Abstract
The way nature evolves and sculpts materials using proteins inspires new approaches to materials engineering but is still not completely understood. Here, we present a cell-free synthetic biological platform to advance studies of biologically synthesized solid-state materials. This platform is capable of simultaneously exerting many of the hierarchical levels of control found in natural biomineralization, including genetic, chemical, spatial, structural, and morphological control, while supporting the evolutionary selection of new mineralizing proteins and the corresponding genetically encoded materials that they produce. DNA-directed protein expression and enzymatic mineralization occur on polystyrene microbeads in water-in-oil emulsions, yielding synthetic surrogates of biomineralizing cells that are then screened by flow sorting, with light-scattering signals used to sort the resulting mineralized composites differentially. We demonstrate the utility of this platform by evolutionarily selecting newly identified silicateins, biomineralizing enzymes previously identified from the silica skeleton of a marine sponge, for enzyme variants capable of synthesizing silicon dioxide (silica) or titanium dioxide (titania) composites. Mineral composites of intermediate strength are preferentially selected to remain intact for identification during cell sorting, and then to collapse postsorting to expose the encoding genes for enzymatic DNA amplification. Some of the newly selected silicatein variants catalyze the formation of crystalline silicates, whereas the parent silicateins lack this ability. The demonstrated bioengineered route to previously undescribed materials introduces in vitro enzyme selection as a viable strategy for mimicking genetic evolution of materials as it occurs in nature.
Keywords: directed evolution, in vitro compartmentalization, DNA shuffling, metal oxide nanoparticles, self-assembly
Although often treated as conceptually distinct, organic and inorganic systems have always been intimately connected in biology. This vital relationship likely originated at reactive mineral surfaces, where many believe that life’s first molecules were formed (1), and persisted through the emergence of ancient unicellular organisms able to process or synthesize minerals (2). The diverse evolutionary products of those initial simple systems, such as bones, teeth, and shells, serve as inspiration for engineers interested in developing new and environmentally benign routes for producing technological materials (3–8). Bioinspired materials engineering also offers the prospects of both combinatorial searches for optimizing material performance (9) and genetic control over biomolecular-mineral interfaces (6, 8, 10, 11). These prospects and recent research suggest that biology and minerals are poised to form new intimate connections now in the realm of human technology. Progress toward this hybrid outcome is highlighted by the recent use of semiconductor arrays to sequence a human genome directly and ultrarapidly (12) and by the fabrication of high-performing batteries that were genetically encoded and templated by viruses (11).
Despite these developments, a challenging divide persists between biological and materials technologies, and the potential for use of biomacromolecules as tools to control the synthesis of solid systems, as exemplified in the precision and selectivity of biomineralization, remains to be fully realized. In contrast to the wide diversity of biomolecules required for natural biomineralization (13), only short stretches of polypeptides (6, 8, 10) or polynucleic acids (14) have been systematically searched by genetic screening for defined mineral interactions in vitro. In contrast to load-bearing (4, 15) or other material properties (16, 17) that are directly selected in nature, laboratory screening has focused on either simple mineral binding (6, 8, 10) or precipitation (14) as the primary selection criterion. The challenge of engineering increasingly complex biomolecular behavior is critically dependent on the development of laboratory platforms in which genetic information can be expressed and analyzed in new ways. This fact has been evident in the progress of enzyme engineering (18, 19), in which emulsion-based cell-free systems have been used as surrogates of living cells to overcome the limitations associated with cellular protein expression (20–22). This strategy of “in vitro compartmentalization,” as it is called, allows excessive background activity (from thousands of cellular proteins) to be avoided; expands opportunities to conduct chemistry that may be toxic or not well supported in cellular environments; and supports the screening of large protein libraries [>107 variants (20–22)] via flow sorting, a technique by which the artificial cells are flowed in single file at high-throughput (>10,000 per second) past one or more laser-based detectors. Protein variants synthesized in specific cell surrogates that produce defined fluorescence or light-scattering signals (as an assay for the proteins’ activity) can be physically sorted into separate channels, after which the encoding genes can be isolated for clonal propagation and further study.
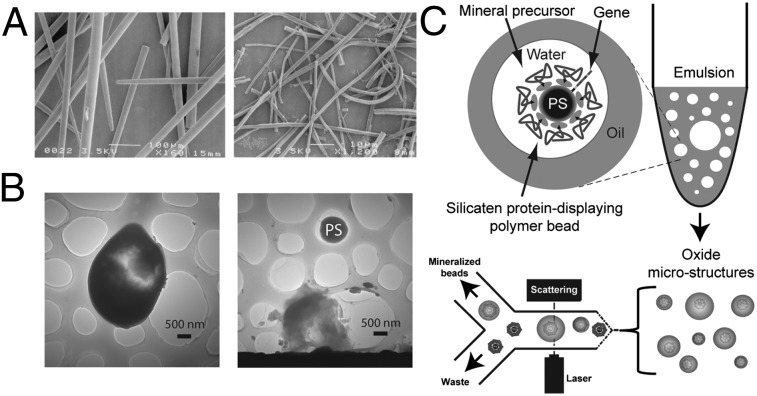
Here, we combine lessons learned from silica biomineralization in marine sponges (23–32) with emulsion-based synthetic cell surrogates (20–22) to select variants of mineralizing enzymes evolutionarily in vitro, thus advancing capabilities for studying genetically encoded inorganic materials. The cell-free enzyme reengineering method we describe is more robust for targeting a wide range of materials than cellular systems that are susceptible to poisoning or damage by certain mineral precursors or the product minerals (24). Silica-synthesizing biomimetic vesicles are first investigated with recombinant silicatein α, an enzyme that catalyzes and templates formation of marine sponge biosilica in vivo. The vesicles are then used evolutionarily to select unique silicatein variants from a combinatorially altered gene library, with reactions targeting either silicon dioxide or titanium dioxide synthesis. In the particular version of in vitro compartmentalization used here (20), the silicatein genes and their product enzymes are displayed and reacted on the surfaces of polystyrene microbeads. This allows us to mimic marine sponge skeletal growth in several ways (Fig. 1), including the use of a silicatein-based scaffold with an enzymatically active surface as a catalytic template for mineralization (20, 23, 25, 30, 32) (Fig. 1A) and the ability to control reactant species within micrometer-scale compartmentalized vesicles (26, 28, 31). Because the amplifiable genes, their encoded silicatein-based catalytic template, and the resulting synthesized minerals all are integrated within the same vesicle-based cell surrogate, evolutionary selection works on these elements as it does in living cells. We show that environmentally selected properties of the mineral and its synthesis system can thus emulate the evolutionary selection of biominerals as it occurs in nature.
