Abstract
Regulation of actin dynamics is key to many cell physiological processes, ranging from protrusion formation and control of cell shape to cellular motility, endocytosis, and vesicle movement. The actin-related protein (ARP)2/3 complex is a major actin nucleator organizing branched filament networks in lamellipodial protrusions and during cell migration downstream of nucleation-promoting factors (NPFs). Although many NPFs have been characterized in detail, only few ARP2/3 inhibitors are known. Here, we identify the trans-Golgi network (TGN)/endosomally localized adaptor protein (AP)-1-associated adaptor protein Gadkin as a negative regulator of ARP2/3 function. Loss of Gadkin is associated with a partial redistribution of ARP2/3 to the plasma membrane and with increased cell spreading and migration, phenotypes that depend on the presence of a functional ARP2/3 complex. Gadkin directly binds to ARP2/3 via a conserved tryptophan-based acidic cluster motif reminiscent of ARP2/3-binding sequences of NPFs but fails to facilitate ARP2/3-mediated actin assembly. Consistent with an inhibitory role of Gadkin on ARP2/3 function, ARP2/3 is found on motile Gadkin-containing endosomal vesicles under migration-inhibiting conditions from where it relocalizes to the plasma membrane following activation of NPFs. Together with the observation that Gadkin-mediated inhibition of cell spreading requires its binding to ARP2/3, these data indicate that Gadkin is a negative regulator of ARP2/3 function present on intracellular membranes.
Regulated actin dynamics play a pivotal role in many cell physiological processes, ranging from intracellular trafficking to cellular motility (1). Accordingly, actin dynamics are under tight spatiotemporal control, involving multiple layers of regulation. Nucleation of actin filaments or branches is kinetically unfavorable and, therefore, requires nucleators such as the heptameric actin-related protein (ARP)2/3 complex (2), an important cellular factor for organizing branched filament networks that partitions between the plasma membrane and intracellular sites. ARP2/3 displays little nucleation activity on its own but can undergo activation by nucleation-promoting factors (NPFs), many of which have been characterized to date (3). Two important NPFs are Wiskott–Aldrich syndrome protein (WASP) and WASP family Verprolin-homologous protein (WAVE) of the WASP family of proteins. Both types of NPFs bind to and stimulate ARP2/3 via their verprolin, cofilin, acidic (VCA) domains following release from auto- or transinhibition by convergent signals involving activated Rho-family GTPases (2, 3). For example, cell migration involves activation of Rac1 (ras-related C3 botulinum toxin substrate 1), resulting in the WAVE-stimulated ARP2/3-mediated formation of lamellipodial protrusions at the leading edge (4). Over the past decade, different groups of NPFs have been identified and characterized in cell biological and mechanistic detail. Most NPFs reside on the plasma membrane, although some of them, most notably WASH, have been found on intracellular structures, including endosomes (5–7). By contrast, our knowledge about the mechanisms that negatively regulate cell motility (8), e.g., via modulating ARP2/3 function is still in its infancy.
Here, we show that the trans-Golgi network (TGN)/endosomally localized peripheral membrane protein Gadkin, a modulator of adaptor protein (AP)-1-mediated endosomal membrane traffic (9–11), negatively regulates cell spreading and motility via inhibition of ARP2/3 function.
Results
Gadkin-Depleted Cells Show Increased Cell Spreading and Migration.
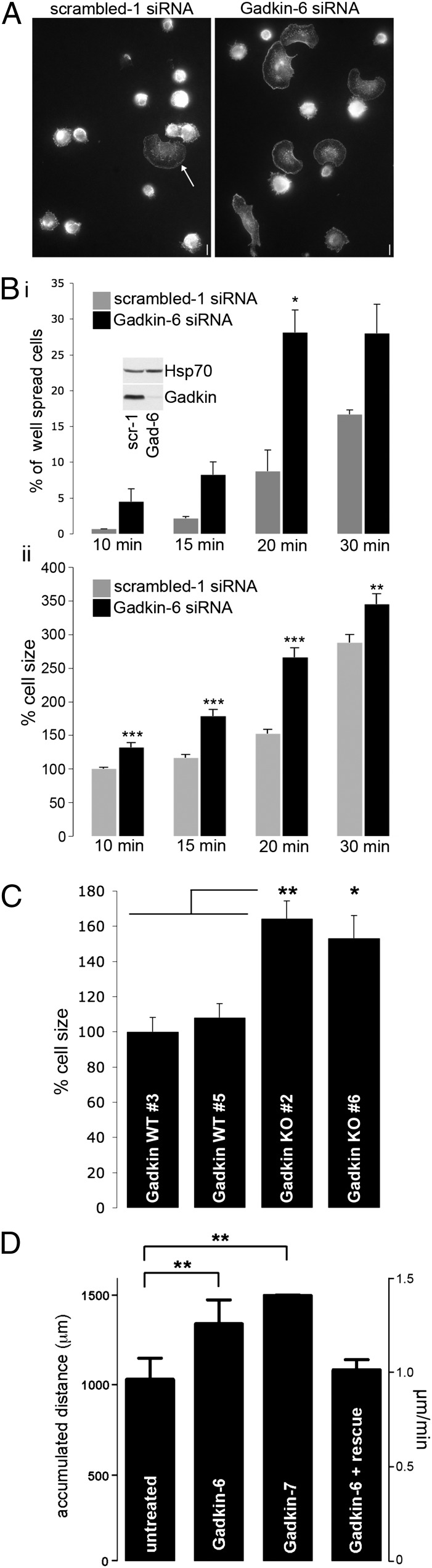
Gadkin overexpression has been reported before to cause the peripheral accumulation of TGN-derived endosomal vesicles, and this phenotype often seemed to correlate with changes in cell morphology (10). To obtain insights into the putative role of Gadkin in modulating cell shape and dynamics, we depleted mouse melanoma B16F1 cells of endogenous Gadkin and assayed the ability of silenced cells to undergo cell spreading. To our surprise, Gadkin-depleted B16F1 cells spread much more efficiently than control cells (Fig. 1A). Cell spreading follows a predictable time course with cells adhering 15–20 min after plating. At this time, they form sheet-like protrusions that are uniformly extended in all directions, while at later time points, cells acquire a more digitated morphology (12). Consistent with this only a very minor fraction (<5%) of B16F1 cells treated with control siRNA had spread and flattened with a circular morphology by 10–15 min, whereas this fraction increased to 9% by 20 min and to 17% by 30 min postplating, with the spreading efficiency also being dependent on extracellular factors, such as coating and FCS. At all of these time points, the fraction of spread cells and their size was significantly increased upon knockdown of Gadkin (Fig. 1B). An enhanced cell-spreading efficiency, as judged by increases in cell area, was also observed in Gadkin-depleted Cos7 and HeLa cells (Fig. S1 A and B) and with several independent Gadkin-specific siRNAs. A role for Gadkin in the regulation of cell spreading is further underscored by our analysis of mouse embryonic fibroblasts (MEFs) isolated from Gadkin knockout (KO) mice (Fig. S2A). Gadkin-deficient MEFs displayed a significant increase in cell size (Fig. 1C) 20–30 min postplating compared with MEFs from WT animals.
