Abstract
Cerebral malaria (CM) is a deadly complication of Plasmodium falciparum infection, but specific interactions involved in cerebral homing of infected erythrocytes (IEs) are poorly understood. In this study, P. falciparum-IEs were characterized for binding to primary human brain microvascular endothelial cells (HBMECs). Before selection, CD36 or ICAM-1–binding parasites exhibited punctate binding to a subpopulation of HBMECs and binding was CD36 dependent. Panning of IEs on HBMECs led to a more dispersed binding phenotype and the selection of three var genes, including two that encode the tandem domain cassette 8 (DC8) and were non-CD36 binders. Multiple domains in the DC8 cassette bound to brain endothelium and the cysteine-rich interdomain region 1 inhibited binding of P. falciparum-IEs by 50%, highlighting a key role for the DC8 cassette in cerebral binding. It is mysterious how deadly binding variants are maintained in the parasite population. Clonal parasite lines expressing the two brain-adherent DC8-var genes did not bind to any of the known microvascular receptors, indicating unique receptors are involved in cerebral binding. They could also adhere to brain, lung, dermis, and heart endothelial cells, suggesting cerebral binding variants may have alternative sequestration sites. Furthermore, young African children with CM or nonsevere control cases had antibodies to HBMEC-selected parasites, indicating they had been exposed to related variants during childhood infections. This analysis shows that specific P. falciparum erythrocyte membrane protein 1 types are linked to cerebral binding and suggests a potential mechanism by which individuals may build up immunity to severe disease, in the absence of CM.
Keywords: cytoadhesion, antigenic variation, parasite ligand
Infection with Plasmodium falciparum may lead to severe disease as a result of infected erythrocyte (IE) binding in brain or placental microvascular blood vessels. P. falciparum-IEs persist in the host and avoid clearance in the spleen by expression of a family of cytoadhesion proteins termed P. falciparum erythrocyte membrane protein 1 (PfEMP1) (1–3). Clonal antigenic variation of var genes modifies the antigenic and binding properties of IEs (2, 4, 5) and is believed to mediate IE tropism for different microvascular sites contributing to organ-specific pathology (6).
Members of the var gene family are classified into three main subfamilies (groups A, B, and C) on the basis of chromosome location, direction of transcription, and upstream promoter sequence (UpsA, UpsB, and UpsC) (7–9). A small number of group B/A genes also have an UpsB promoter and group A coding sequence. This var gene organization is conserved across parasite isolates (10, 11) and may contribute to gene recombination hierarchies that underlie the functional and antigenic specialization of var groups for distinct binding niches (12). Whereas most PfEMP1 proteins bind CD36, group A and the related group B/A have a distinct protein head structure from other var groups (8, 9) and do not bind CD36 (13, 14). Group A and B/A proteins also tend to be among the first PfEMP1 proteins expressed in early childhood infections (15–18) and have been associated with more severe infections (18–21), but whether they have affinity for brain endothelium is unknown. Studies of malaria during pregnancy have demonstrated how a single var gene, VAR2CSA, is primarily responsible for placenta binding (22–24). However, attempts to associate specific PfEMP1 variants with cerebral binding (25) have been complicated by the extensive diversity of the gene family and the lack of animal models for studying binding of P. falciparum-IEs in cerebral malaria.
IEs bind a variety of different host receptors, but the best-understood binding partners are CD36 and intercellular adhesion molecule 1 (ICAM-1) (26). ICAM-1 is efficient at capturing cells from flow and synergizes with CD36 to mediate firm IE binding (27–29). Postmortem studies showed that P. falciparum-IEs colocalize with ICAM-1 in cerebral vessels (30). Because CD36 is weakly expressed on brain endothelium (30, 31), other host receptors would likely need to act in concert with ICAM-1 to mediate cerebral binding. Other candidates, such as HABP1/gC1qR (32) and Fractalkine (33), have been proposed as potential cerebral sequestration receptors, but attempts to link ICAM-1 binding parasites or other binding phenotypes with cerebral malaria have yielded contradictory findings (34–37). In addition, a single P. falciparum genotype can encode multiple ICAM-1–binding PfEMP1 variants (38), but whether all of these are involved in cerebral homing is unknown. Thus, the molecular mechanisms associated with cerebral recruitment remain poorly understood.
In this study, we used several primary human brain microvascular endothelial cell (HBMEC) isolates (39–41) to assess the selectivity of different parasite subpopulations for brain endothelium and to characterize the var gene repertoire and binding characteristics of P. falciparum-IEs that have been selected for increased binding to HBMECs.
Results
Binding of Different P. falciparum Adhesion Types to Resting or Activated HBMECs.
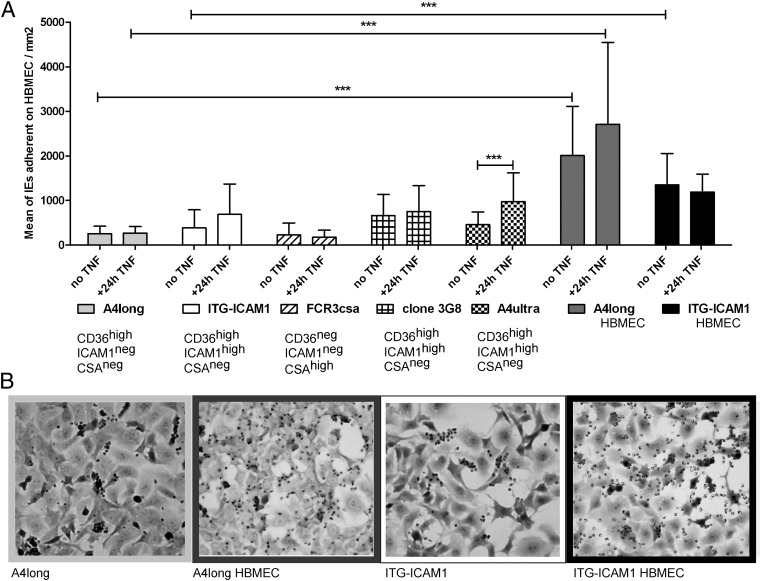
To investigate parasite specificity for primary HBMECs, binding was compared from five parasite lines that were derived from the IT4/FCR3 parasite genotype but expressed different var genes (13). A CSA-binding parasite (FCR3-CSA) displayed the lowest binding level, a CD36 parasite line (A4long) had an intermediate binding level, and three ICAM-1 plus CD36-binding parasite lines (ItG-ICAM-1, 3G8, and A4ultra) bound at similar levels and only slightly higher than the CD36-binding parasite line (Fig. 1).
Fig. 1.
Binding of P. falciparum-IEs to primary HBMECs. (A) Parasites exhibiting different binding phenotypes were compared for binding to resting or TNF-α–activated HBMECs from patient donor 13. The A4longHBMEC and ItG-ICAM-1HBMEC lines were panned three times on HBMECs from donor 13. Results are expressed as mean number of IEs per square millimeter ± SDs from two to three independent experiments (***P < 0.001). (B) The starting A4long and ItG-ICAM-1 parasite lines exhibited a concentrated binding to a subpopulation of HBMECs that became more dispersed after panning of IEs three times on HBMECs.
Notably, all five parasite lines bound in a concentrated pattern to only a subpopulation of HBMECs (Fig. 1 and Fig. S1). Whereas all HBMECs were ICAM-1 surface positive, only a minor subpopulation was CD36 surface positive (∼3%) and CD36 surface levels were ∼10–100× lower than ICAM-1 surface levels (Fig. S2). The binding of both CD36-binding and ICAM-1–binding parasites (A4ultra and ItG-ICAM-1) was nearly abolished by anti-CD36 antibodies (91% and 82% decrease, respectively), whereas anti–ICAM-1 antibodies significantly reduced binding (61% vs. 78%, respectively) (Fig. 2C). Thus, both parasite lines were highly dependent on CD36 for adhesion to “resting” brain endothelial cells.