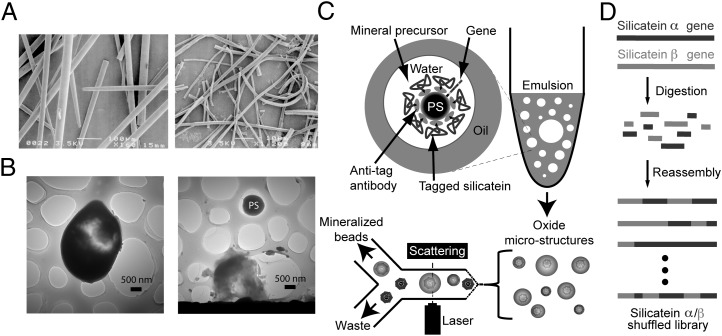
Fig. 1.
Overview of the biomimetic mineralization platform used in this study. (A) SEM images of biosilica spicules (Left) and occluded polymerized silicatein filaments (Right) isolated from the marine sponge T. aurantia. [Reproduced with permission from ref. 29 (Copyright, National Academy of Sciences of the United States)]. (B) TEM images of silicate microstructures formed from biomimetic vesicles, revealing intact bead-silicate composite (Left) and a polystyrene (PS) microbead scaffold released from the fracture of its silicate shell (Right). (C) Polystyrene microbeads coated with silicatein α [by in vitro compartmentalization (20)] are reacted with small metal-containing precursors in water-in-oil emulsions, yielding mineral composites such as those shown in B, which are then isolated from nonmineralized polymer beads by flow sorting, with light scattering used to identify large beads for sorting. (D) Schematic summary of the gene library used for evolutionary selection experiments. Recombinant genes for two natural isoforms of silicatein were digested and reassembled by DNA shuffling (Materials and Methods) to produce a chimeric library of variant silicateins, which were then screened to identify previously undescribed mineralizing variants via the process summarized in C.
Results
In Vitro Silicatein Expression.
Creation of biomimetic vesicles begins by linking silicatein genes and an antibody directed against a defined peptide “tag” sequence to polystyrene microbeads (with a diameter of 0.97 μm). The silicatein genes consist of silicatein-encoding recombinant DNA, whereas the antibody molecules ultimately act as capture agents for the silicatein proteins that will be expressed from the DNA. The DNA molecules are anchored to the beads via their 5′-end, leaving the information-encoding length of nucleic acids accessible to enzymes that can transcribe or replicate the gene. The concentration of DNA in the linkage reaction is diluted to favor the linking of less than one DNA molecule per bead. This DNA-to-bead ratio promotes the attachment of unique DNA molecules (from a mutagenized library of millions of DNA molecules encoding variant silicateins) to the individual microbeads. This ratio was also used when a homogeneous DNA population encoding parent silicatein α was first used to test the biomimetic system. Once linked with DNA and coated with antibodies, the beads are immersed in an aqueous solution, including a suite of biomolecular machinery extracted from bacteria (EcoPro T7 Mix; Novagen) that enzymatically transcribes the DNA on the beads to the corresponding mRNA and translates this mRNA to synthesize multiple copies of the uniquely encoded silicatein protein (either parental or variant, depending on the DNA sequence). Genetic modifications of the original DNA template ensure that the expressed silicateins include a unique polypeptide sequence (i.e., a tag) at their N termini as a handle, and are thus captured by antitag antibodies that were precoated on the microbead surface. In a key step ensuring that proteins expressed from a given bead-gene source become attached to that same bead (instead of diffusing to bind on another bead), the mixture of DNA-linked beads and bacterial extracts is first emulsified in oil, compartmentalizing each bead-gene to its own oil-bound aqueous vesicle, which serves as a surrogate of a living cell. The protein expression reactions yielding silicatein-coated beads occur simultaneously within the population of millions of vesicles contained in the emulsion. The emulsion is then broken by centrifugation, releasing the microbeads that each contain linked to its surface, its display of silicatein proteins, and their corresponding encoding gene. Details of this protein expression method have been described previously by Griffiths and Tawfik (20) and Miller et al. (21), and are summarized in Materials and Methods. In the present study, the presence of recombinant silicatein on the bead surfaces was successfully confirmed by flow cytometry analysis using a silicatein-specific polypeptide (24) as the fluorescent probe (Fig. S1). Antibody-only–coated beads were used for all mineralization control reactions in this study.
Biomimetic Mineralization Reactions and Flow-Sorting Analysis.
To form biomimetic mineralization vesicles, the silicatein-displaying beads (after transcription and translation, as described immediately above) were reemulsified to form a second unique and distinct water-in-oil emulsion that included chemical precursors for mineral synthesis (Fig. 1C), either 100 mM tetraethylorthosilicate [TEOS; as a precursor for silica (23)] or 100 mM titanium bisammonium lactatodihydroxide [TiBALDH; as a precursor for titania (30)], with the emulsion’s aqueous phase buffered at pH 7.4. After incubation to allow mineral synthesis (Materials and Methods), the emulsions were broken and the beads were recovered by centrifugation, as described previously (20, 21). Beads that support mineralization from their surfaces and remain intact through centrifugal bead recovery steps can potentially be detected by flow sorting (using a Becton Dickinson FACSAria flow sorter) through differential light-scattering or fluorescence signals emanating from the mineralized beads. However, the solid oxides targeted in this study do not exhibit intrinsic photoluminescence. Thus, without functionalizing these materials with a fluorophore, only light scattering can be used as a trigger for differential bead sorting. In this study, flow sorting has been used as an assay for mineralization performance. Therefore, to ensure that light-scattering signals were a reliable indicator of mineral growth, we tested our biomimetic system using both fluorescence and scattering analysis, combined with EM characterization of the sorted beads, after using initial reaction conditions that promoted formation of fluorescently labeled silica. This was achieved by using a polypeptide (“Si4,” MSPHPHPRHHHT) that has been previously demonstrated to accelerate silica precipitation from silicic acid (33). Fluorescein-labeled Si4 (Flc-Si4) was included in biomimetic vesicles with silicatein α-displaying beads reacted with TEOS, such that Si4 occluded in the mineralized product would fluorescently label the composite beads.