Fig. 1.
Gadkin negatively regulates cell spreading and migration. (A and B) B16F1 cells treated with scrambled or Gadkin-targeting siRNAs were trypsinized and seeded onto fibronectin-coated glass coverslips. After the indicated time intervals, cells were rapidly washed, fixed, and stained with phalloidin. (A) Representative images of control and Gadkin-depleted cells 20 min postplating. (Scale bar: 10 μm.) (Bi) Percentage of spread cells at different time points (two experiments; n = 128–299 cells per condition per experiment; *P < 0.05; unpaired Student’s t test). Symmetrically flattened out cells with a smooth plasmalemmal rim (see arrow in A) were classified as well spread. (Inset) Knockdown verified by immunoblot analysis. (Bii) Percentage cell size evaluated for the same experiment normalized to the size of control cells at 10 min (***P < 0.0001; unpaired Student’s t test). (C) Percentage cell size of two independent Gadkin WT and KO MEF cell lines 20–30 min postplating normalized to the size of Gadkin WT no. 3 cells (two experiments; n = 221–293 cells per cell line; *P < 0.01, **P < 0.001; one-way ANOVA plus Tukey’s post test). (D) B16F1 cells treated with Gadkin-targeting siRNAs and control cells were seeded onto laminin. Analysis of time lapse movies revealed that Gadkin-depleted cells migrate more, an effect that can be rescued by overexpression of Gadkin-WT-EGFP (n = 2–3; n = 30 cells per experiment; **P < 0.001; unpaired Student’s t test). Error bars always represent SEM.
Because the formation of sheet-like protrusions is involved in both cell spreading and migration (1, 2), we assayed the ability of Gadkin-depleted B16F1 cells to randomly migrate. Quantification of the accumulated distance migrated over time revealed that Gadkin depletion was associated with a significant gain in migration compared with control cells. This effect was specific because it was rescued by reexpression of siRNA-resistant Gadkin-EGFP (Fig. 1D). These data identify Gadkin as a negative regulator of cell spreading and migration and prompted us to dissect the underlying molecular mechanism.
Gadkin Directly Interacts with ARP2/3 via a Tryptophan-Containing Acidic Cluster Motif.
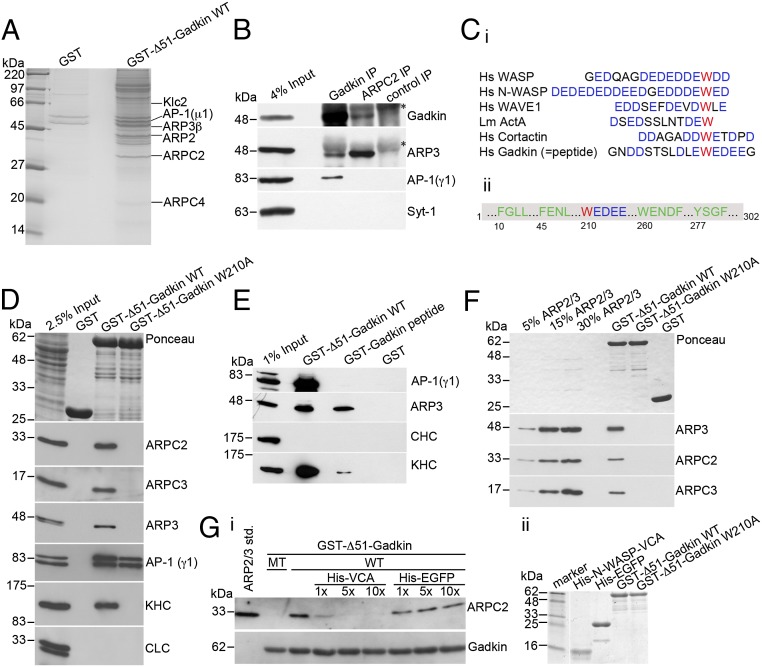
Because Gadkin avidly binds to the TGN/endosomally localized clathrin adaptor AP-1 and to kinesin1 [kinesin family motor protein 5/kinesin light chain 2 (KIF5/KLC2)], we first assessed the effect of silencing AP-1γ or kinesin1 heavy chain (KIF5) on cell spreading. However, neither AP-1 nor kinesin1-depleted cells showed alterations in spreading efficiency (see below). To identify Gadkin-binding proteins that might explain the above phenotypes, we performed affinity chromatography experiments from rat brain extracts using GST-Δ51-Gadkin immobilized on Sepharose beads. Tandem mass spectrometry (MS/MS)-based proteomic approaches identified, in addition to AP-1 and kinesin1, ARP2, ARP3β, ARP2/3 complex subunit 2 (ARPC2), and ARPC4, four subunits of the heptameric ARP2/3 actin-nucleation complex (3) as specific binding partners of Gadkin (Fig. 2A). Complex formation of native endogenous proteins in vivo was verified by reciprocal coimmunoprecipitation experiments using Gadkin- or ARPC2-specific antibodies (Fig. 2B). ARP2/3 regulates the assembly of sheet-like protrusions during the initial phase of cell spreading and migration (12) by promoting actin branch nucleation downstream of nucleation promoting factors (NPFs) such as WAVE and WASP proteins. Most NPFs harbor an acidic region bearing a conserved tryptophan (W) residue required for ARP2/3 binding (3, 13). Gadkin contains a sequence closely resembling W-bearing acidic domains of NPFs (Fig. 2C). To test the importance of this putative interaction motif for Gadkin complex formation with ARP2/3, we mutated W210 to A. As expected, GST-Δ51-Gadkin associated not only with AP-1 and kinesin1 but also with the ARP2/3 complex (detected via its ARP3, ARPC2, and