Abstract
Telomeres are repeating DNA sequences at the ends of chromosomes that protect and buffer genes from nucleotide loss as cells divide. Telomere length (TL) shortens with age in most proliferating tissues, limiting cell division and thereby contributing to senescence. However, TL increases with age in sperm, and, correspondingly, offspring of older fathers inherit longer telomeres. Using data and samples from a longitudinal study from the Philippines, we first replicate the finding that paternal age at birth is associated with longer TL in offspring (n = 2,023, P = 1.84 × 10−6). We then show that this association of paternal age with offspring TL is cumulative across multiple generations: in this sample, grandchildren of older paternal grandfathers at the birth of fathers have longer telomeres (n = 234, P = 0.038), independent of, and additive to, the association of their father’s age at birth with TL. The lengthening of telomeres predicted by each year that the father’s or grandfather’s reproduction are delayed is equal to the yearly shortening of TL seen in middle-age to elderly women in this sample, pointing to potentially important impacts on health and the pace of senescent decline in tissues and systems that are cell-replication dependent. This finding suggests a mechanism by which humans could extend late-life function as average age at reproduction is delayed within a lineage.
Keywords: adaptation, epigenetics, evolution, parental effects, transgenerational plasticity
Telomeres are repeating DNA sequences at the ends of chromosomes that protect and buffer genes from nucleotide loss as cells divide (1). In many tissues, telomere lengths (TL) are shortened by cellular proliferation, and as a result TL tends to decline with age (2–5). As cell replication generally requires a minimal TL, shortened TL is thought to contribute to senescence (6). Consistent with this, elderly persons with shorter telomeres (in blood) for their age have reduced survival (7–13).
Although it is well established that TL shortens with age in most proliferating tissues (e.g., 4, 5), sperm TL is an exception—older men have sperm with longer telomeres (4, 14, 15). This may be explained by the fact that the activity of telomerase (an enzyme that extends TL) is high in testes (16, 17). Consistent with the fact that offspring inherit half their chromosomes from sperm, offspring of older fathers tend to have longer telomeres (4, 18, 19). In contrast, because the pool of ova is established in utero, TL in ova are thought to be stable with age, and there is no evidence for a maternal age effect on TL in offspring (e.g., 4, 20).
We recently hypothesized that the age-related TL increase in sperm could lead to cumulative, and thus more biologically significant, multigenerational lengthening or shortening of TL in response to population trends in reproductive scheduling (21). If average reproductive age of recent patrilineal ancestors cumulatively influences TL, this might also lead to changes in TL of sufficient magnitude to influence late-life function and life expectancies (17). To test the hypothesis that a man’s age at reproduction influences TL in grandchildren, we used a large longitudinal, multigeneration sample from Cebu, Philippines (22) in which we measured TL (23) of DNA extracted from venous blood. We related TL in mothers (aged 36–69 y at blood collection) to their fathers’ ages when the mothers were born. In their offspring (21–23 y at the time of blood collection), we related TL to their fathers’ ages when the offspring were born and to their grandfathers’ ages when their parents were born.
Results
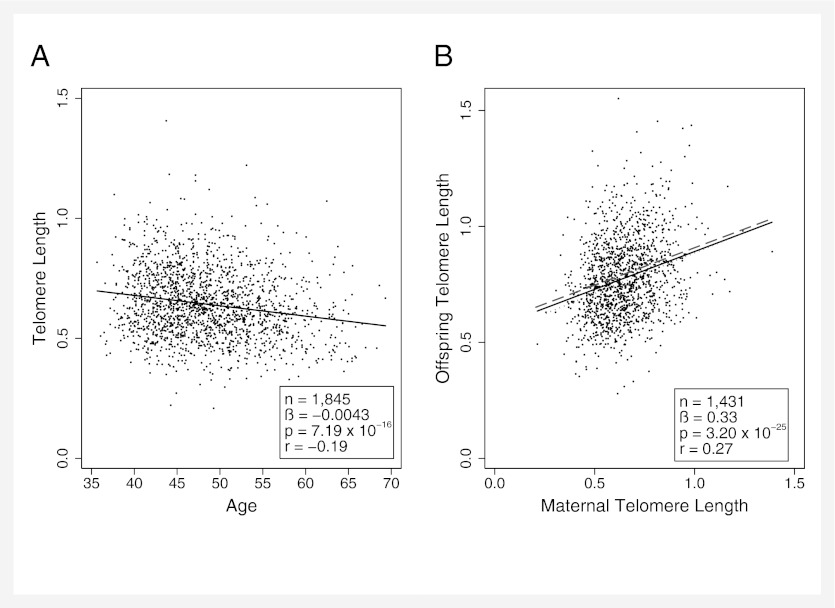
Consistent with the expectations of age-related decline in TL, blood TL was inversely associated with age in the 36- to 69-y-old mothers (Fig. 1A) with a similar magnitude of effect as seen in previous comparable studies (4, 24). Longer age-adjusted maternal TL predicted longer offspring TL (Fig. 1B). This mother–offspring TL correlation was similar in magnitude to previous studies (25–28) and did not vary depending on the sex of the offspring (maternal age adjusted TL × offspring sex interaction, P = 0.81). The TL of male offspring (n = 902, mean TL = 0.763 ± 0.006) was on average shorter than the TL of female offspring (n = 820, mean TL = 0.792 ± 0.006, t = 3.52, df = 1,720, P = 0.0005) (Fig. 1B).
