Abstract
Chromosomal translocations involving the MALT1 gene are hallmarks of mucosa-associated lymphoid tissue (MALT) lymphoma. To date, targeting these translocations to mouse B cells has failed to reproduce human disease. Here, we induced MALT1 expression in mouse Sca1+Lin− hematopoietic stem/progenitor cells, which showed NF-κB activation and early lymphoid priming, being selectively skewed toward B-cell differentiation. These cells accumulated in extranodal tissues and gave rise to clonal tumors recapitulating the principal clinical, biological, and molecular genetic features of MALT lymphoma. Deletion of p53 gene accelerated tumor onset and induced transformation of MALT lymphoma to activated B-cell diffuse large-cell lymphoma (ABC-DLBCL). Treatment of MALT1-induced lymphomas with a specific inhibitor of MALT1 proteolytic activity decreased cell viability, indicating that endogenous Malt1 signaling was required for tumor cell survival. Our study shows that human-like lymphomas can be modeled in mice by targeting MALT1 expression to hematopoietic stem/progenitor cells, demonstrating the oncogenic role of MALT1 in lymphomagenesis. Furthermore, this work establishes a molecular link between MALT lymphoma and ABC-DLBCL, and provides mouse models to test MALT1 inhibitors. Finally, our results suggest that hematopoietic stem/progenitor cells may be involved in the pathogenesis of human mature B-cell lymphomas.
Keywords: molecular-directed therapy, transgenic mice, cancer stem cells, tumor reprogramming
Mucosa-associated lymphoid tissue (MALT) lymphoma is a distinct clinicopathological entity that accounts for 8% of all non-Hodgkin lymphomas (1, 2). MALT lymphomas can be distinguished from other lymphomas because they occur in various extranodal locations, primarily in the stomach, where they are preceded by Helicobacter pylori infection, and also in the ocular adnexa, lung, salivary glands, intestinal tract, skin, thyroid, and genitourinary tract, and are associated with chronic microbial infections or autoimmune disorders. MALT lymphomas show a typical histopathological picture composed of a heterogeneous neoplastic B-cell population that arises from the marginal zone of reactive B-cell follicles, extends to the interfollicular region, and infiltrates the epithelium, forming the characteristic lymphoepithelial lesions. Additionally, these lymphomas frequently show a prominent plasmacytic differentiation (1–3).
Genetically, MALT lymphomas are mainly associated with two chromosomal translocations involving the MALT1 gene. The t(11;18)(q21;q21), which generates an API2-MALT1 fusion transcript, occurs in up to 30% of the cases, whereas the t(14;18)(q32;q21) results in the immunoglobulin H (IGH)-MALT1 translocation and is detected in 15–20% of the cases (4–6). A third translocation, the t(1;14)(p22;q32), forms a B-cell BCL10-IGH fusion and is present in ∼1% of MALT lymphomas. All of these translocations lead to the activation of NF-κB, which regulates target genes involved in immune responses to foreign antigens (7). In B lymphocytes, MALT1 is part of the CARD11/BCL10/MALT1 (CBM) complex, a mediator of the stimulation of the B-cell antigen receptor (BCR). MALT1 mediates the activation of the IκB kinase (IKK) complex, leading to the release and nuclear translocation of NF-κB. Remarkably, the caspase-like domain of MALT1 shows proteolytic activity and can cleave BCL10 and several NF-κB inhibitors, resulting in NF-κB activation (7–9). In MALT lymphomas, it is assumed that underlying chronic infection/inflammation causes persistent BCR-mediated NF-κB activation that promotes B-lymphocyte expansion and accumulation in extranodal tissues, ultimately resulting in clonal lymphoma development (1, 3).
Emerging data also implicate the CBM complex in the pathogenesis of diffuse large cell lymphoma of activated B cells (ABC-DLBCL), a distinct lymphoma subtype that can be distinguished from the germinal center B-cell-like (GCB)-DLBCL (10, 11). Like in MALT lymphomas, a hallmark of ABC-DLBCL is the constitutive signaling of the NF-κB pathway, which is caused by mutations in various genes regulating NF-κB, including activating mutations of CARD11, CD79A, CD79B, and MyD88, and inactivating mutations of TNFAIP3 (11). Notably, mutations in MyD88 and TNFAIP3 genes have been also found in MALT lymphomas, suggesting a molecular link between ABC-DLBCL and MALT lymphoma (11). However, this relationship has not been proved experimentally.
Despite these evidences, a causative role for MALT1 in the development of MALT lymphoma has not been demonstrated. Indeed, the expression of API2-MALT1 or BCL10 in B lymphocytes did not induce lymphoma in mice (12, 13), suggesting that NF-κB activation in B cells may not be sufficient to promote malignant transformation. The lack of genetically engineered human-like MALT lymphoma models has hampered a better understanding of the disease pathogenesis and the development of MALT1-targeted therapies, the relevance of which to the treatment of ABC-DLBCL, an aggressive lymphoma that responds poorly to current immunochemotherapies, has already been highlighted using in vitro cell models (14, 15).