ARPC3 subunits) in affinity chromatography experiments. By contrast, mutant GST-Δ51-Gadkin-W210A failed to associate with either ARP2/3 or kinesin1 but retained the ability to bind to AP-1 (Fig. 2D). Loss of kinesin1 binding of Gadkin-W210A is consistent with earlier results (10) and suggests that Gadkin associates with kinesin1 and the ARP2/3 complex via an overlapping site centered around W210. To determine whether the W-containing acidic cluster motif is not only required but also sufficient for binding to ARP2/3, we assayed the ability of a GST-fused peptide containing this motif (Fig. 2C) to associate with ARP2/3. As seen in Fig. 2E, the immobilized Gadkin-derived peptide was sufficient to retain ARP2/3 and albeit much more weakly also kinesin1, whereas it failed to bind to AP-1. To test whether the interaction between Gadkin and ARP2/3 was direct, we performed binding assays with purified proteins. ARP2/3 specifically bound to GST-Δ51-Gadkin-WT coupled to beads, whereas no binding was detected for GST-Δ51-Gadkin-W210A (Fig. 2F). We conclude that Gadkin directly and specifically associates with the ARP2/3 complex via a W-containing acidic cluster motif similar to those found in NPFs.
Fig. 2.
Gadkin directly interacts with ARP2/3 via a tryptophan-containing acidic cluster motif. (A) Coomassie blue-stained gel of material isolated by affinity chromatography from 12.5 mg of rat brain extract using 1.5 mg of GST or GST-fused Δ51-Gadkin cross-linked to glutathione Sepharose beads as bait. The labeled bands were identified by MALDI-TOF mass spectrometry. (B) Endogenous ARP2/3 and Gadkin coimmunoprecipitate. Solubilized rat brain membranes were subjected to immunoprecipitation using affinity-purified antibodies against Gadkin, ARPC2, or nonspecific rabbit IgGs as control. Samples were analyzed by immunoblotting for Gadkin, ARP3, AP-1, and synaptotagmin 1 (Syt-1). Asterisks denote nonspecific bands originating from Ig-heavy chains. (Ci) Sequence alignment of ARP2/3-interacting A domains of classic NPFs and Gadkin. Acidic residues are shown in blue, and the critical W is in red. (Cii) Scheme of Gadkin primary structure highlighting amino acid positions of motifs involved in binding to AP-1 (green) or ARP2/3 (red/blue). (D) Immunoblot analysis of proteins isolated by affinity chromatography from 1.6 mg of rat brain extract using 20 μg of GST-tagged Δ51-Gadkin-WT or its W210A mutant as bait. Samples were analyzed by immunoblotting for ARPC2, ARPC3, ARP3, AP-1, kinesin heavy chain (KHC), and clathrin light chain (CLC). (E) Immunoblot analysis of material affinity-purified using GST, GST-Δ51-Gadkin, or a GST-Gadkin-peptide as described in C as baits. (F) Direct binding of purified ARP2/3 complex to GST-Δ51-Gadkin. No binding is seen for Gadkin-W210A or GST. Samples were analyzed by immunoblotting for ARP3, ARPC2, and ARPC3. (Gi) Binding of purified ARP2/3 to GST-Δ51-Gadkin is inhibited by addition of a 1–10× molar excess of His6-N-WASP-VCA domain. Addition of His6-EGFP as a control did not affect binding. Samples were analyzed by immunoblotting for ARPC2 and Gadkin. (Gii) Integrity of purified proteins was verified by SDS/PAGE and Coomassie staining.
The fact that Gadkin and NPFs appear to associate with ARP2/3 via similar determinants suggests that both factors may use a common overlapping interface on ARP2/3 and, hence, compete with each other for binding. To analyze whether Gadkin and NPFs associate with ARP2/3 in a mutually exclusive manner, we carried out direct binding studies using GST-Δ51-Gadkin and purified ARP2/3 to which we added increasing amounts of the VCA domain of N-WASP, a potent NPF bearing a W-containing acidic motif (3). Addition of equimolar amounts of His6-VCA substantially decreased binding of ARP2/3 to Gadkin, and excess His6-VCA completely abrogated Gadkin-ARP2/3 complex formation (Fig. 2G). We conclude that Gadkin and NPFs such as N-WASP compete for the same binding site on ARP2/3.