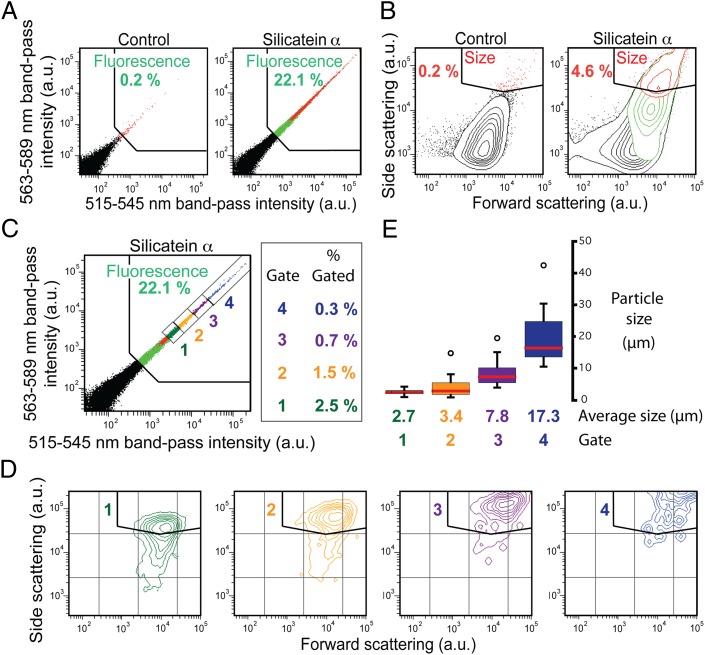
During flow sorting, beads recovered from an emulsion mineralization reaction are hydrodynamically guided past the instrument’s laser and interrogated at a defined excitation wavelength (488 nm). A representative population of 100,000 beads is measured; emission intensities through defined band-pass filters, as well as light-scattering intensities, are acquired from each bead; and the results from all 100,000 measured events are plotted by the instrument. Showing these plots as 2D histograms as in Fig. 2, where the intensities through different acquisition channels are shown on respective axes, provides a useful method for representing the data and readily identifying subpopulations of interest (34). On generating these intensity histograms from a control population (i.e., reacted beads lacking silicatein on their surfaces), “gates” are drawn within the plots to isolate regions of interest. The limits of each gate are fixed arbitrarily and defined by the maximum intensities exhibited by the control beads. Data from the experimental samples are analyzed using the same gates as established for the control. The instrument calculates the fraction of all (100,000) events from the control and experimental samples, respectively, that are binned within each gate. For example, the gate shown in Fig. 2A, (“Fluorescence”) was set against 0.2% of the control beads (i.e., at an intensity limit beneath which 99.8% of the control beads fluoresced; Fig. 2A, Left). By contrast, as indicated in Fig. 2A (Right), 22.1% of the reacted silicatein α bead population fell within this same gate (Fluorescence), showing that a significantly higher amount of Flc-Si4 was associated with the silicatein α-generated reaction products vs. the control products. Similarly, the gate “Size” in the light-scattering plots of Fig. 2B was set against the 0.2% most intensely scattering beads from the control population.
Fig. 2.
Flow-sorting analysis (from a FACSAria instrument) and TEM characterization of sorted protein-coated beads after reaction with the silica precursor TEOS in biomimetic vesicles in the presence of Flc-Si4 (main text). (A) 2D histogram maps show the distribution of fluorescence emission intensities (integrated over the indicated band-pass wavelengths) from reacted antibody-coated control beads (Left) or silicatein α-coated beads (Right). Percentages of “Fluorescence” gated beads are shown (green). (B) Light-scattering histograms from the same bead samples as in A. Percentages of “Size” gated events are shown (red). (C) Gated subpopulations representing the 5% most highly fluorescing beads from the reacted silicatein α-coated beads. One thousand beads were sorted from each subpopulation. (D) Light-scattering distributions from the subpopulations of C. (E) Distribution of average diameters of mineral composite structures sorted in C, as measured by TEM (representative TEM images are shown in Fig. S2). a.u., arbitrary units.
The subpopulations gated in one plot (e.g., Fluorescence) can be highlighted as intensity distributions in another plot (e.g., a light-scattering histogram) by color-coded mapping. In this way, Fig. 2 A and B shows that the largest beads (gated in Size and mapped red) were also the most fluorescent (gated in Fluorescence and mapped green). Further, when four additional gates were applied that binned intensely fluorescent subpopulations (Fig. 2C), it was seen that intensity increases in light-scattering signatures correlated positively with fluorescence intensities (Fig. 2 C and D). These results indicated that the largest beads were the most fluorescent as a result of probe incorporation into the enzymatically synthesized mineral structures.
Analysis of Sorted Silicatein α-Catalyzed Silicate Composites.