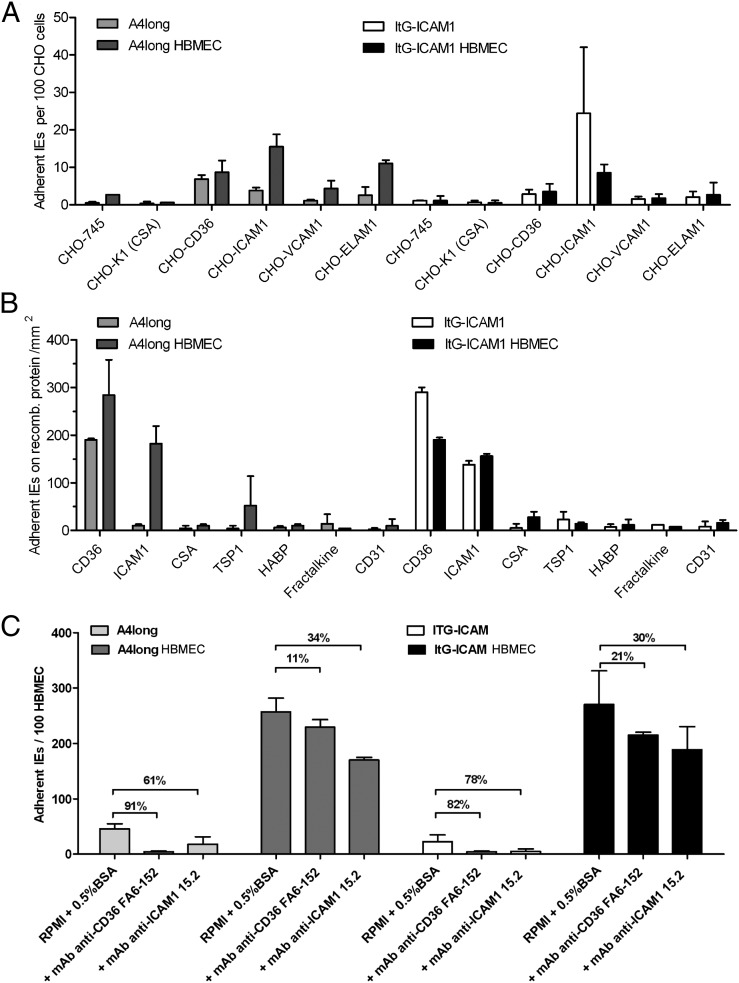
Fig. 2.
The binding specificity of P. falciparum-IEs changed after selection on HBMECs. (A) The initial and HBMEC-panned parasite lines were compared for binding to CHO cells that differed in CSA, CD36, ICAM-1, VCAM-1, or ELAM-1 surface expression. (B) The initial and HBMEC-panned parasite lines were compared for binding to recombinant proteins. (C) The ability of anti-CD36 or anti-ICAM-1 antibodies to block IE binding to primary HBMECs (donor 13) was compared between the initial and HBMEC-panned parasite lines. Percentage of inhibition (%) is indicated between bars. Binding results in A–C are expressed as means ± SDs from two to three independent experiments.
The inflammatory cytokine TNF-α is produced during malaria infections and causes widespread endothelial activation (30). TNF-α activated HBMECs displayed ∼10-fold higher ICAM-1 surface levels, but CD36 surface levels did not change (Fig. S2). TNF-α activation did not enhance binding of CD36- or CSA-binding parasite lines, but binding increased ∼2-fold for two of the three ICAM-1–binding parasite lines (Fig. 1A). However, IEs continued to exhibit a concentrated binding to a subpopulation of cells, except that the A4ultra and 3G8 parasites displayed slightly more dispersed binding to other HBMECs in the culture (Fig. S1). Thus, CD36-binding or ICAM-1–binding parasites were primarily adherent to a small subpopulation of resting or TNF-α–activated HBMECs that were dually positive for CD36 and ICAM-1.
Selection of IEs on Primary HBMECs Increases Binding Activity to all of the Cells in the Culture.
To investigate whether higher-affinity parasites could be selected on brain endothelium, A4long and ItG-ICAM-1 parasite lines were selected on HBMECs. Following panning, binding levels increased 5- to 10-fold for both the selected A4longHBMEC and ItG-ICAM-1HBMEC parasite lines and the pattern of binding became more dispersed to all HBMECs in the culture (Fig. 1). Whereas the ItG-ICAM-1HBMEC parasite line displayed similar binding to resting or TNF-α–activated HBMECs, A4longHBMEC binding was slightly increased to TNF-α–activated HBMECs (Fig. 1), but the increase did not reach significance.
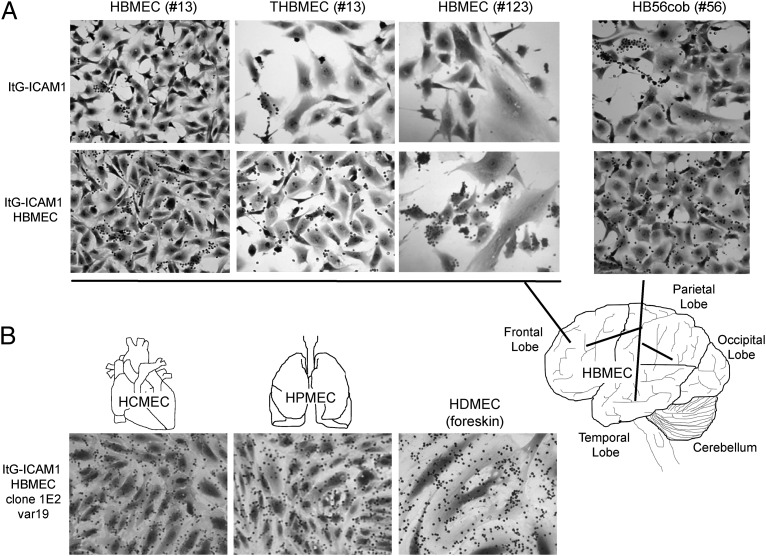
To investigate whether this unique dispersed binding pattern was restricted to particular donor endothelial cells, starting and panned parasite lines were assayed against HBMECs collected from three different donors and an immortalized HBMEC line (THBMEC) that was derived from one of the primary isolates by SV40-LT virus transformation (42). All four HBMEC cultures were ICAM-1 surface positive and expressed low CD36 surface levels on only a minor subpopulation of cells, as determined by live cell immunofluorescence (Fig. S3). In each case, the panned ItG-ICAM-1HBMEC and A4longHBMEC parasite lines had significantly greater binding activity and exhibited a more dispersed binding to HBMECs than the starting parasite populations (Fig. 3 and Fig. S4). Consequently, panning of IEs on brain endothelium broadened IE binding to all of the brain endothelial cells in the culture.
Fig. 3.
HBMEC-panned IEs adhere to brain microvascular endothelial cells from different patient sources. (A) Binding was compared between the starting (ItG-ICAM-1) and selected (ItG-ICAM-1HBMEC) parasite lines on HBMECs harvested from donors 13, 56, and 123 and a transformed HBMEC line (THBMEC) from donor 13. The anatomic origin of HBMEC cultures is illustrated in the brain schematic. (B) A DC8-var-expressing clonal parasite line derived from ItG-ICAM-1HBMEC bound to primary microvascular cells from heart, lung, and dermis.
Selection of Parasites on HBMECs Up-Regulates a Limited Subset of var Genes.