In this work, we show that human MALT lymphoma pathogenesis can be modeled in mice by targeting MALT1 expression to hematopoietic stem/progenitor cells, demonstrating the oncogenic role of MALT1 in lymphomagenesis. Furthermore, our study establishes a molecular link between MALT lymphoma and ABC-DLBCL and provides mouse models to test therapies targeting MALT1.
Results
MALT1 Shows Oncogenic Properties in Primitive Hematopoietic Cells.
Given that the expression of the MALT lymphoma-related genes in mouse B lymphocytes does not induce lymphoma development, we first explored whether MALT1, BCL10, or API2-MALT1 could be tumorigenic in more primitive hematopoietic cells. Human full-length MALT1 and BCL10 genes and the API2-MALT1 fusion gene were stably transfected into murine IL3-dependent hematopoietic BaF3 cells activated with anti-mouse IgM, anti-mouse CD40 antibody, and recombinant mIL4. Isolated single-cell clones expressing MALT1, BCL10, or API2-MALT1 genes exhibited higher NF-κB activation with respect to control cells, but only MALT1-expressing cells grew independently of IL3 (SI Appendix, Fig. S1 A and B). Accordingly, i.v. injection of 1 × 106 cells expressing MALT1, BCL10, and API2-MALT1 into BALB/c nude mice revealed that only MALT1-expressing cells generated tumors that rapidly killed the mice (SI Appendix, Fig. S1C). However, the MALT1-driven leukemias were composed of highly proliferating large lymphoid cells with a CD19−B220weakCD5−IgM− phenotype that did not reproduce the clinicopathological features of MALT lymphoma (SI Appendix, Fig. S1 D and E).
In Vivo Model of Ectopic Expression of Human MALT1 Protein in Hematopoietic Stem/Progenitor Cells.
Having demonstrated that MALT1 can transform primitive hematopoietic cells, we generated transgenic mice in which the mouse stem cell antigen 1 (Sca1, Ly-6E.1) gene promoter was used to drive the expression of the human MALT1 gene in the hematopoietic stem/progenitor cell compartment of C57BL/6 × CBA mice (Fig. S2A) (16). Southern blot analysis of the three founders obtained showed transgene copy numbers ranging from ∼2 to 4 (SI Appendix, Fig. S2B). Measurement of MALT1 mRNA expression levels by RT-PCR showed expression of the exogenous human MALT1 transgene in Sca1+Lin− hematopoietic/stem cells purified from the bone marrow (BM) of Sca1-MALT1 mice, but not in wild-type (WT) littermates (Fig. S2C). In addition, Sca1-driven human MALT1 expression was detected in Sca1+Lin− cells but not in other lymphoid cell subpopulations, as determined by quantitative real-time PCR analysis performed in cells isolated from BM, spleen and lymph nodes (SI Appendix, Fig. S2D). Likewise, immunofluorescence and Western blot studies detected MALT1 protein in transgenic BM Sca1+Lin− cells but not in splenic CD19+IgM+ mature B-lymphocytes, B220+CD21highCD23− marginal zone B cells or thymic T cells (Fig. 1C and SI Appendix, Fig. S2 F and G). These results document that human MALT1 expression is restricted to Sca1+Lin− cells of Sca1-MALT1 mice, validating the model for the functional analysis of MALT1 expression in hematopoietic stem/progenitor cells in vivo.
Fig. 1.
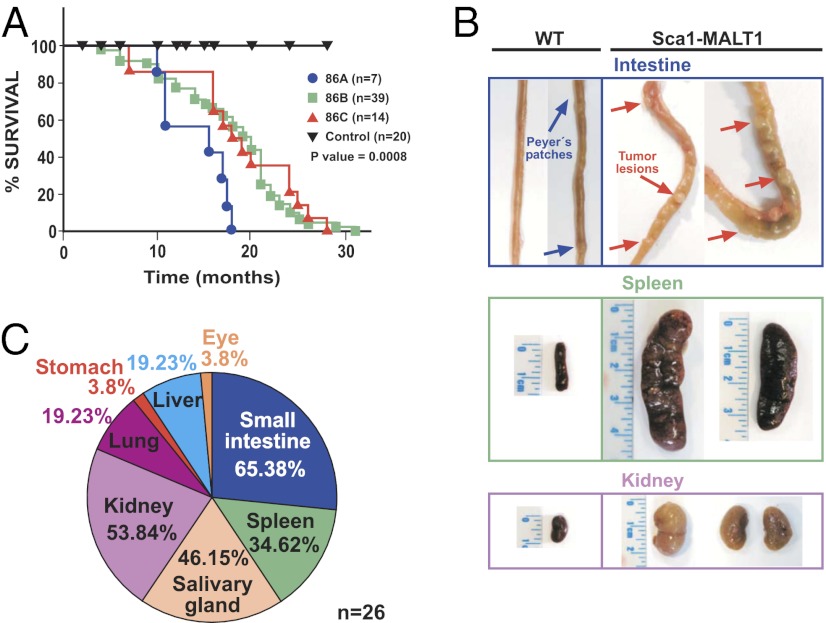
Sca1-MALT1 mice develop extranodal human-like MALT lymphomas. (A) Kaplan–Meier overall survival plots of Sca1-MALT1 mice and WT littermates. Differential survival was analyzed using the log-rank (Mantel–Cox) test: line 86A vs. WT, P = 0.0007; line 86B vs. WT, P = 0.0026; and line 86C vs. WT, P = 0.0173). (B) Macroscopic aspect of the tumors in small intestine, spleen, and kidney of Sca1-MALT1 mice. (C) Tumor location of MALT lymphomas in 26 Sca1-MALT1 mice; note that 65% of mice developed tumors at more than one site.