Gadkin Colocalizes with a Subpool of ARP2/3 on Endosomes.
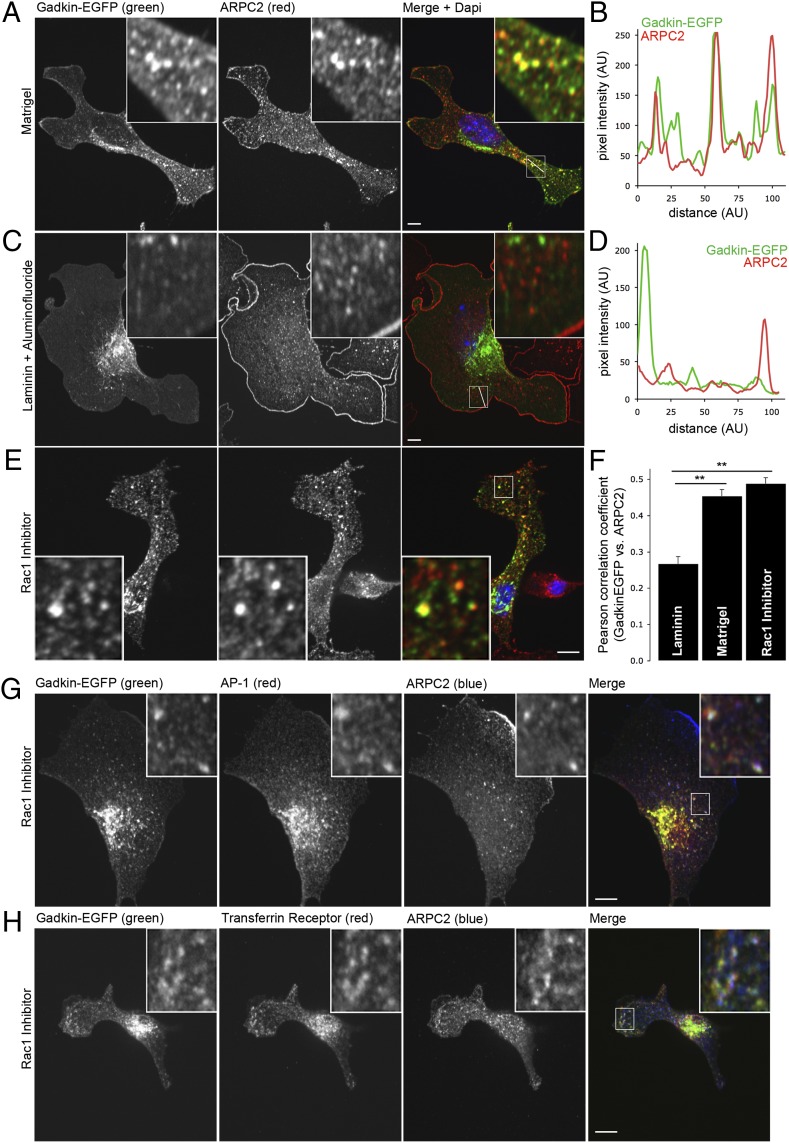
Gadkin restricts cell spreading and migration and directly associates with ARP2/3, a key component of lamellipodia formation and a positive regulator of cell motility. We, therefore, hypothesized that Gadkin may act as an inhibitor of ARP2/3 function. To examine this possibility, we analyzed the subcellular distribution of Gadkin and ARP2/3 in cells plated on different substrates and under promigratory or migration prohibiting conditions. In lamellipodia-free cells plated on Matrigel, endogenous ARP2/3 was mostly distributed intracellularly, where it partially colocalized with a pool of Gadkin-EGFP (Fig. 3 A, B, and F). Treatment of cells grown on laminin with aluminum fluoride, a chemical activator of NPFs stimulating small GTPases, including Rac1 and Cdc42 (cell division control protein 42 homolog), induced the formation of numerous lamellipodia as expected (14), and these changes correlated with a redistribution of ARP2/3 from intracellular pools to the plasma membrane (Fig. 3 C, D, and F). Conversely, when Rac1 activation was blocked, ARP2/3 redistributed from the plasma membrane to intracellular sites including Gadkin-containing endosomes (Fig. 3E), as evidenced by a significant increase in colocalization under these conditions (Fig. 3F). Gadkin- and ARP2/3-positive puncta also contained AP-1 and transferrin receptor (Fig. 3 G and H), identifying these structures as endosomal vesicles. In agreement with these data on fixed cells, a subpool of ARP2/3 was present on mobile Gadkin-positive endosomes observed by live imaging of Gadkin-mRFP- and EGFP-ARPC5B-expressing cells (Fig. S3 A and B). Finally, overexpression of FLAG-Gadkin or Gadkin-EGFP in HeLa cells resulted in the kinesin1-driven accumulation of endogenous ARP2/3 on peripheral Gadkin-positive spots (extensively characterized in ref. 10) beneath the plasma membrane (Fig. S3 C and D), whereas mutant Gadkin-W210A-EGFP was unable to recruit ARP2/3 (Fig. S3C). Collectively, these data suggest that Gadkin in the absence of promigratory signals colocalizes with a subpool of ARP2/3 on AP-1- and transferrin receptor-containing endosomes.
Fig. 3.