The flow sorter can be set to sort beads whose intensities fall within a specified gate. We flow-sorted beads through the Fluorescence subgates “1,” “2,” “3,” and “4” of Fig. 2C to examine the relationship between fluorescence and scattering within the population further, and to investigate the mineralized materials produced from this biomimetic system. One thousand beads were sorted through each subgate into respective collection tubes, washed, and evaporated onto grids for analysis by transmission electron microscopy (TEM). Measured sizes of intact silicate structures confirmed that both fluorescence intensity (Fig. 2C) and light-scattering signatures (Fig. 2D) positively correlated with bead size (Fig. 2E). TEM images of representative beads sorted through respective gates (Fig. S2) suggest that mineralization is initiated at the bead surface and that the resultant pseudospherical composite morphology (e.g., Fig. 1B) is influenced by the geometry of the biomimetic mineralization vesicle. Electron-dispersive spectroscopy (EDS) of several different mineral-coated beads confirmed the presence of silicon and oxygen while also revealing a heterogeneous distribution of additional metals, including Al, Ca, Cl, K, Fe, and Na (Fig. S2). The various reagents used for transcription, translation, and mineralization likely were the source of these additional metals. [Reaction components included mineral oil (Sigma), aqueous buffers, commercially synthesized Flc-Si4 polypeptide (Sigma–Genosys), commercial streptavidin-coated polystyrene beads (Bangs Laboratories), PCR-amplified DNA, commercial antibody (Roche), the silica precursor TEOS (Sigma), and a proprietary solution of bacterial extracts for in vitro transcription/translation (EcoPro T7 Mix; Novagen) (Materials and Methods).] Selected area electron diffraction (SAED) showed that the mineralized structures were crystalline (Fig. S2); although attributable to the heterogeneous chemistry within these crystalline products, the patterns could not be unequivocally indexed to known silica or silicate polymorphs.
The sizes of the confined reaction environments, which vary among vesicles within the emulsion used for this study (21), can have a significant impact on both enzymatic activity (21) and mineralization (35). Such variable effects, in combination with the heterogeneous distribution of metal impurities, may contribute to the compositional chemical diversity of the crystalline phases. TEM and EDS analyses of a sorted fraction of the TEOS-reacted antibody-only control beads revealed no Si in the sorted materials and showed only precipitated salts (Fig. S3). The presence of surface-displayed recombinant silicatein is thus critical for initiating directed growth of stable, higher order silicate microstructures. The sorted silicatein-containing composites exhibited crystalline mineral products both immediately adjacent to and relatively distant from the bead surfaces (Fig. S2). This suggests that after mineralization is enzymatically initiated at the bead surface, subsequent mineral growth can occur through concerted mechanisms involving the TEOS precursor, the Flc-Si4 polypeptide, the mineral surface, surfactants at the oil-water vesicle interface, and metal and molecular additives within the compartmentalized reaction environment. Although elucidation of the detailed mechanisms controlling mineral formation and growth under these conditions requires further investigation, this biomimetic system represents a highly tractable laboratory model relative to living organisms for such studies (i.e., for investigating general aspects of the concerted genetic, chemical, spatial, and interfacial mechanisms that operate during biomineralization).
Silicatein Gene Library Creation.
A wide variety of bioengineering approaches are available for genetically altering recombinant DNA (18–20, 36). We used two approaches that are commonly utilized for producing diverse gene libraries for evolutionary enzyme selection experiments. The first is error-prone PCR, in which the encoded amino acid sequence of a length of DNA is randomly mutated through errors in replication made by DNA polymerases. The second method is DNA shuffling (or “molecular sex”), in which two parent genes encoding related proteins with similar functions and shared sequence identity are randomly recombined with one another (by enzymatic scission and rejoining), yielding gene chimeras as progeny. Each progeny DNA molecule has the same overall length as the parents but randomly recombined segments of genetic information within its gene sequence (Fig. 1D). The silicateins α and β from the marine sponge Tethya aurantia are compatible with the generation of a shuffled library because they share >70% sequence identity at the gene level (29). The DNA shuffling approach we used, which was a variation of family shuffling with ssDNA as described by Zha et al. (37) (Fig. S4), also introduced random point mutations into the gene library. Sequencing of 10 randomly selected variants confirmed that the library contained silicatein genes exhibiting both gene-to-gene crossover points and random point mutations (Fig. S5).
Genetic Screening Using Biomimetic Vesicles.
The silicatein library produced as described above yields a diverse gene pool encoding millions of unique silicatein variants. Partitioning these genes to different biomimetic mineralization vesicles and screening them via the mineralization activity of their encoded proteins (as assayed by flow sorting) allowed us to select previously undescribed functional silicatein enzymes. In vitro expression for display of silicatein proteins on bead surfaces was conducted as described above. Here, each set of DNA and corresponding proteins displayed from a given microbead represented a unique silicatein variant, and 2 × 108 genes were applied to beads for screening in a single experiment (20). In initial screening attempts, biomimetic mineralization reactions were carried out with TEOS as the precursor and in the presence of polypeptide Flc-Si4. This resulted in flow-sorting fluorescence and light-scattering gate enrichments for the library-displaying beads compared with the control beads (Fig. S6), similar to the effect produced by silicatein α-displaying beads (Fig. 2). TEM analyses showed that the majority of the reacted control beads remained uncoated by mineral, whereas the reacted silicatein library-displaying beads were embedded in a matrix of inorganic material (Fig. S6). Also similar to the results with silicatein α, in some product regions, crystalline materials were observed by SAED (Fig. S6). Collectively, these results confirmed that the diverse gene pool, when subjected to evolutionary selection via biomimetic reactions and flow sorting, included functional (i.e., synthetically active) silicatein variants.
Just as gene replication is a necessary requirement for biological evolution in nature, in vitro evolutionary selection requires that successful genes be replicated (amplified) so that they may be identified and further studied. In bead-based in vitro compartmentalization (20), gene replication of the DNA attached to the individually selected (flow-sorted) beads is accomplished by PCR, in which the genes displayed from the individual bead surfaces are enzymatically amplified. We found PCR amplification of sorted mineralizing genes to be experimentally challenging as a result of the presence of occluding mineral on the bead surfaces and the very low (femtomolar) concentration of sorted DNA available for PCR. We attempted a variety of PCR conditions and bead pretreatments to liberate the mineral-occluded encoding DNA, including mechanical agitation and silica etching with hydrofluoric acid or NaOH. We also explored various reaction and sorting conditions to minimize mineral synthesis and coverage of the beads (e.g., reduced reaction times) and to identify surface-exposed genes (e.g., sorting based on binding of fluorescently labeled cDNA). Using silica as a model system, we found the most reliable approach to achieving significant gate enrichment during flow sorting, while also promoting successful gene amplification, involved conducting mineralization in the absence of the fluorescently labeled polypeptide Flc-Si4 to reduce oxide growth subsequent to initial mineralization at the bead surfaces and then mechanically agitating the sorted beads before PCR. This selection process, which facilitates use of light scattering alone (instead of fluorescence) as a signal for flow sorting, allowed us to select beads with significant yet still disruptible mineral oxides without interference from the labeled polypeptide on the mineralization reactions. Further, these experimental constraints introduced a unique set of “environmental pressures” in which the genetically encoded mineral composites were subjected to mechanical stresses, thus experiencing at the microscale an analog of the structural pressures that select for naturally evolved biominerals.