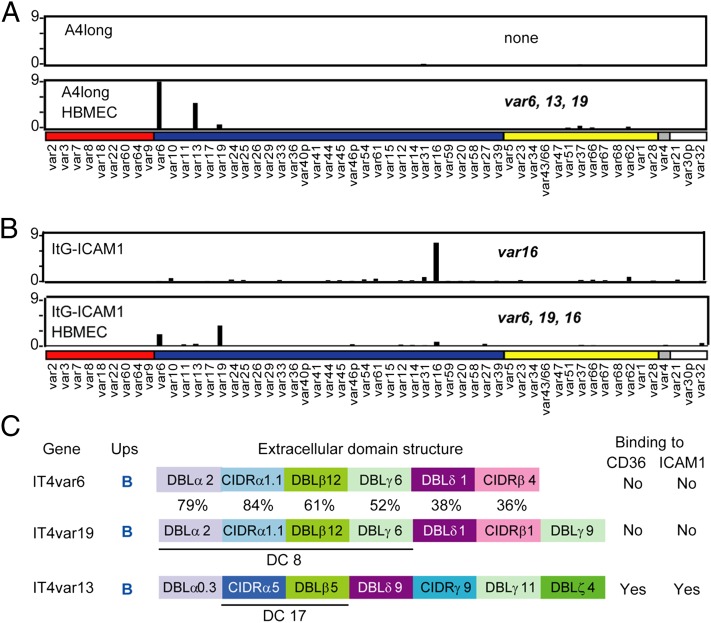
To investigate whether HBMEC panning selected for unique parasite adhesion traits, the parasite lines were characterized for var gene transcription by quantitative (Q)-RT-PCR with specific primers to family members (13). Before selection, the starting ItG-ICAM-1 predominantly transcribed IT4var16, but after panning the IT4var16 transcript was strongly decreased and two new var transcripts (It4var6 and IT4var19) were selected (Fig. 4B). Significantly, the same two transcripts and a third transcript (IT4var13) were highly increased in A4longHBMEC (Fig. 4A).
Fig. 4.
New var genes are up-regulated in parasite lines panned on primary HBMECs. (A and B) Transcription of var genes was compared from ring-stage parasites before and after panning on primary HBMECs (donor 13). Results were normalized to the housekeeping control gene adenylosuccinate lyase (asl). Genes are organized by Ups category, UpsA (red), UpsB (blue), UpsC (yellow), and UpsE (gray), as well as three genes for which the Ups type has not been determined (white). The names of parasite lines are indicated at the left, and var genes that expressed onefold or more of asl are indicated. (C) The extracellular domain architecture and predicted binding features of the three main genes up-regulated on HBMECs are shown. The DC8 cassette in IT4var6 and IT4var19 and the DC17 cassette in IT4var13 are underlined. The percentage of amino acid domain identity is shown for the two DC8-var products. Binding predictions are from repertoire-wide analysis of PfEMP1 recombinant proteins between CIDR-CD36 (14) and DBLβ-ICAM-1 (38).
Two of the selected genes, IT4var6 and IT4var19, belong to the group B/A (Fig. 4C) and were not predicted to bind to the host receptors CD36 or ICAM-1 (14, 38). Both encode a tandem domain cassette 8 (DC8) (DBLα2-CIDRα1.1–DBLβ12-DBLγ4/6) (11) and have higher sequence identity within this region (Fig. 4C). The third up-regulated var gene, ITvar13, has a domain cassette 17 (DC17) (CIDRα5-DBLβ5) that is predicted to bind both CD36 and ICAM-1 (14, 38). Consistent with these binding predictions (Fig. 4C), the A4longHBMEC parasite line (IT4var6hi, IT4var13hi, IT4var19lo) underwent a dramatic increase in ICAM-1 binding from the starting A4long parasite line (Fig. 2 A and B), likely due to up-regulation of the IT4var13 transcript (Fig. 4A). By comparison, ItG-ICAM-1HBMEC (IT4var6hi, IT4var19hi, IT4var16lo) was slightly decreased in CD36 and ICAM-1 binding from the starting parasite culture, as would be expected from the reduction of IT4var16 transcription and gain of two non-CD36/non–ICAM-1 binding var products. Moreover, both HBMEC panned parasite lines were less sensitive to inhibition with anti-CD36 or anti–ICAM-1 mAbs than the starting populations (Fig. 2C). The residual low level of antibody inhibition would be expected because each of the panned lines retained a subpopulation of parasites that encode ICAM-1 plus CD36 binding activity (IT4var13 and IT4var16, respectively).
Selection on HBMECs Enriches for Unique Parasite Adhesion Types.
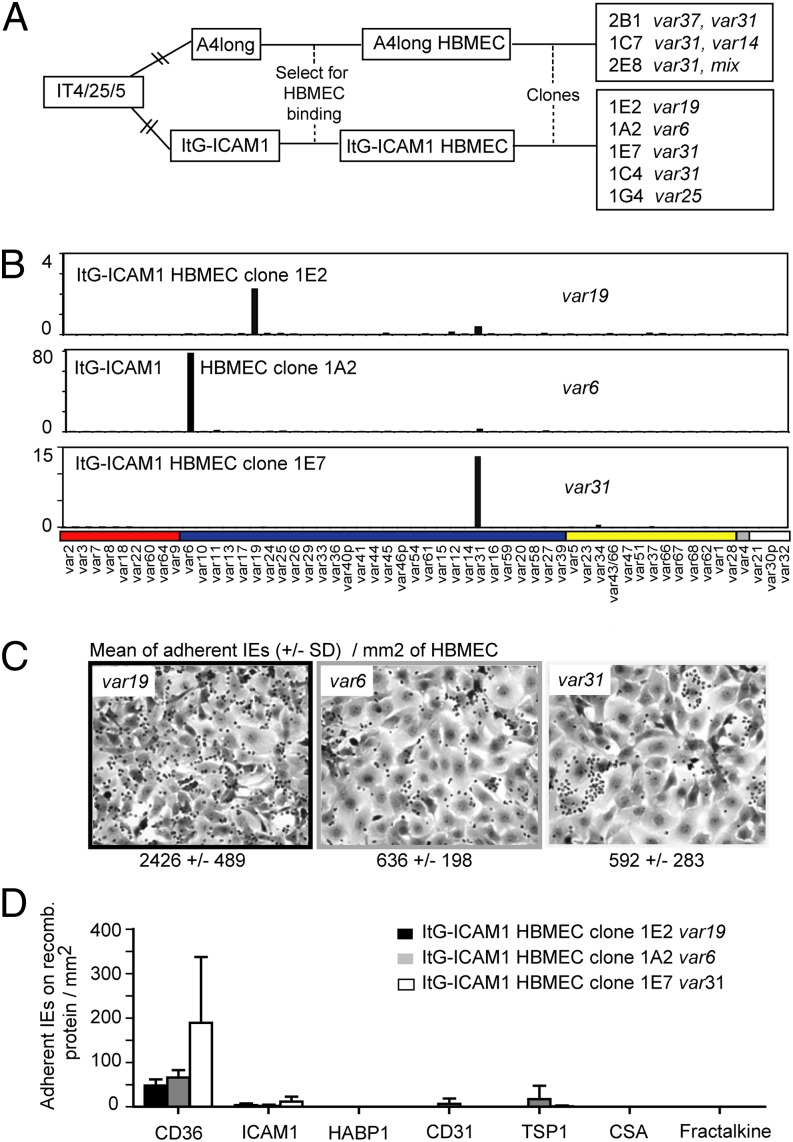
To further investigate cerebral binding interactions, HBMECs were analyzed by immunofluorescence assay for known cytoadhesion receptors other than CD36 and ICAM-1. All of the HBMEC cultures and the THBMEC line were positive for the endothelial cell markers, von Willebrand Factor (VWF), and CD31. In contrast, they were mostly negative for ICAM-2, endothelial-leukocyte adhesion molecule 1 (ELAM-1), vascular cell adhesion molecule 1 (VCAM-1), and the brain endothelial candidate CX3CL1/fractalkine (33) (Fig. S3). In addition, clonal parasite lines were isolated from A4longHBMEC and ItG-ICAM-1HBMEC by dilution cloning. The five parasites lines isolated from ItG-ICAM1HBMEC each expressed a unique predominant var transcript (IT4var19, IT4var6, IT4var25, or IT4var31, isolated twice) (Fig. 5 and Fig. S5). In contrast, the three parasite lines isolated from A4longHBMEC each expressed a mixture of var transcripts including IT4var31 (Fig. 5 and Fig. S5), which has also been observed to be a frequent switch event in previous limited dilution clonings (13). A clonal parasite line expressing IT4var13 was not isolated.
