biogenesis is a core function of Blastocystis MROs. Components of the CIA machinery have also been identified in the Blastocystis transcriptome. More unexpectedly, a partial sequence of a putative sufC gene was also identified in the Blastocystis transcriptomic data, suggestive of the existence of alternative and/or complementary Fe/S cluster biosynthetic machinery in this organism.
Here we investigate in detail the makeup, cellular locations, and functions of the various Fe/S cluster biogenetic systems in Blastocystis by using protein structural prediction, phylogenetic analyses, cellular localization, gene expression, genetic complementation, and/or biochemical investigations of members of the ISC machinery along with the putative SUF protein.
Results and Discussion
The Blastocystis SUF System Is a Unique Acquisition.
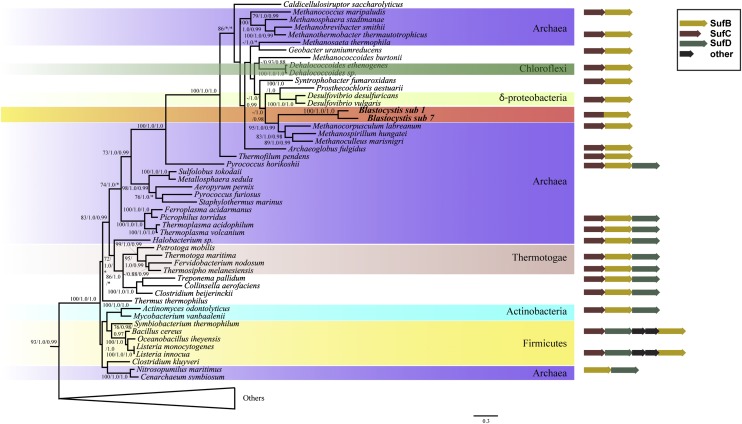
We identified a Blastocystis SufCB fused protein (SI Appendix, Fig. S1), herein referred to as Bh-SufCB, with domains homologous to the proteins encoded by the sufC and sufB genes of the sufCB operon of some bacteria and archaea (SI Appendix, Fig. S2). Phylogenetic analyses (SI Appendix, Supplementary Results) demonstrated that both domains of the fused Bh-SufCB protein cluster within a distinct group of homologues from the anaerobic or thermophilic archaea and bacteria (SI Appendix, Figs. S3 and S4), with closest affinities to the Methanomicrobiales (Fig. 1 and SI Appendix, Fig. S5). The Bh-SufCB protein shares a number of specific sequence features with these organisms (SI Appendix, Fig. S5), and hence it seems that Blastocystis has acquired its sufCB gene by lateral gene transfer (LGT) from a relative of the Methanomicrobiales. Recent studies have shown a rich phylogenetic diversity of these methane-producing anaerobic archaea in the gastrointestinal tract of humans (16), a habitat also occupied by Blastocystis. Consequently, it is likely that, within such an environment, an ancestor of Blastocystis acquired a sufCB operon from a methanoarchaeon gastrointestinal tract inhabitant and incorporated it into its genome; the transfer was then followed by point mutations and/or deletions that converted the two genes into a single ORF. Fusions of genes acquired by LGT corresponding to operons in bacteria have been documented in eukaryotes previously (17–19), and it may turn out to be a common mode of LGT-mediated adaptation. An extensive genome/transcriptome-wide analysis of multiple Blastocystis genomes is needed to determine if other gene(s) have been laterally acquired from this division of Archaea, as LGT was recently suggested to contribute to the adaptation of Blastocystis to its parasitic lifestyle (13).
Fig. 1.
Phylogeny of a concatenated SufCB alignment estimated by maximum likelihood (RAxML) and Bayesian analysis based on available sequences from Bacteria, Archaea, and Eukaryotes. The tree was generated from 532 aligned amino acids (111 taxa) using the LG+Gamma model. Blastocystis SufCB sequences cluster within a group of predominantly anaerobic prokaryotes, and with strong bootstrap support (95%) and posterior probability (1.0/0.99) as a sister group to the Methanobacteriales. Support values are shown above branches in the following order: maximum likelihood bootstrap support (LG model, RAxML), posterior probability (LG+G+F model, MrBayes), and posterior probability (C20 model, PhyloBayes). Only bipartitions that received >65% ML bootstrap support are labeled.
The Bh-SufCB Protein Is Localized in Cytosol and Functions in Fe/S Protein Maturation.