Gadkin colocalizes with a subpool of ARP2/3 on endosomes. (A–D) Gadkin and ARPC2 colocalize under conditions that do not stimulate lamellipodia formation. Localization of Gadkin-EGFP and ARPC2 in B16F1 cells grown on Matrigel (A) or on laminin in the presence of aluminum fluoride (C) or in the presence of Rac1 inhibitor (E). Twenty-four hours after seeding, cells were fixed and immunostained with ARPC2-specific antibodies. (B and D) Fluorescence-intensity profiles along lines indicated within boxed areas in (A and C). (F) Pearson correlation coefficients illustrating the degree of colocalization between Gadkin-EGFP and ARPC2 under different conditions (40–67 cells per condition from 2 to 4 experiments; error bars represent SEM; **P < 0.01; one-way ANOVA plus Dunnett’s post test). (G and H) Gadkin-EGFP-expressing B16F1 cells were treated with Rac1 inhibitor. Twenty-four hours after seeding, cells were fixed and costained with antibodies specific for the indicated protein. (Scale bar: 5 μm.) (Insets) Magnifications: 5× (A–E); 4× (G and H).
ARP2/3 is an abundant protein complex with concentrations up to 2–10 μM, depending on cell type (15). Negative regulation of ARP2/3 function by association with Gadkin would require the latter to be present at similarly high concentrations. We, therefore, determined the approximate concentrations of ARP2/3 and Gadkin in B16F1 cells by quantitative immunoblotting [assuming an average cell volume of 2,780 μm3 (16)]. Gadkin and ARPC2 were present at near equimolar ratios of about 2.3 and 2.6 μM, respectively (Fig. S4A). These data not only fit well with published values for ARP2/3 but are also consistent with the possibility that Gadkin regulates ARP2/3 via intracellular sequestration. This hypothesis is in agreement with the observation that Gadkin and ARPC2 are both widely expressed (Fig. S4B). Gadkin may affect ARP2/3 function either by sequestration or by direct inhibition of NPF-stimulated ARP2/3-dependent actin polymerization. To test the latter possibility, we used established in vitro actin polymerization assays based on the incorporation of fluorescent pyrene-actin. Incubation of either actin alone or together with purified ARP2/3 only led to a moderate rate of actin polymerization, consistent with the known requirement for NPFs to activate ARP2/3. Addition of Gadkin did not alter the rate of actin polymerization, indicating that Gadkin does not act as an NPF. This is in line with the fact that Gadkin binds ARP2/3 via a W-based acidic cluster motif but lacks the V and C domains of typical NPFs required for ARP2/3 activation. By contrast, addition of the WASP-VCA domain promoted actin assembly irrespective of the presence of Gadkin added in up to 250× molar excess to the assay (Fig. S4C). These results, together with in vitro binding competition experiments (Fig. S4D), indicate that Gadkin neither acts as an NPF nor is able to compete off active VCA domain from ARP2/3, at least in vitro.
To assess whether Gadkin regulates the partitioning of ARP2/3 between the cell surface and intracellular sites, we analyzed plasma membrane-associated surface proteins isolated from WT or Gadkin-deficient MEFs by quantitative stable isotope labeling (SILAC)-based MS/MS. As seen in Table S1, this analysis revealed a clear about 1.7-fold enrichment of all subunits of the ARP2/3 complex at the plasma membrane of Gadkin KO MEFs. By contrast, the surface levels of platelet-derived growth factor receptor, an important regulator of cell shape and migration, were not increased. A similar enrichment of the ARP2/3 subunit ARPC2 was also observed in immunostainings of Gadkin KO vs. WT MEFs with ARPC2 specific antibodies (Fig. S1B). Collectively, our data identify Gadkin as an intracellular regulator of ARP2/3 function and distribution.
Gadkin Regulates Cell Spreading and Size via Modulating ARP2/3 Function.
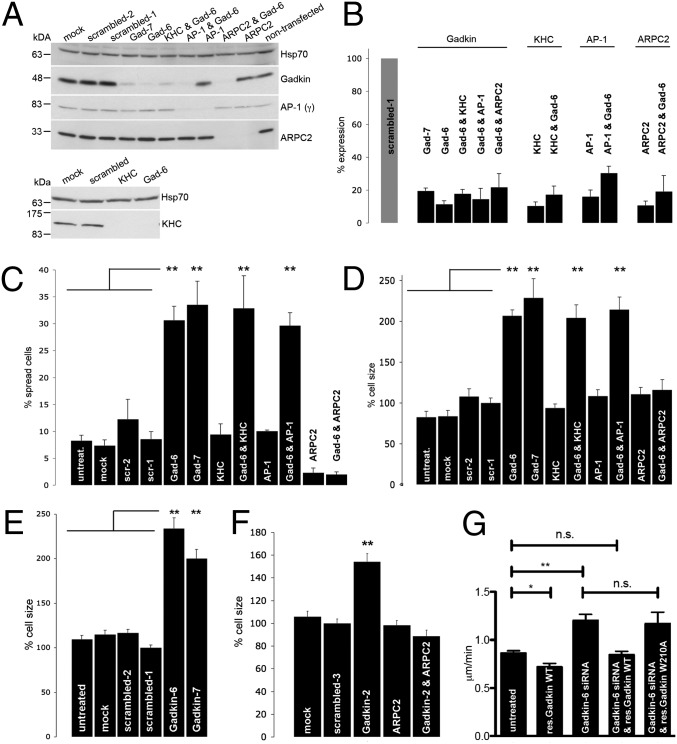
Gadkin, in addition to ARP2/3, also interacts with AP-1 (9, 11) and kinesin1 (KIF5B) (10). To mechanistically dissect the role of Gadkin in regulating lamellipodia-mediated cell spreading and to identify the responsible effectors, we depleted cells of endogenous Gadkin, KIF5B, AP-1(γ1), or ARPC2 [a subunit required for ARP2/3 complex function (17, 18)], either alone or in combination (Fig. 4 A and B). Depletion of Gadkin using two independent siRNAs resulted in profoundly increased cell spreading, whereas depletion of either AP-1(γ1) or KIF5B had no effect (Fig. 4C). Conversely, silencing of ARPC2 strongly impaired cell spreading 15–20 min after plating (Fig. 4C) (as described in ref. 12). Consistent with the proposed function of Gadkin as a negative regulator of ARP2/3, Gadkin silencing did not enhance cell spreading in the absence of ARPC2 (Fig. 4C). Cells depleted of both Gadkin and KIF5B or Gadkin and AP-1(γ1) behaved identical to those treated with Gadkin siRNA alone (Fig. 4C). Hence, the ability of Gadkin to regulate cell spreading depends on an intact ARP2/3 complex but not on kinesin1 or on AP-1(γ1).