Fig. 5.
Cloned parasite lines expressing IT4var6 or IT4var19 var products were weak CD36 binders and did not bind ICAM-1. (A) A panel of eight clonal parasite lines was generated from A4longHBMEC and ItG-ICAM-1HBMEC, using limited dilution cloning. Five parasite lines from the ItG-ICAM1HBMEC line express a unique predominant var transcript IT4var19, IT4var6, IT4var31, or IT4var25, whereas three parasite lines from A4longHBMEC express a mixture of var transcripts including the frequent-switch event IT4var31. (B) Profiling of var transcription, performed by Q-RT-PCR, for the clonal parasite lines expressing IT4var6, IT4var19, or IT4var31. (C) Parasite lines expressing IT4var19 and IT4var6 exhibit the dispersed HBMEC binding of the parental line, whereas the parasite line expressing IT4var31 binds to only a subpopulation of HBMECs. (D) Parasite lines were compared for binding to recombinant proteins. Results are expressed as means ± SDs from two independent experiments.
In HBMEC binding assays the IT4var19-expressing clonal line had higher binding activity than the IT4var6-expressing clonal line, but both DC8-var parasites displayed a dispersed binding pattern similar to that of the HBMEC-panned parental line (Fig. 5C). In contrast, cloned CD36-binding parasite lines expressing IT4var31, IT4var25, or a mixture of IT4var14 and IT4var31 all bound in a punctate fashion to a subpopulation of HBMECs (Fig. 5C and Fig. S5). In recombinant protein binding assays, the IT4var6 and IT4var19 parasite clones exhibited little or no adhesion to CD36 or other known host cytoadhesion receptors (Fig. 5D), including the three brain endothelial candidate receptors, ICAM-1, HABP1/gC1qR, or Fractalkine (30, 32, 33). They also did not form rosettes with uninfected red blood cells or autoagglutinate. Although the selected A4HBMEC parasite line exhibited a slight increase in CHO-ELAM-1 binding (Fig. 2A), this was not observed in the ItG-ICAM-1HBMEC parasite line that primarily expressed the two DC8-var genes (IT4var6 and IT4var19) (Figs. 2 and 4).
Some PfEMP1 variants adhere to heparin sulfate (HS)-like glycosaminoglycans on endothelial cells or red blood cells (43–46). To investigate the role of glycans in cerebral binding, HBMECs were pretreated with chondroitinase ABC (Case ABC), neuraminidase, hyaluronidase, or heparinase III. Whereas neuraminidase pretreatment slightly increased binding of A4longHBMEC and ItG-ICAM-1HBMEC parasite lines and hyaluronidase pretreatment slightly increased binding of ItG-ICAM-1HBMEC, none of these treatments reduced IE binding (Fig. S6). Taken together, HBMEC selection up-regulated a subfamily of DC8-var–encoded products that were not dependent on the primary microvascular receptor CD36 or on ICAM-1 or other established receptors, suggesting a unique host receptor(s) is involved in cerebral endothelial binding.
DC8-var–Encoded Proteins Bind Brain Endothelium and Can Inhibit IE Binding.
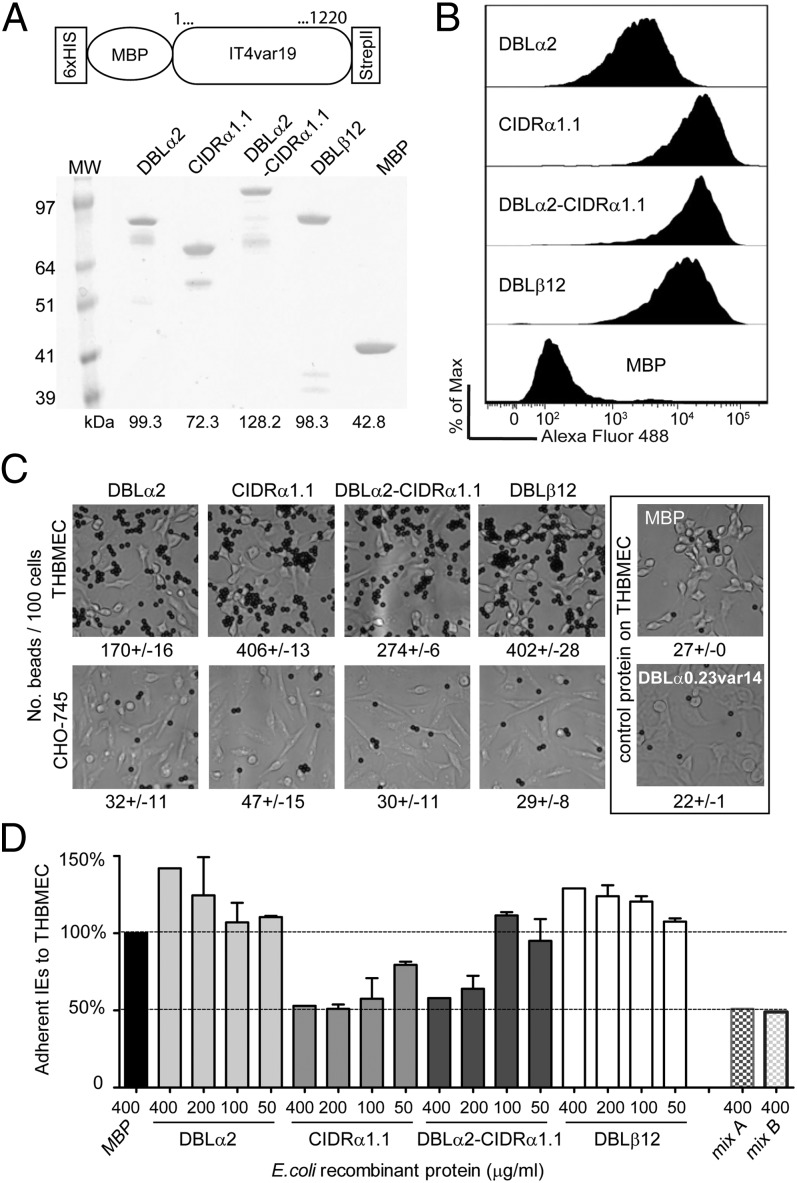
To investigate the role of the DC8 cassette in cerebral binding, the first three individual domains of the IT4var19 DC8 cassette (NTS-DBLα2, CIDRα1.1, and DBLβ12) were produced as MBP-fusion proteins, as well as an NTS-DBLα2-CIDRα1.1 “head structure” recombinant protein. As controls, an MBP-alone recombinant protein was made and an IT4var14 NTS-DBLα recombinant protein was generated from the A4ultra parasite that exhibited a punctate binding pattern (Fig. S1). All recombinant proteins migrated at the expected molecular weight in SDS/PAGE gels (Fig. 6A).
Fig. 6.
