Abstract
MicroRNA-155 (miR-155) is an oncogenic microRNA that regulates several pathways involved in cell division and immunoregulation. It is overexpressed in numerous cancers, is often correlated with poor prognosis, and is thus a key target for future therapies. In this work we show that overexpression of miR-155 in lymphoid tissues results in disseminated lymphoma characterized by a clonal, transplantable pre-B-cell population of neoplastic lymphocytes. Withdrawal of miR-155 in mice with established disease results in rapid regression of lymphadenopathy, in part because of apoptosis of the malignant lymphocytes, demonstrating that these tumors are dependent on miR-155 expression. We show that systemic delivery of antisense peptide nucleic acids encapsulated in unique polymer nanoparticles inhibits miR-155 and slows the growth of these “addicted” pre-B-cell tumors in vivo, suggesting a promising therapeutic option for lymphoma/leukemia.
Keywords: oncogene, oncomiR addiction, nanotechnology
MicroRNAs (miRNAs) govern nearly every biological process investigated (1–8). MicroRNA-155 (miR-155) in particular is one of the most salient miRNAs, and although its role in immune function has been the subject of much attention (9–14), manifold studies also implicate it in cancer pathways, particularly those of hematopoietic origin (11, 15–21). miR-155 is induced in several lymphomas, such as diffuse large B-cell lymphomas, Hodgkin lymphomas, and subsets of Burkitt lymphomas (16, 18, 22), and ectopic expression of miR-155 in a transgenic mouse model leads to B-cell malignancy (15, 23). Furthermore, miR-155 induction in hematopoietic cells of myeloid origin results in a myeloproliferative pathology characterized by granulocyte/monocyte expansion in lymphoid tissues (24, 25). At the molecular level, miR-155 directly targets SH2-containing inositol phosphatase (SHIP1), a negative regulator of myeloid cell proliferation and survival. Furthermore, in diffuse large B-cell lymphomas, miR-155 induction and subsequent SHIP1 repression are TNF-α-dependent (23, 24, 26). Although these studies provide compelling evidence for miR-155 involvement in lymphoproliferative disease, the degree to which the survival of these cancers depend on maintained miR-155 expression has not been established, a crucial point to ascertain in determining whether or not inhibition of miR-155 has therapeutic promise.
It is known that several protein-coding oncogenes such as Egfr, Myc, Ras, and Her2 (among others) exhibit oncogene addiction and that tumors can become dependent on maintained expression of these genes in mouse models (27–29). Recently, we described a miR-21-induced mouse model of lymphoma in which tumors were dependent on maintained expression of miR-21, demonstrating that miRNAs are a unique class of genes that mediate oncogene addiction (30).
Toward cancer therapy, anti-miR molecules could potentially be used to attenuate oncogenic miRNAs, ultimately exploiting the miRNA dependence exhibited by certain tumors. In recent years, anti-miRs have emerged as useful tools for inhibiting the function of miRNAs. Chemically modified anti-miRs, such as cholesterol-conjugated “antagomirs,” locked nucleic acid oligonucleotides, and polylysine-conjugated peptide nucleic acids (PNAs) have demonstrated effectiveness in vivo (31–33). From a sterics perspective, the range of functional modifications achievable for anti-miRs is limited. On the contrary, nanoparticulate delivery systems (< 1-μm diameter) are well suited to support a multitude of delivery-enhancing modifications. Recently, Su et al. described a nanocomplex that delivers anti-miR-122 to the liver (34). Furthermore, nanoparticles composed of poly(lactic-co-glycolic acid) (PLGA) are safe, biodegradable, and have a long history of success in enhancing the delivery of therapeutic agents.
Here, we have generated a sophisticated in vivo system to induce and withdraw miR-155 expression in mouse tissues of interest using Cre-loxP and tTA technologies and show that anti-miR-155 molecules introduced to tumors using a unique nanoparticulate-delivery strategy have therapeutic efficacy.
Results
Generation of mir-155LSLtTA Mice.
We designed a miR-155 Cre-loxP tetracycline-controlled knockin mouse model, mir-155LSLtTA, in which mir-155 is targeted to the ROSA26 locus. ROSA26 is ubiquitously expressed in mouse tissues and its disruption does not exhibit a phenotype (35), allowing for broad regulation of our system in tissues of interest. The construct contains a loxP-floxed stop cassette that prevents transcription until the stop cassette is removed by Cre recombinase, allowing for tissue-specific expression of miR-155 depending on the promoter-Cre line into which the animal is crossed (Fig. 1A). Furthermore, this system contains a tTA element that deactivates miR-155 induction upon exposure to doxycycline (DOX), providing temporal control of miR-155 expression (36) (Fig. 1A).
Fig. 1.
Controlled expression of miR-155 in mir-155LSLtTA mice. (A) Schematic of controlled gene expression in mir-155LSLtTA mice. miR-155 expression is prevented until exposure to Cre recombinase, which excises a stop cassette allowing transcription to proceed. Transcription can subsequently be suppressed by exposure to doxycycline. Modified from (36). (B) Recombination and stop cassette removal detected by PCR in the spleen, thymus, and bone marrow of a 2-wk-old NesCre8; mir-155LSLtTA mouse (n = 20). (C) miR-155 levels in the spleens of wild-type mice and NesCre8; mir-155LSLtTA mice with and without doxycycline. Graph represents averages of three mice per group ± SD. (D) Percentage of mice that showed signs of lymphadenopathy and/or paresis (median time 115 d); p was calculated with a log-rank (Mantel-Cox) test (n = 20).
To determine whether induction of miR-155 in various tissues is sufficient for tumorigenesis, we activated recombination in brain and lymphoid tissues by crossing these mice with a NesCre8 driver (30) to generate double heterozygote NesCre8; mir-155LSLtTA mice, and withdrawing doxycycline from their chow at birth. Recombination was evident in the spleen, thymus, and bone marrow of 2-wk-old NesCre8; mir-155LSLtTA mice (Fig. 1B), and in the absence of doxycycline, miR-155 expression was increased approximately 20-fold in spleen and approximately 120-fold in bone marrow of 2-wk-old mice (Fig. S1 A and B). miR-155 expression was increased by approximately 70-fold in adult brain (Fig. S1C) and approximately 45-fold in adult spleen (Fig. 1C). This is within the range of miR-155 induction reported in human lymphoma lines (16). Feeding doxycycline after induction reduced miR-155 levels to less than fivefold above normal in brain (Fig. S1C) and less than twofold in spleen (Fig. 1C), confirming that the elevated levels of miR-155 were dependent on the transgene.
Induction of miR-155 in Lymphoid Tissues Causes Disseminated Lymphoma.