Expression of MALT1 in Sca1+ Cells Induces NF-κB Signaling, Promotes Hematopoietic Stem/Progenitor Cell Expansion, and Enhances B-Cell Lymphopoiesis in Young Mice.
Flow cytometric analysis of the different hematopoietic cell compartments in 2-mo-old Sca1-MALT1 mice did not detect major abnormalities in myeloid or T-cell subpopulations (as determined by Gr1, Mac1, CD4, or CD8 staining) in BM, peripheral blood (PB), spleen, and thymus (SI Appendix, Fig. S2H). However, an expansion of the hematopoietic stem/progenitor Sca1+Kit+Lin− cells was detected in the BM of transgenic mice in comparison with WT littermates (1.24% vs. 0.44%) (Fig. S2I). This expansion was accompanied by increased numbers of pro-B and pre-B lymphoid cells in BM (Fig. S2I) and with moderate accumulation of mature B cells in PB and spleen but not in lymph nodes (Fig. S2I). To investigate the molecular mechanisms underlying this enhanced B-cell lymphopoiesis, the transcriptional profiles of BM-sorted Sca1+Lin− cells from transgenic and WT mice were compared using gene-expression microarrays (SI Appendix, Fig. S2J). Linear Models of Microarray Data Analysis (LIMMA) identified 110 genes differentially expressed between transgenic and WT cells [false discovery rate (FDR) < 0.19; P < 0.0006), defining the Sca1+Lin− MALT1 signature. This included underexpression of genes encoding essential regulators of early B-cell development [Sox4, E2A (Tcef2a), Vpreb1, Vpreb2, and Rag1/Rag2], and up-regulation of differentiated B-cell markers (Prdm1, Xbp1, and Ig genes) (Fig. S2K and SI Appendix, Table S1). Gene Set Enrichment Analysis (GSEA) determined that the Sca1+Lin− MALT1 dataset was significantly enriched in NF-κB target genes (FDR = 0.009; http://www.bu.edu/nf-kb/gene-resources/target-genes), which is consistent with the activation of NF-κB pathway in Sca1+Kit+Lin− cells (Fig. S2L). Additionally, Ingenuity Pathway Analysis (IPA) (Ingenuity Systems) identified that the Sca1+Lin− MALT1 dataset was enriched in inflammatory response genes [FDR (Benjamini–Hochberg) < 1 × 10−10] (SI Appendix, Fig. S2M). These results indicate that MALT1-driven NF-κB activation induced the expansion of mouse BM hematopoietic stem/progenitor cells, which showed an abnormal early expression of B-cell markers and were selectively directed toward B-cell differentiation, promoting the accumulation of mature B lymphocytes in extranodal tissues.
Sca1-MALT1 Mice Develop Human-Like MALT Lymphomas.
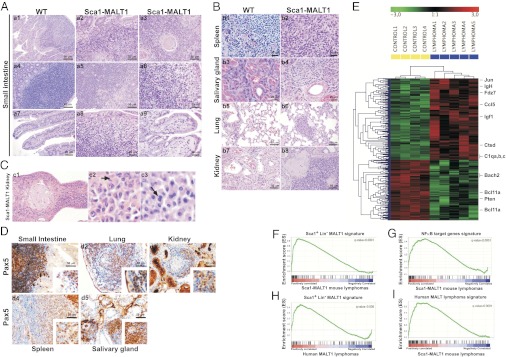
Adult Sca1-MALT1 transgenic mice developed clinical signs of disease, having a shorter lifespan in comparison with their WT littermates (Fig. 1A and SI Appendix, Table S2A). Systematic necropsies in transgenic mice (n = 60) showed consistent macroscopic tumors (Fig. 1B) and the presence of normocytic anemia (SI Appendix, Fig. S2N). Tumors involved different extranodal sites such as the small intestine, salivary glands, kidneys, lungs, liver, stomach, ocular adnexa, and spleen but did not affect the lymph nodes (Fig. 1C). More than one site of involvement was observed in 17 of 26 (65%) Sca1-MALT1 mice with macroscopic abnormalities (SI Appendix, Table S2B). Histological examination of the tumors revealed an extranodal cell infiltration comprising a morphologically heterogeneous lymphoid population, predominantly composed of marginal zone (centrocyte-like) cells mixed up with small lymphocytes, scattered large blasts, and plasma cells (Fig. 2 A and B). The neoplastic lymphoid infiltrate was located in the marginal zone of the B follicles, surrounding reactive germinal centers and extending into the interfollicular region. In epithelial tissues, the cells infiltrated the epithelium and formed lymphoepithelial lesions (Fig. 2 A and B and SI Appendix, Fig. S3 A–E). Prominent plasma cells were observed, particularly surrounding blood vessels, showing pleomorphic features with occasional binuclei, which suggests the neoplastic nature of at least a proportion of these cells (Fig. 2C). Immunohistochemistry (IHC) analysis showed that the lymphoid-infiltrating tumor cells expressed the B-cell-specific markers CD20 and Pax5, with moderate to strong IgM expression and weak IgD expression, whereas CD3 highlighted a few scattered T cells within the tumor (Fig. 2D and SI Appendix, Fig. S3F). Most tumors were oligoclonal or clonal, according to PCR and Southern blot analyses of V(D)J rearrangements. PCR amplification and sequencing of Ig-rearranged genes, however, did not show somatic hypermutation in Sca1-MALT1 mouse lymphomas (SI Appendix, Fig. S3 G and H). As in humans, mouse Sca1-MALT1 lymphomas frequently spread to the spleen, and a spontaneous transformation to human-like ABC-DLBCLs with a CD20+, Foxp1+, CD10−, Gcet1−, Mum1/Irf4−, Bcl6− phenotype was observed in 4 of 26 (15%) mice (17, 18) (SI Appendix, Fig. S3I). Taken together, Sca1-MALT1 mice developed mature B-cell lymphomas that involved extranodal sites and displayed clinical, histological, immunophenotypic, and genetic features resembling human MALT lymphoma.