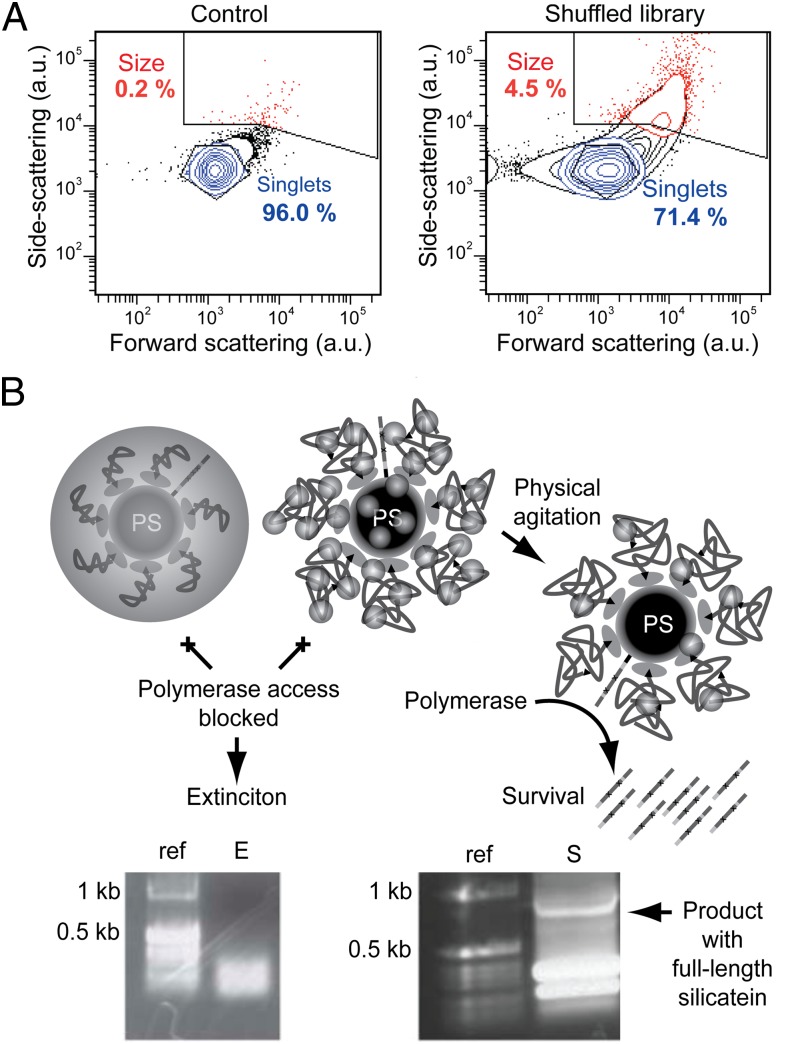
In parallel screening experiments, TEOS was used as the precursor for silica-targeting reactions and TiBALDH was used as the mineral precursor for titania-targeting reactions. TEM analyses confirmed that Si- and Ti-containing materials were formed under screening reaction conditions (Fig. S6). After recovering the beads from emulsion-based biomimetic mineralization reactions and immediately before flow sorting, the beads were physically agitated by immersion in a sonication bath (5 min) to disrupt bead agglomerates and background salt precipitates from control bead surfaces; this treatment (in contrast to Flc-Si4–assisted TEOS reaction conditions) was required to yield a significant differential light-scattering signal from the silicatein library displaying beads relative to control beads. The majority of the control population then appeared as bead singlets by flow-sorting analysis (Fig. 3A and Fig. S6). To collect sorted beads for PCR amplification, the entirety of each reacted silicatein bead sample was subjected to flow sorting, thereby resulting in circa 600,000 TEOS-reacted and 185,000 TiBALDH-reacted beads, respectively, sorted through the Size gate (Fig. 3A and Fig. S6). Gate enrichment and sorting yield were lower in the titania-targeting selection experiment vs. the silica-targeting selection experiment. To disrupt the encasing mineral and liberate the encoding DNA, the sorted beads were subjected to more intense physical agitation consisting of three cycles of a 5-min sonication bath, 1 min on a Baxter SP vortexer at maximum speed, and 5 min of centrifugation at 20,800 × g. After each centrifugation step, small particulates became increasingly visible by eye above the beads along the length of the test tube sidewall; these particulates were removed along with the supernatant and replaced with fresh buffer before the subsequent agitation cycle. The agitated and washed beads were finally applied as gene template carriers to a PCR after dislodged mineral particles were removed by filtration through 0.22-μm filter units (retaining the beads with a diameter of ∼1 μm). This final filtration step was critical for successful gene amplification. Amplified full-length genes (Fig. 3B) were then cloned and sequenced.
Fig. 3.
Mineralization performance of the silicatein library in biomimetic vesicles. (A) Light-scattering signatures of bead populations reacted with TEOS, after agitation by immersion in a sonication bath to disrupt bead agglomeration. Relative population percentages of “Size”-gated events (red) and “Singlets”-gated events (blue) are shown. The control bead populations are disrupted into bead singlets (i.e., uncoated and nonagglomerated polystyrene beads), whereas the silicatein-library bead population maintains an intensely scattering subpopulation, indicating the presence of mechanically robust silica-polymer composite structures. Similar results were obtained from reactions using the titania precursor TiBALDH (Fig. S6). (B) Schematic illustrations (Upper) and 1% agarose gels of electrophoresed DNA products from PCR reactions (Lower) show that mineralizing enzymes that catalyze synthesis of uniform or extensively agglomerated mineral coatings block gene amplification (E), whereas silicatein variants that self-assemble and catalyze synthesis of relatively dispersed oxide nanoparticles meet flow-sorting criteria and are also amenable to postsorting mechanical disassembly, allowing the encoding gene to be exposed for amplification (S). ref., reference DNA ladder.
