Abstract
The conversion of recalcitrant plant-derived cellulosic biomass into biofuels is dependent on highly efficient cellulase systems that produce near-quantitative levels of soluble saccharides. Similar to other fungal and bacterial cellulase systems, the multienzyme cellulosome system of the anaerobic, cellulolytic bacterium Clostridium thermocellum is strongly inhibited by the major end product cellobiose. Cellobiose-induced inhibition can be relieved via its cleavage to noninhibitory glucose by the addition of exogenous noncellulosomal enzyme β-glucosidase; however, because the cellulosome is adsorbed to the insoluble substrate only a fraction of β-glucosidase would be available to the cellulosome. Towards this end, we designed a chimeric cohesin-fused β-glucosidase (BglA-CohII) that binds directly to the cellulosome through an unoccupied dockerin module of its major scaffoldin subunit. The β-glucosidase activity is thus focused at the immediate site of cellobiose production by the cellulosomal enzymes. BglA-CohII was shown to retain cellobiase activity and was readily incorporated into the native cellulosome complex. Surprisingly, it was found that the native C. thermocellum cellulosome exists as a homooligomer and the high-affinity interaction of BglA-CohII with the scaffoldin moiety appears to dissociate the oligomeric state of the cellulosome. Complexation of the cellulosome and BglA-CohII resulted in higher overall degradation of microcrystalline cellulose and pretreated switchgrass compared to the native cellulosome alone or in combination with wild-type BglA in solution. These results demonstrate the effect of enzyme targeting and its potential for enhanced degradation of cellulosic biomass.
Keywords: biomass conversion, enzyme inhibition, enzyme synergy, alternative energy
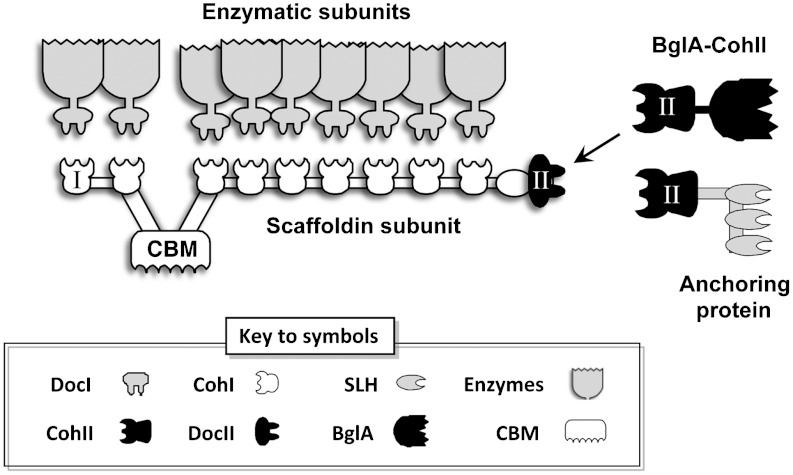
Cellulose, the most abundant biopolymer on earth, is an excellent source of energy for polysaccharide-degrading microorganisms and an unexploited potential for converting this renewable biomass into biofuels (1–6). Due to the highly ordered, insoluble, crystalline nature of the cellulose, very few microorganisms possess the necessary enzymatic system to efficiently degrade cellulosic substrates to soluble sugar (7, 8). Highly efficient cellulose degradation has been demonstrated by a multienzyme complex termed cellulosome produced by the anaerobic, thermophilic, cellulolytic bacterium, C. thermocellum (9–12). The cellulosome (Fig. 1) contains a noncatalytic subunit called scaffoldin that binds the insoluble substrate via a cellulose-specific carbohydrate-binding module (CBM). The C. thermocellum scaffoldin also contains a set of nine subunit-binding modules coined cohesins that mediate the specific incorporation and organization of the catalytic subunits through a complementary binding module (dockerin) that is carried by each enzymatic subunit. The scaffoldin contains another type of dockerin (type II) at its C terminus that mediates the attachment of the cellulosome to the cell wall through a selective binding interaction with a set of cell-anchoring proteins. The assembly of the enzymes into the complex ensures their collective targeting to a specific region of the substrate thereby facilitating stronger synergism among the catalytic components (13–15).
Fig. 1.
Schematic view of the C. thermocellum cellulosome and the proposed attachment of the chimeric β-glucosidase-fused type-II cohesin (BglA-CohII). The type-I cohesin-dockerin interaction integrates the dockerin-containing enzymatic subunits into the complex via interaction with the scaffoldin-borne type-I cohesins, whereas the carbohydrate-binding module (CBM) binds the complex to the insoluble substrate. C. thermocellum can produce over 70 different dockerin-bearing enzymes that are integrated into the cellulosome through interaction with any of the nine cohesins of the scaffoldin subunit. In the cell surface-attached state, the cellulosomal DocII module binds selectively to the CohII of an anchoring protein. In the cell-free state, unoccupied DocII positions can be used for specific incorporation of BglA-CohII into the purified cellulosome.
The synergistic degradation of the different enzymes comprising the cellulosome results in the formation of large concentrations of the major soluble disaccharide end product cellobiose. In the native environment, the cellobiose and other oligodextrins are transported directly into the cell by ATP binding cassette transporter systems (16) during which they are hydrolyzed to glucose by periplasmic β-glucosidases (17). The assimilation of oligodextrins can be accomplished by various additional microorganisms in the environment, and cellobiose is rapidly removed from the medium (14, 18). In the native ecosystem, cellobiose serves in a regulatory capacity and acts as a strong inhibitor, mainly for exocellulases; near-complete inhibition of the C. thermocellum cellulosome occurs at a concentration of 2% cellobiose (19, 20). Therefore, in a cell-free system, removal of the inhibitory cellobiose is essential for constant degradation of the lignocellulose substrate.