Fig. 4.
Gadkin modulates cell spreading and size via regulation of ARP2/3. (A) Representative immunoblots of lysates from cells treated with the indicated siRNAs incubated with antibodies against the proteins indicated on the right. (B) Quantification of knockdown efficiency based on immunoblotting (n = 2–4). (C and D) B16F1 cells treated with the indicated siRNAs were trypsinized and seeded onto glass coverslips. Fifteen to 20 min postplating, the fraction (%) of spread cells (C), as well as the cell size normalized to cells treated with scrambled-1 siRNA (D), was determined (n = 2–4 experiments; n = 105–312 cells per condition per experiment; **P < 0.001; one-way ANOVA plus Tukey’s post test). (E and F) B16F1 (E) respectively A431 (F) cells treated with the indicated siRNAs were fixed 24 h after plating and stained with phalloidin. Cell size was quantified and normalized to the size of cells treated with scrambled-1 respectively scrambled-3 siRNA (A431: n = 2, n = 131–266 cells per condition; B16F1: n = 3–4, n = 217–270 cells per condition; **P < 0.001; one-way ANOVA plus Tukey’s post test). (G) B16F1 cells treated with control or Gadkin-specific siRNAs transiently expressing siRNA resistant WT or ARP2/3 binding defective mutant (W210A) Gadkin-EGFP were assayed for their ability to randomly migrate. Reexpression of WT but not W210A mutant Gadkin rescued enhanced migration seen in Gadkin-depleted cells (n = 3; n = 30 cells per experiment; **P < 0.001; *P < 0.05; unpaired Student’s t test; mean ± SEM).
As an additional parameter, we analyzed cell size 15–20 min after plating. Similar to the results seen for the morphological analysis of cell spreading, depletion of Gadkin but not of KIF5B or AP-1(γ1) led to a significant increase in cell size. Knockdown of ARPC2 did not alter cell size, consistent with the observation that the ability of cells to form lamellipodia is not directly linked to the control of plasma membrane area (19, 20). However, ARPC2 silencing completely abrogated increases in cell size elicited by knockdown of Gadkin, whereas depletion of KIF5B or AP-1(γ1) had no effect (Fig. 4D). Gadkin-depleted cells had an increased size not only early after plating but remained significantly larger even after 24 h in culture (Fig. 4E). Similar increases in plasma membrane area were observed in A431 Gadkin knockdown cells, a phenotype rescued by cosilencing of ARPC2 expression (Fig. 4F). The increased size of Gadkin-depleted cells correlated with an elevation in the number of protrusion containing cells compared with control (Fig. S5), which was not caused by general alterations in the expression level of WASH or AP-1 or in F-actin distribution (Fig. S6).
Finally, we wanted to know whether the function of Gadkin as a negative regulator of cell migration (compare Fig. 1) depends on its ability to directly associate with ARP2/3, as expected from the above results. To this aim, we carried out rescue experiments by reexpressing siRNA-resistant WT or ARP2/3-binding defective mutant versions of Gadkin-EGFP in cells depleted of the endogenous protein. As expected, WT-Gadkin-EGFP rescued the increased motility of cells depleted of endogenous Gadkin (Figs. 1D and 4G) and even decreased the migration rate of nondepleted control cells (Fig. 4G), whereas ARP2/3-binding defective Gadkin-EGFP (W210A) failed to rescue enhanced migration (Fig. 4G).
Our collective data thus indicate that Gadkin negatively regulates cell spreading, size, and motility via sequestration of ARP2/3.
Discussion
Here, we identify Gadkin as a negative regulator of ARP2/3 function, modulating ARP2/3-dependent processes such as cell spreading and motility. We demonstrate that Gadkin via a W-based acidic motif directly associates with ARP2/3 and that both proteins colocalize on motile endosomal vesicles under migration-inhibiting conditions. Loss of Gadkin facilitates the partial redistribution of ARP2/3 from internal pools to the plasma membrane and leads to increased cell spreading, size, and migration via a mechanism that depends on ARP2/3 activity. Hence, Gadkin represents a negative regulator of ARP2/3 function present on intracellular membranes.
Gadkin differs in several important aspects from previously identified inhibitors of ARP2/3-mediated actin nucleation. The PDZ-BAR domain protein Pick1 (protein interacting with C kinase 1) (13), the actin-binding protein coronin 1B (21), and the cofilin homolog glia maturation factor (GMF) (22) directly interact with both ARP2/3 and actin to inhibit NPF-stimulated ARP2/3-mediated actin polymerization. Others, including tropomyosin (23), caldesmon (24), and EPLIN (epithelial protein lost in neoplasm) (25), mainly act via tight association with F-actin, thereby competing with ARP2/3 binding to the mother actin filament. In all of these cases, negative regulation on ARP2/3-mediated branch nucleation is imparted directly at the site of ARP2/3 function. For example, Pick1 (13) acts at sites of glutamate receptor internalization in neuronal dendrites, whereas coronin 1B coordinates ARP2/3-mediated filament dynamics at the leading edge of migrating cells (21). There is no report that loss of function of any of these inhibitors is associated with increased cell motility. To the contrary, expression of mutant coronin with elevated ARP2/3 binding caused increased cell migration (26). Our biochemical and cell biological data indicate that Gadkin regulation of ARP2/3 function occurs via a different mechanism that likely involves sequestration of ARP2/3 at intracellular sites without directly inhibiting the activity of the complex. Support for this model comes from our observations that (i) excess Gadkin is incapable of outcompeting active VCA domain from ARP2/3; (ii) Gadkin does not affect VCA-domain stimulated ARP2/3-dependent actin polymerization; and (iii) Gadkin KO cells display elevated plasma membrane levels of ARP2/3 and increased cell spreading. Based on these data, we favor a role for Gadkin as a regulator of an intracellular reservoir of ARP2/3. Release of ARP2/3 from this pool might be triggered by signaling cascades, resulting in the activation and localized recruitment of NPFs to their site of action. Such activated NPFs via their VCA domains likely are capable of retrieving ARP2/3 from Gadkin-complexed intracellular pools. Consistent with this hypothetical model, we have been unable to observe lamellipodia formation in Gadkin-depleted cells in the absence of promigratory signals.