Evolved Mineralizing Genotypes.
From a sample of 30 sequenced genes from each selection procedure (targeting silica and titania, respectively), the majority of genes were found to encode a truncated polypeptide sequence that also was found present in high numbers in the starting library (SI Text and Fig. S5). However, within each sampled population, we found one full-length silicatein gene (Fig. 4A) different in character from both the parent silicateins and the representative genes from the starting library (Fig. S5). We name these variants screened for silica and titania synthesis, respectively, silicatein X1 and silicatein XT. Each variant exhibits several unique DNA recombinations and point mutations. Collectively, almost all (22 of 24) of these changes in protein sequence involve hydroxyl residues, either directly (Fig. 4A, stars) or indirectly by occurring immediately adjacent to a hydroxyl moiety (Fig. 4A, asterisks). We consider these changes likely to be important because hydroxyls have been implicated previously as surface functional groups capable of guiding mineral oxide deposition (25, 29, 30, 38).
Fig. 4.
Silicatein sequences and modeled active sites. (A) Multiple protein sequence alignment of silicatein parents (α and β) and progeny (X1 and XT). Stars indicate sequence changes that directly involve hydroxyl residues; asterisks indicate changes occurring immediately adjacent to hydroxyl residues. Yellow and blue highlights denote residues structurally conserved in a putative catalytic triad configuration as determined by homology modeling; gray highlights denote residues in the distorted active site region of silicatein XT (Fig. S7). (B) Homology-modeled active site of silicatein α overlaid with that of silicatein X1, showing the shared putative catalytic triad configuration. Native silicatein α has been previously proposed to operate through an SN2-type attack by the active site serine-25/26 on a silicon precursor center, leading to precursor hydrolysis and subsequent polycondensation (23).
For Si-O-protein bond formation to occur, pentacoordinated Si intermediates are required (23, 32, 39). This suggests a biochemical role for hydroxyl-adjacent amino acids, whose importance is further underscored by a previous study showing that genetic alterations adjacent to a silicon-interacting hydroxyl critically affected silica precipitation activity in a cysteine hydrolase enzyme (40). Additionally, silicatein surface biochemistry has been suggested to be critical for its self-assembly into polymeric filaments in vivo (27, 41). Energy-minimization modeling using cathepsin L [a structurally closely related homolog of silicatein with a known X-ray structure (27, 32)] as the structural template showed that the majority of point mutations are located on the solvent-exposed protein surfaces (Fig. S7). The models further suggest that the active site residues of silicatein α (Ser26, His165, and Asn185) are structurally conserved in silicatein X1 (Fig. 4B) but partially reconfigured in silicatein XT (Fig. S7).
Silicatein has been proposed to operate through an SN2-type attack by the active site serine-26 on the silicon precursor center, leading to the release of ethanol and a metal-protein intermediate (3, 23, 25, 32). Subsequent attack on the metal center by water releases the hydrolyzed precursor, which can then condense with other precursor species. In silicatein α, the active site histidine-165 hydrogen bonds with the active site serine-26 hydroxyl to increase the nucleophilicity of the serine oxygen and activate it for metal-center attack (Fig. 4B). Asparagine-185 is hydrogen-bonded with His-165 on its opposite site (away from Ser-26) to withdraw electron density and increase the strength of the imidazole-hydroxyl hydrogen bond. In the model of silicatein XT, only Ser-26 appears to be structurally conserved (Fig. S7). Although this suggests that the catalytic triad of silicatein α is energetically strained in silicatein XT, it is presently unclear whether this reconfiguration represents a functionally adjusted active site or whether a catalytic triad is not necessary for silicatein-mediated titania formation. The role of active site chemistry in enzymatic mineralization with different metallorganic substrates is currently a subject of ongoing investigations (42, 43).
Evolved Mineralized Phenotypes.
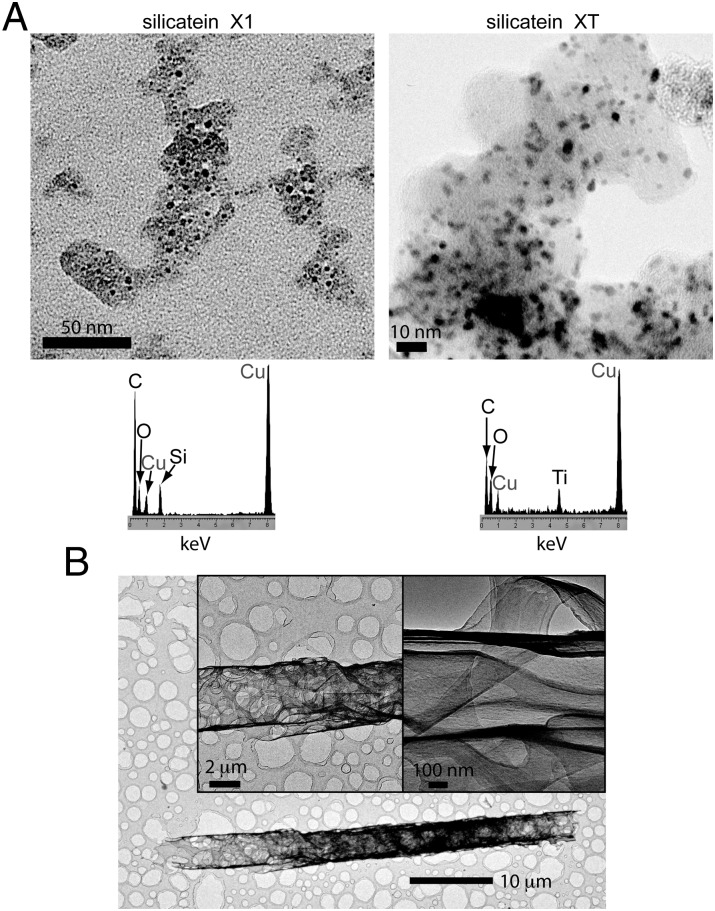
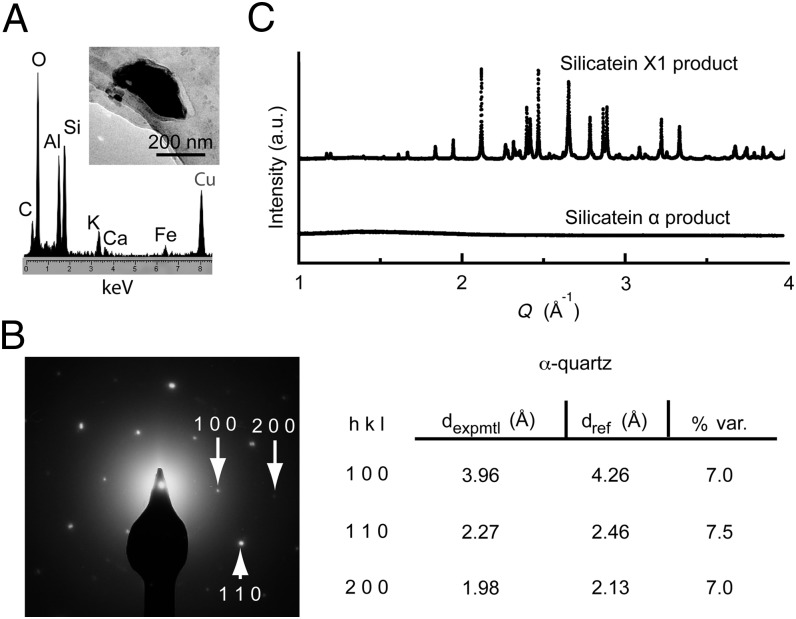