Fig. 1.
(A) Telomere length (relative T/S ratio) in blood decreases with age in 36- to 69-y-old mothers. (B) Telomere length in mothers (adjusted for age) predicts offspring telomere length (adjusted for offspring age, sex, and age × sex). Statistics and black line show maternal–offspring TL association (maternal TL adjusted for maternal age and offspring TL adjusted for offspring age, sex, and age × sex). Dashed gray line shows maternal–daughter association and dotted gray line shows maternal–son association (unadjusted offspring TL values).
As observed in previous studies, we found that longer TL was predicted by older paternal age at reproduction in both the offspring cohort (Table 1, model 1, n = 1,681, P = 4.00 × 10−5) and their mothers (Table 2, model 1, n = 342, P = 0.003; mother and offspring combined: n = 2,023, P = 1.84 × 10−6). The association of paternal age with offspring TL was little changed by controlling for birth order, household income, or body mass index (BMI) at the time of blood collection in both the offspring (Table 1, model 2) and their mothers (Table 2, model 2). The paternal age association did not differ in sons versus daughters (paternal age × sex interaction, P = 0.844). Consistent with a linear association, paternal age did not exhibit a quadratic relationship with offspring TL (paternal age2, P = 0.689 for offspring and P = 0.995 for mothers).
Table 1.
Linear regression models relating paternal age and grandparental age to telomere lengths (relative T/S ratios) in 21- to 23-y-old offspring (both sexes pooled)
| Model 1 | Model 2 | Model 3 | Model 4 | Model 5 | Model 6 | Model 7 | Model 8 | |
| Age (self)† | −0.048** | −0.056** | −0.048** | 0.0068 | −0.010 | −0.0013 | 0.022 | 0.023 |
| Male | −0.031*** | −0.035*** | −0.031*** | −0.055* | −0.061** | −0.054* | −0.063*** | −0.073*** |
| Male × age (self)† | −0.030 | −0.025 | −0.030 | −0.071 | −0.062 | −0.086 | −0.13* | −0.11* |
| Father’s age | 0.0027*** | 0.0030** | 0.0029** | 0.0035‡ | 0.0034 | 0.0041 | ||
| Birth order | −0.0019 | |||||||
| Family income (log) | 0.0041 | −0.00074 | ||||||
| BMI | 0.0012 | 0.0058 | ||||||
| Mother's age | −0.00030 | |||||||
| Father's father's age | 0.0029* | 0.0026‡ | 0.0029 | |||||
| Father's mother's age | −0.000028 | |||||||
| Mother's father's age | 0.0011 | −0.00061 | ||||||
| Mother's mother's age | 0.0025 | |||||||
| Constant | 0.72*** | 0.67*** | 0.72*** | 0.62*** | 0.52*** | 0.61*** | 0.76*** | 0.74*** |
| Observations | 1,681 | 1,602 | 1,681 | 234 | 218 | 194 | 342 | 288 |
| Adjusted R2 | 0.032 | 0.034 | 0.031 | 0.045 | 0.048 | 0.038 | 0.047 | 0.059 |
Values are β-coefficients; ‡P < 0.10; *P < 0.05; **P < 0.01; and ***P < 0.001.
†Age is mean centered.
Table 2.
Linear regression models relating paternal age to telomere lengths (relative T/S ratios) in 36- to 69-y-old mothers
| Model 1 | Model 2 | Model 3 | |
| Age (self) | −0.0032* | −0.0031 | −0.0018 |
| Father’s age | 0.0032** | 0.0033* | 0.0024 |
| Family income (log) | −0.0082 | ||
| BMI | 0.0016 | ||
| Mother’s age | 0.0026 | ||
| Constant | 0.70*** | 0.70*** | 0.60*** |
| Observations | 342 | 339 | 290 |
| Adjusted R2 | 0.030 | 0.030 | 0.041 |
Values are β-coefficients; ‡P < 0.10 (none); *P < 0.05; **P < 0.01; and ***P < 0.001.
The association of paternal age with longer TL is thought to be due to direct inheritance from longer TL in sperm (4). To test an alternative hypothesis that offspring of older fathers have a slower age-related attrition rate of TL in adulthood, we examined if the association between offspring age and TL was reduced among offspring of older fathers, but found no support for this hypothesis (paternal age × offspring age interaction term in offspring, P = 0.341 and P = 0.472 in mothers).