Fig. 2.
Characterization of human-like MALT lymphomas arising in Sca1-MALT1 mice. (A) Representative hematoxylin–eosin (HE) staining analysis of the mouse lymphomas. A typical lymphoid cell population is present in the small intestine of Sca1-MALT1 mice, infiltrating the lamina propria and the epithelium in small groups, giving rise to lymphoepithelial lesions. (B) Lymphoid tumor cells also infiltrate other tissues. (C) Lymphomas developed in the kidneys showed prominent plasma cells surrounding blood vessels, which were pleomorphic and showed occasional binuclei (marked with arrows). (D) IHC analysis of Pax5 expression in Sca1-MALT1 lymphomas. (E) Gene-expression microarray analysis defined the Sca1-MALT1 lymphoma transcriptional signature. Five Sca1-MALT1 splenic lymphomas and four WT spleen were studied. (F–I) Bioinformatic studies of Sca1-MALT1 murine lymphoma transcriptional signature using GSEA.
Sca1-MALT1 Murine Lymphomas Share Abnormal Transcriptional and Genomic Profiles with Human MALT Lymphomas.
The transcriptional signature of human MALT lymphoma has been incompletely reported. Therefore, to compare the molecular profiles of mouse Sca1-MALT1 lymphomas and human MALT lymphomas, gene-expression microarrays were applied to diagnostic biopsies from patients with MALT lymphoma and with other common B-cell lymphoma subtypes (SI Appendix, Tables S2 C and D and Fig. S4A). Bioinformatic analysis of microarray data using the Prediction Analysis for Microarrays (PAM) software identified 132 genes that distinguished MALT lymphoma from other B-cell lymphomas, thereby defining the human MALT lymphoma transcriptional signature (SI Appendix, Table S3A and Fig. S4B). A representative selection of deregulated genes (FCRL4-Irta1, LTF, MEIS1, PBX1, ETV6, FZD7, and FOXO3) and proteins [LTF and EGF receptor (EGFR)], selected using biological and functional criteria because of their special interest for lymphoma, was studied by quantitative real-time PCR and IHC analyses in normal tissue samples and lymphoma cells, validating the microarray data (SI Appendix, Fig. S4 C and D). LIMMA comparison of the gene-expression profiles between splenic Sca1-MALT1 lymphomas and WT mouse spleens defined the Sca1-MALT1 lymphoma transcriptional signature, composed of 246 differentially expressed genes (FDR < 0.058, P < 0.00063) (Fig. 2E and SI Appendix, Table S3B). Bioinformatic studies using GSEA revealed that the Sca1+Lin−MALT1 signature (see Fig. S2K) was strongly conserved in the Sca1-MALT1 murine lymphomas (FDR < 0.0001) (Fig. 2F). In addition, the transcriptional profiles of both the Sca1-MALT1 lymphomas and human MALT lymphomas were significantly enriched in NF-κB target genes (GSEA, FDR < 0.0001) (Fig. 2G and SI Appendix, Fig. S4E) and inflammatory response genes (Ingenuity function enrichment analysis, FDR (Benjamini–Hochberg) <5%; P < 1 × 10−10) (SI Appendix, Fig. S4 F and G). Moreover, the Sca1+Lin− MALT1 signature was over-represented in human MALT lymphomas, but not in the other B-cell lymphoma subgroups (Fig. 2H). To gain further insight, we focused on the genes that were over-expressed in the Sca1+Lin− MALT1 signature, such as Prdm1, Xbp1 and Ig genes, which are involved in plasmacytic differentiation. This process is defined at the molecular level by an XBP1-target gene signature (19), which was significantly enriched in the transcriptional profiles of Sca1+Lin− BM-sorted cells (FDR < 0.001), Sca1-MALT1 mouse lymphomas (FDR < 0.001), and human MALT lymphomas (FDR < 0.05), but not in the other B-cell lymphoma subgroups (FDR > 0.1) (SI Appendix, Fig. S4H). In addition, the human MALT lymphoma transcriptional signature was significantly enriched in the Sca1-MALT1 mouse lymphomas (FDR < 0.0001) (Fig. 2I). Finally, high-resolution microarray-based comparative genomic hybridization (aCGH) analysis revealed that all nine Sca1-MALT1 mouse lymphomas analyzed displayed genomic changes, including gains of chromosome 14D3-qE3 (n = 8) and 17A1-E5 (n = 2), and loss of chromosome 12A1-F2 (n = 2) (SI Appendix, Fig. S4 I and J). These abnormalities are syntenic with the gains of chromosome 3p24 and 18p11 and with the loss of chromosome 7q21-q31, which are common in human marginal-zone B-cell lymphomas (1). These results indicate that human and mouse lymphomas display similar genomic profiles and common molecular signatures, sharing MALT1-mediated NF-κB activation, proinflammatory signaling and XBP1-induced plasmacytic differentiation.