Previous works have shown that addition of the cellobiose-degrading enzyme β-glucosidase can enhance the rate and degree of solubilization of crystalline cellulose by the C. thermocellum cellulosome (21, 22). It does so by converting cellobiose to two molecules of noninhibitory glucose; however, in the process of crystalline cellulose degradation, the cell-free cellulosome binds to the insoluble cellulosic substrate and, therefore, only a fraction of the soluble β-glucosidase can be involved directly in digestion of cellobiose that accumulates in the immediate environment of the substrate-attached cellulosome. A mechanism that would bind the β-glucosidase to the cellulosome would therefore be expected to cleave the inhibitory cellobiose at greater efficiency and enhance the overall cellulose degradation.
The Lego-like architecture of the C. thermocellum cellulosome holds great potential for creating “designer cellulosomes,” artificial assemblies comprising hybrid forms of cellulosomal components for improved hydrolysis of cellulosic substrates (14). To date, most of the designer cellulosome experiments try to mimic the enzymatic synergism observed for native cellulosome systems by fabricating complexes composed of an artificial chimeric cohesin-containing scaffoldin and a set of matching dockerin-containing cellulases (23–25).
In this work, we examined an alternative approach by which the cellulosomal type-II cohesin-dockerin interaction was employed for specific incorporation of exogenous β-glucosidase into the native cellulosome (Fig. 1). As the main function of the interaction between the terminal scaffoldin-borne type-II dockerin (DocII) and the type-II cohesin (CohII) is cell-surface attachment, in a cell-free system this interaction should not affect the enzymatic content of the native cellulosomes. Therefore, in cases where the terminal scaffoldin-borne type-II dockerin (DocII) module is unoccupied, it could be used for attachment of a CohII-bearing component. Towards this end, we created a chimeric enzyme (BglA-CohII) containing the C. thermocellum β-glucosidase (GenBank Acc. No. X60268) (26) fused at its C terminus to a type-II cohesin from the C. thermocellum anchoring protein Orf2p (27). The chimeric enzyme was assayed to determine its ability to bind to the native cellulosome and its contribution towards the enhancement of enzymatic degradation of cellulosic substrates.
Results
Properties of BglA-CohII.
A clone expressing the CohII-fused BglA produced an approximately 72 kDa N-terminal (His)6-tagged polypeptide. Ni-NTA affinity purification of the soluble fraction resulted in a > 90% purified enzyme as detected by SDS-PAGE and β-glucosidase activity assay. As fusion of a binding module has been shown to alter the activity and the thermal stability of the catalytic domain (28, 29), the kinetic parameters (Km and kcat), thermal stability and the optimal pH and temperature profiles of BglA-CohII were determined and compared to those of the wild-type enzyme (WT BglA). The thermal stability assay revealed that BglA-CohII retains 80% of its initial activity after 3 h at 60 °C as compared to 91% retention of the activity shown by WT BglA thus indicating a decrease of 13%. After 43 h at 60 °C, the fusion protein retained 29% of its original activity under these conditions vs. 34% for the wild-type protein. The kcat/Km ratio of BglA-CohII was about 9% lower than WT BglA (52.8 and 57.7 s-1 mM-1, respectively). Optimal activity for both enzymes was observed at 60 °C and pH 6.5.
Incorporation of BglA-CohII into the Cellulosome Complex.
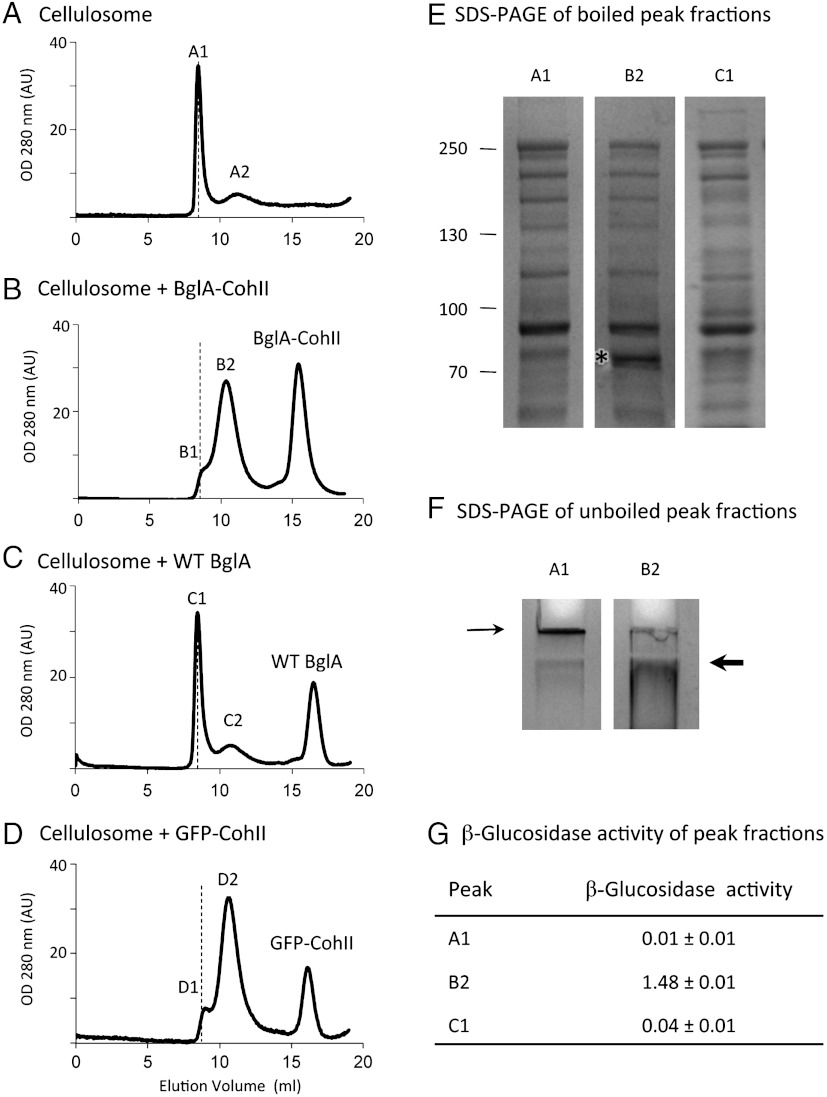