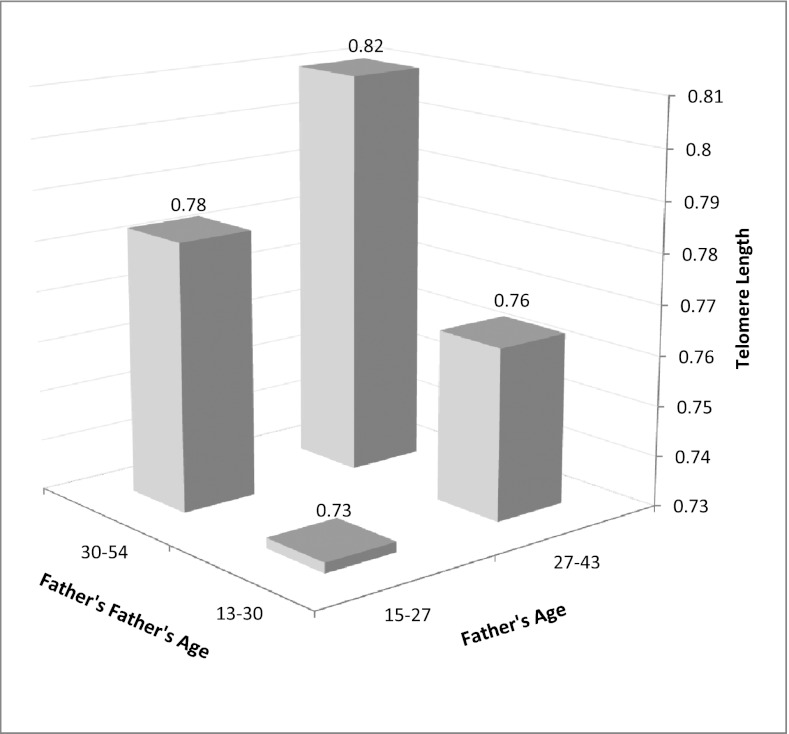
Consistent with our hypothesis, we found that, independent of the association of TL with the father’s age at reproduction, the paternal grandfather’s age at the father’s birth predicted longer TL in grandchildren (Table 1, model 4, n = 234, P = 0.038). The association between TL and grandpaternal age was nearly identical in size to the association with paternal age (Fig. 2; Table 1, models 1 and 4; Table 3), and was little changed after adjusting for household income and BMI at the time of blood collection (Table 1, model 5). As was found for the association of paternal age with TL, the relationship between paternal grandfather’s age and TL did not vary by the age of the grandchild (grand-paternal age × grandchild age interaction, P = 0.920) or by the sex of grandchild (grand-paternal age × sex interaction, P = 0.370) and did not exhibit a quadratic relationship (grand-paternal age2 term, P = 0.809).
Fig. 2.
Telomere length (relative T/S ratio) increases with both father’s age at offspring birth and father’s father’s age at father’s birth (age groups are defined by median splits). Bar heights represent mean telomere length adjusted for offspring age, sex, and age × sex. Median paternal age was 23.59 (interquartile range: 21.33–27.29) and grandpaternal age 30.44 (24.85–36.61).
Table 3.
Comparing effect sizes of paternal and grandpaternal age associations with telomere length (relative T/S ratios) to the age-related decline in telomere length in mothers (36–69 y old)
| Description | β (/y) | 95% CI | P |
| Age-related decline in mothers | −0.0032 | −0.0058 to −0.0005 | 0.018 |
| Paternal age effect on offspring | +0.0027 | +0.0014 to +0.0040 | 4 × 10−5 |
| Paternal age effect on mothers | +0.0032 | +0.0011 to +0.0053 | 0.003 |
| Paternal grandfather age effect on offspring | +0.0029 | +0.0002 to +0.0057 | 0.038 |
| Maternal grandfather age effect on offspring | +0.0011 | −0.0017 to +0.0038 | 0.442 |
CI, confidence interval.
The age of the maternal grandfather at the birth of the mother was also positively associated with TL, but this relationship was not significant (Table 1, model 7). In accord with the fact that male gametes continue to divide throughout life, whereas female gametes are produced prenatally, TL did not vary in relation to maternal age at the time of birth: after adjusting for paternal and grandpaternal age, maternal and grandmaternal ages were not significantly associated with the child’s or the grandchild’s TL (Table 1, models 3, 6, and 8; Table 2, model 3).
To place the magnitude of the paternal age associations with TL in a biological context, we compared the increase in TL predicted by each year that the father or grandfather delayed reproduction to the yearly shortening of TL in the mothers (age range: 36–69 y). The yearly decline in TL in the mothers was −0.0032/y (Table 2, model 1; Table 3), which is equal in magnitude, but opposite in direction, to the increase in TL predicted by a 1-y delay in reproduction by the father or paternal grandfather.
Discussion
In this large sample from the Philippines, we found that telomeres measured in blood were longer in individuals whose paternal grandfathers were older at their father’s birth. This effect was equal in size, and additive to, the longer TL predicted by a delay in the father’s age at their own birth, suggesting that paternal age at reproduction has effects on TL that are transmitted with high fidelity and cumulatively across at least two generations. The longer TL predicted by each year that paternal or grandpaternal reproduction was delayed was comparable to the yearly shortening in TL seen in older individuals in this population, suggesting that these intergenerational changes may be biologically important. These findings point to a mechanism by which shifts in the age of paternal reproduction within a population can lead to cumulative, multigenerational changes in telomere length in descendants.