Brains from NesCre8; mir-155LSLtTA mice were harvested from induced mice euthanized at ages 4–5 mo. Complete gross and histopathological analysis was performed on all tissues. Microscopic examinations did not reveal any abnormal brain pathology, suggesting that the high levels of miR-155 alone are insufficient to induce brain tumors in our model in the timeframe evaluated. Similarly, necropsy and histopathologic analysis on lung tissues harvested from intranasal Adenovirus-Cre-treated mir-155LSLtTA mice expressing elevated miR-155 (Fig. S1D) euthanized at 6 mo post-infection, did not reveal any histopathologic changes indicative of neoplasia, and lung histology appeared similar to control mice. In contrast, NesCre8; mir-155LSLtTA mice reared without doxycycline developed clinical signs of lymphoid pathology as early as age 2 wk, at which time there was marked expansion of the splenic white pulp by a monotypic population of lymphocytes (Fig. S1E). As the mice developed postnatally (age 2–4 mo), they presented with a scruffy fur coat, hunched posture, and, occasionally, ataxia. By age 5 mo the majority of mice developed pronounced lymphadenopathy (Fig. 1D and Fig. 2A and B), splenomegaly (Fig. 2B), and labored breathing (in some mice, the ataxia progressed to severe hind-limb paresis), while all NesCre8; mir-155LSLtTA mice developed splenomegaly. Complete gross necropsy revealed that NesCre8; mir-155LSLtTA mice without doxycycline had marked splenomegaly (Fig. 2B, spleen indicated by triple asterisk), severe diffuse lymphadenopathy (Fig. 2B, submandibular lymph nodes indicated by arrows; axillary lymph nodes by arrowheads), and, in some cases, hepatomegaly (Fig. 2B, double asterisk) and marked thymic hyperplasia (Fig. 2B, single asterisk) suggestive of lymphoma. Enlarged spleens averaged approximately 10 times the weight of corresponding littermate controls (Fig. S1F). Complete histopathologic examination was performed on tissues from these mice. Wild-type mice exhibit normal splenic and thymic architecture (Fig. 2C and D, respectively). In contrast, histopathology of NesCre8; mir-155LSLtTA mice revealed the presence of a monotonous population of lymphoblasts that effaced and replaced the normal splenic and thymic architecture (Fig. 2E and F). Malignant lymphocytes were found in high numbers around hepatic central veins (Fig. 2G, arrow) and within the submandibular lymph nodes (Fig. 2H, asterisk). Mice with posterior paresis had inappropriate accumulations of lymphoblasts within the meninges of the brain (Fig. 2I, asterisk) and spinal cord (Fig. 2J, arrowhead) in addition to greatly enlarged paravertebral lymph nodes (Fig. 2J, arrows). Furthermore, complete blood count analysis was performed on three mice. Mice with lymphoma tended to be anemic [RBC count 3.3·106 RBC/cmm (reference range 5.5–10·106 cells/cmm); hemoglobin 5.9 g/dL (reference range 13–15 g/dL); hematocrit 18% (reference range 33–35%)] and leukopenic [3.17·103 WBC/cmm (reference range 5.5–10.5·103 WBC/cmm)]. Neither wild-type littermates nor NesCre8; mir-155LSLtTA mice maintained on doxycycline developed any signs of lymphoma (Fig. 1D), indicating that miR-155 induction is necessary for disease pathology in this model.
Fig. 2.
mir-155LSLtTA mice develop disseminated lymphoma. (A) NesCre8; mir-155LSLtTA mouse with ruffled fur and massively enlarged peripheral and axillary lymph nodes (arrowheads) (n = 20). (B) Gross anatomical level of enlarged spleen (arrow) and lymph nodes (arrowheads) (n = 20). (C, D) Representative H&E sections from wild-type mice reveal distinct periarteriolar lymphocyte sheaths (PALS) in spleen and normal cortex and medullary in thymus; (n = 20). (E, F) Representative H&E sections from NesCre8; mir-155LSLtTA mouse spleen and thymus. Spleen and thymic parenchyma are effaced and replaced by a monotonous population of lymphoblasts(n = 20). (G–J) Lymphocyte infiltration into liver, submandibular lymph nodes (SMLN), meninges of the brain (I, asterisk), paravertebral lymph nodes (J, arrows), and meninges of the spinal cord (J, arrowhead). Scale bars, 500 μ (n = 20). (K) Lymphadenopathy is observed in nude mice when injected subcutaneously with NesCre8; mir-155LSLtTA spleen cells (n = 5). (L) Representative flow cytometry plot reveals that NesCre8; mir-155LSLtTA spleens (p = 0.0006) and thymuses (p = 0.0002) are highly enriched with a B220+, IgM population of cells (Upper Left) (n = 3). The following channels were used for gating: FL1 channel to detect FITC-conjugated antibodies, FL2 to detect PE-conjugated antibodies, FL3 to detect APC-conjugated antibodies, and FL4 to detect PI.
Furthermore, given our previous report that miR-21 induction leads to lymphoma (30), we performed quantitative real-time PCR (qPCR) to investigate whether or not miR-21 is induced in spleens of NesCre8; mir-155LSLtTA mice and found that it was not (Fig. S1G). This indicated that the miR-155 phenotype was not merely the result of overexpressing miR-21.
Interestingly, in a few instances we observed that NesCre8; mir-155LSLtTA mice reared without doxycycline did not develop the severe signs of lymphoma, but instead lived for over a year before they were euthanized. These mice presented with splenomegaly (as did all examined NesCre8; mir-155LSLtTA mice reared without doxycycline) but did not exhibit the characteristic signs of disseminated lymphoma or pathologic changes observed in the majority of NesCre8; mir-155LSLtTA mice. qPCR revealed that miR-155 induction in spleens of these mice was only approximately 12-fold above wild-type levels (Fig. S1H), which is roughly half the induction seen in mice that developed aggressive disease, and is borderline insignificant (p = 0.06). Although we are uncertain why we observed variability in the levels of miR-155 induction between animals, we find it interesting that levels seemingly need to reach a certain threshold to induce aggressive disease pathology.
Tumor Cells are Clonal, Transplantable, and Migratory Pre-B cells.
We sought to determine the clonality status of our model by taking advantage of a PCR strategy developed to detect different diversity (D) and joining (J) rearrangements in splenic B-cell Ig heavy chain genes (37, 38). Primers flanking the D and J.4 regions of genomic DNA were used to analyze samples; in the case of a clonal population, we would expect at most two bands representing a subset of possible rearrangements for the two alleles in one cell. In contrast to control spleens, which are comprised of a polyclonal population of cells, we found that NesCre8; mir-155LSLtTA spleens were expanded with a clonal or oligoclonal population of cells (Fig. S2). This suggests that our mouse model, unlike some other lymphoma models, may be particularly relevant for understanding human B-cell malignancy, which is almost invariably clonal (39, 40). Interestingly, two mice with reduced miR-155 expression and less aggressive disease (age > 1 year) had a polyclonal population of cells (Fig. S2). These results suggest that cells in mice with aggressive disease may have acquired secondary mutations, conferring them with a selective growth advantage.
We sought to examine the tumorigenic potential of lymphoma cells from NesCre8; mir-155LSLtTA mice. Affected mice showing lymphadenopathy or paresis were euthanized and spleens were harvested and dispersed into single-cell suspensions, which were injected subcutaneously into nude mice. At an average of 10 d, palpable tumors were observed (Fig. 3C, Upper), and mice were subsequently euthanized for humane reasons after another 3–4 d because of aggressive tumor growth. Intriguingly, we noticed that nude mice injected subcutaneously with cells from NesCre8; mir-155LSLtTA spleens variably developed splenomegaly and peripheral lymphadenopathy (Fig. 2K, arrowheads). PCR from the spleen and lymph nodes from these nude mice confirmed that the cells originated from the NesCre8; mir-155LSLtTA mice, indicating that these cells had migrated from the original site of subcutaneous tumor cell injection and colonized these organs.
Fig. 3.