Deletion of p53 Accelerates Tumor Onset and Induces ABC-DLBCL in Sca1-MALT1 Mice.
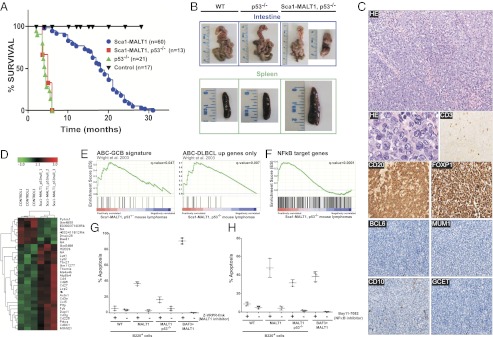
Previous studies have shown that P53 inactivation is associated with the transformation of MALT lymphoma to DLBCL (20). Based on these data, Sca1-MALT1 and p53−/− mice were crossed to generate Sca1-MALT1,p53−/− mice (21). Loss of p53 decreased the overall survival of Sca1-MALT1 mice (P < 0.0001, Log-rank (Mantel-Cox) test), which was similar to that of p53−/− animals (P = 0.53) (Fig. 3A). However, while p53−/− mice were killed by thymic T-cell lymphomas (21), Sca1-MALT1,p53−/− mice developed aggressive lymphomas characterized by a diffuse infiltrate of large B-lymphocytes with nuclei with dispersed chromatin, deeply basophilic cytoplasm and multiple mitotic figures, resembling human DLBCL (Fig. 3 B and C and SI Appendix, Fig. S5A). IHC studies revealed moderate expression of CD20 and strong expression of Foxp1, with negative expression for Bcl6, Gcet1, Mum1 (Irf4) and CD10, and with few scattered CD3+ T cells, allowing their classification as human-like ABC-DLBCL (17) (Fig. 3C). Notably, over-expression of FOXP1 is a common feature of aggressive human ABC-DLBCL (22). PCR and Southern blot analyses of the Sca1-MALT1,p53−/− lymphomas revealed their oligoclonal and clonal nature in most cases (SI Appendix, Fig. S3 G and H). Gene expression profiling analysis identified 34 genes differentially expressed between B cells isolated from splenic Sca1-MALT1,p53−/− lymphomas and from WT spleens (LIMMA; FDR < 0.3; P < 0.0004) (Fig. 3D and SI Appendix, Table S3C). GSEA using a gene-expression-based predictor to classify human DLBCL (10) revealed that the ABC-DLBCL transcriptional signature was highly enriched in the transcriptional profile of Sca1-MALT1,p53−/− lymphomas when using the entire classifier (FDR < 0.047) and specially when selecting the up-regulated genes (FDR < 0.007) (Fig. 3E). Additionally, GSEA showed a significant enrichment in NF-κB target genes in the Sca1-MALT1,p53−/− lymphomas (FDR < 0.0001), which is one of the main features of human ABC-DLBCL (7, 10, 11, 18) (Fig. 3F). Finally, aCGH studies of clonal lymphomas developed in Sca1-MALT1,p53−/− mice identified a complex pattern of DNA copy number changes that were syntenic with the gain/amplification of human chromosomes 18q21 (including the BCL2 gene locus) and 3q25-q26.2, both of which are frequently observed in ABC-DLBCL but not in GBC-DLBCL (23) (SI Appendix, Fig. S5 B and C). These results demonstrate that p53 loss is associated with the transformation of MALT lymphoma to human-like ABC-DLBCL in Sca1-MALT1 mice, further corroborating the parallelism between mouse and human disease.