Consistent with previous studies (4, 18, 19), the increase in TL predicted by each year that the father’s reproduction was delayed was nearly identical in size to the yearly age-related decline in TL in middle- to old-age adults. The lack of interaction between paternal age and offspring age in models predicting offspring TL, as well as the similar effect sizes of paternal age on TL in the younger and older cohorts (offspring and their mothers, respectively), suggests that the effect of paternal age on TL does not act by changing the rate of telomere shortening in adulthood. However, because both the cohorts were already in adulthood, we are not able to rule out the possibility that the pace of telomere shortening in somatic tissues varies in relation to paternal or grandpaternal age at conception for those ages younger than that of the mothers and offspring in this study. Some (4, 19), but not all (18) previous studies have suggested that the father’s age at conception has greater effects on TL in male offspring. In Cebu, we found that the change in TL predicted by a delay in the father’s or grandfather’s ages at birth did not vary by offspring sex.
A standard model of genetic inheritance leads to the prediction that each offspring receives half of his or her telomere DNA from their father and one quarter of their telomere DNA from each of their paternal and maternal grandfathers. On the basis of this, we expected the effect of delaying reproduction among paternal and maternal grandfathers to be equal in size and half as strong as the effect of a comparable delay in paternal age at reproduction. Contrary to this expectation, we found that the effect size of the paternal grandfather’s age on their grandchildren’s TL was larger than the effect of the maternal grandfather’s age at the mother’s birth on the grandchild’s TL. In addition, the paternal age association with offspring’s TL and the paternal grandfather age association with the grandchild’s TL were equal. Although it is not certain what accounts for these results, both findings might be explained by the fact that heritability of TL is substantially greater from father to offspring than from mother to offspring (25–27), which recent work suggests could relate to telomere protection proteins that are specific to sperm (29, 30).
The longer TL among offspring of older fathers has been explained by the fact that sperm TL are longer in older men (4, 14, 15), with high telomerase activity in testes being the most common explanation for the increase in sperm TL seen with age (16, 31). Because men produce an estimated 100 million sperm cells daily (32–34), telomere maintenance mechanisms are required to avoid rapid telomere shortening (17). It is presently less clear why high testicular telomerase expression should lead to gradual and progressive lengthening of sperm telomeres with age, rather than simply maintaining a stable length. If testicular telomerase expression extends short and long telomeres equally, the distribution of sperm TL in young and old men would be expected to have the same shape, but centered on a longer mean length in older men. However, Kimura and colleagues noted that, compared with younger men, older men have a TL distribution skewed slightly in favor of long telomeres (4). Telomerase preferentially extending long telomeres might explain this, but to the extent that telomerase has effects that vary based upon the length of telomeres, past work suggests that it is likely to have the strongest effects on short telomeres (reviewed in ref. 21). Kimura and colleagues interpreted the skewed distribution of sperm TLs in older men as evidence that sperm stem cells with shorter telomere lengths disproportionately die out with age (4). It is presently not clear if this selective cellular attrition would be sufficient to account for the age-related rate of telomere lengthening observed in sperm.
To our knowledge, no study has used longitudinal sampling of sperm or studied full siblings to firmly establish whether the longer TL in offspring of older male patrilineal ancestors is caused by increases in sperm TL as a man ages. Although longer sperm TL in older men compared with younger men (4, 14, 15) provides convergent evidence for this prominent hypothesis, it is also known that sperm donors represent nonrandom samples of the population (35, 36). It is possible that older men who volunteer to donate sperm have better reproductive health and longer sperm TL and that a similar selection bias results in men with longer sperm TL tending to reproduce at later ages. Such a strong and persistent selection bias would imply important effects of TL on male reproductive health or that some unmeasured factor influences both sperm TL and reproductive health (which could make sperm TL an important biomarker).
Regardless of the mechanism underlying the longer telomeres in descendants of older fathers and grandfathers, there is evidence that TL is a predictor of health and mortality, suggesting that intergenerational lengthening or shortening of TL could have effects on health and the pace of senescence (17). Several human studies report higher mortality among elderly individuals with shorter blood telomere lengths (7–13). There are converging lines of evidence that telomere lengths per se are at least partial causes of health and survival. Because the mechanics of DNA replication lead to incomplete replication of chromosome ends (the “end-replication problem”), the buffer of repeating telomere sequence can be depleted after multiple rounds of cell division. If replication continues past this point, genes may be progressively lost. To prevent gene loss, shortened telomeres generally activate damage response pathways that cause cell death or impairment of cellular function (2, 3). Evidence that cell replication is limited by shorter TL has been reported in vitro (37–40) through in vivo experimentation in animal models (41–43) and in cross-sectional and longitudinal observational studies (7–13, 44–48). On the other hand, whether inherited TL alters the risks of developing cancer is unsettled (20, 21, 49–53), so the implications of the association of paternal ages at reproduction with TL for cancer risk are unclear. Direct assessment of the health implications of paternal-age effects on descendants’ TL is lacking and awaits future study.