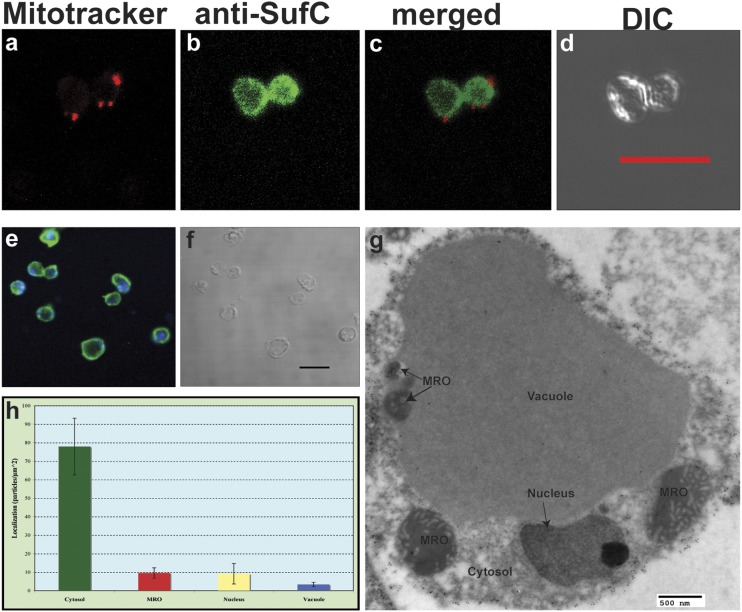
Not only does the Bh-SufCB protein have a genetic organization (SI Appendix, Fig. S2) and an origin that is completely distinct from the SUF system of photosynthetic eukaryotes, it also functions in a different cellular context. Western blot analysis of subcellular fractions (SI Appendix, Fig. S10E), immunofluorescence microscopy (Fig. 2 A–F), and immunogold EM (Fig. 2 G and H and SI Appendix, Fig. S11) experiments show that Bh-SufCB functions within the parasite’s cytosol, not within plastids like in photosynthetic eukaryotes (20–22). This is particularly intriguing because Blastocystis also encodes homologues of components of the CIA machinery (also likely localized in the cytosol) that, in yeast, are responsible for the maintenance of the cytosolic and nuclear Fe/S proteins (6).
Fig. 2.
Cellular localization of SufCB in Blastocystis cells. (A) MitoTracker red localizing to discrete structures corresponding to mitochondrion-related organelles of Blastocystis. (B) Rabbit anti-E. chrysanthemi SufC antiserum (1:500; green) shows a cytosolic localization corresponding to Bh SufCB. (C) Localization of MitoTracker and of the SufCB protein merged in one image. (D) Differential interference contrast (DIC) images of the cells used for immunofluorescence. (E) Rabbit anti-E. chrysanthemi SufC antisera (1:500; green) show a cytosolic localization corresponding to Bh SufCB and the blue corresponds to the dsDNA staining by using DRAQ5 dye. (F) DIC image of the cells. (Scale bar: 10 μm.) (G) Localization of SufCB in Blastocystis cell by transmission EM shows cytosolic localization. Two additional figures can be found in SI Appendix, Fig. S11H. Densities of labeling in different compartments of blastocystis cells suggest that SufCB is mainly localized in the cytosol of the parasite.
It is currently impossible to manipulate Blastocystis genetically. Thus, to determine if the Bh-SufCB could function in the maturation of Fe/S proteins in vivo, we employed two E. coli Fe/S cluster dependent transcriptional regulators, IscR and NsrR, as reporters for Fe/S protein maturation in E. coli. A vector expressing the Bh-SufCB protein was introduced in two E. coli strains: one carrying the lacZ reporter gene fused to a gene whose expression is repressed by the Fe/S bound form of IscR, iscR (piscR::lacZ); and the second one carrying the lacZ reporter gene fused to hmpA gene (phmpA::lacZ), whose expression is repressed by the Fe/S-bound form of NsrR (SI Appendix) (23, 24). Both strains were defective for ISC (ΔiscAU) and SUF (ΔsufCD) biogenesis pathways, and contain the eukaryotic mevalonate-dependent pathway that does not depend on Fe/S enzymes (25). These strains are incapable of synthesizing Fe/S clusters, and thus IscR and NsrR both remain in their “apo” form (i.e., lacking Fe/S cluster), and thus they do not block the transcription of piscR::lacZ and phmpA::lacZ fusions, respectively (SI Appendix, Fig. S6, empty vector). Transformation of these strains with the pBAD vector expressing the E. coli sufABCDSE operon caused a drastic decrease in piscR::lacZ and phmpA::lacZ fusions expression (∼80% for each), showing that Fe/S biogenesis (and thus maturation of IscR and NsrR) was restored by the SUF system (SI Appendix, Fig. S6). When the analogous experiment was performed by introducing the pBAD Bh-sufCB, decreased expression of the phmpA::lacZ (30%) and the piscR::lacZ (19%) lacZ fusions was observed (SI Appendix, Fig. S6). Therefore, in the absence of functional native ISC and SUF scaffolds, Bh-SufCB is able to partially restore NsrR and IscR maturation by assembling their Fe/S clusters, albeit less efficiently than the native E. coli SUF system.
The Bh-SufCB Protein Displays Similar Properties in Vitro to E. coli SufBC2D.