Fig. 3.
Sca1-MALT1-p53−/− mice develop human-like ABC-DLBCL. (A) Kaplan–Meier survival plots of Sca1-MALT1, p53−/−, p53−/−, and control WT mice. (B) Macroscopic aspect of the intestine and spleen in Sca1-Malt1,p53−/− mice vs. WT and p53−/− age-matched mice. (C) Representative hematoxylin–eosin (HE) staining and IHC analysis for CD20, Foxp1, Mum1, Gcet1, CD10, and Bcl6 in Sca1-Malt1,p53−/− mice. Appropriate positive and negative control tissues for each antibody were used (see Materials and Methods). (D) Heat-map image showing the Sca1-MALT1,p53−/− lymphoma transcriptional signature. B220+ cells isolated from three Sca1-MALT1,p53−/− lymphomas and three WT spleens were studied. (E) GSEA showed a significant positive enrichment of the human ABC-DLBCL transcriptional signature in Sca1-MALT1,p53−/− lymphomas. (F) Sca1-MALT1,p53−/− lymphomas were significantly enriched in NF-κB target genes. (G and H) B220+ splenic B lymphoma cells isolated from three Sca1-MALT1 and three Sca1-MALT1,p53−/− mice were incubated with (+) or without (−) z-VRPR-fmk (75 μM) (G) or Bay11-7082 (5 μM) (H) for 48 h. Induction of apoptosis was assessed by flow cytometry using annexin V/PI staining. Data represent the means ± SD percentages of apoptotic cells from three independent experiments.
Sca1-MALT1 and Sca1-MALT1,p53−/− Mice Are Useful Models to Test MALT1 Inhibitors.
Previous publications have reported that the MALT1 caspase-like domain shows proteolytic activity that can be inhibited by the peptidic inhibitor, z-VRPR-fmk (8, 9). Measurement of Malt1 mRNA with murine-specific primers did not identify higher Malt1 mRNA expression in Sca1-MALT1 lymphomas than in WT splenocytes (SI Appendix, Fig. S5D). However, endogenous Malt1 showed proteolytic activity in the murine lymphomas and cleaved Bcl10 (SI Appendix, Fig. S5E). To evaluate whether our models may be suitable tools for testing MALT1 inhibitors (14, 15), B220+ lymphoma B cells isolated from transgenic mice were treated with the specific MALT1 inhibitor z-VRPR-fmk and with the NF-κB inhibitor Bay11-7082 (9). Both, Sca1-MALT1 and Sca1-MALT1,p53−/− lymphoma cells showed a statistically significant increase in the apoptotic rates upon incubation with z-VRPR-fmk (P < 0.05) (Fig. 3G) or with Bay11-7082 (P < 0.05) (Fig. 3H and SI Appendix, Fig. S5F). These data indicate that constitutive Malt1-induced NF-κB signaling is required for lymphoma cell survival, suggesting that these mice can be useful for testing MALT1 inhibitors.
Bone Marrow CD34+ Cells Isolated from Patients with MALT Lymphoma Show an Aberrant Transcriptional Profile.
To investigate whether hematopoietic/stem cells could be involved in the pathogenesis of MALT lymphoma, bone marrow CD34+ cells (with purity of >95%) were isolated from six patients with MALT lymphoma and of five healthy donors (SI Appendix, Table S4A). Using FISH or semiquantitative RT-PCR, the t(11;18)(q21;q21) was detected in the mature B-cell lymphoma cells of two patients but not in the CD34+ cells (SI Appendix, Fig. S6 A and B). Gene-expression-profiling analysis of CD34+ cells isolated from MALT lymphoma patients vs. healthy individuals identified 743 gene probes differentially expressed (LIMMA; FDR < 0.017; P < 0.001) (SI Appendix, Fig. S6C). The human CD34+ MALT lymphoma transcriptional signature revealed among the top biological functions “inflammatory response” and “antigen presentation” (SI Appendix, Fig. S6D). In addition, overexpression of 23 gene probes that corresponded to Ig genes was detected in MALT lymphoma CD34+ cells (SI Appendix, Table S4B). GSEA identified that the human CD34+ MALT lymphoma dataset was positively enriched in NF-κB target genes (FDR < 0.0001) (SI Appendix, Fig. S6E). Moreover, the human CD34+ MALT lymphoma transcriptional signature was significantly enriched in the previously defined human MALT lymphoma dataset (FDR < 0.012) (SI Appendix, Fig. S6F) but not in the datasets from patients with DLBCL (FDR = 0.059), follicular lymphoma (FDR = 0.113), or splenic marginal-zone lymphoma (FDR = 0.357) (SI Appendix, Fig. S6 G–I). Finally, the molecular signatures of both Sca1+Lin− cells and mature B-lymphoma cells isolated from the Sca1-MALT1 mice were enriched in the human CD34+ MALT lymphoma dataset (FDR < 0.0001 and FDR = 0.005, respectively) (SI Appendix, Fig. S6 J and K). These results show that NF-κB and inflammatory signaling are present in human MALT lymphoma CD34+ cells, along with the overexpression of a set of Ig genes, which is compatible with early lymphoid priming. Because these Ig genes are not normally expressed in CD34+ cells but rather in committed B lymphocytes, our findings suggest that the hematopoietic stem/progenitor cells may be involved in the pathogenesis of human lymphoma.