Together with this evidence, our findings lead us to the prediction that being born into a lineage in which recent male ancestors, particularly patrilineal ancestors, reproduced at later ages could have long-term health benefits. In particular, we expect changes in TL inherited at conception to have the largest effects on tissues and biological functions that are especially dependent upon rapid cellular proliferation, such as the immune system, the gastrointestinal tract, and skin (21). In contrast, cells that are not as dependent on proliferation, such as neurons or cardiomyocytes (21), might be less impacted by the age of reproduction in recent ancestors. Although some negative outcomes, such as miscarriages (54), are more common in offspring born to fathers of advanced age, the age-related lengthening of TL that we note here is present across the range of paternal and grandpaternal ages—not just at advanced ages.
The finding that telomere length may be extended cumulatively when multiple generations of male ancestors delay their ages of reproduction has potentially important evolutionary implications. Reproductive life span is a key evolutionary life-history parameter (e.g., 55–58), and this is particularly true in humans owing to the importance of late-life intergenerational transfers to reproduction and survival of offspring and grand-offspring (56, 59–61). In light of past work showing that blood TL predicts late-life survival (7–13), we believe that our findings point to a possible mechanism of plasticity in the pace of aging and senescent functional decline. Notably, as paternal ancestors delay reproduction, longer TL will be passed to offspring, which could allow life span to be extended as populations survive to reproduce at older ages. Having been born to an older father could signal that an individual is likely to grow up in a social and ecological context within which mortality rates are low and reproduction is likely to occur later in life, thus placing more of a premium on a durable long-lived body (21). However, the age at reproduction of one’s father alone is likely to be an imprecise signal of demographic conditions because factors like birth order will introduce additional variability. The high fidelity and cumulative multigenerational nature of the paternal age effect that we document are important in this regard because they suggest that TL could serve as a more robust signal of average paternal ages at reproduction over longer time frames (21). By integrating information about the average age at reproduction across multiple generations of ancestors, we speculate that the paternal age effect on TL could allow a unique form of transgenerational plasticity that modifies physiologic function in response to a relatively stable cue of recent ancestral experience and behavior.
In sum, we find that grandpaternal age at the father’s birth is associated with longer TL in grandchildren, which is independent of, and additive to, the association of TL with the father’s age at the offspring’s birth. This evidence for cumulative, multigenerational lengthening of telomeres hints at a capacity for age-related changes in sperm TL to be transmitted with high fidelity across at least two generations. The implications of these findings for understanding heterogeneity in health and the pace of aging is an important question to be addressed by future research. We speculate that an effect of the age of paternal ancestors on TL could allow increases in life expectancy under demographic conditions of low mortality and delayed reproduction, when investment in a more durable and long-lived body is likely to reap higher fitness returns.
Materials and Methods
Data Collection.
Data are from the Cebu Longitudinal Health and Nutrition Survey, a birth cohort study in Cebu City, the Philippines, that began with enrollment of 3,327 pregnant mothers in 1983–1984 (22). Mothers came from randomly selected rural and urban neighborhoods. Ages of the ancestor of the offspring were retrieved from household rosters (data available at http://www.cpc.unc.edu/projects/cebu), and as such there is missing age information reflecting ancestors who never lived with surveyed families. Venous blood samples were collected in 2005, when the offspring were 20.8–22.5 y old and the mothers were 35.7–69.3 y old. Written informed consent was obtained from all participants, and data and sample collection were conducted with approval and oversight from the Institutional Review Boards of the University of North Carolina and Northwestern University. Telomere measurement and analysis in de-identified samples and data were not considered human subject research by Northwestern University’s Institutional Review Board.
Telomere Length Measurement.
Automated and manual DNA extraction (Puregene, Gentra) was conducted on venous blood from 1,893 mothers and 1,779 offspring. DNA isolated in this fashion from blood is widely considered to be of leukocyte origin. Although this is no doubt mostly true, there are other possible cellular and noncellular sources of DNA found in blood (21, 62–66) that preclude our referring to the telomere lengths measured here as exclusively leukocyte in origin. Telomere lengths were measured using the monochrome multiplex quantitative PCR assay (23) with the following modifications. Reactions were run with telomere primers (telg/telc) at 500 nM each and albumin (single-copy control) primers (albd/albu) at 300 nM each on a Bio-Rad iCycler iQ thermocycler with a modified thermo-profile: internal well factor collection for 1.5 min at 95 °C; denaturation and Taq activation for 13.5 min at 95 °C; two repeats of 2 s at 98 °C followed by 30 s at 49 °C; 34 repeats of 2 s at 98 °C; 30 s at 59 °C; 15 s at 74 °C with signal acquisition; 30 s at 84 °C; and 15 s at 85 °C with signal acquisition, followed by a melt curve for PCR product verification. Data were analyzed with a per-well efficiency calculation method (7, 67, 68) using LinRegPCR version 12.7 (69, 70). All telomere to single copy gene (T/S) ratios were normalized to (divided by) the same control sample run with six replicates per 96-well plate. Interrun correlations of the same samples (r = 0.92; n = 705; P < 0.0001) were comparable to a previous analysis (71). Samples with coefficients of variation (CVs) greater than 15% were rerun at least once, and 2.0% of all samples were dropped from the analysis due to CV <15% not being achieved. Geometric mean CV of T/S ratios before exclusion was 5.7% and 5.6% after high-CV exclusion.