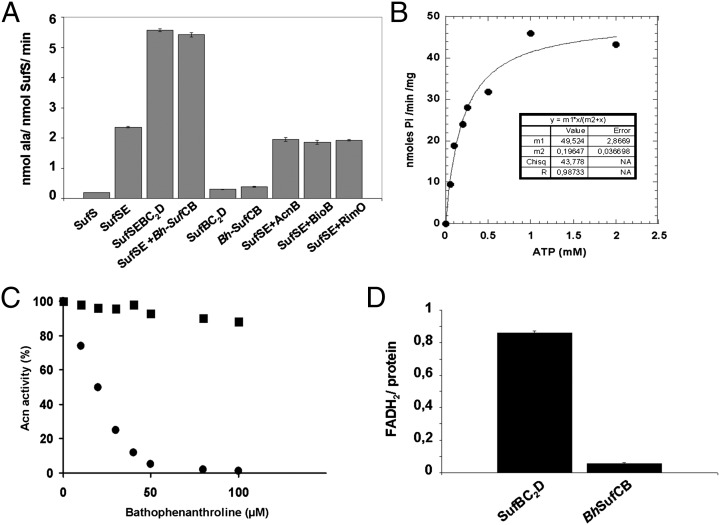
To provide further evidence that Bh-SufCB is involved in Fe/S protein maturation, we performed in vitro studies on the purified Bh-SufCB protein. Bh-SufCB expression in E. coli cells led to the production of insoluble protein in inclusion bodies, regardless of the growth conditions used. Therefore, we purified Bh-SufCB from inclusion bodies onto an Ni-NTA column and checked that the protein was correctly refolded by circular dichroism (SI Appendix, Fig. S7). In SDS/PAGE electrophoresis, the unfolded pure protein migrates at a size close to 77 kDa, in agreement with the predicted molecular weight. The refolded protein, that exists in a dimeric form (∼154 kDa) as determined by analytical size exclusion chromatography, was then assayed for several properties that were previously used to characterize the E. coli SufBC2D complex (26). The cysteine desulfurase activity of SufSE from E. coli in the presence of Bh-SufCB was first measured (Fig. 3A). The E. coli SufBC2D complex and Bh-SufCB protein enhanced the cysteine desulfurase activity of E. coli SufSE complex by a factor of two (Fig. 3A), and, as expected, the Bh-SufCB, like SufBC2D, displays no cysteine desulfurase activity in the absence of SufSE (26). In a control experiment, no enhancement of the SufSE cysteine desulfurase activity was detected when Bh-SufCB was replaced by proteins, such as apo-aconitase B, apo-biotin synthase (i.e., BioB), or the apo-methylthiotransferase RimO from E. coli, all of which, as Fe/S cluster-binding proteins, might potentially increase cysteine desulfurase activity. Thus, like the E. coli SufBC2D complex, the refolded Bh-SufCB protein specifically recognizes SufSE and stimulates its cysteine desulfurase activity.
Fig. 3.
(A) Enhancement of SufSE cysteine desulfurase activity. Cysteine desulfurase activity assays were performed as described in Materials and Methods. All assay mixtures contained 0.5 μM SufS from E. coli, 1.5 μM of SufE from E. coli, and 100 μM of L-cysteine. Either SufBC2D from E. coli or SufCB from Blastocystis (Bh-SufCB) were added at a final concentration of 1.5 μM. (B) Characterization of ATPase activity of purified Bh-SufCB protein. Activity was tested at pH 7.5 and 30 °C, in the presence of 5 mM MgCl2 and 1 mM ATP, in 25 mM Tris-HCl with a protein concentration of 10 μM in a final volume of 60 μL. Kinetic activities of SufCB were tested over a range of ATP concentrations (0–2 mM) for 5 min, allowing Vmax and Km values to be determined. The ATPase activity as a function of ATP was then plotted. Data have been fitted by regression to a saturation hyperbola according to Eq. 1. (C) Intact Fe/S cluster transfer from Bh-SufCB to apo-AcnB. Apo-AcnB (0.2 nmol) pretreated with 1 mM DTT and desalted was incubated anaerobically with either Bh-SufCB-(Fe/S) (0.2 nmol protein containing 4.1 nmol of Fe and 4.7 nmol of S) (■) or fivefold molar excess of Fe2+, S2−, and 5 mM DTT (●) in 100 μL of 50 mM Tris-HCl, pH 7.6, with increasing amounts of bathophenanthroline. After 20 min incubation, the activity of AcnB was measured by monitoring the absorption at 340 nm. For this, 300 μM MnCl2, 25 mM citrate, 0.5 unit of isocitrate dehydrogenase, and 0.25 mM NADP+ was added to the protein mixture in a final volume of 100 μL. (D) Protein-bound FADH2. SufBC2D from E. coli and SufCB from Blastocystis (in nmole) were incubated anaerobically for 2 h with fivefold molar excess of FAD, in the presence of 10 mM DTT and catalytic amount of DAF under illumination. After desalting, proteins were air-oxidized (O/N), boiled for 5 min, and centrifuged, and the corresponding clear supernatants were analyzed for their FAD content.
Bh-SufCB was found to exhibit an ATPase activity at pH 7.5 in the presence of MgCl2 measured by the release of Pi (SI Appendix, Supplementary Results). A steady-state kinetic analysis of the production of Pi catalyzed by Bh-SufCB is shown in Fig. 3B. Kinetic parameters could be estimated because the experimental values fit the following equation, indicating that the enzyme displays canonical Michaelis–Menten kinetics.
For Bh-SufCB, the Vmax value was 0.049 μmol of inorganic phosphate per minute per milligram protein, and the Km value for ATP was 196 μM (Fig. 3B). In comparison, the E. coli SufBC2D complex has a Vmax of 0.035 μmol of inorganic phosphate per minute per milligram protein and a Km of 200 μM. Collectively, these results show that the refolded Bh-SufCB protein possesses comparable ATP-hydrolyzing properties to the E. coli SufBC2D complex.