Doxycycline-induced withdrawal of miR-155 leads to rapid tumor regression. (A) Regression of enlarged lymph nodes in NesCre8; mir-155LSLtTA mice exposed to doxycycline (n = 10). (B) H&E spleens from NesCre8; mir-155LSLtTA mice without doxycycline or on doxycycline for 48 h or 2 wk. In recovered mice, spleen architecture is restored similar to wild-type mice (n = 10). (C) Nude mice injected subcutaneously with lymphoma spleen cells develop aggressive tumors. Tumors regress rapidly when exposed to doxycycline (n = 3 mice per group). (D) Change in tumor volume, V = (length × width2)/2 (n = 3 mice per group, represented as mean ± SD); t test was used for statistical analysis. (E) Kaplan-Meier survival curve. After developing conspicuous signs of disease, mice were fed with doxycycline food for recovery or normal food (n = 10 mice per group).
We investigated the specific cell type involved in the disseminated lymphoma by performing flow cytometry on NesCre8; mir-155LSLtTA spleens, thymuses, and lymph nodes. We found that the tumor cells were B220+, IgM-, CD3-, and CD43+, revealing a pre-B-cell lymphoblastic lymphoma/leukemia (41) (Fig. 2L).
Doxycycline-Induced Withdrawal of miR-155 Causes Rapid Tumor Regression.
Our system allowed us to test whether miR-155-induced pre-B-cell lymphoma cells become addicted to miR-155 expression. Mice that presented with clear clinical symptoms of disseminated lymphoma (lymphadenopathy and/or paresis) were fed doxycycline-impregnated food and monitored. Strikingly, mice fed with doxycycline food rapidly recovered from disease-within 24 h, regression of lymphadenopathy occurred. By 48 h, lymph node size was markedly reduced and disease virtually undetectable (Fig. 3A, Lower), and by 1 wk, lymph nodes were visibly the same size as control mice. Within 1 wk the gross symptoms of disease-including the scruffy coat, hunched posture, paresis, and labored breathing (likely caused by the markedly enlarged submandibular lymph nodes)-had also disappeared. In contrast to mice not fed doxycycline (Fig. 3B, Upper), histopathological analysis of the spleen at 48 h in mice given doxycycline revealed partial recovery of the splenic architecture (Fig. 3B, Middle). By 2 wk after doxycycline exposure, the spleen (Fig. 3B, Lower) and other lymphoid organs displayed normal architecture. Additionally, tumors from nude mice injected with NesCre8; mir-155LSLtTA tumor cells derived from spleen or lymph nodes rapidly recovered (within days) when exposed to doxycycline (Fig. 3C and D).
To determine differences in overall survival between mice that received doxycycline and mice that did not, we collected data for a survival curve. Twenty NesCre8; mir-155LSLtTA mice that developed clear clinical signs of disseminated lymphoma (paresis and/or lymphadenopathy, which occurred between age 3–5 mo) were divided into two arms: One arm received doxycycline and the other did not. Mice died between age 3–5 mo if they were not fed doxycycline, while mice that were fed doxycycline recovered and survived until at least 10 mo, when we stopped monitoring them (Fig. 3E).
Tumor Regression Occurs in Part via Apoptosis.
Given the rapid nature of tumor regression in NesCre8; mir-155LSLtTA mice fed with doxycycline, we hypothesized that recovery occurs, at least in part, via apoptosis. Supporting this hypothesis, in a genome-scale screen to identify miRNAs that could modulate tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis, miR-155 could suppress apoptosis in MDA-MB-453 cells and T-cell leukemia Jurkat cells (42). To investigate the degree to which apoptosis occurs in doxycycline-fed mice, we compared TUNEL-stained sections of spleen-derived tumors from nude mice with and without doxycycline. Histopathologic analysis revealed greater numbers of TUNEL-positive cells in regressing tumors in doxycycline-fed mice than in tumors from mice not exposed to doxycycline (Fig. 4A). Furthermore, we investigated the earlier stages of apoptosis by examining annexin-V binding in recovering tumors (43). We found that tumors exposed to doxycycline had higher levels of apoptosis compared to nondoxycycline tumors as measured by annexin-V binding (Fig. 4B and C). To determine if withdrawal of miR-155 affected cell differentiation, we removed spleen-derived tumors undergoing regression from nude mice fed on doxycycline and stained the tumor cells for annexin-V and CD43, a marker for immature B cells (44). We found that the proportion of CD43-positive tumor cells remained high in doxycycline-fed mice (Fig. 4C). Together, these results suggest that miR-155 expression inhibits apoptosis in NesCre8; mir-155LSLtTA tumors and that miR-155 withdrawal does not cause tumor cell differentiation.
Fig. 4.
Tumor regression cccurs in part via apoptosis. (A) TUNEL staining of tumors from nude mice fed with doxycycline for 48 h or without doxycycline (n = 3). (B. C) Nude tumors from mice fed with or without doxycycline for 48 h and analyzed for annexin-V binding and/or CD43 staining via flow cytometry (n = 3). (D) Western blot of spleens from wild-type and NesCre8; mir-155LSLtTA mice treated with doxycycline for 72 h with or without doxycycline (n = 3). (E) AlamarBlue assay to detect relative viability of cells with and without doxycycline (n = 3, data represented as mean ± SD).
SHIP1 is a miR-155 target gene (23, 24) and has been reported to be a positive regulator of apoptosis (45–47). We found that SHIP1 protein levels were decreased in splenic cells from NesCre8; mir-155LSLtTA mice and increased slightly during the recovery period, 72 h after doxycycline feed (Fig. 4D). Additionally, we tested whether miR-155 withdrawal reduced tumor cell viability via Alamar Blue assay. We found that viability in NesCre8; mir-155LSLtTA spleen-derived cells not exposed to doxycycline was highly increased; doxycycline exposure led to a dramatic reduction in viability in these cells but not in Toledo B cells (Fig. 4E).
Unique Nanoparticles Deliver Anti-miRs to Lymphocytes.
The involvement of deregulated miRNA expression in promoting tumor initiation and progression prompts the exploitation of miRNAs as therapeutic targets. Several studies have demonstrated the utility of inhibiting miRNAs using complementary anti-miR molecules; chemically modified antagomirs and PNAs show particular promise in vivo (31, 32, 48, 49).
Because of their stability and excellent binding affinity, PNAs are ideal anti-miRs. However, current methods of delivering PNAs require chemical modifications and extremely high doses to achieve miRNA-inhibition effects (31). We hypothesized that the delivery efficacy of anti-miR PNAs could be enhanced by loading the agents into PLGA polymer nanoparticles. Previously we showed that PLGA nanoparticles could safely and effectively silence genes in vivo when loaded with siRNA (50) and could effectively deliver recombination-inducing PNAs in vitro when loaded with PNA and DNA (51). Unlike most nucleic acids, PNAs have a charge-neutral backbone, which makes their encapsulation into polymer nanoparticles uniquely independent of electrostatic interactions between cargo and delivery vehicle. We have shown that less than 5% of naked siRNA is encapsulated into PLGA nanoparticles by standard fabrication methods; however, precomplexing siRNA with a polycation can improve loading efficiency into nanoparticles to approximately 50% (50). In this work we observed that without the aid of a polycation, inherently charge-neutral PNAs displayed a similar encapsulation efficiency (approximately 50%) when loaded into nanoparticles-we mainly attribute this favorable loading to less charge repulsion during the nanoparticle synthesis process. In addition, although polycations are commonly exploited for nucleic acid delivery, because of their charge density, they may contribute adverse side effects in physiological and cellular environments (52). Loading charge-neutral PNAs into nanoparticles presents a nucleic acid–delivery strategy that is potentially more benign and more effective than other delivery systems.