In order to determine whether BglA-CohII can bind to the cellulosome, the cellulosome was allowed to interact with WT BglA, BglA-CohII, and GFP-CohII (as a nonenzymatic CohII-bearing control protein) followed by size exclusion chromatography (Fig. 2). As can be seen in Fig. 2A, the majority of the native cellulosome emerged in the void volume of the column (peak A1) with a smaller peak (A2) that eluted later. When BglA-CohII was added to the cellulosome, the second peak (B2), presumably representing the cellulosome-complexed chimeric β-glucosidase, was increased at the apparent expense of the void-volume peak, and a third peak appeared representing excess free, uncomplexed BglA-CohII consistent with its apparent molecular mass. In contrast, the addition of WT BglA failed to affect the position of the cellulosome peak (C1) and the free enzyme eluted in an uncomplexed state. Interestingly, the presence of the cohesin-bearing control protein (GFP-CohII) caused a delay in the elution pattern of the major peak (D2) very similar to that observed for BglA-CohII thus indicating its incorporation into the cellulosome complex. Indeed, BglA-CohII was clearly incorporated into the cellulosome complex as evident from the denaturing SDS-PAGE data (Fig. 2E). A strong approximately 72-kDa protein band was observed in the major peak (B2) in accordance with the estimated molecular mass of the BglA-CohII chimera. This band was absent from the native cellulosome in the absence or presence of WT BglA (peaks A1 and C1). Surprisingly, when the samples were not boiled prior to SDS-PAGE (Fig. 2F), the native cellulosome failed to penetrate the stacking gel indicating an aggregation or oligomerization of cellulosome complexes under these conditions. In contrast, upon interaction of the cellulosome with BglA-CohII the sample clearly entered the stacking and separating gels. The fact that BglA-CohII is unambiguously integrated into the cellulosome complex is evident from the β-glucosidase activity displayed by the relevant peak (Fig. 2G). The gel-filtration data combined with the nondenaturing SDS-PAGE results suggest that the purified cellulosome exists largely in a homooligomeric state that dissociates upon binding of CohII.
Fig. 2.
Incorporation of BglA-CohII into the purified native cellulosome and its effect on the oligomeric state of the complex. (A–D). Size-exclusion chromatographic profiles of the cellulosome alone (A), after incubation with BglA-CohII (B), WT BglA (C), or GFP-CohII (D) using a Superdex 200 (HL 10/30) column. The proteins were allowed to interact overnight prior to chromatography. (Void volume is indicated by the dashed lines in A–D). (E) SDS-PAGE analysis of fully denatured samples of the indicated cellulosome-containing fractions. Samples were incubated for 10 min at 100 °C prior to SDS-PAGE. The 72-kD band corresponding to the incorporated BglA-CohII (B2) is indicated by an asterisk. Note the absence of this band in the native cellulosome (peak A1) and the cellulosome following interaction with WT BglA and chromatographic separation (peak C1). (F) SDS-PAGE analysis of nondenatured samples of the native cellulosome and cellulosome/BglA-CohII fractions (peaks A1 and B2, respectively). The samples were incubated at 25 °C prior to electrophoresis under conditions where the complex does not dissociate extensively into its component parts. Note that the native cellulosome alone (A1) fails to penetrate the 3% stacking gel (thin arrow), whereas the cellulosome-incorporated BglA-CohII complex accumulates as a major band immediately after entering the 6% separating gel (thick arrow). (G) The presence of BglA in the cellulosome-containing fractions was detected by β-glucosidase (pNPGase) activity assay. Triplicates of each reaction were carried out, and standard errors are indicated.
Enhancement of Cellulolytic Activity by the Cellulosome-Bearing BglA-CohII.