Inclusion bodies obtained from Bh-SufCB–expressing cells were red-brown in color, and, during purification, a slight pale pink color remained, suggesting the presence of an Fe/S cluster within Bh-SufCB. However, iron and sulfur composition analysis revealed only 0.1 ± 0.05 Fe and 0.08 ± 0.03 S per protein suggesting that an Fe/S cluster was lost during purification as is often observed. Therefore, we tested the ability of Bh-SufCB to assemble a full Fe/S cluster by incubating the isolated refolded Bh-SufCB protein in the apo-form with a sixfold molar excess of ferrous iron and sodium sulfide under anaerobic conditions for 5 h. After desalting to remove unbound iron and sulfide, the protein solution was analyzed by UV-visible spectroscopy and for its iron and sulfur content. The UV spectrum of the chemically reconstituted Bh-SufCB [Bh-SufCB-(Fe/S)] is characteristic of a (4Fe-4S)2+ cluster containing protein with a unique absorption band at 420 nm (SI Appendix, Fig. S8). Iron and sulfur composition analysis showed an average of 4.2 ± 0.2 Fe atoms per dimer and a slightly higher amount of S (4.5 ± 0.3 sulfur atoms per dimer), as previously observed for E. coli SufBC2D (26). Reduction with dithionite under anaerobic conditions led to a rapid bleaching of the solution. During this reaction, the initial electron paramagnetic resonance silent protein was converted to a S=1/2 species, characterized by an axial electron paramagnetic resonance signal with g values at 2.04(1) and 1.92(9) (SI Appendix, Fig. S8, Inset). Temperature dependence and microwave power saturation properties of the signal were in agreement with the presence of a (4Fe-4S)+1 center that accounts for 40% of total iron.
We then investigated the ability of Bh-SufCB to transfer its Fe/S cluster to E. coli aconitase B (AcnB), a (4Fe-4S) protein. A stoichiometric amount of Bh-SufCB-(Fe/S) was incubated with apo-AcnB to provide a sufficient amount of Fe/S to fully maturate AcnB. As shown in Fig. 3C, aconitase activation was observed when the apo-protein was incubated with (Fe/S)-Bh-SufCB for 20 min. Addition of increasing concentration of a strong chelator, bathophenanthroline, to the reaction mixture had very little effect on the Fe/S cluster transfer from Bh-SufCB to apo-AcnB as AcnB still remains fully active. On the contrary, the chelator completely inhibited the chemical reconstitution of apo-AcnB performed by using 5× molar excess of iron and sulfide in the presence of DTT. This experiment unambiguously demonstrates that the Bh-SufCB protein can function as a scaffold Fe/S protein.
Finally, we analyzed the flavin-binding properties of the refolded Bh-SufCB. When E. coli SufBC2D complex is anaerobically purified from E. coli, it has a bound FADH2 molecule (0.8 ± 0.1 mol FADH2/mol of complex). Alternatively, the same complex can be reconstituted from an apo-form by incubation with an excess of FADH2 and desalting under anaerobic conditions, leading to a complex containing 0.86 ± 0.1 equivalent of FADH2 (26). Under similar conditions, Bh-SufCB failed to bind FADH2 (Fig. 3D), consistent with the absence of the flavin-binding residues observed in the sequence analyses of Bh-SufCB (SI Appendix, Fig. S9).
The ISC System Is Localized in Blastocystis MROs.
In addition to the Blastocystis SUF machinery, we sought to investigate the localization and function of its putative mitochondrial ISC system previously suggested to function within its biochemically unique anaerobic MRO (12, 13). The unusual properties of the Blastocystis MRO raises questions about the localization and functionality of its ISC machinery, especially because recent studies have shown that other parasitic anaerobic protists with MROs, including the microsporidian Trachipleistophora hominis and the amoebozoan Entamoeba histolytica, may have relocated some components of their Fe/S cluster biosynthetic machinery into the cytosol (7, 27). Our investigations reveal that Blastocystis possesses homologues of nearly all components of the mitochondrial ISC machinery in yeast (SI Appendix, Table S1). Moreover, in the Blastocystis transcriptome, we identified components of the CIA machinery such as Cia1, Nbp35, and Nar1. Although targeting prediction programs suggested that some of the ISC components are targeted to Blastocystis MROs (SI Appendix, Table S1), the limited predictive power of these programs when applied to diverse lineages of microbial eukaryotes led us to conduct thorough experimental investigations.