Statistical Analysis.
Regression models were run with robust SEs because some of the regression models were heteroscedastic using the Breusch–Pagan/Cook–Weisberg test (“hettest” command in Stata). The household income variable, reflecting a probable multiplicative rather than additive effect, was logged. P values were combined using Fisher’s method (“metap” command in Stata). All tests were two-tailed with α = 0.05. All analyses were conducted using Stata 11.2.
Acknowledgments
DNA extracts were provided by Karen Mohlke. Richard Cawthon, Justin Tackney, Katarina Nordfjäll, Klelia Salpea, Christine Ackerman, and Margrit Urbanek provided laboratory advice; and Jared Bragg, Yarrow Axford, and Zaneta Thayer gave feedback on earlier drafts of this manuscript. We thank the many researchers at the Office of Population Studies, University of San Carlos, for their central role in study design and data collection and the Filipino participants who provided their time and samples for this study. Laboratory analysis was funded by the National Science Foundation (Doctoral Dissertation Improvement Grant BCS-0962282) and the Wenner-Gren Foundation (Grant 8111). Institutional support was received from Northwestern University. Data and sample collection were funded by National Institutes of Health Grants RR20649 and ES10126. D.T.A.E. was supported by a National Science Foundation Graduate Research Fellowship.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- 2.Olovnikov AM. Principle of marginotomy in template synthesis of polynucleotides (Translated from the Russian) Dokl Akad Nauk SSSR. 1971;201:1496–1499. [PubMed] [Google Scholar]
- 3.Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 4.Kimura M, et al. Offspring’s leukocyte telomere length, paternal age, and telomere elongation in sperm. PLoS Genet. 2008;4:e37. doi: 10.1371/journal.pgen.0040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishii A, et al. Telomere shortening with aging in the human pancreas. Exp Gerontol. 2006;41:882–886. doi: 10.1016/j.exger.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 6.Harley C. Telomere loss: Mitotic clock or genetic time bomb? Mutat Res. 1991;256(2–6):271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 7.Ehrlenbach S, et al. Influences on the reduction of relative telomere length over 10 years in the population-based Bruneck Study: Introduction of a well-controlled high-throughput assay. Int J Epidemiol. 2009;38:1725–1734. doi: 10.1093/ije/dyp273. [DOI] [PubMed] [Google Scholar]
- 8.Cawthon RM, Smith KR, O’Brien E, Sivatchenko A, Kerber RA. Association between telomere length in blood and mortality in people aged 60 years or older. Lancet. 2003;361:393–395. doi: 10.1016/S0140-6736(03)12384-7. [DOI] [PubMed] [Google Scholar]
- 9.Bakaysa SL, et al. Telomere length predicts survival independent of genetic influences. Aging Cell. 2007;6:769–774. doi: 10.1111/j.1474-9726.2007.00340.x. [DOI] [PubMed] [Google Scholar]
- 10.Kimura M, et al. Telomere length and mortality: A study of leukocytes in elderly Danish twins. Am J Epidemiol. 2008;167:799–806. doi: 10.1093/aje/kwm380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honig LS, Schupf N, Lee JH, Tang MX, Mayeux R. Shorter telomeres are associated with mortality in those with APOE epsilon4 and dementia. Ann Neurol. 2006;60(2):181–187. doi: 10.1002/ana.20894. [DOI] [PubMed] [Google Scholar]
- 12.Martin-Ruiz C, et al. Telomere length predicts poststroke mortality, dementia, and cognitive decline. Ann Neurol. 2006;60(2):174–180. doi: 10.1002/ana.20869. [DOI] [PubMed] [Google Scholar]
- 13.Fitzpatrick AL, et al. Leukocyte telomere length and mortality in the Cardiovascular Health Study. J Gerontol A Biol Sci Med Sci. 2011;66:421–429. doi: 10.1093/gerona/glq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baird DM, et al. Telomere instability in the male germline. Hum Mol Genet. 2006;15:45–51. doi: 10.1093/hmg/ddi424. [DOI] [PubMed] [Google Scholar]
- 15.Allsopp RC, et al. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet. 1996;18(2):173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Zalenskaya IA, Zalensky AO. Telomeres in mammalian male germline cells. Int Rev Cytol. 2002;218:37–67. doi: 10.1016/s0074-7696(02)18011-9. [DOI] [PubMed] [Google Scholar]
- 18.De Meyer T, et al. Asklepios investigators Paternal age at birth is an important determinant of offspring telomere length. Hum Mol Genet. 2007;16:3097–3102. doi: 10.1093/hmg/ddm271. [DOI] [PubMed] [Google Scholar]