Another key advantage of nanoparticulate delivery systems is their amenability to surface modification with molecules that can modulate nanoparticle function. To further enhance the ability of PLGA nanoparticles to delivery cargo intracellularly, we decorated the particles (with a 100–200-nm diameter measured by light scattering) with a cell-penetrating peptide, penetratin (53), using an effective surface attachment strategy that allows for a high density of ligand deposition and conformational flexibility (54). Interestingly, we found that pre-B cells uniquely favored uptake of nanoparticles coated with penetratin (ANTP-NP) as opposed to other cell-penetrating peptides (e.g., TAT and polyarginine) (Fig. S3 A and B). We then tested whether ANTP-NP-delivering anti-miR PNAs would have therapeutic potential in B-cell tumor models. Because lymphocytes are usually refractory to most transfection methods (55), we first used a miR-155 luciferase sensor to verify that nanoparticles could effectively deliver PNAs to B cells (Fig. 5A). Anti-miR-155 delivery by ANTP-NP into human Toledo B cells elicited an approximately fourfold increase in luciferase expression, likely a direct result of miR-155 inhibition. This increase was significantly greater than that achieved by nanoparticles without surface enhancements and ANTP-NP loaded with scramble (scrm) control anti-scrm (Fig. 5A). Notably, to ensure target specificity, preconditioning cells with pre-miR-155 resulted in loss of detectable miR-155 inhibition after ANTP-NP anti-miR-155 delivery (Fig. 5A).
Fig. 5.
Nanoparticle delivery of anti-miR-155 PNAs inhibits miR-155 in vitro. (A) miR-155 dual luciferase sensor demonstrates feasibility of nanoparticle-mediated anti-miR delivery to Toledo B cells (n = 3, data represented as mean ± SD); t test was used for statistical analysis relative to ANTP-NP anti-155 group. (B) Dose response of cultured Toledo B cells to anti-miR treatment with (siSHIP1) and without knockdown of SHIP1. After 48 h nanoparticle incubation, cell viability was measured using CellTiter-Blue (Promega) (n = 3, data represented as mean ± SD). Two-way ANOVA was used for statistical analysis relative to ANTP-NP anti-155-treated SHIP1 knockdown group. (C) miR-155 levels in miR-155-addicted pre-B cells treated with anti-miRs (n = 3, data represented as mean ± SD); t test was used for statistical analysis relative to ANTP-NP anti-scrm group; (D) Western blot of pre-B cells treated with anti-miRs demonstrates SHIP1 response to miR-155 inhibition. Shown is a representative blot of n = 3 independent experiments per group. (E) Dose response of cultured pre-B cells to anti-miR treatment. Cell viability measured using CellTiter-Blue (Promega) (n = 3, data represented as mean ± SD). Two-way ANOVA was used for statistical analysis.
B-cell lymphomas with elevated levels of miR-155 have previously been shown to be sensitive to reduction in miR-155 levels (26); specifically, Toledo xenograft tumors treated with an anti-TNF-α regimen to decrease the expression of miR-155 showed reduced cell proliferation and delayed tumor growth. Toward the realization of miRNA-inhibition therapy, nanoparticles loaded with anti-miRs have more applicability than gene-specific therapeutic strategies that attenuate miR expression (e.g., anti-TNF-α), which may also have undesirable off-target effects. Concordantly, ANTP-NP-delivering anti-miR-155 was effective at diminishing Toledo cell viability with an IC50 of 6.1 μM (Fig. 5B). This efficacy was decreased by more than 10-fold (IC50 of 90.1 μM) when SHIP1 was knocked down prior to treatment, which further demonstrates the involvement of miR-155 in regulating apoptosis and the relevance of SHIP1 as a proapoptotic target of miR-155 (Fig. 5B). However, it is important to note that after knockdown of SHIP1, lymphoma B cells still exhibited some sensitivity to anti-miR-155 treatment. Therefore, SHIP1 is likely one of several miR-155 targets that are functionally relevant for maintenance of lymphoma.
Demonstrating therapeutic efficacy in this genetically complex Toledo model of human B-cell lymphoma has implications for the widespread applicability of this anti-miR technology. However, it is unclear whether these B cells developed their cancer phenotype as a direct result of miR-155, so inhibition of miR-155 in this model may only have ancillary therapeutic effects. On the contrary, our miR-addicted pre-B cells present a less convoluted model in which cancer was induced and maintained specifically by miR-155. To investigate exclusively the involvement of miR-155 in tumor maintenance, we utilized miR-155-addicted pre-B cells isolated from NesCre8; mir-155LSLtTA mice as a therapeutic model. In cultured cells, ANTP-NP elicited 65% reduction of functional miR-155 levels, and uncoated nanoparticles yielded 23% reduction (Fig. 5C). We note that miRNA degradation is not the primary outcome of anti-miR binding; reduction of miRNA level is attributed to isolation and detection artifacts inherent to anti-miR-miRNA hybrids (56). Nevertheless, this reduction of miR-155 indicates effective delivery and binding of complementary PNAs resulting in loss of endogenous miRNA activity. To further demonstrate miR specificity, inhibition of miR-155 using ANTP-NP relieved the suppression of the miR-155 target, SHIP1; anti-miR-treated pre-B cells displayed elevated levels of SHIP1 relative to both scrambled anti-miR and untreated controls (Fig. 5D). Thus, PNA binding causes miR-155 inhibition.
Our model capitalizes on the involvement of miR-155 in tumor maintenance. Accordingly, we observed that neoplastic pre-B cells derived from NesCre8; mir-155LSLtTA mice were sensitive to ANTP-NP-mediated anti-miR-155 treatment with an IC50 of 1.1 μM (Fig. 5E), which was over fivefold more effective than similar treatment in Toledo cells. Anti-miR-155-loaded nanoparticles without penetratin modification showed an ineffective dose response that was similar to anti-scrm-loaded ANTP-NP (Fig. 5E), which further illustrates the utility of enhancing the nanoparticle surfaces with functional ligands. Thus, nanoparticles have been shown to be effective anti-miR delivery vehicles to lymphocytes in vitro.
Anti-miR-155 Therapy in miR-155-Addicted Tumors.