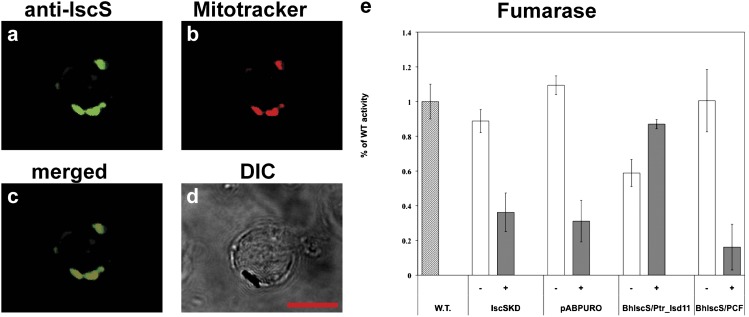
Immunofluorescence analyses (SI Appendix, Supplementary Results) showed that heterologous antibodies against Blastocystis IscS, IscU, and frataxin (SI Appendix, Fig. S10 B–D) colocalized with the mitochondrial marker dye MitoTracker (Fig. 4 A–D and SI Appendix, Fig. S12), demonstrating that core elements of the ISC biosynthetic machinery function within the Blastocystis MROs, similar to mitochondria and mitochondrion-related organelles of most eukaryotes (3, 28). Although the primary sequences of these proteins have conserved sequence features associated with canonical eukaryotic ISC functions (SI Appendix, Figs. S13–S15), we sought more direct evidence of their activities. To characterize the above proteins, full-length copies of Blastocystis frataxin, iscU, and iscS genes were amplified from cDNA and were cloned into the pABPURO vector for transfection into Trypanosoma brucei (Tb). Upon linearization, the constructs were introduced into the 29–13 procyclic Tb cells, in which the expression of the Tb frataxin (29), or IscU or IscS (30), was ablated by inducible RNAi. The empty pABPURO vector and Tb-frataxin, Tb-IscU, or Tb-IscS RNAi knock-down cells were used as controls. Although the Bh-frataxin and Bh-IscS transfections were successful, repeated attempts to transfect the Tb procyclic cells with the pABPURO vector containing Bh-IscU were unsuccessful. To overcome this problem, we used different strategies involving alternative targeting sequences at the N-terminal sequence of Bh-IscU [an approach that has been successful in Tb rescued with human frataxin (31)], which also failed. After the addition of tetracycline, which triggers RNAi, growth of the Tb-frataxin and Tb-IscS knock-down controls virtually stopped at day 5 postinduction (SI Appendix, Figs. S16 and S17), as, in the absence of these proteins, the cells cease dividing and eventually die (29, 30). However, the procyclic trypanosomes in which RNAi-ablated Tb-frataxin was replaced by Bh-frataxin in trans retained almost the same growth as their noninduced counterparts (SI Appendix, Fig. S16), suggesting that Bh frataxin can replace the Tb homologue. In contrast, no obvious rescue of growth was observed for Bh-IscS introduced into the inducible Tb-IscS RNAi background (SI Appendix, Fig. S17), likely a consequence of the failure of Bh-IscS to function in the context of the heterologous Tb ISC machinery. Specifically, IscS is known to closely interact with the ISC protein Isd11 in yeast (32, 33), humans (33), and probably other eukaryotes (34), including Tb (35). Therefore, it is possible that Bh-IscS is unable to form a functional complex with Tb-Isd11. If this hypothesis were correct, growth of the RNAi-induced Tb-IscS knock-down may be restored by the parallel introduction of Bh-IscS and its probable native binding partner (as detailed later). However, no Isd11 homologue could be identified among the Blastocystis ESTs, nor could we amplify it by degenerate PCR. Fortunately, in the recently sequenced genome of the diatom Phaeodactylum tricornutum, a stramenopile relative of Blastocystis, we identified a full-length Isd11 homologue (Pt-Isd11). On the assumption that Pt-Isd11 may more closely resemble the Bh orthologue than the Tb-Isd11 (SI Appendix, Fig. S18), we amplified full-length Pt-Isd11 and cloned it into the Tb pCF-4 vector. Tb-IscS RNAi procyclic trypanosomes expressing Bh-IscS were transfected with Pt-Isd11 or an empty PCF-4 vector used as a control. Cells depleted for Tb-IscS and coexpressing Bh-IscS and Pt-Isd11 showed partial growth rescue relative to their noninduced counterparts (SI Appendix, Fig. S17), whereas no rescue was observed in cells transfected with Bh-IscS and the empty pCF-4 vector control (SI Appendix, Fig. S17).
Fig. 4.
Cellular localization of IscS protein in Blastocystis and characterization in Tb knock-downs. (A) Rabbit anti-Trichomonas IscS antibodies (1:200) detect Blastocystis IscS. (B) MitoTracker red labels discrete structures corresponding to the MROs of Blastocystis. (C) Colocalization of MitoTracker red with the IscS proteins. (D) DIC images of the cells used for immunofluorescence. (Scale bar: 10 μm.) (E) Activities of fumarase in total Tb-IscS cell lysates after 5 d of RNAi induction. White bars (−), noninduced knock-down cells, gray bars (+), RNAi-induced knock-down cells; vector, Tb knock-down cells transfected with the empty pABPURO vector as a control; Bh, Tb knock-down cells transfected with the Bh IscS gene; WT, 29–13 WT strain of Tb; PCF, empty vector as control.
The rescued growth phenotype (SI Appendix, Figs. S16 and S17) indicates that Bh-frataxin and Bh-IscS plus Pt-Isd11 can partially replace the function of Tb-frataxin and Tb-IscS in the Tb mitochondrion. As previous studies have shown that the down-regulation of Tb-frataxin and Tb-IscS causes a disruption of Fe/S cluster assembly (29, 36), it is likely that this function is provided in trans by the Blastocystis homologue. To verify this assumption, we measured activities of several previously described Fe/S cluster-containing enzymes with mitochondrial and/or cytosolic localization (29, 37). Activity of the mitochondrial Fe/S cluster-containing aconitase dropped to less than 20% of the WT activity in the RNAi-induced Tb-frataxin (SI Appendix, Fig. S16) and in the RNAi-induced Tb-IscS knock-down cells (SI Appendix, Fig. S17) or in the corresponding cells transfected with an empty pABPURO vector (SI Appendix, Figs. S16 and S17). However, in cells expressing Bh-frataxin or Bh-IscS plus Pt-Isd11, respectively, aconitase activity increased to approximately 80% to 90% of the WT level (SI Appendix, Figs. S16 and S17). Similarly, the activities of the mitochondrial enzymes fumarase and succinate dehydrogenase (respiratory complex II) in the RNAi-induced cells transfected with either of these constructs were three to four times higher than in their absence (Fig. 4E and SI Appendix, Figs. S16 and S17). Surprisingly, in the RNAi-induced Tb-IscS cells coexpressing Bh-IscS plus Pt-Isd11, the enzymatic activities reached ∼125% of those from the noninduced cells. The lowered activity of the noninduced cells relative to vector-only controls is likely a result of nonfunctional hybrid IscS-Isd11 complexes formed between Trypanosoma and Blastocystis or Phaeodactylum proteins. In any case, as expected, the activity of the control Fe/S cluster-lacking enzyme threonine dehydrogenase was similar in the studied cell lines and the controls (SI Appendix, Figs. S16 and S17). In summary, these data suggest that Blastocystis ISC homologues (IscS and frataxin) are conserved, located in MROs, and can functionally replace the Trypanosoma brucei homologues in Fe/S cluster biogenesis. Our data also tentatively supports the possibility that Blastocystis IscS functions through interaction with an Isd11 homologue in its MROs. Because we were unable to find an Isd11 homologue in the transcriptomic data from this organism or in the published Blastocystis subtype 7 genome (13), the possibility of an IscS–Isd11 interaction in Blastocystis will require further experimental investigation.