Although the proposed function of Gadkin as an intracellular safeguard for ARP2/3 activity is consistent with our data and with previous work (2, 3), we cannot rule out that Gadkin-associated endosomal ARP2/3 plays additional roles in cell physiology apart from restricting cell migration. ARP2/3 via N-WASP (27) and WASP homolog associated with actin, membranes, and microtubules (WHAMM) (28) has been implicated in membrane traffic at the Golgi and the TGN and via WASH at endosomes (5–7). It is, therefore, conceivable that the restrictive role of Gadkin in regulating ARP2/3 function at the cell surface may extend to intracellular membranes. Future studies will need to address this possibility. The role of Gadkin as a negative regulator of ARP2/3-mediated cell spreading and migration parallels the recent identification of Sharpin as an inhibitor of integrin activity (8). Loss of Sharpin, similar to loss of Gadkin, is associated with an elevated propensity of cell to migrate, suggesting that signal-dependent control of cell shape and motility is under spatiotemporal control imparted by activators and inhibitors acting at multiple stages of the pathway.
Materials and Methods
Descriptions of the following are available in SI Materials and Methods: plasmids, antibodies, reagents, cell culture and transfection, preparation of cell and tissue lysates, protein quantification, actin polymerization assay, binding and competition experiments, generation of Gadkin KO mice and derived MEFs, and surface biotinylation followed by SILAC-based MS analysis.
Immunocytochemistry.
Cells grown on glass coverslips were washed and fixed with 4% (wt/vol) paraformaldehyde/4% (wt/vol) sucrose in PBS. Following permeabilization and blocking in goat serum dilution buffer (GSDB) [25% (vol/vol) goat serum, 0.3% Triton X-100 in PBS], cells were incubated with primary and secondary antibodies in GSDB. Washed cells were mounted in ImmuMount (Thermo Electron) supplemented with 1 μg/mL DAPI.
Cell-Spreading Assay.
Cells, if necessary, were transfected 2× with siRNA (days 1 and 3), trypsinized, and seeded in full growth medium onto glass coverslips. Cells were incubated at 37 °C in the presence of 5% CO2 for 10–30 min. Immediately afterward, cells were washed and processed for immunocytochemistry.
Cell-Motility Assay.
B16F1 cells were transfected 2× with siRNA (days 1 and 3), trypsinized, and seeded sparsely into 6-cm dishes coated with laminin. Cells were imaged at low magnification every 3 min for 18 h. Tracks were identified from recorded movies using the manual tracking plugin of Image J. The “chemotaxis and migration“ plugin (Ibidi) was used for further analysis.
Fluorescence Microscopy.
Epifluorescent images for the quantification of cell size, cell shape and fluorescence intensities were acquired on a motorized Zeiss Axiovert 200M inverted microscope equipped with the Stallion System (Intelligent Imaging Innovations). Data were processed using Slidebook 5 software (Intelligent Imaging Innovations). Confocal images and live cell imaging were carried out with a Zeiss Axiovert 200M-based Perkin-Elmer UltraView ERS dual spinning disk system. Data were processed using Volocity software (Improvision) and ImageJ.
Affinity Chromatography.
GST-fusion proteins were purified from overexpressing Escherichia coli using GST-bind resin (Novagen) according to standard protocols. Affinity chromatography experiments were carried out as described in ref. 10. Samples were analyzed by SDS/PAGE and immunoblotting or MS/MS-based MS.
Direct-Binding Assay.
ARP2/3 complex (tebu-bio) was diluted to 0.1 μg/μL in 10 mM Hepes (pH 7.4), 100 mM KCl, 1 mM MgCl2, and 0.1 mM EDTA supplemented with 1 mM PMSF and mammalian protease inhibitor mixture (Sigma). GST-fusion protein (10 μg) on GST-bind resin was incubated for 2 h at 4 °C under gentle agitation with 6.7 μg of ARP2/3 in 500 μL of the same buffer supplemented with 0.1% Tween-20. Following extensive washes, bound ARP2/3 was eluted in sample buffer. Samples were analyzed by SDS/PAGE and immunoblotting.
Immunoprecipitation.