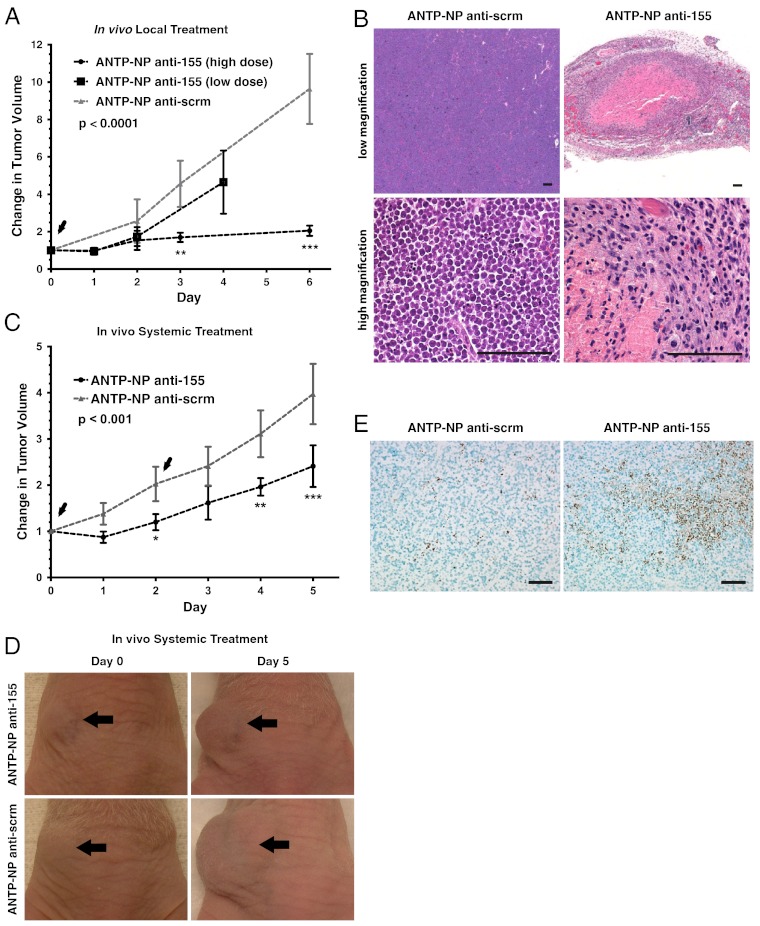
Given the dramatic tumor regression seen in response to doxycycline treatment (Fig. 3C), we investigated the anti-tumor potency of anti-miR-155 treatment in vivo. As with our previous studies (Fig. 3C), cells derived from the spleens of NesCre8; mir-155LSLtTA mice were inoculated into the flanks of nude mice to generate an aggressive subcutaneous tumor model of miR-155 addiction. A single local injection of anti-miR-155 loaded in ANTP-NP significantly diminished tumor growth over the course of 4–6 d (Fig. 6A). Anti-scrm-treated tumors displayed an approximately 10-fold increase in volume relative to starting volume, while anti-miR-155-treated tumors were limited to less than an approximately twofold increase at a dose of 1.5 mg/kg (high dose) (Fig. 6A). Anti-tumor efficacy was dependent on the dose of anti-miR-155; tumors treated with 0.5 mg/kg (low dose) showed less significant growth delay than the high dose-treated tumors. Notably, confocal microscopy of FITC-labeled PNAs showed that our nanoparticle delivery system could effectively deliver anti-miR PNAs to cells throughout tumor tissue and that nanoparticles were retained by the tumor for at least 2 d (Fig. S3C). Cell-penetrating peptides, such as penetratin, facilitate uptake into all types of cells, so it is possible that nanoparticles are taken up by various cells comprising the tumor; however, ANTP-NPs exhibit a proclivity for uptake by pre-B cells derived from the spleens of NesCre8; mir-155LSLtTA mice (Fig. S3 A and B). Histopathological analysis of treated tumors identified regions of hemorrhage and necrosis surrounded by infiltrative inflammatory cells (Fig. 6B). Control-treated tumors comprised a relatively homogeneous population of neoplastic lymphocytes with no areas of cell death (Fig. 6B).
Fig. 6.
Nanoparticle-based inhibition of miR-155 induces therapeutic effects. (A) Fold change in tumor growth in response to locally administered anti-miRs. Arrowheads, nanoparticle injection (n = 3 mice for control group, n = 4 mice for high dose treatment group, n = 3 for low dose treatment group); on day 1, one high dose anti-miR-155-treated mouse was removed for early time point analysis. Data represented as mean ± SD. Statistics were done using two-way ANOVA. (B) H&E analysis of locally treated tumors. Scale bar, 100 µm for both high and low magnification images. Representative images shown for n = 3 tumors. (C) Fold change in tumor growth in response to systemically administered anti-miRs. Arrowheads indicate nanoparticle injection (n = 3 mice per group, data represented as mean ± SD). Two-way ANOVA was used for statistical analysis. (D) Representative tumor images before and after systemic treatment, n = 3 mice per group. In accordance with proper animal care regulations, mice were euthanized when tumors reached a volume of 2,000 mm3. (E) TUNEL staining of systemically treated tumors. Scale bar, 50 µm. Representative images shown for n = 3 tumors.
An important benefit of this nanoscale delivery system is its size-dependent exploitation of the enhanced permeability and retention effect (EPR) for passive targeting of the nanoparticles to tumors (57)-current methods for miRNA inhibition (i.e., antagomirs and other chemically modified oligonucleotides) cannot capitalize on this phenomenon because of size constraints. Effective and prolonged tumor targeting and accumulation of systemically administered nanoparticles for up to 5 d was monitored by near-IR fluorescent imaging (Fig. S3D). ANTP-NP showed preferential targeting to tumor tissue compared to the lungs, liver, kidneys, spleen, stomach, heart, and brain (Fig. S3E). In agreement with these targeting results, tail-vein delivery of 1.5 mg/kg anti-miR-155 PNAs loaded in ANTP-NP for two treatments over the course of 3 d significantly delayed tumor growth; after 5 d of treatment, tumors had an approximately 50% decrease in growth relative to control-treated tumors (Fig. 6 C and D). As with the local injection studies, histopathological analysis confirmed the therapeutic response in systemically treated tumors (Fig. S3F). Treated tumors showed positive TUNEL staining in distinct regions throughout the tissue (Fig. 6E), consistent with the implicated involvement of miR-155 in regulating apoptosis. Quantification of miR-155 levels in treated tumors revealed that control-treated tumors had approximately 80,000 molecules of miR-155 per tumor cell, while DOX treatment reduced miR-155 levels to approximately 3,000 molecules per cell (Fig. S3G), which is consistent with previously reported miR-155 levels in B-cell lymphomas (16). Both intratumoral (resulting in approximately 15,000 molecules per cell) and systemic (resulting in approximately 50,000 molecules per cell) ANTP-NP anti-miR-155 treatment down-regulated miR-155 levels in tumors. In accordance with the anti-tumor response, inhibition of miR-155 was more effective in intratumoral-treated tumors (Fig. S3G). We have shown that the therapeutic response is proportional to the anti-miR-155 dose delivered to the tumor site (Fig. 6A); therefore, we expect that improved efficacy for systemic treatment would result at higher doses. We also note that the systemic dose used here was approximately 25-fold less than anti-miR doses that have been used by other groups to achieve therapeutic miRNA in vivo (32). Thus, anti-miR PNAs delivered by nanoparticles represent a therapeutically viable approach to miRNA inhibition, which can be a particularly potent treatment strategy for oncomiR-addicted tumors.
Discussion
Results from our targeted mouse model, in which miR-155 induction can be controlled both spatially and temporally, indicate that miR-155 induction in brain and lung is insufficient alone to drive tumorigenesis at these sites, at least during the timeframe investigated, but is sufficient for tumor initiation and survival in the hematopoietic system. Our results do not exclude a role for miR-155 in initiating solid tumor formation at these sites, however, because miR-155 may act cooperatively with other oncogenes to drive tumorigenesis. We show that miR-155 withdrawal can lead to rapid tumor regression in vivo, demonstrating that malignant lymphocytes can become dependent on elevated miR-155 levels. This is just the second known example of oncomiR addiction.