Oxygen-Stress Up-Regulates the SUF System.
Collectively, our results demonstrate that at least two (and likely three, including the CIA system) conserved and functional Fe/S cluster biosynthetic systems coexist in the Blastocystis cell (SI Appendix, Fig. S19). This leads naturally to the question of why such apparent redundancy in Fe/S biogenesis systems has evolved in this anaerobic microbial eukaryote. We hypothesize that Blastocystis acquired the SUF machinery by LGT as a unique adaptation to support the metabolic requirements of its distinctive anaerobic lifestyle. For example, the hydrogenosome-like metabolism in the Blastocystis MRO involves the activities of oxygen-sensitive Fe/S enzymes such as pyruvate:ferredoxin oxidoreductase, [FeFe]-hydrogenase, and ferredoxins (12). It is possible that Blastocystis, like some prokaryotes, uses its alternative SUF system to support and/or repair its Fe/S clusters under stress conditions (20, 38), a hypothesis supported by our observation that Bh-sufCB is highly up-regulated under oxygen stress (SI Appendix, Fig. S20), whereas the expression level of ISC components and several Fe/S proteins remained constant under these conditions. Enhanced expression of the Bh-sufCB gene could mitigate the degradation of Fe/S clusters damaged by oxidative stress; Bh-SufCB could thus provide the necessary substrate for the repair of the damaged Fe/S clusters and subsequent recovery of the Fe/S proteins (SI Appendix, Fig. S19). If so, it is possible that similar SUF systems could be discovered in other poorly studied anaerobic protistan lineages. In addition, as this machinery is unique among plastid-lacking eukaryotes, it could be used as a potential drug-target for Blastocystis, which has been linked to gastrointestinal disease in humans (39).
Conclusions
We have demonstrated that, uniquely among plastid-lacking eukaryotes, Blastocystis has acquired a sufCB gene by LGT from a Methanobacteriales archaeon that was likely part of the gut microbiome of its host. We hypothesize this served as a protective adaptation to exposure to atmospheric oxygen, a stressful environment that the parasite encounters during its transmission to a new host. Unlike photosynthetic eukaryotes in which components of the SUF machinery function within their plastids, the unique Bh-SufCB fusion protein is localized in the cytosol, is capable of functioning in Fe/S biogenesis in vivo in E. coli and in vitro, and may aid in the repair of Fe/S proteins after exposure to oxygen.
Materials and Methods
Western blot analyses were used to determine the specificity of antibodies raised against IscS of Trichomonas vaginalis (36), IscU of Giardia intestinalis (40), Frataxin of Tb (29), and SufC of Erwinia chrysanthemi (41) in Blastocystis protein extracts as previously described (42). To determine the cellular localization of these proteins, we used confocal immunofluorescence analyses, and, additionally, in the case of Bh-SufCB, immunogold transmission EM. For the Tb complementation studies, full-length Blastocystis iscs and frataxin genes were cloned into the pABPURO vector. After linearization, the vectors were transfected into Tb knockdown strains. The rescued strains were tested for complementation by measuring the activities of mitochondrial and cytosolic Fe/S enzymes as demonstrated earlier (29). In the case of Bh-SufCB, functional characterization of the protein was performed to determine the FADH2 binding affinities of the protein, the presence of an ATPase activity, and its capacity to interact with a cysteine desulfurase as described for E. coli homologues (26). Genetic complementation of E. coli ISC and SUF KO strains by Bh-SufCB was assessed through reporter gene repression assays (43). For the phylogenetic analyses, selected sequences of the orthologous genes were obtained from GenBank and other databases and aligned. The alignments were manually trimmed and subsequently analyzed. To assay transcriptional expression, total RNA was isolated using TRIzol (Invitrogen) and reverse-transcribed. Quantitative reverse transcriptase-PCR assays were performed. Detailed procedures are described in the SI Appendix.