DC8-var19–encoded recombinant proteins exhibit binding capacity for brain endothelium. (A) Schematic of the protein construct. Protein boundaries are the following: DBLα2, M1-V484; CIDRα1.1, C485-C732; DBLα2-CIDRα1.1, M1-C732; and DBLβ12, P733-C1220. Proteins were analyzed on SDS/PAGE gel and visualized with GelBlue code. (B) Recombinant proteins binding to unfixed THBMEC cells were determined by flow cytometry. Recognition via the anti-strepII tag antibody tag is shown. (C) Protein-coupled Dynal Bead binding assays to THBMEC cells or CHO-745 cells as a negative control. Results are expressed as mean of beads binding per 100 cells ± SDs. (D) Binding of the IT4var19-expressing parasite clone (1E2) to transformed HBMECs in the presence of each of the E. coli fusion recombinant proteins, from 0.4 mg/mL to 0.05 mg/mL final concentration. Mix A corresponds to 0.4 mg/mL of three single domains DBLα2, CIDRα1.1, and DBLβ12 combined and mix B to 0.4 mg/mL of the tandem domain plus the DBLβ12 domain combined. The percentage of binding is expressed relative to binding in the presence of control protein MBP. Results are expressed as mean ± SDs from two independent experiments.
All four IT4var19 recombinant proteins bound to the THBMEC line, both in a flow cytometry assay (Fig. 6B) and after coating onto Dynal beads (Fig. 6C). In contrast, neither MBP alone nor the negative control IT4var14 NTS-DBLα recombinant protein bound to THBMECs, and none of the IT4var recombinant proteins bound to a negative control CHO-745 cell line (Fig. 6C). In addition, both the CIDRα1.1 recombinant protein and the NTS-DBLα2-CIDRα1.1 were able to inhibit binding of P. falciparum-IEs to brain endothelial cells by up to 50% in a dose-dependent manner (Fig. 6D). Binding inhibition was not increased beyond 50% when the three single domains (DBLα2, CIDRα1.1, and DBLβ12) or the protein head structure plus DBLβ12 domain were combined (Fig. 6D). This analysis suggests that multiple N-terminal domains in IT4var19 encode binding activity for brain endothelial cells and the CIDRα1.1 domain confers a critical IE-binding interaction.
DC8-var–Encoded Products Are Not Restricted to Brain Endothelium.
It is paradoxical that the parasite encodes potentially deadly adhesive properties, such as cerebral binding, which could reduce parasite transmission. To investigate whether DC8-var–encoded products may have alternative sites of sequestration, parasites were examined for binding to microvascular endothelial cells from different organs/tissues. Compared with the starting parasite populations, both of the HBMEC-panned parasites exhibited increased and more dispersed binding to primary human dermal microvascular endothelial cells (HDMECs) (Fig. S4). Furthermore, the DC8-var–expressing IT4var19 clonal parasite line also bound avidly to primary microvascular endothelial cells from dermis, lung, or heart (Fig. 3), confirming that expression of this unique adhesion receptor(s) is not limited to brain endothelium.
HBMEC-Panned Parasite Isolates Are Commonly Recognized by Malaria Endemic Plasma from Young Children.
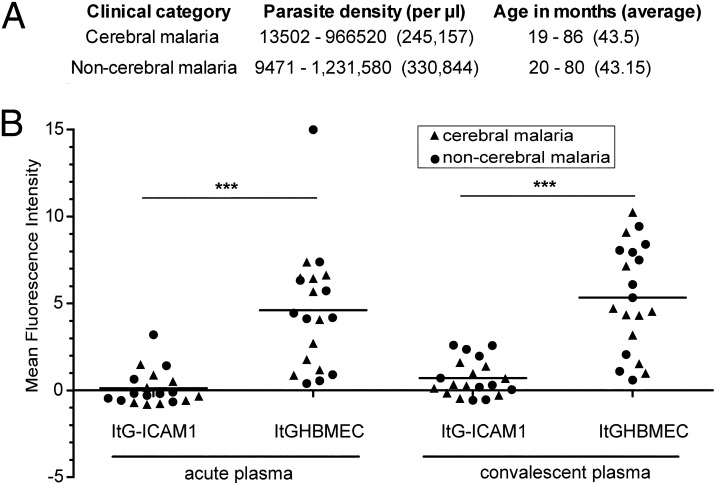
To investigate whether young African children are exposed to related parasite variants, antibody reactivity was compared between the prepanned and the HBMEC-panned parasite lines. Plasma was compared from children with cerebral malaria (CM) or age-matched children with a non-CM episode collected at the time of hospital arrival (acute sample) or 3–4 wk after hospitalization (convalescent sample). This analysis showed that panned ItG-ICAM-1HBMEC was better recognized by acute and convalescent plasma samples than the starting highly homogeneous ItG-ICAM-1 parasite line [Fig. 7; acute sample ItG-ICAM-1, median = 0, interquartile range (IQR) = 0–3.2; acute sample ItG-ICAM-1HBMEC, median = 4.3, IQR = 0.4–15.0; convalescent sample, ItG-ICAM-1, median = 0.3, IQR = 0–2.6; convalescent sample ItG-ICAM-1HBMEC, median = 5.0, IQR = 0.6–10.3]. Of interest, HBMEC-panned parasite lines were equally well recognized by plasma from both the CM and the non-CM children groups and antibody reactivity increased in convalescent plasma of specific individuals for both groups (Fig. 7). Previous studies have found a general boost in antimalaria antibodies accompanying malaria infections (47). These results suggest that young African children are exposed to parasites that are antigenically related to the HBMEC-panned parasite lines, but this exposure is not necessarily associated with a CM episode.
Fig. 7.
Endemic sera recognition of HBMEC-panned parasite lines. (A) Acute and convalescent sera pairs were collected from 10 children with cerebral malaria and 10 age-matched children with nonsevere malaria. (B) Surface recognition of infected erythrocytes was analyzed by flow cytometry. The mean fluorescence intensity (MFI) of uninfected erythrocytes was subtracted from the MFI of the infected erythrocyte population to give a specific MFI of the infected erythrocytes. The specific MFI reported in the graph was further corrected by subtracting antibody reactivity of a nonimmune European control plasma, which had an MFI of 1 on ItG-ICAM-1HBMEC and no reactivity on ItG-ICAM-1. Antibody reactivity between starting and selected parasite lines was significantly different at the time of acute disease and at convalescence (***P < 0.001, Mann–Whitney test).
Discussion
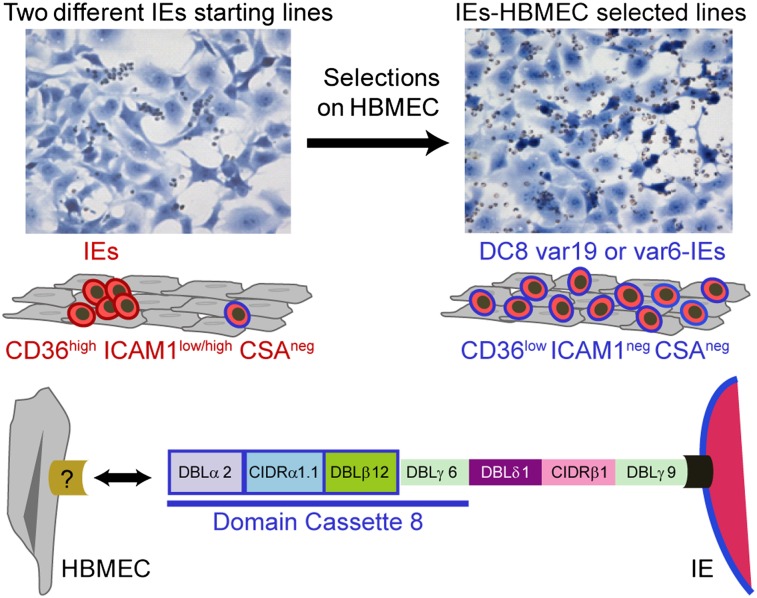
A hallmark of CM is sequestration of P. falciparum-IEs in cerebral microvasculature (48–50), but there is still limited understanding of how IEs sequester in brain. In this study, we show that CD36- and CSA-binding parasites adhere in low numbers to primary HBMECs and that binding of three different CD36-binding plus ICAM-1–binding parasite lines was largely restricted to a subpopulation of HBMECs and highly dependent on CD36. Panning of IEs on HBMECs led to a more dispersed binding phenotype and the up-regulation of two unique var genes associated with CD36low/ICAM-1−–binding parasite variants and a unique CD36+/ICAM-1+–binding variant. This analysis indicates that IEs were not dependent on CD36 for adhesion to primary HBMECs and that ICAM-1 may be one factor in cerebral homing, but ICAM-1 is not required for broadly dispersed binding to all HBMECs. Of interest, the unique host receptor(s) involved in HBMEC binding were not restricted to cerebral vasculature and were also present on primary microvascular cells from dermis, heart, and lung.