- 19.Unryn BM, Cook LS, Riabowol KT. Paternal age is positively linked to telomere length of children. Aging Cell. 2005;4(2):97–101. doi: 10.1111/j.1474-9728.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- 20.Arbeev KG, Hunt SC, Kimura M, Aviv A, Yashin AI. Leukocyte telomere length, breast cancer risk in the offspring: The relations with father’s age at birth. Mech Ageing Dev. 2011;132(4):149–153. doi: 10.1016/j.mad.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisenberg DTA. An evolutionary review of human telomere biology: The thrifty telomere hypothesis and notes on potential adaptive paternal effects. Am J Hum Biol. 2011;23(2):149–167. doi: 10.1002/ajhb.21127. [DOI] [PubMed] [Google Scholar]
- 22.Adair LS, et al. Cohort profile: The Cebu longitudinal health and nutrition survey. Int J Epidemiol. 2011;40:619–625. doi: 10.1093/ije/dyq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37:e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svenson U, et al. Breast cancer survival is associated with telomere length in peripheral blood cells. Cancer Res. 2008;68:3618–3623. doi: 10.1158/0008-5472.CAN-07-6497. [DOI] [PubMed] [Google Scholar]
- 25.Nordfjäll K, Larefalk A, Lindgren P, Holmberg D, Roos G. Telomere length and heredity: Indications of paternal inheritance. Proc Natl Acad Sci USA. 2005;102:16374–16378. doi: 10.1073/pnas.0501724102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nordfjäll K, Svenson U, Norrback K-F, Adolfsson R, Roos G. Large-scale parent-child comparison confirms a strong paternal influence on telomere length. Eur J Hum Genet. 2010;18:385–389. doi: 10.1038/ejhg.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Njajou OT, et al. Telomere length is paternally inherited and is associated with parental lifespan. Proc Natl Acad Sci USA. 2007;104:12135–12139. doi: 10.1073/pnas.0702703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nawrot TS, Staessen JA, Gardner JP, Aviv A. Telomere length and possible link to X chromosome. Lancet. 2004;363:507–510. doi: 10.1016/S0140-6736(04)15535-9. [DOI] [PubMed] [Google Scholar]
- 29.Gao G, Cheng Y, Wesolowska N, Rong YS. Paternal imprint essential for the inheritance of telomere identity in Drosophila. Proc Natl Acad Sci USA. 2011;108:4932–4937. doi: 10.1073/pnas.1016792108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gineitis AA, Zalenskaya IA, Yau PM, Bradbury EM, Zalensky AO. Human sperm telomere-binding complex involves histone H2B and secures telomere membrane attachment. J Cell Biol. 2000;151:1591–1598. doi: 10.1083/jcb.151.7.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yashima K, et al. Expression of the RNA component of telomerase during human development and differentiation. Cell Growth Differ. 1998;9:805–813. [PubMed] [Google Scholar]
- 32.Freund M. Effect of frequency of emission on semen output and an estimate of daily sperm production in man. J Reprod Fertil. 1963;6:269–285. doi: 10.1530/jrf.0.0060269. [DOI] [PubMed] [Google Scholar]
- 33.Amann RP, Howards SS. Daily spermatozoal production and epididymal spermatozoal reserves of the human male. J Urol. 1980;124:211–215. doi: 10.1016/s0022-5347(17)55377-x. [DOI] [PubMed] [Google Scholar]
- 34.Gao J, et al. Semen quality in a residential, geographic and age representative sample of healthy Chinese men. Hum Reprod. 2007;22:477–484. doi: 10.1093/humrep/del383. [DOI] [PubMed] [Google Scholar]
- 35.Muller A, et al. Selection bias in semen studies due to self-selection of volunteers. Hum Reprod. 2004;19:2838–2844. doi: 10.1093/humrep/deh521. [DOI] [PubMed] [Google Scholar]
- 36.Handelsman DJ. Sperm output of healthy men in Australia: Magnitude of bias due to self-selected volunteers. Hum Reprod. 1997;12:2701–2705. doi: 10.1093/humrep/12.12.2701. [DOI] [PubMed] [Google Scholar]
- 37.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: The path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 38.Counter CM, et al. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 40.Allsopp RC, Harley CB. Evidence for a critical telomere length in senescent human fibroblasts. Exp Cell Res. 1995;219(1):130–136. doi: 10.1006/excr.1995.1213. [DOI] [PubMed] [Google Scholar]
- 41.Ilmonen P, Kotrschal A, Penn DJ. Telomere attrition due to infection. PLoS ONE. 2008;3:e2143. doi: 10.1371/journal.pone.0002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahin E, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bize P, Criscuolo F, Metcalfe NB, Nasir L, Monaghan P. Telomere dynamics rather than age predict life expectancy in the wild. Proc Biol Sci. 2009;276(1662):1679–1683. doi: 10.1098/rspb.2008.