Notably, our results also suggest that tumors in NesCre8; mir-155LSLtTA mice may acquire secondary mutations that render one or a few clonal cell population(s) able to outcompete others. Recently, Croce and coworkers reported that miR-155 regulates genes in the DNA mismatch repair (MMR) pathway (58). Furthermore, the same group reported that miR-155 enhances mutation rates via regulation of pathways other than MMR (59). Accordingly, high expression of miR-155 may lead to a cellular context in which secondary activating mutations are more likely to occur, especially when combined with increased proliferation rates. Thus, we show that our model closely resembles human B-cell malignancies in which secondary mutations arise and confer monoclonal status to the tumor (60). We also find in our model that tumor regression involves apoptosis and that miR-155 may directly regulate apoptosis via SHIP1. As Hanahan and Weinberg describe in their landmark review (61), cancer cells must overcome numerous obstacles in order to persist. By regulating pathways involved in proliferation and apoptosis, miR-155 may provide cancer cells with the “multiple hits” they need to overcome sentinel pathways and survive.
While the levels of miR-155 overexpression in our diseased animals is in line with levels of overexpression in human lymphomas, we also note that some of our animals had relatively less induction of miR-155 and that these animals did not develop aggressive disease. This data suggest a threshold of expression is required for aggressive disease in our mouse model. We do not understand this phenomenon, but it could be related to the increased mutation rate at high levels of miR-155 mentioned above. To our knowledge, the observation of a human threshold for miR-155-induced lymphoma has not been established, but we believe our study and mouse model provide a basis for further investigations in humans.
In recent years, studies using anti-miRNA molecules with various chemical modifications have shown some promise. Nonetheless, the potential of utilizing miRNA inhibition as cancer therapy has so far been unrealized. Effective transport of anti-miRs into hard-to-transfect lymphocytes presents a technical challenge (55). Cholesterol-conjugated antagomirs can effectively enter certain cells, but cellular internalization of lipophilic oligonucleotide conjugates is dependent on lipoprotein transport and, putatively, the presence of the transmembrane protein SID1 (62). The low expression of SID1 in leukocytes may make them refractory to antagomir treatment (63). On the contrary, we show that PLGA nanoparticles surface-endowed with penetratin can effectively deliver anti-miRs to neoplastic human B cells and murine pre-B cells. Improved intracellular delivery imparted by cell-penetrating peptides is just one of many potential enhancements for ligand-coated nanoparticles. For example, nanoparticles can be coated with targeting molecules that improve delivery to specific, inflicted tissues and cells; also, nanoparticles can be coated with bioactive polymers like PEG to increase circulation time when administered systemically (64). Coating nanoparticles with targeting ligands may improve selective uptake into tumor cells over nonhematopoietic cells, and thus improve therapeutic efficacy. However, we observed that size-dependent passive targeting of ANTP-NPs was sufficient to achieve accumulation in lymphoid tumors that comprised a relatively homogeneous population of neoplastic lymphocytes. Furthermore, it has been shown that passive targeting of nanotherapeutics can have a greater impact on biodistribution than active targeting ligands (65). Notably, in this study we attached penetratin to the nanoparticle surface via a PEG linker, which may have improved the passive delivery of the nanoparticles to the hypervascularized pre–B-cell tumors. Long-circulating nanoparticles can better capitalize on the EPR effect by which systemically administered nanoscale particles (with a selective size range of 10–200 nm) target to and accumulate in tumors that are characterized by leaky vasculature and poor lymphatic drainage. Through such modifications, nanoparticles can be engineered to overcome most any conceivable physiological or cellular delivery barrier.
While antagomirs and other chemically modified oligonucleotides can be effective anti-miRs, their utility in achieving miRNA inhibition in vivo may be limited without a delivery vehicle-when administered systemically, antagomirs show substantial targeting to the liver (32). Additionally, antagomir treatments generally require higher concentrations to achieve an effect compared to the concentration of nanoparticle-encapsulated PNAs that we used in our study. For example, in their seminal study testing systemic delivery of antagomirs, Krutzfeldt and coworkers used 80 mg/kg concentration; this has set the standard for numerous other studies, many of which have used even higher concentrations: e.g., 120 mg/kg (49) or 300 mg/kg (48). In the present study, we observed that a systemic dose of 1.5 mg/kg was sufficient to achieve a significant therapeutic response. Nanoparticles and liposomes have been successfully used to deliver DNA and RNA; however, our work is a previously undescribed in vivo demonstration employing a nanoscale delivery vehicle to encapsulate charge-neutral nucleic acids (i.e., PNAs) without the use of a cationic component. Through interactions with cells and proteins as well as altered nucleic acid release dynamics, electrostatic interactions can confound most delivery systems. Efficiently loading charge-neutral nucleic acids into PLGA nanoparticles represents a less convoluted strategy for nanoparticulate gene therapy. To our knowledge, our study uniquely employs a systemically delivered nanoparticulate delivery system to show that inhibition of oncomiRs in vivo can lead to lymphoid tumor growth delay, and suggests a possible therapeutic route for lymphoma/leukemia.
Materials and Methods
Generation of Mice.
miR-155 was PCR-amplified from mouse genomic DNA using the following primers: 5′ CTAGCTAGCGAACTATGAACCGTGGCTG and 5′ CCGCTCGAGGAGATGTTGTTTAGGATACTGC. This product was cloned into the pTET-BigT vector containing the tTA “Tet-off” control element (kindly provided by Andrew McMahon, Harvard University, Cambridge, MA) (36) using NheI and XhoI restriction sites. We subsequently subcloned this “tTA-miR-155” construct into the pROSA26PAS vector using PacI and AscI restriction sites. The Yale Transgenic Facility targeted this final “tTA-miR-155-ROSA26” construct to the ROSA26 locus in B6;129-hybrid ES cells. Mice were generated, mated to B6 mice, screened for the mir-155LSLtTA transgene, and propagated to form a colony. Once a colony was established, mice were crossed into NesCre8 mice from a FVB/NJ background. miR-155-positive mice were identified and genotyped via PCR using the following sets of primers: knockin allele: P1 + P2, P3 + P4: wild-type allele: P2 + P3; P1: 5′ GGGAGGATTGGGAAGACAAT; P2: 5′ GGCGGATCACAAGCAATAAT; P3: 5′ TCCCAAAGTCGCTCTGAGTT; P4: 5′ GAAAGACCGCGAAGAGTTTG.
Mice were maintained at Yale University in accordance with Yale Animal Resource Center and Institutional Animal Care and Use Committee guidelines. Mice were maintained on doxycycline-impregnated food (with 200-mg DOX per kg of chow, Bio-Serv). To induce miR-155 overexpression, mice were fed standard chow from birth.
Histopathology.
Mice were euthanized by CO2 asphyxiation or, when blood collection was desired, by terminal cardiac puncture. Hematologic analysis was performed by ANTECH Diagnostics East. Tissues were immersion fixed in either 10% natural buffered formalin or Bouin’s fixative (Ricca Chemical Company), processed, embedded, sectioned at 5 μm, and stained by hematoxylin and eosin (H&E) by routine methods. TUNEL staining was performed as previously described (66). Tissue sections were evaluated blind to experimental manipulation (CJB) and digital light microscopic images were recorded with a Zeiss Axio Imager A1 microscope, AxioCam MRc5 camera, and AxioVision 4.7.1 imaging software (Carl Zeiss Microimaging).
Western Blot.