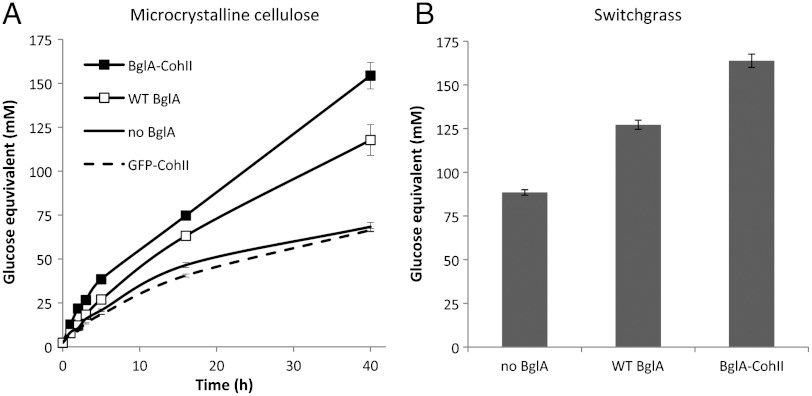
Because BglA-CohII binds specifically to the cellulosome complex in an enzymatically active form, it was thus of interest to determine whether the overall cellulolytic activity of the cellulosome would be enhanced accordingly. In this context, the combined cellulosome and BglA-CohII were examined for their ability to degrade microcrystalline cellulose or pretreated switchgrass vs. those of the various controls. Thus, the cellulosome was combined with the cellulosic substrate, and the production of soluble reducing sugars was assessed. In parallel samples, the substrate-adsorbed cellulosome was combined with BglA-CohII, WT BglA (added at equivalent specific activity), or GFP-CohII (at equivalent molar concentration to BglA-CohII). As can be seen in Fig. 3, complexation of BglA-CohII to the cellulose-adsorbed cellulosome enhanced the hydrolysis of microcrystalline cellulose and pretreated switchgrass by 2.3- and 1.9-fold, respectively, over that of the native cellulosome. Combination of WT BglA and cellulosome resulted in a corresponding 1.7- and 1.4-fold enhancement. The difference in enhancement was even more pronounced if the reaction mixtures were not agitated and their contents allowed to settle. Because the WT BglA and BglA-CohII enzymes were added on the basis of specific activity, the main difference between the enzymes appears to be their ability to bind to the cellulosome. The addition of GFP-CohII failed to affect cellulosome-mediated cellulolytic activity indicating that the presence of CohII by itself and consequent disruption of cellulosome oligomers is not directly responsible for the observed enhanced hydrolysis of the cellulosic substrates. These findings suggest that localization of BglA-CohII onto the cellulosome focuses the cellobiose-degradation activity at the general site of cellobiose accumulation where the enzyme would be more effective in reducing the inhibitory action of the disaccharide product on the cellulosomal enzymes.
Fig. 3.
Degradation of cellulosic substrates by cellulosome combined with BglA-CohII. Microcrystalline cellulose (A) and pretreated switchgrass (B) by cellulosome alone or combined with BglA-CohII, WT BglA, or GFP-CohII. The reaction mixture contained substrate (12.5% microcrystalline cellulose or 4.2% pretreated switchgrass) and 0.475 mg/mL cellulosome. The β-glucosidase-containing samples comprised 0.75 μM WT BglA or BglA-CohII in an equivalent specific activity. The effect of CohII on cellulosome-mediated cellulolytic activity was determined using 0.94 μM GFP-CohII. The level of degradation was determined at 60 °C by the reducing sugar assay. The level of degradation of the pretreated switchgrass samples was determined after 40 h. Triplicates of each reaction were carried out, and standard errors are indicated.
Discussion
Improvement of enzymatic deconstruction of lignocellulosic biomass is an essential step for effective production of plant-derived biofuels (1, 7, 9). Previous studies have shown that various cellulases are inhibited by cellobiose, the major enzyme-mediated degradation product of cellulosic substrates (19, 30–32). In contrast to natural environments where the cellobiose is removed from the medium by microbial assimilation, efficient cellulolysis by cell-free enzymatic systems would benefit by the removal of cellobiose (22, 33). In this context, Kadam and Demain (21) used a combined system of cloned β-glucosidase (BglB) from C. thermocellum with the crude cellulase system from the same strain. A 96-h reaction resulted in 10-fold enhancement of crystalline cellulose degradation. Lamed et al. (22) achieved similar results using a purified β-glucosidase from the fungus Aspergillus niger combined with purified preparations of the C. thermocellum cellulosome. Nevertheless, neither of these approaches address the fact that the cellulosome is bound to the insoluble substrate; thus, cellobiose accumulates in close proximity to the cellulosome-cellulose interface and only a fraction of soluble β-glucosidase would be available to convert the inhibitory cellobiose to the noninhibitory glucose. The possible synergistic effect of coupling the cellulosome and β-glucosidase together was not examined.
The results reported here show that the efficiency of one of nature's most potent cellulolytic machinery—the cellulosome of C. thermocellum—can be enhanced using a chimeric type-II cohesin-fused β-glucosidase (BglA-CohII). The CohII module mediates the specific high-affinity binding of the chimeric enzyme to the exposed cellulosomal DocII module. Consequently, the cellobiose-degrading enzyme is targeted to the precise sites on the insoluble cellulosic substrate where cellulosome-mediated degradation to cellobiose occurs. The emplacement of this critical enzyme close to the site of cellobiose accumulation would thus be expected to reduce its inhibitory effect on the native cellulosome-borne enzymes. In contrast, the free β-glucosidase (lacking the CohII) is distributed homogeneously throughout the solution phase and thus diluted in the immediate vicinity of the cellulosome-borne enzymes with respect to the concentration of BglA-CohII.
It has been claimed that organization of appropriate cellulolytic enzymes into a complex of specified composition and spatial arrangement can enhance cellulolysis (34). Fierobe et al. (24), thus, prepared a designer cellulosome composed of three cellulases arranged into an artificial CBM-bearing scaffoldin that enhanced the degradation of microcrystaline cellulose by 3.8 fold (compared to the noncomplexed enzymes). A similar design composed of two xylanases, two cellulases, and a xylan-binding module enhanced the degradation of a wheat straw substrate by 2.6 fold (25). Similar levels of enhanced ethanol production were achieved on phosphoric acid-swollen cellulose (PASC) by mini cellulosomes composed of two cellulases and β-glucosidase attached to yeast cells (35); however, to date, the efficiency of artificial cellulosome systems has been inferior to that of the native cellulosome possibly due to the larger variety of enzymes and preferred conformation of the native complex (24).