Two of the three var transcripts selected on brain endothelium (IT4var6 and IT4var19) encode a unique tandem domain cassette called the DC8 (11). Multiple N-terminal domains in the DC8 cassette encoded binding activity for brain endothelial cells and the CIDR domain in this cassette could inhibit binding of P. falciparum-IEs by 50%, highlighting a critical role for the DC8 cassette in cerebral binding. The DC8 cassette is usually found in zero to four var copies per parasite genotype and is typically associated with an UpsB promoter, although sometimes it is associated with an UpsA promoter (11). The fact that most DC8 var genes have mixed features (UpsB promoter and group A coding features) could limit gene recombination with other group A or group B genes and favor the evolution of specialized adhesion properties within group B/A genes. Significantly, DC8 var products tend to be among the first PfEMP1s expressed in early childhood infections (15, 16), indicating they may encode unique adhesion properties that confer a growth advantage in malaria-naive children. This study demonstrates that DC8 var products can encode binding activity for brain endothelium.
It has been a paradox how deadly adhesive traits, such as cerebral binding, are maintained in the parasite population. In the accompanying articles by Claessens et al. (51) and Lavstsen et al. (52), they found that DC8 and DC13 var products from different parasite strains were enriched on immortalized brain endothelial cells and that DC8 var products were enriched in children with CM, hyperparasitemia, or severe malaria anemia. Taken together, these findings suggest that DC8 var products may be more broadly associated with severe malaria, possibly because of their ability to bind to diverse endothelium. We hypothesize that although DC8 var products have high binding activity for brain endothelium, they are likely not under evolutionary selective pressure to bind in brain because it can lead to lethal infections. Instead, DC8 variants must have other attributes that favor parasite growth in individuals with limited malaria immunity, and they may be under even greater selection for a nonbrain endothelial niche, which maintains them in the parasite population. Thus, unlike placental binding variants, which cause limited deaths through acute severe disease in pregnant women in high malaria transmission regions and have their major impact on the developing fetus (53), there must be sufficient transmission advantage to the parasite that it maintains potentially deadly adhesion properties in DC8 var products (e.g., cerebral binding), even though some people may die without transmitting the parasite. If cerebral binding variants have alternative sequestration sites, this result could explain how young children can rapidly develop immunity against this rare subset, even in the absence of cerebral malaria symptoms. It is also consistent with seroepidemiological studies showing that severe malaria isolates are relatively antigenically restricted and commonly recognized by plasma from young African children (54–56), even though severe malaria infections are relatively rare. We found that young African children with CM or nonsevere control cases had antibodies to HBMEC-selected parasites, indicating they had been exposed to related variants during childhood infections. Antibody reactivity differed even within the CM group, indicating that although the variant surface antigens associated with CM may be restricted (54–56), they may still constitute more than a discrete handful of serotypes.
In conclusion, this study suggests a limited subset of var types in a given parasite repertoire is responsible for cerebral binding and has important implications for the molecular basis of CM pathogenesis and natural immunity to severe malaria.
Materials and Methods
Parasite.
P. falciparum parasites were cultured under standard conditions (13) using human O red blood cells (RBCs) in RPMI-1640 medium (Invitrogen), except that parasites were grown in human A red blood cells for rosetting assays (SI Materials and Methods). Cloning by limited dilution of the parasite lines was done as previously described (13).
Human Plasma.
Plasma samples were selected from children attending Kilifi District Hospital with varying levels of malaria severity and who had been recruited as study participants in the Kemri–Wellcome Trust study of integrated development of natural immunity to malaria in children in Kilifi District between 2006 and 2010. Ethical approval for this study was granted by the Kenya Medical Research Institute Ethical Review Committee and informed consent obtained from the parents/guardians of all study participants. CM was defined as malaria with a Blantyre coma score ≤2 and without any sign of respiratory distress and/or severe malaria anemia. Each CM sample was matched to a nonsevere control by age, date of admission, and blood group. The nonsevere controls were defined as children who were either seen in the outpatient department and required no admission or children who were admitted but did not develop any signs of severe disease throughout their admission. Exclusion criteria were a positive blood or cerebrospinal fluid culture or hypoglycemia. An acute plasma sample was collected at the time of admission and the convalescent sample was collected 3–4 wk later.
Endothelial Cell Cultures.
Primary HBMECs were isolated from human brain cortex by surgical resection during surgery for seizure disorders and purified by cloning via fluorescence-activated cell sorting as previously described (57) or by selection using magnetic anti-CD31 beads (Dynal; Invitrogen) according to manufacturer’s instructions (SI Materials and Methods). Primary dermal, lung, or heart microvascular endothelial cells were purchased from ScienCell and cultured according to manufacturer’s recommendation between passages 1 and 5.
Characterization and Phenotypic Analyses of Endothelial Cell Lines.
Human endothelial cell cultures were characterized by the immunofluorescence method with antibodies to vWF/Factor VIII (1/40; Dako A0082) on methanol-fixed cells and by anti–CD31-PE conjugated (PECAM) (1/5; Molecular Probes) on live cells. For TNF-α stimulation, 10 ng/mL of TNF-α (Sigma; T0157) was added for 24 h at 37 °C before analysis. Live cells were labeled with antibodies against ICAM-1, VCAM-1, CD36, E-selectin/CD62E, and ICAM-2 (SI Materials and Methods).
Selection of IEs on HBMECs.
A collagen-coated flask was seeded with HBMECs to reach confluency on the day of the experiment. The culture medium was removed and HBMEC cells were overlaid with either A4long or ItG-ICAM-1 trophozoite-stage gelatin-enriched IEs. After 1 h of incubation, unbound erythrocytes were removed by several gentle washes. The remaining cells were covered with 5 mL of complete Plasmodium culture medium containing 500 μL of 5% hematocrit RBCs and incubated overnight at 37 °C. The next day, the parasite culture was resuspended and transferred to a new flask under standard P. falciparum culture conditions. HBMEC selection was repeated twice, allowing four to five cycles of growth between pannings.
Binding Assays on Human Endothelial Cells.
Endothelial cells were seeded on coated four- or eight-well slides (BD Biocoat) and allowed to grow for 3–4 d before the binding assays to achieve confluency. For TNF-α stimulation, 10 ng/mL of TNF-α (Sigma; T0157) was added to the confluent monolayers of endothelial cells for 24 h before analysis. For IE binding-inhibition assays, HBMECs were preincubated with monoclonal antibodies to CD36 or ICAM-1 or with IT4var19 PfEMP1 recombinant proteins. For PfEMP1 binding assays, IT4var19 recombinant proteins were coated onto Dynal beads and added to an HBMEC monolayer. Alternatively, HBMECs were lifted with 8 mM EDTA and then incubated with monomeric recombinant proteins before visualizing binding by flow cytometry (SI Materials and Methods).
IE Binding Assays to Recombinant Protein or Transfected CHO Cell Lines.
Binding assays to CHO-K1 (CSA surface positive), CHO745 (CSA surface negative), and CHO745 transfectants expressing CD36, ICAM-1, E-selectin, or VCAM-1 were performed as previously described (13). For recombinant protein assays, 10-μL spots at 50 μg/mL of CD36-Fc, ICAM-1-Fc, HABP1/gC1qR-6×-HIS, CX3CL1/Fractalkine-6×-HIS, CD31, TSP-1-10× HIS (R&D Systems), or CSA (Sigma) were applied to bacterial Petri dishes as described (13). Binding was quantified by determining the number of IEs adhering per square millimeter in four random fields under 400× magnification.