Western blotting was performed according to standard protocols. Briefly, spleens were ground by mortar and pestle and lysed using RIPA buffer (50 mM Tris•HCl pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS). Protein extracts were size fractionated by SDS/PAGE and transferred to a nitrocellulose or PVDF membrane. Blotting was performed using antibodies as follows: SHIP1 (D1163); SHIP1 (sc-1964); CASP3 (8G10) (cell signaling); Tubulin (sc-32293) (Santa Cruz Biotechnology); GAPDH (ab9485) (Abcam).
Adenovirus Treatment.
Five 4–8-wk-old mir-155LSLtTA mice (not crossed with NesCre8 mice) were intranasally infected with 5·108 pFU Adenovirus-Cre (Gene Transfer Vector Core). They were monitored for 6 mo after treatment, euthanized, and their lungs analyzed by processing and H&E staining, as described above.
qPCR.
Total RNA was extracted using Trizol according to standard procedures. RT and qPCR were performed using primers from ABI and the 7900HT Fast Real-Time PCR System (ABI) or MyiQ Real-Time Thermal Cycler (Bio-Rad). All RT reactions were performed using 10 ng of total cell RNA. Measured miR-155 levels were normalized to U6 internal control (Ambion) and absolute quantification of miR-155 was performed using a standard curve generated from synthetic miR-155. Values were converted to copy number per cell by assuming 0.67 pg of total RNA per pre-B cell (67).
Nanoparticle Synthesis.
Anti-miR-loaded PLGA nanoparticles were fabricated using a double emulsion solvent evaporation method as previously described (50). Nanoparticle surfaces were modified with the cell-penetrating peptide, penetratin, as previously described (54). After washing unencapsulated PNA and unattached penetratin, nanoparticles were lyophilized with the cryoprotectant, trehalose, at an equal mass ratio of polymer to carbohydrate. Anti-miR PNAs were obtained from Bio-Synthesis, Inc.; the sequences were as follows: anti-miR-155: 5′-ACCCCTATCACAATTAGCATTAA-3′; anti-scrm is a scrambled anti-miR-155 sequence: 5′-ACCCAATCGTCAAATTCCATATA-3′.
Transplantation.
Spleens or lymph nodes were extracted from pathologic NesCre8; mir-155LSLtTA mice and immediately placed on ice in 5% fetal bovine serum (FBS) in PBS. Tissues were dispersed into single-cell suspensions, red blood cells were lysed, and 5·106 cells were subcutaneously injected into nude (CrTac:NCr-Foxn1nu) mice. Tumor volume was calculated as: (length × width2)/2.
PCR To Detect DJ Rearrangements.
DJ rearrangements were detected by PCR as described previously (38). Briefly, the following primers were used to amplify the D and J regions of genomic DNA: DFS primer: 5′AGGGATCCTTGTGAAGGGATCTACTACTGTG3′; JH4 primer: 5′AAAGACCTGCAGAGGCCATTCTTACC3′. A polyclonal population of cells results in a “ladder” corresponding the DJ.1, DJ.2, DJ.3, and DJ.4 rearrangements, while a clonal population results in two or less bands.
Flow Cytometry.
RBC-lysed cells were stained with CD45R (B220)-PE, IgM-FITC, CD3-FITC, and CD43-APC (clone 1b11) (BD) and analyzed using a Calibur Flow cytometer (BD) with CellQuest software. Apoptosis studies were done using the Annexin-V-FITC Apoptosis Detection kit (BD) and following the manufacturer’s instructions. FL4 was used to gate for living cells without PI. Flow cytometric analysis of nanoparticle uptake by pre-B cells was performed by incubating single-cell suspensions derived from NesCre8; mir-155LSLtTA spleens with nanoparticles labeled with the fluorescent tracer dye, coumarin 6. After 1 h, live cells were stained with CD45R (B220)-PE, washed, and analyzed by FACScan (BD). For FACS experiments, the following channels were used: FL1 channel to detect FITC-conjugated antibodies, FL2 to detect PE-conjugated antibodies, FL3 to detect APC conjugated antibodies, and FL4 to detect PI.
Transfection.
Toledo cells were transfected with dual luciferase miR-155 reporter using Lipofectamine LTX with PLUS Reagent (Invitrogen). After 24 h, cells were treated with anti-miR or relevant controls. Cell lysates were measured for luciferase activity 48 h after initial transfection using the Dual-Luciferase Reporter Assay System (Promega). Renilla luciferase signal (containing the miR-155 target sequence within the 3′UTR) was normalized to Firefly luciferase. For SHIP1 knockdown studies, cells were incubated with 1 μM of Accell siRNA (Invitrogen) targeted against SHIP1 per the manufacturer’s protocol 12 h prior to nanoparticle administration.
Viability Assay.
Tumor cells were isolated from nude mouse tumors and cultured in the presence or absence of doxycycline (5 μg/mL); Toledo cells were cultured using standard methods. Alamar Blue (Invitrogen) or CellTiter-Blue (Promega) were used to assay for cell viability in triplicate.
Intra-Tumoral and Systemic Delivery of Anti-miRs.
Prior to injection, nanoparticles loaded with 3.5 μg PNA per mg PLGA were suspended in PBS. When tumors reached an approximate size of 100–300 mm3, nanoparticles were administered intravenously in a volume of 0.2 ml (43 mg/ml) or intratumorally in a volume of 0.1 ml (86 mg/ml for high dose, 29 mg/ml for low dose). Confocal imaging was performed using Leica TCS SP5 Spectral Confocal Microscope. Tumor tissues were harvested and fixed in 10% neutral-buffered formalin (NBF), and then incubated overnight in sucrose/OCT. Tumors were flash frozen in OCT before sectioning and staining. In vivo fluorescence imaging was performed on an IVIS Spectrum (Caliper) system using systemically administered nanoparticles loaded with a near-IR fluorescent dye (IR-780; Sigma). Fluorescence images were acquired using 1-s acquisition, small binning, and 12.5-cm field of view. Live mice were anesthetized using isoflurane during image acquisition. For biodistribution imaging, organs were harvested and fixed in NBF before IVIS visualization.
Supplementary Material
Acknowledgments.