Supplementary Material
Acknowledgments
The authors thank Prof. Jan Tachezy for providing the IscS and IscU antibodies; Dr. Barbara Karten for her help with the quantitative gene expression experiments; Grant C. Stevens for the immunoelectron microscopy experiments; S. Arragain (Laboratoire de Chimie et Biologie des Métaux/Biocatalyse) for providing RimO protein; and Dr. V. Forge (Institut de Recherches en Technologies et Sciences pour le Vivant/Laboratoire de Chimie et Biologie des Métaux/Amyloid Fibres: From Foldopathies to NanoDesign, Commissariat à l'Energie Atomique) for circular dichroism spectrum analysis. This work was supported by Comparative Genomics and Evolutionary Bioinformatics postdoctoral awards from the Tula Foundation (to A.D.T. and E.G.), an EMBO fellowship (to A.D.T.), an International Outgoing Fellowships Marie Curie Fellowship (to A.D.T.), the Canada Research Chairs Program (A.J.R.), the Canadian Institute for Advanced Research Program in Integrated Microbial Biodiversity (to A.J.R. and J.L.), Canadian Institutes for Health Research Operating Grant MOP-62809 (to A.J.R.), Czech Republic Grant Agency Grants 204/09/1667 and P305/11/2179 (to J.L.), a Praemium Academiae Award (to J.L.) and grants from the Centre National de la Recherche Scientifique, the Agence Nationale de la Recherche (Blanc SPV05511), and the Commissariat à l'Energie Atomique (to F.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GeneBank database (accession nos. JN399203–JN399216).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116067109/-/DCSupplemental.
References
- 1.Py B, Barras F. Building Fe-S proteins: Bacterial strategies. Nat Rev Microbiol. 2010;8:436–446. doi: 10.1038/nrmicro2356. [DOI] [PubMed] [Google Scholar]
- 2.Beinert H, Kiley PJ. Fe-S proteins in sensing and regulatory functions. Curr Opin Chem Biol. 1999;3:152–157. doi: 10.1016/S1367-5931(99)80027-1. [DOI] [PubMed] [Google Scholar]
- 3.Lill R, Mühlenhoff U. Iron-sulfur protein biogenesis in eukaryotes: Components and mechanisms. Annu Rev Cell Dev Biol. 2006;22:457–486. doi: 10.1146/annurev.cellbio.22.010305.104538. [DOI] [PubMed] [Google Scholar]
- 4.Xu XM, Møller SG. Iron-sulfur clusters: Biogenesis, molecular mechanisms, and their functional significance. Antioxid Redox Signal. 2011;15:271–307. doi: 10.1089/ars.2010.3259. [DOI] [PubMed] [Google Scholar]
- 5.Frazzon J, Dean DR. Formation of iron-sulfur clusters in bacteria: An emerging field in bioinorganic chemistry. Curr Opin Chem Biol. 2003;7:166–173. doi: 10.1016/s1367-5931(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 6.Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 7.Maralikova B, et al. Bacterial-type oxygen detoxification and iron-sulfur cluster assembly in amoebal relict mitochondria. Cell Microbiol. 2010;12:331–342. doi: 10.1111/j.1462-5822.2009.01397.x. [DOI] [PubMed] [Google Scholar]
- 8.Mi-ichi F, Abu Yousuf M, Nakada-Tsukui K, Nozaki T. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc Natl Acad Sci USA. 2009;106:21731–21736. doi: 10.1073/pnas.0907106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lill R, Mühlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: Mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 10.Tan KS. Blastocystis in humans and animals: new insights using modern methodologies. Vet Parasitol. 2004;126:121–144. doi: 10.1016/j.vetpar.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 11.Zierdt CH. Blastocystis hominis—past and future. Clin Microbiol Rev. 1991;4:61–79. doi: 10.1128/cmr.4.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stechmann A, et al. Organelles in Blastocystis that blur the distinction between mitochondria and hydrogenosomes. Curr Biol. 2008;18:580–585. doi: 10.1016/j.cub.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denoeud F, et al. Genome sequence of the stramenopile Blastocystis, a human anaerobic parasite. Genome Biol. 2011;12:R29. doi: 10.1186/gb-2011-12-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long S, et al. Stage-specific requirement for Isa1 and Isa2 proteins in the mitochondrion of Trypanosoma brucei and heterologous rescue by human and Blastocystis orthologues. Mol Microbiol. 2011;81:1403–1418. doi: 10.1111/j.1365-2958.2011.07769.x. [DOI] [PubMed] [Google Scholar]
- 15.Tsaousis AD, et al. The Blastocystis mitochondrion-like organelles. In: Clark CG, Adam RD, Johnson PJ, editors. Anaerobic Parasitic Protozoa: Genomics and Molecular Biology. Norwich, UK: Horizon Scientific Press; 2010. [Google Scholar]
- 16.Oxley AP, et al. Halophilic archaea in the human intestinal mucosa. Environ Microbiol. 2010;12:2398–2410. doi: 10.1111/j.1462-2920.2010.02212.x. [DOI] [PubMed] [Google Scholar]
- 17.Andersson JO. Gene transfer and diversification of microbial eukaryotes. Annu Rev Microbiol. 2009;63:177–193. doi: 10.1146/annurev.micro.091208.073203. [DOI] [PubMed] [Google Scholar]
- 18.Andersson JO, Roger AJ. A cyanobacterial gene in nonphotosynthetic protists—an early chloroplast acquisition in eukaryotes? Curr Biol. 2002;12:115–119. doi: 10.1016/s0960-9822(01)00649-2. [DOI] [PubMed] [Google Scholar]
- 19.Andersson JO, Sjögren AM, Davis LA, Embley TM, Roger AJ. Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr Biol. 2003;13:94–104. doi: 10.1016/s0960-9822(03)00003-4. [DOI] [PubMed] [Google Scholar]