Determination of var Transcription by Q-RT-PCR.
The var gene transcription profiles were performed using a set of gene-specific primers to the IT4 var repertoire (13). In brief, RNA was extracted in TRIzol LS (Invitrogen) from ring stage parasites at ∼6–12 h postinvasion. Two micrograms of starting total RNA was used to compare the full set of primers. Quantitative real-time PCR reactions were performed on an ABI Prism 7500 thermocycler and the relative transcription was determined by normalization to the control housekeeping gene adenylosuccinate lyase (ASL) (PFB0295w).
Production of DC8-var19–Encoded Recombinant Proteins.
His6-MBP-StrepII–tagged fusion recombinant proteins from IT4var19 (GenBank accession no. EF158075) or IT4var14 (A4var; L42244.1) were produced in Escherichia coli pSHuffle Express (NEB) expression hosts. Inserts were cloned using the Gateway destination vector technique as described in ref. 58 (SI Materials and Methods).
Serological Reactivity of Endemic Sera.
Flow cytometry was performed on P. falciparum-IEs using a previously published procedure (59) (SI Materials and Methods). To adjust for background signal, the mean fluorescence intensity (MFI) of the uninfected cells was subtracted from that of the infected cells to give an adjusted MFI. Further adjustment was performed for all patient plasma by subtracting the median adjusted MFI of a European plasma performed in triplicate against each parasite line.
Statistical Analyses.
Statistical analyses were performed with Prism Software. IE binding data were analyzed by the Krustal–Wallis one-way ANOVA test and endemic sera reactivity by a Mann–Whitney test.
Supplementary Material
Acknowledgments
This paper was submitted with the permission of the director of Kenya Medical Research Institute. Funding for this work was provided by the National Institutes of Health (R01 AI47953 to J.D.S.), the Bloomberg Foundation as Sponsor of the Johns Hopkins Malaria Research Institute (A.K.T., D.J.S., and M.F.S.), the Wellcome Trust Programme Grants (084535 and 077092, to P.C.B.), and a Wellcome Trust Strategic Award (084538 to C.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 10158 (volume 109, number 26).
See Commentary on page 10130.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120534109/-/DCSupplemental.
References
- 1.Baruch DI, et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 2.Smith JD, et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su XZ, et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 4.Biggs BA, et al. Adherence of infected erythrocytes to venular endothelium selects for antigenic variants of Plasmodium falciparum. J Immunol. 1992;149:2047–2054. [PubMed] [Google Scholar]
- 5.Roberts DJ, et al. Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature. 1992;357:689–692. doi: 10.1038/357689a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 7.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kraemer SM, Smith JD. Evidence for the importance of genetic structuring to the structural and functional specialization of the Plasmodium falciparum var gene family. Mol Microbiol. 2003;50:1527–1538. doi: 10.1046/j.1365-2958.2003.03814.x. [DOI] [PubMed] [Google Scholar]
- 9.Lavstsen T, Salanti A, Jensen AT, Arnot DE, Theander TG. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malar J. 2003;2:27. doi: 10.1186/1475-2875-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraemer SM, et al. Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: Comparisons of geographically diverse isolates. BMC Genomics. 2007;8:45. doi: 10.1186/1471-2164-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rask TS, Hansen DA, Theander TG, Gorm Pedersen A, Lavstsen T. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes—divide and conquer. PLoS Comput Biol. 2010;6:6. doi: 10.1371/journal.pcbi.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraemer SM, Smith JD. A family affair: var genes, PfEMP1 binding, and malaria disease. Curr Opin Microbiol. 2006;9:374–380. doi: 10.1016/j.mib.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Janes JH, et al. Investigating the host binding signature on the Plasmodium falciparum PfEMP1 protein family. PLoS Pathog. 2011;7:e1002032. doi: 10.1371/journal.ppat.1002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson BA, Welch TL, Smith JD. Widespread functional specialization of Plasmodium falciparum erythrocyte membrane protein 1 family members to bind CD36 analysed across a parasite genome. Mol Microbiol. 2003;47:1265–1278. doi: 10.1046/j.1365-2958.2003.03378.x. [DOI] [PubMed] [Google Scholar]
- 15.Cham GK, et al. Sequential, ordered acquisition of antibodies to Plasmodium falciparum erythrocyte membrane protein 1 domains. J Immunol. 2009;183:3356–3363. doi: 10.4049/jimmunol.0901331. [DOI] [PubMed] [Google Scholar]
- 16.Cham GK, et al. Hierarchical, domain type-specific acquisition of antibodies to Plasmodium falciparum erythrocyte membrane protein 1 in Tanzanian children. Infect Immun. 2010;78:4653–4659. doi: 10.1128/IAI.00593-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen AT, et al. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J Exp Med. 2004;199:1179–1190. doi: 10.1084/jem.20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warimwe GM, et al. Plasmodium falciparum var gene expression is modified by host immunity. Proc Natl Acad Sci USA. 2009;106:21801–21806. doi: 10.1073/pnas.0907590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falk N, et al. Analysis of Plasmodium falciparum var genes expressed in children from Papua New Guinea. J Infect Dis. 2009;200:347–356. doi: 10.1086/600071. [DOI] [PubMed] [Google Scholar]
- 20.Kyriacou HM, et al. Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia. Mol Biochem Parasitol. 2006;150:211–218. doi: 10.1016/j.molbiopara.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rottmann M, et al. Differential expression of var gene groups is associated with morbidity caused by Plasmodium falciparum infection in Tanzanian children. Infect Immun. 2006;74:3904–3911. doi: 10.1128/IAI.02073-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fried M, Duffy PE. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science. 1996;272:1502–1504. doi: 10.1126/science.272.5267.1502. [DOI] [PubMed] [Google Scholar]
- 23.Salanti A, et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 24.Salanti A, et al. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J Exp Med. 2004;200:1197–1203. doi: 10.1084/jem.20041579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montgomery J, et al. Differential var gene expression in the organs of patients dying of falciparum malaria. Mol Microbiol. 2007;65:959–967. doi: 10.1111/j.1365-2958.2007.05837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowe J, Claessens A, Corrigan R, Arman M. Adhesion of Plasmodium falciparum-infected erythrocytes to human cells: Molecular mechanisms and therapeutic implications. Exp Rev Mol Med. 2009;11:e16. doi: 10.1017/S1462399409001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooke BM, et al. Rolling and stationary cytoadhesion of red blood cells parasitized by Plasmodium falciparum: Separate roles for ICAM-1, CD36 and thrombospondin. Br J Haematol. 1994;87:162–170. doi: 10.1111/j.1365-2141.1994.tb04887.x. [DOI] [PubMed] [Google Scholar]
- 28.Gray C, McCormick C, Turner G, Craig A. ICAM-1 can play a major role in mediating P. falciparum adhesion to endothelium under flow. Mol Biochem Parasitol. 2003;128:187–193. doi: 10.1016/s0166-6851(03)00075-6. [DOI] [PubMed] [Google Scholar]