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heidinger BJ, et al. Telomere length in early life predicts lifespan. Proc Natl Acad Sci USA. 2012;109:1743–1748. doi: 10.1073/pnas.1113306109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salpea KD, Nicaud V, Tiret L, Talmud PJ, Humphries SE. EARS II group The association of telomere length with paternal history of premature myocardial infarction in the European Atherosclerosis Research Study II. J Mol Med (Berl) 2008;86:815–824. doi: 10.1007/s00109-008-0347-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soerensen M, et al. Genetic variation in TERT and TERC and human leukocyte telomere length and longevity: A cross-sectional and longitudinal analysis. Aging Cell. 2012;11:223–227. doi: 10.1111/j.1474-9726.2011.00775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atzmon G, et al. Evolution in health and medicine Sackler colloquium: Genetic variation in human telomerase is associated with telomere length in Ashkenazi centenarians. Proc Natl Acad Sci USA. 2010;107(Suppl 1):1710–1717. doi: 10.1073/pnas.0906191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma H, et al. Shortened telomere length is associated with increased risk of cancer: A meta-analysis. PLoS ONE. 2011;6:e20466. doi: 10.1371/journal.pone.0020466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez-Delgado B, et al. Genetic anticipation is associated with telomere shortening in hereditary breast cancer. PLoS Genet. 2011;7:e1002182. doi: 10.1371/journal.pgen.1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez-Delgado B, et al. Shorter telomere length is associated with increased ovarian cancer risk in both familial and sporadic cases. J Med Genet. 2012 doi: 10.1136/jmedgenet-2012-100807. [DOI] [PubMed] [Google Scholar]
- 52.DePinho RA, Wong KK. The age of cancer: Telomeres, checkpoints, and longevity. J Clin Invest. 2003;111:S9–S14. [PubMed] [Google Scholar]
- 53.Shay JW, Wright WE. Hallmarks of telomeres in ageing research. J Pathol. 2007;211(2):114–123. doi: 10.1002/path.2090. [DOI] [PubMed] [Google Scholar]
- 54.De La Rochebrochard E, McElreavey K, Thonneau P. Paternal age over 40 years: The “amber light” in the reproductive life of men? J Androl. 2003;24:459–465. doi: 10.1002/j.1939-4640.2003.tb02694.x. [DOI] [PubMed] [Google Scholar]
- 55.Tuljapurkar SD, Puleston CO, Gurven MD. Why men matter: Mating patterns drive evolution of human lifespan. PLoS ONE. 2007;2:e785. doi: 10.1371/journal.pone.0000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weismann A. Essays upon Heredity. 2nd Ed. Oxford: Clarendon Press; 1889. Electronic edition at http://www.esp.org/books/weismann/essays/facsimile/ [Google Scholar]
- 57.Kirkwood TBL, Holliday R. The evolution of ageing and longevity. Proc R Soc Lond B Biol Sci. 1979;205:531–546. doi: 10.1098/rspb.1979.0083. [DOI] [PubMed] [Google Scholar]
- 58.Stearns SC. The Evolution of Life Histories. Oxford; New York: Oxford Univ. Press; 1992. pp xii, 249. [Google Scholar]
- 59.Medawar PB. An Unsolved Problem of Biology. London: published for University College London by H. K. Lewis; 1952. p. 24. [Google Scholar]
- 60.Hamilton WD. The moulding of senescence by natural selection. J Theor Biol. 1966;12(1):12–45. doi: 10.1016/0022-5193(66)90184-6. [DOI] [PubMed] [Google Scholar]
- 61.Kaplan HS, Robson AJ. The emergence of humans: The coevolution of intelligence and longevity with intergenerational transfers. Proc Natl Acad Sci USA. 2002;99:10221–10226. doi: 10.1073/pnas.152502899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hermansen MC. Nucleated red blood cells in the fetus and newborn. Arch Dis Child Fetal Neonatal Ed. 2001;84:F211–F215. doi: 10.1136/fn.84.3.F211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stachon A, Holland-Letz T, Krieg M. High in-hospital mortality of intensive care patients with nucleated red blood cells in blood. Clin Chem Lab Med. 2004;42:933–938. doi: 10.1515/CCLM.2004.151. [DOI] [PubMed] [Google Scholar]
- 64.Lo YMD, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: Implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lo YM, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 66.Chan CW, Garruto RM, Lum JK. Paleoparasite populations from archived sera: Insights into chloroquine drug resistance in Papua New Guinea. J Infect Dis. 2006;194:1023–1024; author reply 1024–1025. doi: 10.1086/507433. [DOI] [PubMed] [Google Scholar]
- 67.Ehrlenbach S, et al. Raising the bar on telomere epidemiology. Int J Epidemiol. 2010;39:308–309. doi: 10.1093/ije/dyp383. [DOI] [PubMed] [Google Scholar]
- 68.Willeit P, et al. Telomere length and risk of incident cancer and cancer mortality. JAMA. 2010;304(1):69–75. doi: 10.1001/jama.2010.897. [DOI] [PubMed] [Google Scholar]
- 69.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339(1):62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 70.Ruijter JM, et al. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aviv A, et al. Impartial comparative analysis of measurement of leukocyte telomere length/DNA content by Southern blots and qPCR. Nucleic Acids Res. 2011;39:e134. doi: 10.1093/nar/gkr634. [DOI] [PMC free article] [PubMed] [Google Scholar]