Addition of exogenous components to the native cellulosome has been proposed earlier in the form of a “supercellulosome” where exogenous enzymes are incorporated into the intact cellulosome using bifunctional crosslinking reagents (14, 36); however, the nonspecific chemical nature of crosslinking could impair the activities of the β-glucosidase and the cellulosomal enzymes (37). In contrast, our data show that the use of fused CohII module allows specific incorporation of the enzyme into the cellulosome that preserves its essential cellobiase activity. Moreover, bifunctional crosslinking can be time and resource consuming (37), whereas BglA-CohII can be added directly to the cellulosome. Consequently, the CohII fused module enables self assembly of functional BglA into the complex suggesting that the fused CohII module can be used as a general tool for incorporation of new functions into the native cellulosome.
Conversion of BglA from a free enzyme to a chimeric CohII-fused enzyme and its immobilization might affect its properties. For example, Ong et al. (38) created a chimeric enzyme composed of a CBM from Cellulomonas fimi fused to a β-glucosidase from Agrobacterium sp. The specific activity of the resultant fused enzyme was 14% lower than the original enzyme, though its CBM sustained its binding activity. A similar study reported insignificant differences in the fused and native enzyme (39). Chimeric proteins that contained type-I or -II cohesins as binding modules were shown to sustain their binding activity (40, 41). These data are consistent with our results that the BglA-CohII had but a mild negative effect on the kcat/Km ratio, and the fusion protein formed a stable complex with the cellulosome. Previous studies regarding immobilization of β-glucosidase were unclear regarding its effect on activity and thermal stability (42); however, the minor decrease (13%) in thermal stability of the immobilized BglA-CohII seems to be negligible as the localization of the enzyme into the cellulosome enhances the degradation rate of microcrystalline cellulose by 2.3-fold (35% higher than the combination of cellulosome and WT BglA).
The reduced size of the cellulosome:BglA-CohII complexes vs. that of the native cellulosome likely reflects dissociation of cellulosome oligomers by the high-affinity binding of the CohII-fused proteins. Adams et al. (43) showed that, upon calcium binding, the cellulosomal DocII module undergoes a conformational change that results in homodimerization. As the affinity of the type-II cohesin-dockerin interaction is several orders of magnitude stronger than that of homodimerization (Kd of 1.8·10-9 and 4·10-5 M, respectively), the binding of the CohII-bearing protein to the DocII module dissociates the oligomerized complex into a discrete cellulosome:BglA-CohII complex. In this respect, the DocII modules in most of the cellulosome molecules appear to be unoccupied and available for interaction with CohII indicating that most of the cell-free cellulosome molecules are not attached to complementary anchoring proteins.
The combined system of cellulosome with BglA-CohII was shown to enhance the degradation rate of insoluble substrates (microcrystalline cellulose and pretreated switchgrass) to a higher degree than the parallel system with the soluble enzyme (WT BglA) suggesting that this method could be useful for other types of natural complex cellulosic substrates (e.g., wheat straw, sugar-cane bagasse and other lignocellulosic wastes).
The importance of substrate targeting to cellulosome efficiency has been discussed previously in the context of CBM-mediated attachment of cellulosomal cellulases to the insoluble substrate (23–25). Nevertheless, our data suggest that in a cell-free system another type of targeting mechanism can play an important role, i.e., the targeting of the β-glucosidase to the cellobiose-susceptible cellulases. These findings could be used in future studies for development of improved designer cellulosomes.
To conclude, a fusion protein containing the prominent C. thermocellum β-glucosidase, BglA together with type-II cohesin was examined as a tool for targeting the enzyme to the cellulosome. Its effect on the cellulosomal system and cellobiose inhibition constituted an important precedence for the possible use of the unoccupied type-II dockerin site on the cellulosome for the incorporation of new and/or improved functions to the native cellulosome by other types of cohesin-fused components. The localization of cohesin-fused BglA to the cellulosome was shown to provide exogenous cellobiase activity to the cellulosome and enhance the degradation of insoluble substrates to a higher level than that observed for soluble wild-type BglA. This system can be a powerful tool for industrial solubilization of natural cellulosic substrates and for designing improved cellulolytic machineries.
Materials and Methods
Cellulosome Preparation.
The cellulosome used in this work was prepared from 3 d growth culture media of C. thermocellum ATCC 27405 by the affinity purification method (44).
Cloning, Expression, and Purification.
DNA encoding the BglA was amplified from C. thermocellum strain ATCC 27405 genomic DNA by PCR using 5′ CAGTCCATGGCAAAGATAAC, 3′ CACGCTCGAGGAAACCGTTGTTTTTGATTAC, and 3′ CATGGGTACCGAAACCGTTGTTTTTGATTAC (NcoI, XhoI and KpnI sites in boldface type, respectively). DNA encoding the type-II cohesin module from the Orf2p anchoring protein was amplified from previously described CohII-CBD construct (45) using 5′ GTTTCGGTACCTTACCGGACGATGCACACAT and 3′ TGGTGCTCGAGAATCACAGTAATT (KpnI and XhoI sites in boldface type, respectively). DNA encoding the GFP was amplified from previously described GFP-wtDoc construct (46) using 5' CACCTCATGAGTAAAGGAGAAGAACTT and 3′ GGTAAGGTACCTTTGTAGAGCTCATCCATGC (BspHI and KpnI sites in boldface type, respectively). The PCR amplified bglA gene was digested by NcoI/XhoI and ligated into pET28a resulting in the final vector pBglA. PCR amplified bglA and CohII were digested (using NcoI/KpnI and KpnI/XhoI, respectively) and ligated into pET28a resulting in the final vector pBglA-CohII. pGFP-CohII vector was constructed as follows: The PCR amplified GFP was digested with BspHI/KpnI and ligated into NcoI/KpnI, digested, and dephosphorylated (Shrimp alkaline phosphatase, Roche Applied Science, Indianapolis, IN) pBglA-CohII. Expression of the proteins was achieved by adding isopropyl β-D-thiogalactopyranoside (0.1 mM final concentration) to midexponential phase cultures of E. coli BL21(DE3) harboring target plasmids with incubation for a further 3 h at 37 °C. The His-tagged recombinant proteins were purified from cell-free extracts by immobilized metal ion affinity chromatography as described previously (47). Yields for WT BglA are typically between 70 and 90 mg per liter culture medium, and the yield of purified BglA-CohII was 25.2 mg per liter of culture medium.