Two p1 rat brains were homogenized in 5 mL of 25 mM Hepes (pH 7.5), 125 mM NaCl, and 2 mM MgCl2 supplemented with 1 mM PMSF and mammalian protease inhibitor mixture (Sigma). Cellular debris was removed by spinning 2× 10 min at 1,000 × g at 4 °C. The supernatant was centrifuged for 30 min at 180,000 × g at 4 °C. The resulting membrane pellet was resuspended in 1 mL of 25 mM Hepes (pH 7.4), 125 mM NaCl, 2 mM MgCl2, and 0.5% TritonX-100 supplemented with 1 mM PMSF and mammalian protease inhibitor mixture (Sigma). Following solubilization on ice for 15 min, lysates were precleared by ultracentrifugation (15 min at 180,000 × g at 4 °C). Lysate (1.2 mg; 3 mg/mL) was subjected to immunoprecipitation for 4 h at 4 °C using antibodies [6 μg of rabbit anti-ARPC2; 12 μg of rabbit anti-Gadkin (no. 11); 12 μg of rabbit immunoglobulins] coupled to protein A/G agarose (Santa Cruz Biotechnology). Beads were washed extensively and eluted with sample buffer. Samples were analyzed by SDS/PAGE and immunoblotting.
Supplementary Material
Acknowledgments
We thank Drs. Theresia Stradal (University Münster) and Klemens Rottner (University Bonn) for cells and reagents and Dr. Shawn Ferguson (Yale University) for sharing siRNA sequences. We thank Fabian Feutlinske for help with sample preparation for MS/MS. This work was supported by Deutsche Forschungsgemeinschaft (DFG) Grants HA2686/1-1 and 1-2 (to V.H. and T.M.); L.M.M. and T.Z. were funded by Cancer Research UK.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206468109/-/DCSupplemental.
References
- 1.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campellone KG, Welch MD. A nucleator arms race: Cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goley ED, Welch MD. The ARP2/3 complex: An actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 4.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 5.Derivery E, et al. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zech T, et al. The Arp2/3 activator WASH regulates α5β1-integrin-mediated invasive migration. J Cell Sci. 2011;124:3753–3759. doi: 10.1242/jcs.080986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rantala JK, et al. SHARPIN is an endogenous inhibitor of β1-integrin activation. Nat Cell Biol. 2011;13:1315–1324. doi: 10.1038/ncb2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neubrand VE, et al. Gamma-BAR, a novel AP-1-interacting protein involved in post-Golgi trafficking. EMBO J. 2005;24:1122–1133. doi: 10.1038/sj.emboj.7600600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt MR, et al. Regulation of endosomal membrane traffic by a Gadkin/AP-1/kinesin KIF5 complex. Proc Natl Acad Sci USA. 2009;106:15344–15349. doi: 10.1073/pnas.0904268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maritzen T, et al. A novel subtype of AP-1-binding motif within the palmitoylated trans-Golgi network/endosomal accessory protein Gadkin/gamma-BAR. J Biol Chem. 2010;285:4074–4086. doi: 10.1074/jbc.M109.049197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholson-Dykstra SM, Higgs HN. Arp2 depletion inhibits sheet-like protrusions but not linear protrusions of fibroblasts and lymphocytes. Cell Motil Cytoskeleton. 2008;65:904–922. doi: 10.1002/cm.20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rocca DL, Martin S, Jenkins EL, Hanley JG. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat Cell Biol. 2008;10:259–271. doi: 10.1038/ncb1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahne P, Sechi A, Benesch S, Small JV. Scar/WAVE is localised at the tips of protruding lamellipodia in living cells. FEBS Lett. 2001;492:215–220. doi: 10.1016/s0014-5793(01)02239-6. [DOI] [PubMed] [Google Scholar]
- 15.Blanchoin L, Pollard TD, Mullins RD. Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr Biol. 2000;10:1273–1282. doi: 10.1016/s0960-9822(00)00749-1. [DOI] [PubMed] [Google Scholar]
- 16.McMillan TJ, Rao J, Hart IR. Enhancement of experimental metastasis by pretreatment of tumour cells with hydroxyurea. Int J Cancer. 1986;38:61–65. doi: 10.1002/ijc.2910380111. [DOI] [PubMed] [Google Scholar]
- 17.Kempiak SJ, Yip SC, Backer JM, Segall JE. Local signaling by the EGF receptor. J Cell Biol. 2003;162:781–787. doi: 10.1083/jcb.200303144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferguson SM, et al. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell. 2009;17:811–822. doi: 10.1016/j.devcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Nardo A, et al. Arp2/3 complex-deficient mouse fibroblasts are viable and have normal leading-edge actin structure and function. Proc Natl Acad Sci USA. 2005;102:16263–16268. doi: 10.1073/pnas.0508228102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vidali L, Chen F, Cicchetti G, Ohta Y, Kwiatkowski DJ. Rac1-null mouse embryonic fibroblasts are motile and respond to platelet-derived growth factor. Mol Biol Cell. 2006;17:2377–2390. doi: 10.1091/mbc.E05-10-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007;128:915–929. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gandhi M, et al. GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr Biol. 2010;20:861–867. doi: 10.1016/j.cub.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol. 2001;11:1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- 24.Yamakita Y, Oosawa F, Yamashiro S, Matsumura F. Caldesmon inhibits Arp2/3-mediated actin nucleation. J Biol Chem. 2003;278:17937–17944. doi: 10.1074/jbc.M208739200. [DOI] [PubMed] [Google Scholar]
- 25.Maul RS, et al. EPLIN regulates actin dynamics by cross-linking and stabilizing filaments. J Cell Biol. 2003;160:399–407. doi: 10.1083/jcb.200212057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai L, Holoweckyj N, Schaller MD, Bear JE. Phosphorylation of coronin 1B by protein kinase C regulates interaction with Arp2/3 and cell motility. J Biol Chem. 2005;280:31913–31923. doi: 10.1074/jbc.M504146200. [DOI] [PubMed] [Google Scholar]
- 27.Anitei M, et al. Protein complexes containing CYFIP/Sra/PIR121 coordinate Arf1 and Rac1 signalling during clathrin-AP-1-coated carrier biogenesis at the TGN. Nat Cell Biol. 2010;12:330–340. doi: 10.1038/ncb2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.