We thank T. Nottoli and D. Scavone (Animal Genomic Services, Section of Comparative Medicine, Yale University) for help generating the knockin mice; G. Terwilliger (Yale Mouse Research Pathology, Section of Comparative Medicine, Yale University) for necropsy of mice; M. Bonk, K. McLaughlin, and D. Clark (Yale Animal Resource Center) for animal care; V. Horsley for assistance with flow cytometry; C. Weller for assistance with nanoparticle experiments; Jo Handelsman, Jonathan Holt, and Jing Du (Department of Molecular, Cellular, and Developmental Biology, Yale University) for assistance with in vivo imaging of nanoparticles; P. Medina for sharing protocols and advice; A. Kasinski and P. Trang for critical reading of the manuscript and stimulating discussions; and A. Brooks in Research Histology (Pathology Department, Yale University) for immunohistochemistry. I.A.B. received a Gilliam Fellowship (Howard Hughes Medical Institute). This work was supported by grants from The James S. McDonnell Foundation and National Institutes of Health to F.J.S. and J.B.W., a pilot grant from the Yale Cancer Center to F.J.S., and two grants from the National Institutes of Health to W.M.S.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Author Summary on page 10140 (volume 109, number 26).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201516109/-/DCSupplemental.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Babar IA, Slack FJ, Weidhaas JB. miRNA modulation of the cellular stress response. Future Oncol. 2008;4:289–298. doi: 10.2217/14796694.4.2.289. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 4.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 5.Chen X. MicroRNA biogenesis and function in plants. FEBS Lett. 2005;579:5923–5931. doi: 10.1016/j.febslet.2005.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du T, Zamore PD. microPrimer: The biogenesis and function of microRNA (Translated from English) Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 7.Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Wienholds E, Plasterk RH. MicroRNA function in animal development (Translated from English) FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 9.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: New regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 10.Calame K. MicroRNA-155 function in B Cells. Immunity. 2007;27:825–827. doi: 10.1016/j.immuni.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: A typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792:497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 15.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eis PS, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 18.Kluiver J, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 19.Tam W, Dahlberg JE. miR-155/BIC as an oncogenic microRNA. Genes Chromosomes Cancer. 2006;45:211–212. doi: 10.1002/gcc.20282. [DOI] [PubMed] [Google Scholar]
- 20.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 22.Kluiver J, et al. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer. 2006;45:147–153. doi: 10.1002/gcc.20273. [DOI] [PubMed] [Google Scholar]
- 23.Costinean S, et al. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood. 2009;114:1374–1382. doi: 10.1182/blood-2009-05-220814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedersen IM, et al. Onco-miR-155 targets SHIP1 to promote TNFalpha-dependent growth of B cell lymphomas. EMBO Mol Med. 2009;1:288–295. doi: 10.1002/emmm.200900028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Politi K, et al. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma SV, Settleman J. Oncogene addiction: Setting the stage for molecularly targeted cancer therapy. Genes Dev. 2007;21:3214–3231. doi: 10.1101/gad.1609907. [DOI] [PubMed] [Google Scholar]
- 29.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 30.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 31.Fabani MM, et al. Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acids Res. 2010;38:4466–4475. doi: 10.1093/nar/gkq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 33.Lennox KA, Behlke MA. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011;18:1111–1120. doi: 10.1038/gt.2011.100. [DOI] [PubMed] [Google Scholar]
- 34.Su J, Baigude H, McCarroll J, Rana TM. Silencing microRNA by interfering nanoparticles in mice. Nucleic Acids Res. 2011;39:e38. doi: 10.1093/nar/gkq1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zambrowicz BP, et al. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci USA. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao J, Barrow J, McMahon J, Vaughan J, McMahon AP. An ES cell system for rapid, spatial and temporal analysis of gene function in vitro and in vivo. Nucleic Acids Res. 2005;33:e155. doi: 10.1093/nar/gni146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang Y, Paige CJ, Wu GE. Enumeration and characterization of DJH structures in mouse fetal liver. EMBO J. 1992;11:1891–1899. doi: 10.1002/j.1460-2075.1992.tb05241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pennycook JL, Chang Y, Celler J, Phillips RA, Wu GE. High frequency of normal DJH joints in B cell progenitors in severe combined immunodeficiency mice. J Exp Med. 1993;178:1007–1016. doi: 10.1084/jem.178.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy R, Warnke R, Dorfman RF, Haimovich J. The monoclonality of human B-cell lymphomas. J Exp Med. 1977;145:1014–1028. doi: 10.1084/jem.145.4.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelicci PG, et al. Multiple monoclonal B cell expansions and c-myc oncogene rearrangements in acquired immune deficiency syndrome-related lymphoproliferative disorders Implications for lymphomagenesis. J Exp Med. 1986;164:2049–2060. doi: 10.1084/jem.164.6.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morse HC, 3rd, et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 2002;100:246–258. doi: 10.1182/blood.v100.1.246. [DOI] [PubMed] [Google Scholar]
- 42.Ovcharenko D, Kelnar K, Johnson C, Leng N, Brown D. Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res. 2007;67:10782–10788. doi: 10.1158/0008-5472.CAN-07-1484. [DOI] [PubMed] [Google Scholar]
- 43.Koopman G, et al. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood. 1994;84:1415–1420. [PubMed] [Google Scholar]
- 44.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 45.Liu Q, et al. SHIP is a negative regulator of growth factor receptor-mediated PKB/Akt activation and myeloid cell survival. Genes Dev. 1999;13:786–791. doi: 10.1101/gad.13.7.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu L, et al. The Src homology 2 (SH2) domain of SH2-containing inositol phosphatase (SHIP) is essential for tyrosine phosphorylation of SHIP, its association with Shc, and its induction of apoptosis. J Biol Chem. 1997;272:8983–8988. doi: 10.1074/jbc.272.14.8983. [DOI] [PubMed] [Google Scholar]
- 47.Boer AK, Drayer AL, Vellenga E. Effects of overexpression of the SH2-containing inositol phosphatase SHIP on proliferation and apoptosis of erythroid AS-E2 cells. Leukemia. 2001;15:1750–1757. doi: 10.1038/sj.leu.2402261. [DOI] [PubMed] [Google Scholar]
- 48.Ma L, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fontana L, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woodrow KA, et al. Intravaginal gene silencing using biodegradable polymer nanoparticles densely loaded with small-interfering RNA. Nat Mater. 2009;8:526–533. doi: 10.1038/nmat2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McNeer NA, et al. Nanoparticles deliver triplex-forming PNAs for site-specific genomic recombination in CD34+ human hematopoietic progenitors. Mol Ther. 2010;19:172–180. doi: 10.1038/mt.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 53.Derossi D, et al. Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J Biol Chem. 1996;271:18188–18193. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 54.Cheng CJ, Saltzman WM. Enhanced siRNA delivery into cells by exploiting the synergy between targeting ligands and cell-penetrating peptides. Biomaterials. 2011;32:6194–6203. doi: 10.1016/j.biomaterials.2011.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson ML, Spandidos DA, Coggins JR. Electroporation of lymphoid cells: factors affecting the efficiency of transfection. J Biochem Biophys Methods. 1991;22:207–222. doi: 10.1016/0165-022x(91)90069-9. [DOI] [PubMed] [Google Scholar]
- 56.Davis S, et al. Potent inhibition of microRNA in vivo without degradation (Translated from English) Nucleic Acids Res. 2009;37:70–77. doi: 10.1093/nar/gkn904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peer D, et al. Nanocarriers as an emerging platform for cancer therapy (Translated from English) Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 58.Valeri N, et al. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci USA. 2010;107:6982–6987. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tili E, et al. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc Natl Acad Sci USA. 2011;108:4908–4913. doi: 10.1073/pnas.1101795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 62.Wolfrum C, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 63.Zhang H. Reversal of HIV-1 latency with anti-microRNA inhibitors. Int J Biochem Cell Biol. 2009;41:451–454. doi: 10.1016/j.biocel.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beletsi A, Panagi Z, Avgoustakis K. Biodistribution properties of nanoparticles based on mixtures of PLGA with PLGA-PEG diblock copolymers. Int J Pharm. 2005;298:233–241. doi: 10.1016/j.ijpharm.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 65.Schmidt MM, Wittrup KD. A modeling analysis of the effects of molecular size and binding affinity on tumor targeting. Mol Cancer Ther. 2009;8:2861–2871. doi: 10.1158/1535-7163.MCT-09-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu B, et al. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4 (Translated from English) Proc Natl Acad Sci USA. 2010;107:21635–21640. doi: 10.1073/pnas.1016814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]