β-Glucosidase Specific Activity Assay.
β-Glucosidase activity was measured using 4-nitrophenyl-β-D-glucopyranoside (pNPG, Sigma Chemical Co., St. Louis, MO) as a substrate. Samples were incubated for appropriate time with 200 μL solution containing 2 mM pNPG, 50 mM citrate buffer, and 15 mM CaCl2 (pH 6) at 60 °C. The reaction was terminated upon addition of 50 μL of 1 M Na2CO3 and the absorbance at 405 nm was measured. Initial rates were monitored by measuring the formation of p-nitrophenol at 405 nm (ε = 3404.8 M-1 cm-1) using a spectrophotometer (Cary 5 UV-Vis-IR double-beam, Cary-5, Australia). Determination of kinetics parameters were accomplished using the Graphpad prism 5 program (www.graphpad.com).
β-Glucosidase Thermal Stability.
The thermal stability was determined as the ratio between the remaining activity measured using pNPG, after 3 h at 60 °C and the initial activity. The samples were preincubated for 2 h with cellulosome (3.14 mg/mL) prior to activity measurement.
Pretreatment of Switchgrass.
Switchgrass was pretreated with 2% HCl for 1 h at 100 °C (HCl∶Switchgrass ratio was 10∶1 wt/wt). The acid pretreatment was followed by extensive washing steps using 100 volumes of water per volume of biomass. The second pretreatment stage included boiling of the biomass (cellulolignin) in 2% NaOH for one h at 100 °C (NaOH∶switchgrass ratio was 10∶1 wt/wt). The cellulose enriched biomass was washed extensively with water (pH approximately 6). Samples of double pretreated biomass were oven dried overnight at 70 °C, and the dry mass was calculated.
Cellulase Activity Assay.
The activity was tested in an 800 μL final volume containing substrate (12.5% microcrystalline cellulose) [MCC PH301 (Avicel), FMC, Philadelphia] or 4.2% pretreated switchgrass (SG NA10, Designer Energy, Ltd., Rehovot, Israel) and cellulosome 0.475 mg/mL in a 50 mM Citrate buffer (pH 6.0) and 15 mM CaCl2. The β-glucosidase combined samples contained 0.75 μM WT BglA or BglA-CohII in an equivalent specific activity (measured by pNPG in the presence of 0.475 mg/mL cellulosome). The GFP-CohII combined sample contained 0.94 μM GFP-CohII. The reaction mixture was carried out at 60 °C with constant shaking, and the reactions were terminated at predetermined time points by transferring the tubes to an ice water bath. After a centrifugation step (5 min at 14,000 rpm), supernatant samples (20 μL) composed mainly of cellobiose and glucose were transferred into reaction mixtures containing 0.5 μM WT BglA in 50 mM citrate buffer (pH 6) supplemented with 15 mM CaCl2. The reaction was incubated at 60 °C for 3 h, sufficient to convert all cellobiose into glucose. Reducing sugars measurement (DNS) was performed as described previously (25).
Binding of CohII to the Cellulosome.
The cellulosome and assayed proteins were allowed interaction overnight in Tris-buffered saline (TBS) (137 mM NaCl, 2.7 mM KCl, 25 mM Tris•HCl, pH 7.4) supplemented with 15 mM CaCl2 (pH 7.4) at 4 °C. Analytical size-exclusion chromatography of the samples was carried out at 24 °C on a Superdex 200 (HL 10/30) column connected to an AKTA HPLC system (GE Healthcare, Piscataway, NJ). The running buffer was composed of TBS supplemented with 15 mM CaCl2. Proteins were eluted at a flow rate of 1 mL/ min with 500 μL fractions being collected and the optical density of the eluent being monitored at 280 nm. Fractions corresponding to the cellulosome peak were pooled, analyzed by SDS-PAGE, and assayed for β-glucosidase activity using pNPG.
ACKNOWLEDGMENTS.