We overexpressed the selected variant silicateins and parent silicatein α in Escherichia coli, and purified and refolded the recombinant proteins using column chromatography (Materials and Methods and Fig. S7). When freshly refolded silicatein X1, silicatein α, or heat-denatured silicatein X1 was each reacted with 100 mM TEOS at pH 7.4, no product was observed from the denatured silicatein X1 reaction. Insoluble products generated by silicatein α and silicatein X1 were collected by centrifugation, washed, and analyzed by TEM. Whereas silicatein α produced agglomerated, interconnected silica nanoparticles (Fig. S8), silicatein X1 synthesized relatively dispersed silica nanoparticles (Fig. 5A). Analogous results were obtained with silicatein α and silicatein XT from reactions targeting titania synthesis from TiBALDH (Fig. 5A and Fig. S8). Under certain TEOS reaction conditions (static instead of agitated incubation and over an extended incubation time; Materials and Methods), silicatein X1 produced uniquely ordered silica-protein composite filaments in the form of folded sheets (Fig. 5B and Fig. S8). In some cases, silicatein X1 or silicatein XT (but not silicatein α) was observed to produce diverse crystalline inorganic products from these aqueous reactions (Fig. 6 and Fig. S8). These crystals were larger in size than the amorphous nanoparticles produced from the same mineralization reactions, and EDS analysis revealed impurity metals (Al, K, Ca, and/or Fe) to be associated with these crystalline species (Fig. 6A and Fig. S8). Only C, O, and Si (or Ti) were found in the amorphous product regions (Fig. 5A). Interestingly, some of these mineral crystals appeared to be closely related to well-known crystalline polymorphs of silica, such as α-quartz, as measured by SAED (Fig. 6B). Synchrotron X-ray diffraction analysis (44) of the Si-containing products obtained from reactions with silicatein α and silicatein X1 confirmed that silicatein X1 produces a heterogeneous population of crystals, whereas the products from silicatein α are completely amorphous (Fig. 6C). The chemical information from EDS suggests the crystals are mixed silicates (Fig. 6A), but the heterogeneous nature of the products precluded precise structure determination.
Fig. 5.
(A) TEM image and associated EDS spectrum of mineralization products from bacterially expressed silicatein X1 reacted with 100 mM TEOS under stirring conditions (Left) or from bacterially expressed silicatein XT reacted with 100 mM TiBALDH (Right; also from stirring conditions) (Materials and Methods), showing that both reactions produce relatively dispersed nanoparticles on an apparent protein template. (B) TEM images of products from bacterially expressed silicatein X1 reacted with 100 mM TEOS under static incubation conditions (Materials and Methods), showing that uniquely ordered filament and sheet structures are formed. The structures are composites that include silicon, and silicatein α yields less ordered structures under the same conditions (Fig. S8).
Fig. 6.
Silicatein variants were evolutionarily selected to catalyze synthesis of crystalline oxides from buffered aqueous solution, pH 7.4. (A) TEM analysis with EDS on products from silicatein X1 reacted with TEOS shows that crystals are formed, which are composed of silicon with additive metals; analogous results were found from titania-targeting silicatein XT reactions (Fig. S8). Additional diverse crystals in each system exhibited a variable range of metal additives in their EDS spectra, whereas the amorphous nanoparticulate product regions (e.g., Fig. 5) showed no additive metals. (B) Crystal structure of the example presented in A closely resembles that of α-quartz as determined by SAED. dexpmtl, experimentally measured d-spacings; dref, reference d-spacings for alpha-quartz; % var., % variation between the experimentally measured and reference d-spacing values. (C) Synchrotron X-ray diffraction confirms that silicatein X1 uniquely catalyzes a mixture of crystalline silicates; the heterogeneous nature of the products precluded precise structure determination.
Discussion
This study demonstrates multiscale mimicry of skeletal silicification in marine sponges by integrating several reaction strategies used in natural biomineralization. During in vivo formation of spicules (sponge skeletal elements; Fig. 1A), silicatein protein filaments are initially assembled intracellularly before being exported to an extracellular vesicular compartment (26, 28, 31). In both the in vivo and in vitro processes, silicatein enzymatically hydrolyzes precursor molecules to initiate mineralization at the silicatein filament (or silicatein-bead) surface; mineralization is templated along the silicatein scaffold, and the mineral grows radially from the scaffold surface within a shape-constraining compartment as the templating polymer becomes axially occluded within a spicule (23, 28) (Fig. 1A) or centrally occluded within a pseudospherical mineral shell (Fig. 1B). In vivo, additional biomolecules, including silicatein and other proteins, may guide silica growth within the vesicle once the filament is covered by mineral (28). The Si4 polypeptide added to the biomimetic mineralization reactions served to mimic this guided mineral growth while also fluorescently labeling the silica product. This labeling, combined with polydisperse emulsion vesicle sizes (20) (SI Text), was useful for demonstrating by flow-sorting and TEM analysis that both fluorescence and light-scattering intensities positively correlate with composite microstructure diameter.