- 20.Rangachari K, et al. SufC hydrolyzes ATP and interacts with SufB from Thermotoga maritima. FEBS Lett. 2002;514:225–228. doi: 10.1016/s0014-5793(02)02369-4. [DOI] [PubMed] [Google Scholar]
- 21.Shen B, Goldbeck JH. Assembly of the bound iron-sulfur clusters in photosystem I. In: Goldbeck JH, editor. Photosystem I: The Light-Driven Plastocyanin:Ferredoxin Oxidoreductase. Dordrecht, The Netherlands: Springer; 2006. pp. 529–547. [Google Scholar]
- 22.Xu XM, Møller SG. AtNAP7 is a plastidic SufC-like ATP-binding cassette/ATPase essential for Arabidopsis embryogenesis. Proc Natl Acad Sci USA. 2004;101:9143–9148. doi: 10.1073/pnas.0400799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bodenmiller DM, Spiro S. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J Bacteriol. 2006;188:874–881. doi: 10.1128/JB.188.3.874-881.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartz CJ, et al. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc Natl Acad Sci USA. 2001;98:14895–14900. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loiseau L, et al. ErpA, an iron sulfur (Fe S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:13626–13631. doi: 10.1073/pnas.0705829104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wollers S, et al. Iron-sulfur (Fe-S) cluster assembly: the SufBCD complex is a new type of Fe-S scaffold with a flavin redox cofactor. J Biol Chem. 2010;285:23331–23341. doi: 10.1074/jbc.M110.127449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldberg AV, et al. Localization and functionality of microsporidian iron-sulphur cluster assembly proteins. Nature. 2008;452:624–628. doi: 10.1038/nature06606. [DOI] [PubMed] [Google Scholar]
- 28.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 29.Long S, et al. Ancestral roles of eukaryotic frataxin: Mitochondrial frataxin function and heterologous expression of hydrogenosomal Trichomonas homologues in trypanosomes. Mol Microbiol. 2008;69:94–109. doi: 10.1111/j.1365-2958.2008.06260.x. [DOI] [PubMed] [Google Scholar]
- 30.Smíd O, et al. Knock-downs of iron-sulfur cluster assembly proteins IscS and IscU down-regulate the active mitochondrion of procyclic Trypanosoma brucei. J Biol Chem. 2006;281:28679–28686. doi: 10.1074/jbc.M513781200. [DOI] [PubMed] [Google Scholar]
- 31.Long S, Jirku M, Ayala FJ, Lukes J. Mitochondrial localization of human frataxin is necessary but processing is not for rescuing frataxin deficiency in Trypanosoma brucei. Proc Natl Acad Sci USA. 2008;105:13468–13473. doi: 10.1073/pnas.0806762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiedemann N, et al. Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 2006;25:184–195. doi: 10.1038/sj.emboj.7600906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi Y, Ghosh MC, Tong WH, Rouault TA. Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Hum Mol Genet. 2009;18:3014–3025. doi: 10.1093/hmg/ddp239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards TA, van der Giezen M. Evolution of the Isd11-IscS complex reveals a single alpha-proteobacterial endosymbiosis for all eukaryotes. Mol Biol Evol. 2006;23:1341–1344. doi: 10.1093/molbev/msl001. [DOI] [PubMed] [Google Scholar]
- 35.Paris Z, et al. The Fe/S cluster assembly protein Isd11 is essential for tRNA thiolation in Trypanosoma brucei. J Biol Chem. 2010;285:22394–22402. doi: 10.1074/jbc.M109.083774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutak R, et al. Mitochondrial-type assembly of FeS centers in the hydrogenosomes of the amitochondriate eukaryote Trichomonas vaginalis. Proc Natl Acad Sci USA. 2004;101:10368–10373. doi: 10.1073/pnas.0401319101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horváth A, et al. Downregulation of the nuclear-encoded subunits of the complexes III and IV disrupts their respective complexes but not complex I in procyclic Trypanosoma brucei. Mol Microbiol. 2005;58:116–130. doi: 10.1111/j.1365-2958.2005.04813.x. [DOI] [PubMed] [Google Scholar]
- 38.Mettert EL, Outten FW, Wanta B, Kiley PJ. The impact of O(2) on the Fe-S cluster biogenesis requirements of Escherichia coli FNR. J Mol Biol. 2008;384:798–811. doi: 10.1016/j.jmb.2008.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zierdt CH. Blastocystis hominis, a long-misunderstood intestinal parasite. Parasitol Today. 1988;4:15–17. doi: 10.1016/0169-4758(88)90049-x. [DOI] [PubMed] [Google Scholar]
- 40.Tovar J, et al. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature. 2003;426:172–176. doi: 10.1038/nature01945. [DOI] [PubMed] [Google Scholar]
- 41.Nachin L, Loiseau L, Expert D, Barras F. SufC: An unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 2003;22:427–437. doi: 10.1093/emboj/cdg061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsaousis AD, et al. A functional Tom70 in the human parasite Blastocystis sp.: Implications for the evolution of the mitochondrial import apparatus. Mol Biol Evol. 2011;28:781–791. doi: 10.1093/molbev/msq252. [DOI] [PubMed] [Google Scholar]
- 43.Vinella D, Brochier-Armanet C, Loiseau L, Talla E, Barras F. Iron-sulfur (Fe/S) protein biogenesis: Phylogenomic and genetic studies of A-type carriers. PLoS Genet. 2009;5:e1000497. doi: 10.1371/journal.pgen.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.