The authors appreciate the assistance of Dr. Yoav Barak (Chemical Research Support, Weizmann Institute) for invaluable assistance in constructing the BglA-CohII and GFP-CohII chimaeras. This research was supported by grants from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel, by the Weizmann Institute of Science Alternative Energy Research Initiative (AERI); a grant from the Israel Strategic Alternative Energy Foundation (I-SAEF) (to E.A.B.), a grant from the Israel Ministry of Science (IMOS) (966/09 to E.A.B. and 159/07 and 24/11 to RL, respectively) by the Israel Science Foundation and establishment of an Israeli Center of Research Excellence (I-CORE Center No. 152/11 to E.A.B. and Y.S.) managed by the Israel Science Foundation. E.M. and E.A.B. are also pleased to acknowledge funding within the framework of the NOFAR program (issued by the MAGNET directorate in the Israeli Ministry of Industry, Trade and Labor). E.A.B. is the incumbent of The Maynard I. and Elaine Wishner Chair of Bioorganic Chemistry.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Lynd LR, et al. How biotech can transform biofuels. Nat Biotechnol. 2008;26:169–172. doi: 10.1038/nbt0208-169. [DOI] [PubMed] [Google Scholar]
- 2.Ragauskas AJ, et al. The path forward for biofuels and biomaterials. Science. 2006;311:484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- 3.Sims RE, Mabee W, Saddler JN, Taylor M. An overview of second generation biofuel technologies. Bioresour Technol. 2010;101:1570–1580. doi: 10.1016/j.biortech.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 4.Bayer EA, Lamed R, Himmel ME. The potential of cellulases and cellulosomes for cellulosic waste management. Curr Opin Biotech. 2007;18:237–245. doi: 10.1016/j.copbio.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Himmel ME, Bayer EA. Lignocellulose conversion to biofuels: Current challenges, global perspectives. Curr Opin Biotech. 2009;20:316–317. doi: 10.1016/j.copbio.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Wall JD, Harwood CS, Demain A, editors. Bioenergy. Washington, DC: ASM Press; 2008. p. 454. [Google Scholar]
- 7.Himmel ME, et al. Biomass recalcitrance: Engineering plants and enzymes for biofuels production. Science. 2007;315:804, 807. doi: 10.1126/science.1137016. and erratum (2007) 316:982. [DOI] [PubMed] [Google Scholar]
- 8.Reiter WD. Biosynthesis and properties of the plant cell wall. Curr Opin Plant Biol. 2002;5:536–542. doi: 10.1016/s1369-5266(02)00306-0. [DOI] [PubMed] [Google Scholar]
- 9.Demain AL, Newcomb M, Wu JH. Cellulase, clostridia, and ethanol. Microbiol Mol Biol Rev. 2005;69:124–154. doi: 10.1128/MMBR.69.1.124-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doi RH, Kosugi A. Cellulosomes: Plant-cell-wall-degrading enzyme complexes. Nat Rev Microbiol. 2004;2:541–551. doi: 10.1038/nrmicro925. [DOI] [PubMed] [Google Scholar]
- 11.Bayer EA, Belaich J-P, Shoham Y, Lamed R. The cellulosomes: Multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 12.Himmel ME, et al. Microbial enzyme systems for biomass conversion: Emerging paradigms. Biofuels. 2010;1:323–341. [Google Scholar]
- 13.Fontes CM, Gilbert HJ. Cellulosomes: Highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu Rev Biochem. 2010;79:655–681. doi: 10.1146/annurev-biochem-091208-085603. [DOI] [PubMed] [Google Scholar]
- 14.Bayer EA, Morag E, Lamed R. The cellulosome—A treasure-trove for biotechnology. Trends Biotechnol. 1994;12:379–386. doi: 10.1016/0167-7799(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 15.Shoham Y, Lamed R, Bayer EA. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 1999;7:275–281. doi: 10.1016/s0966-842x(99)01533-4. [DOI] [PubMed] [Google Scholar]
- 16.Nataf Y, et al. Cellodextrin and laminarbiose ABC transporters in Clostridium thermocellum. J Bacteriol. 2009;191:203–209. doi: 10.1128/JB.01190-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strobel HJ. Growth of the thermophilic bacterium Clostridium thermocellum in continuous culture. Curr Microbiol. 1995;31:210–214. [Google Scholar]
- 18.Zhang YH, Lynd LR. Cellulose utilization by Clostridium thermocellum: Bioenergetics and hydrolysis product assimilation. Proc Natl Acad Sci USA. 2005;102:7321–7325. doi: 10.1073/pnas.0408734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamed R, Kenig R, Setter E, Bayer EA. Major characteristics of the cellulolytic system of Clostridium thermocellum coincide with those of the purified cellulosome. Enzyme Microb Technol. 1985;7:37–41. [Google Scholar]
- 20.Andric P, Meyer AS, Jensen PA, Dam-Johansen K. Effect and modeling of glucose inhibition and in situ glucose removal during enzymatic hydrolysis of pretreated wheat straw. Appl Biochem Biotechnol. 2010;160:280–297. doi: 10.1007/s12010-008-8512-9. [DOI] [PubMed] [Google Scholar]
- 21.Kadam SK, Demain AL. Addition of cloned β-glucosidase enhances the degradation of crystalline cellulose by the Clostridium thermocellum cellulose complex. Biochem Biophys Res Commun. 1989;161:706–711. doi: 10.1016/0006-291x(89)92657-0. [DOI] [PubMed] [Google Scholar]
- 22.Lamed R, et al. Efficient cellulose solubilization by a combined cellulosome-b-glucosidase system. Appl Biochem Biotechnol. 1991;27:173–183. [Google Scholar]
- 23.Fierobe H-P, et al. Degradation of cellulose substrates by cellulosome chimeras: Substrate targeting versus proximity of enzyme components. J Biol Chem. 2002;277:49621–49630. doi: 10.1074/jbc.M207672200. [DOI] [PubMed] [Google Scholar]