Discussion
Understanding the biology underlying MALT lymphoma has been hampered by the lack of genetically modified mouse models that develop human-like tumors (1–3, 7). Here, we show that the expression of human MALT1 in Sca1+Lin− hematopoietic stem/progenitor cells induced an abnormal NF-κB signaling that promoted expansion and early priming of these cells toward B-cell differentiation, eventually resulting in the accumulation of mature B lymphocytes in extranodal tissues and in progressive lymphoma development. These tumors accurately reflected most clinical, histopathological, genetic, and molecular features of human MALT lymphomas (1–3). Further paralleling the natural history of human disease, spontaneous transformation of mouse MALT lymphoma to ABC-DLBCL was observed in a fraction of Sca1-MALT1 mice, which was accelerated after the constitutive deletion of the p53 gene (20). Therefore, human lymphoma pathogenesis can be modeled in mice by targeting MALT1 expression to the hematopoietic stem/progenitor cells. Our results also show that constitutive Malt1 signaling is required for the survival of murine lymphoma cells, indicating that our transgenic models may be suitable tools for developing and testing MALT1 inhibitors (24).
Additionally, our data suggest that hematopoietic/stem progenitor cells may be involved in the development of mature B-cell lymphomas. In support of this hypothesis, one recent report demonstrated that patients can harbor inactivating mutations of TET2 gene in hematopoietic stem cells that give rise to B-cell lymphomas (25). Similarly, in B-cell chronic lymphocytic leukemia (CLL), the propensity to generate clonal B cells had been already acquired at the hematopoietic stem cell stage (26). Thus, CLL-derived hematopoietic stem cells showed an abnormal expression of lymphoid-related genes, presumably reflecting their cell-intrinsic priming into the lymphoid lineage (26). Although our results indicate that hematopoietic stem/progenitor cells may play a role in human mature B-cell lymphoma development, it cannot be excluded, however, that the abnormal gene-expression profile of human MALT lymphoma CD34+ cells could be consequence of having cancer.
In summary, our study shows that human MALT lymphoma pathogenesis can be modeled in mice by targeting MALT1 expression to hematopoietic stem/progenitor cells, demonstrating the oncogenic role of MALT1 in lymphomagenesis. Furthermore, our findings establish a molecular link between MALT lymphoma and ABC-DLBCL and provide mouse models to test therapies targeting MALT1 proteolytic activity.
Materials and Methods
Generation of Transgenic Mice.
The Sca1-MALT1 vector was generated by inserting the human MALT1 cDNA into the ClaI site of the pLy6 vector. The transgene fragment was excised from its vector by restriction digestion with NotI, purified, and injected (2 ng/mL) into CBA × C57BL/6J fertilized eggs (16). Heterozygous p53+/− mice (21) were bred to Sca1-MALT1 mice to generate null p53−/− mice hemizygous for Sca1-MALT1. The study was performed in accordance with Consejo Superior de Investigaciones Científicas, University of Salamanca and University of Navarra Institutional Animal Care and Use Committees and the Spanish National Institute of Health guidelines.
Patient Samples.
Biopsies from patients with MALT lymphoma (n = 75), DLBCL (n = 26), follicular lymphoma (n = 15), and splenic marginal B-cell lymphoma (n = 12) were included in the study. For BM CD34+ cell isolation, Spanish patients newly diagnosed with MALT lymphoma and healthy donors were included. Informed consent was obtained from the patients in accordance with the Declaration of Helsinki. The study was approved by Consejo Superior de Investigaciones Científicas, University of Salamanca and University of Navarra Institutional Research Ethics Committees.
Microarray Data.
Affymetrix microarray data files are available at GEO (accession numbers GSE25636, GSE25637, GSE25638, GSE25639, and GSE34015).
Additional materials and methods are presented in SI Appendix.
Supplementary Material
Acknowledgments
We thank E. Dzierzak for the Sca1 promoter, M. J. Dyer for MALT1 and BCL10 cDNAs, M. Baens for the API2-MALT1 cDNA, L. Ortiz and C. Gomez-Abad for support with microarray analyses, and M. Morente (Spanish Tumor Bank Network) and A. Fortuño (University of Navarra Tumor Bank) for providing lymphoma biopsies. This work was funded by Spanish Ministry of Science and Innovation Grant PI080164 and Consejo Superior de Investigaciones Cientificas (CSIC) P.I.E.200920I055 (to C.C.), SAF2009-08803 and CSD2007-0017 (to I.S.-G.), FIS-PI081878, and RTICCRD06/0020/0088-0103-0111-0006 and PPT-300000-2008-4 (to J.A.M.-C.); by Junta de Castilla y León SA060A09 (to C.C.), CSI007A11-2 (to I.S.-G.), and SAN/39/2010 (to C.V.-D.); by Fundación Ramón Areces (to C.C.); and by an Affymetrix Collaboration in Cancer Program grant (to J.A.M.-C.). I.R.-C., F.A.-J., E.C.-S., M.B.-D., L.F., J.I.M.-F., and M.A.A. were supported by Spanish Ministry of Science and Innovation fellowships. E.C.-S. was a “Residencia de Estudiantes” Fellow. C.B. was supported by the Navarra Government. All Spanish funding is cosponsored by the European Union Fondo Europeo de Desarrollo Regional (FEDER) program. The laboratory of I.S.-G. is an Associate Principal Investigators (API) laboratory of the EuroSyStem project.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE25636–GSE25639 and GSE34015).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204127109/-/DCSupplemental.