- 29.Ho M, Hickey MJ, Murray AG, Andonegui G, Kubes P. Visualization of Plasmodium falciparum-endothelium interactions in human microvasculature: Mimicry of leukocyte recruitment. J Exp Med. 2000;192:1205–1211. doi: 10.1084/jem.192.8.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner GD, et al. An immunohistochemical study of the pathology of fatal malaria. Evidence for widespread endothelial activation and a potential role for intercellular adhesion molecule-1 in cerebral sequestration. Am J Pathol. 1994;145:1057–1069. [PMC free article] [PubMed] [Google Scholar]
- 31.Wassmer SC, et al. Vascular endothelial cells cultured from patients with cerebral or uncomplicated malaria exhibit differential reactivity to TNF. Cell Microbiol. 2011;13:198–209. doi: 10.1111/j.1462-5822.2010.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biswas AK, et al. Plasmodium falciparum uses gC1qR/HABP1/p32 as a receptor to bind to vascular endothelium and for platelet-mediated clumping. PLoS Pathog. 2007;3:1271–1280. doi: 10.1371/journal.ppat.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatabu T, Kawazu S, Aikawa M, Kano S. Binding of Plasmodium falciparum-infected erythrocytes to the membrane-bound form of Fractalkine/CX3CL1. Proc Natl Acad Sci USA. 2003;100:15942–15946. doi: 10.1073/pnas.2534560100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayor A, et al. Association of severe malaria outcomes with platelet-mediated clumping and adhesion to a novel host receptor. PLoS ONE. 2011;6:e19422. doi: 10.1371/journal.pone.0019422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newbold C, et al. Receptor-specific adhesion and clinical disease in Plasmodium falciparum. Am J Trop Med Hyg. 1997;57:389–398. doi: 10.4269/ajtmh.1997.57.389. [DOI] [PubMed] [Google Scholar]
- 36.Ochola LB, et al. Specific receptor usage in Plasmodium falciparum cytoadherence is associated with disease outcome. PLoS ONE. 2011;6:e14741. doi: 10.1371/journal.pone.0014741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogerson SJ, et al. Cytoadherence characteristics of Plasmodium falciparum-infected erythrocytes from Malawian children with severe and uncomplicated malaria. Am J Trop Med Hyg. 1999;61:467–472. doi: 10.4269/ajtmh.1999.61.467. [DOI] [PubMed] [Google Scholar]
- 38.Howell DP, et al. Mapping a common interaction site used by Plasmodium falciparum Duffy binding-like domains to bind diverse host receptors. Mol Microbiol. 2008;67:78–87. doi: 10.1111/j.1365-2958.2007.06019.x. [DOI] [PubMed] [Google Scholar]
- 39.Tripathi AK, Sullivan DJ, Stins MF. Plasmodium falciparum-infected erythrocytes increase intercellular adhesion molecule 1 expression on brain endothelium through NF-kappaB. Infect Immun. 2006;74:3262–3270. doi: 10.1128/IAI.01625-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tripathi AK, Sullivan DJ, Stins MF. Plasmodium falciparum-infected erythrocytes decrease the integrity of human blood-brain barrier endothelial cell monolayers. J Infect Dis. 2007;195:942–950. doi: 10.1086/512083. [DOI] [PubMed] [Google Scholar]
- 41.Tripathi AK, Sha W, Shulaev V, Stins MF, Sullivan DJ., Jr Plasmodium falciparum-infected erythrocytes induce NF-kappaB regulated inflammatory pathways in human cerebral endothelium. Blood. 2009;114:4243–4252. doi: 10.1182/blood-2009-06-226415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stins MF, Badger J, Sik Kim K. Bacterial invasion and transcytosis in transfected human brain microvascular endothelial cells. Microb Pathog. 2001;30:19–28. doi: 10.1006/mpat.2000.0406. [DOI] [PubMed] [Google Scholar]
- 43.Carlson J, et al. Disruption of Plasmodium falciparum erythrocyte rosettes by standard heparin and heparin devoid of anticoagulant activity. Am J Trop Med Hyg. 1992;46:595–602. doi: 10.4269/ajtmh.1992.46.595. [DOI] [PubMed] [Google Scholar]
- 44.Rowe A, Berendt AR, Marsh K, Newbold CI. Plasmodium falciparum: A family of sulphated glycoconjugates disrupts erythrocyte rosettes. Exp Parasitol. 1994;79:506–516. doi: 10.1006/expr.1994.1111. [DOI] [PubMed] [Google Scholar]
- 45.Vogt AM, et al. Heparan sulfate on endothelial cells mediates the binding of Plasmodium falciparum-infected erythrocytes via the DBL1alpha domain of PfEMP1. Blood. 2003;101:2405–2411. doi: 10.1182/blood-2002-07-2016. [DOI] [PubMed] [Google Scholar]
- 46.Vogt AM, Winter G, Wahlgren M, Spillmann D. Heparan sulphate identified on human erythrocytes: A Plasmodium falciparum receptor. Biochem J. 2004;381:593–597. doi: 10.1042/BJ20040762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kinyanjui SM, Mwangi T, Bull PC, Newbold CI, Marsh K. Protection against clinical malaria by heterologous immunoglobulin G antibodies against malaria-infected erythrocyte variant surface antigens requires interaction with asymptomatic infections. J Infect Dis. 2004;190:1527–1533. doi: 10.1086/424675. [DOI] [PubMed] [Google Scholar]
- 48.MacPherson GG, Warrell MJ, White NJ, Looareesuwan S, Warrell DA. Human cerebral malaria. A quantitative ultrastructural analysis of parasitized erythrocyte sequestration. Am J Pathol. 1985;119:385–401. [PMC free article] [PubMed] [Google Scholar]
- 49.Pongponratn E, Riganti M, Punpoowong B, Aikawa M. Microvascular sequestration of parasitized erythrocytes in human falciparum malaria: A pathological study. Am J Trop Med Hyg. 1991;44:168–175. doi: 10.4269/ajtmh.1991.44.168. [DOI] [PubMed] [Google Scholar]
- 50.Taylor TE, et al. Differentiating the pathologies of cerebral malaria by postmortem parasite counts. Nat Med. 2004;10:143–145. doi: 10.1038/nm986. [DOI] [PubMed] [Google Scholar]
- 51.Claessens A, et al. A subset of group A-like var genes encode the malaria parasite ligands for binding to human brain endothelial cells. Proc Natl Acad Sci USA. 2012;109:E1772–E1781. doi: 10.1073/pnas.1120461109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lavstsen T, et al. Plasmodium falciparum erythrocyte membrane protein 1 domain cassettes 8 and 13 are associated with severe malaria in children. Proc Natl Acad Sci USA. 2012;109:E1791–E1800. doi: 10.1073/pnas.1120455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brabin BJ, et al. The sick placenta-the role of malaria. Placenta. 2004;25:359–378. doi: 10.1016/j.placenta.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 54.Bull PC, Lowe BS, Kortok M, Marsh K. Antibody recognition of Plasmodium falciparum erythrocyte surface antigens in Kenya: Evidence for rare and prevalent variants. Infect Immun. 1999;67:733–739. doi: 10.1128/iai.67.2.733-739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bull PC, et al. Plasmodium falciparum-infected erythrocytes: agglutination by diverse Kenyan plasma is associated with severe disease and young host age. J Infect Dis. 2000;182:252–259. doi: 10.1086/315652. [DOI] [PubMed] [Google Scholar]
- 56.Nielsen MA, et al. Plasmodium falciparum variant surface antigen expression varies between isolates causing severe and nonsevere malaria and is modified by acquired immunity. J Immunol. 2002;168:3444–3450. doi: 10.4049/jimmunol.168.7.3444. [DOI] [PubMed] [Google Scholar]
- 57.Stins MF, Gilles F, Kim KS. Selective expression of adhesion molecules on human brain microvascular endothelial cells. J Neuroimmunol. 1997;76:81–90. doi: 10.1016/s0165-5728(97)00036-2. [DOI] [PubMed] [Google Scholar]
- 58.Nallamsetty S, Austin BP, Penrose KJ, Waugh DS. Gateway vectors for the production of combinatorially-tagged His6-MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli. Protein Sci. 2005;14:2964–2971. doi: 10.1110/ps.051718605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bejon P, et al. Analysis of immunity to febrile malaria in children that distinguishes immunity from lack of exposure. Infect Immun. 2009;77:1917–1923. doi: 10.1128/IAI.01358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]