- 24.Fierobe H-P, et al. Action of designer cellulosomes on homogeneous versus complex substrates: Controlled incorporation of three distinct enzymes into a defined tri-functional scaffoldin. J Biol Chem. 2005;280:16325–16334. doi: 10.1074/jbc.M414449200. [DOI] [PubMed] [Google Scholar]
- 25.Moraïs S, et al. Cellulase-xylanase synergy in designer cellulosomes for enhanced degradation of a complex cellulosic substrate. MBio. 2010;1:e00285–10. doi: 10.1128/mBio.00285-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grabnitz F, Seiss M, Rucknagel K, Staudenbauer W. Structure of the b-glucosidase gene bglA of Clostridium thermocellum. Sequence analysis reveals a superfamily of cellulases and b-glycosidases including human lactase/phlorizin hydrolase. Eur J Biochem. 1991;200:301–309. doi: 10.1111/j.1432-1033.1991.tb16186.x. [DOI] [PubMed] [Google Scholar]
- 27.Leibovitz E, Béguin P. A new type of cohesin domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J Bacteriol. 1996;178:3077–3084. doi: 10.1128/jb.178.11.3077-3084.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gundllapalli SB, Pretorius IS, Cordero Otero RR. Effect of the cellulose-binding domain on the catalytic activity of a β-glucosidase from Saccharomycopsis fibuligera. J Ind Microbiol Biotechnol. 2007;34:413–421. doi: 10.1007/s10295-007-0213-9. [DOI] [PubMed] [Google Scholar]
- 29.Kataeva IA, Blum DL, Li XL, Ljungdahl LG. Do domain interactions of glycosyl hydrolases from Clostridium thermocellum contribute to protein thermostability? Protein Eng. 2001;14:167–172. doi: 10.1093/protein/14.3.167. [DOI] [PubMed] [Google Scholar]
- 30.Johnson EA, Reese ET, Demain AL. Inhibition of Clostridium thermocellum cellulase by end products of cellulolysis. J Appl Biochem. 1982;4:64–71. [Google Scholar]
- 31.Morag E, Halevy I, Bayer EA, Lamed R. Isolation and properties of a major cellobiohydrolase from the cellulosome of Clostridium thermocellum. J Bacteriol. 1991;173:4155–4162. doi: 10.1128/jb.173.13.4155-4162.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kristensen JB, Felby C, Jørgensen H. Yield-determining factors in high-solids enzymatic hydrolysis of lignocellulose. Biotech Biofuels. 2009;2:11. doi: 10.1186/1754-6834-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chauve M, et al. Comparative kinetic analysis of two fungal β-glucosidases. Biotech Biofuels. 2010;3:3. doi: 10.1186/1754-6834-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang YH. Substrate channeling and enzyme complexes for biotechnological applications. Biotechnol Adv. 2011;29:715–725. doi: 10.1016/j.biotechadv.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 35.Tsai SL, Oh J, Singh S, Chen R, Chen W. Functional assembly of minicellulosomes on the Saccharomyces cerevisiae cell surface for cellulose hydrolysis and ethanol production. Appl Environ Microbiol. 2009;75:6087–6093. doi: 10.1128/AEM.01538-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bayer EA, Lamed R. The cellulose paradox: Pollutant par excellence and/or a reclaimable natural resource? Biodegradation. 1992;3:171–188. doi: 10.1007/BF00129082. [DOI] [PubMed] [Google Scholar]
- 37.Rao SV, Anderson KW, Bachas LG. Oriented immobilization of proteins. Mikrochim Acta. 1998;128:127–143. [Google Scholar]
- 38.Ong E, Gilkes NR, Warren RAJ, Miller RC. Enzyme immobilization using the cellulose-binding domain of a Cellulomonas fimi exoglucanase. Nat Biotechnol. 1989;7:604–607. [Google Scholar]
- 39.Ong E, Gilkes NR, Miller RCJ, Warren RAJ, Kilburn DG. Enzyme immobilization using a cellulose-binding domain: Properties of a b-glucosidase fusion protein. Enzyme Microb Technol. 1991;13:59–65. doi: 10.1016/0141-0229(91)90189-h. [DOI] [PubMed] [Google Scholar]
- 40.Carrard G, Koivula A, Soderlund H, Béguin P. Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc Natl Acad Sci USA. 2000;97(19):10342–10347. doi: 10.1073/pnas.160216697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barak Y, et al. Matching fusion-protein systems for affinity analysis of two interacting families of proteins: The cohesin-dockerin interaction. J Mol Recognit. 2005;18:491–501. doi: 10.1002/jmr.749. [DOI] [PubMed] [Google Scholar]
- 42.Woodward J. Immobilized cellulases for cellulose utilization. J Biotechnol. 1989;11:299–311. [Google Scholar]
- 43.Adams JJ, Webb BA, Spencer HL, Smith SP. Structural characterization of type II dockerin module from the cellulosome of Clostridium thermocellum: Calcium-induced effects on conformation and target recognition. Biochemistry. 2005;44:2173–2182. doi: 10.1021/bi048039u. [DOI] [PubMed] [Google Scholar]
- 44.Morag E, Bayer EA, Lamed R. Affinity digestion for the near-total recovery of purified cellulosome from Clostridium thermocellum. Enzyme Microb Technol. 1992;14:289–292. [Google Scholar]
- 45.Haimovitz R, et al. Cohesin-dockerin microarray: Diverse specificities between two complementary families of interacting protein modules. Proteomics. 2008;8:968–979. doi: 10.1002/pmic.200700486. [DOI] [PubMed] [Google Scholar]
- 46.Demishtein A, Karpol A, Barak Y, Lamed R, Bayer EA. Characterization of a dockerin-based affinity tag: Application for purification of a broad variety of target proteins. J Mol Recognit. 2010;23:525–535. doi: 10.1002/jmr.1029. [DOI] [PubMed] [Google Scholar]
- 47.Vazana Y, Moraïs S, Barak Y, Lamed R, Bayer EA. Interplay between Clostridium thermocellum family-48 and family-9 cellulases in the cellulosomal versus non-cellulosomal states. Appl Environ Microbiol. 2010;76:3236–3243. doi: 10.1128/AEM.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]