References
- 1.Farinha P, Gascoyne RD. Molecular pathogenesis of mucosa-associated lymphoid tissue lymphoma. J Clin Oncol. 2005;23:6370–6378. doi: 10.1200/JCO.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 2.Isaacson PG, Du MQ. MALT lymphoma: From morphology to molecules. Nat Rev Cancer. 2004;4:644–653. doi: 10.1038/nrc1409. [DOI] [PubMed] [Google Scholar]
- 3.Sagaert X, De Wolf-Peeters C, Noels H, Baens M. The pathogenesis of MALT lymphomas: Where do we stand? Leukemia. 2007;21:389–396. doi: 10.1038/sj.leu.2404517. [DOI] [PubMed] [Google Scholar]
- 4.Dierlamm J, et al. The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood. 1999;93:3601–3609. [PubMed] [Google Scholar]
- 5.Sanchez-Izquierdo D, et al. MALT1 is deregulated by both chromosomal translocation and amplification in B-cell non-Hodgkin lymphoma. Blood. 2003;101:4539–4546. doi: 10.1182/blood-2002-10-3236. [DOI] [PubMed] [Google Scholar]
- 6.Streubel B, et al. T(14;18)(q32;q21) involving IGH and MALT1 is a frequent chromosomal aberration in MALT lymphoma. Blood. 2003;101:2335–2339. doi: 10.1182/blood-2002-09-2963. [DOI] [PubMed] [Google Scholar]
- 7.Thome M, Charton JE, Pelzer C, Hailfinger S. Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harb Perspect Biol. 2010;2:a003004. doi: 10.1101/cshperspect.a003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coornaert B, et al. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat Immunol. 2008;9:263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- 9.Rebeaud F, et al. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat Immunol. 2008;9:272–281. doi: 10.1038/ni1568. [DOI] [PubMed] [Google Scholar]
- 10.Wright G, et al. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci USA. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2:a000109. doi: 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baens M, et al. Selective expansion of marginal zone B cells in Emicro-API2-MALT1 mice is linked to enhanced IkappaB kinase gamma polyubiquitination. Cancer Res. 2006;66:5270–5277. doi: 10.1158/0008-5472.CAN-05-4590. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, et al. Emu-BCL10 mice exhibit constitutive activation of both canonical and non-canonical NF-kappaB pathways generating marginal zone (MZ) B cell expansion as a precursor to splenic MZ lymphoma. Blood. 2009;114:4158–4168. doi: 10.1182/blood-2008-12-192583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferch U, et al. Inhibition of MALT1 protease activity is selectively toxic for activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2009;206:2313–2320. doi: 10.1084/jem.20091167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hailfinger S, et al. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc Natl Acad Sci USA. 2009;106:19946–19951. doi: 10.1073/pnas.0907511106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez-Caro M, et al. Cancer induction by restriction of oncogene expression to the stem cell compartment. EMBO J. 2009;28:8–20. doi: 10.1038/emboj.2008.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi WW, et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res. 2009;15:5494–5502. doi: 10.1158/1078-0432.CCR-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Visco C, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: A report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012 doi: 10.1038/leu.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaffer AL, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Du M, Peng H, Singh N, Isaacson PG, Pan L. The accumulation of p53 abnormalities is associated with progression of mucosa-associated lymphoid tissue lymphoma. Blood. 1995;86:4587–4593. [PubMed] [Google Scholar]
- 21.Jacks T, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 22.Sagaert X, et al. Forkhead box protein P1 expression in mucosa-associated lymphoid tissue lymphomas predicts poor prognosis and transformation to diffuse large B-cell lymphoma. J Clin Oncol. 2006;24:2490–2497. doi: 10.1200/JCO.2006.05.6150. [DOI] [PubMed] [Google Scholar]
- 23.Lenz G, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci USA. 2008;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunleavy K, et al. Differential efficacy of bortezomib plus chemotherapy within molecular subtypes of diffuse large B-cell lymphoma. Blood. 2009;113:6069–6076. doi: 10.1182/blood-2009-01-199679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quivoron C, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Kikushige Y, et al. Self-renewing hematopoietic stem cell is the primary target in pathogenesis of human chronic lymphocytic leukemia. Cancer Cell. 2011;20:246–259. doi: 10.1016/j.ccr.2011.06.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.