The results from studies of purified, refolded silicateins showed that the evolutionarily selected silicatein variants acquired several specific mineralizing phenotypes as a result of the genetic screen, including the abilities to synthesize dispersed metal or metalloid oxide nanoparticles, to self-assemble into higher ordered structures under certain conditions, and to catalyze synthesis of multimetallic oxide crystals from near-neutral aqueous solution at ambient temperature. The similar flow-sorting and light-scattering signatures, biochemical mutations, and mineral characteristics observed from both the silica- and titania-targeting selections suggest that acquisition of the unique phenotypes was directly governed by the imposed selection environment. To “survive,” a given silicatein gene variant displayed on a bead would need to (i) code for an enzyme that yields a light-scattering signature (via mineralization) of sufficient intensity to be included in the flow-sorting gate (Fig. 3A and Fig. S6) and (ii) be sufficiently accessible for postsorting extraction and polymerase-catalyzed DNA amplification (Fig. 3B). Self-assembly of the active silicatein enzyme variant on the bead surface would effectively expand the bead diameter, thereby providing a larger polymeric template for biomimetic mineralization. Formation of dispersed oxide nanoparticles would then introduce mechanical stabilization while providing a rough surface, leading to an intense side-scattering signal for flow sorting. Such a composite architecture would be susceptible to postsorting mechanical disruption to expose the encoding DNA (Fig. 3B) yet would be stable enough to resist presorting physical agitation (Fig. 3A and Fig. S6).
Interestingly, multimetallic silicate crystals have been identified in natural sponge biosilica (26), suggesting that recombinant silicatein activity in these in vitro mineralization vesicles exhibits this natural activity as well. Our results from genetic screening showed that this crystal-forming ability could be (re)acquired via evolutionary selection for expression in a range of in vitro environments. It is unknown whether the crystal-forming ability of the evolved silicateins was a requirement for selection or a secondary trait indirectly coselected by the screening reaction environment. All observed silicatein α-catalyzed composites that remained intact through centrifugal recovery and flow sorting also exhibited a crystalline mineral structure (Fig. S2), providing indirect evidence that crystal formation may promote composite strength. Completely aqueous (i.e., nonemulsified) mineralization reactions with bacterially expressed, purified, and refolded enzymes, which yielded only amorphous silica from parental silicatein α (Fig. 6C), demonstrated that the variant silicatein X1 retained its crystal-forming behavior as an intrinsic activity (Fig. 6), even under conditions in which metal additives were scarce and vesicle interfaces were absent. This confirmed that crystal-forming activity was uniquely acquired through evolutionary selection from the biomimetic mineralization vesicles. By random chance alone, it is unlikely that either variant (silicatein X1 or silicatein XT) would exhibit the number of hydroxyl-associated changes that we found as mutations in these two selected protein variants (Fig. 4A). We thus infer that the observed mutations are mechanistically important in mineral formation. Collectively, the above findings demonstrate that laboratory selection pressures imposed by an evolutionary reaction environment enabled the detection of mutationally produced unique or unexpected materials not formed by conventional chemical syntheses or by reaction with unaltered parental protein sequences as obtained directly from nature.
This study adds to growing evidence that enzymatic routes to new materials, as exploited in biomineralization, may be effectively harnessed to produce technological materials (3, 23, 24, 32, 40, 43, 45, 46). A typical enzyme, in a properly folded conformation, presents both a chemically catalytic active site and a solution-exposed surface with specific surface chemistry and atomic-scale topological features. Through genetic evolution, driven by specific selective conditions, both chemical activity and surface definitions of an enzyme are tunable. For inorganic materials synthesis in any context, the dynamic states of both of these parameters, local chemical changes and molecular-scale interactions at heterogeneous interfaces, are critically influential in the nucleation and growth of specific mineral phases. Thus, the essential and genetically tunable features inherent to enzymes make these functional biomolecules potentially useful agents for mediating and controlling mineral synthesis from aqueous solution.
In this study, enzymes have been evolutionarily reengineered to synthesize newly identified inorganic materials catalytically. Thus far, we have produced only a single generation of silicatein progeny to investigate salient features of the screening process. Our ability to recover unique functional genes after a single round of selection was facilitated by our use of a shuffled gene library (18, 36). Our findings show that enzymes can indeed be engineered to synthesize new material structures through mutation and evolutionary selection, but the mineralization reaction environment during screening must be tightly controlled before more quantitative studies can be conducted to define the role of enzymes in achieving specific crystalline products. The observed problem of contamination is analogous to similar problems faced in electronic materials fabrication, in which clean-room environments are required for fabricating high-purity, highly controlled semiconducting crystals. However, although impurity elements pose a concern in elucidating the enzymatic mechanisms of mineralization, the structural complexity of biominerals in nature suggests that DNA-driven systems can accommodate and functionally adapt such chemical additives toward achieving specific new materials properties and performance. The in vitro study presented here supports this conclusion and suggests that we may begin harnessing this flexibility for anthropogenic materials fabrication.
Conclusion
We have reported the acquisition of previously undescribed enzymatic biomineralizing phenotypes through a single generation of in vitro evolution, including the synthesis of crystalline silicates from near-neutral aqueous solution. We have thus shown it is possible to evolve solid-state materials genetically using an approach that mimics natural biomineral evolution, in which the fittest materials systems are selected. In principle, the targeted mineral characteristics could be extended to any material property (e.g., photoluminescence, magnetization) for which an effective selection can be designed. Tunable parameters in the demonstrated platform for further studies include vesicle size; vesicle and other mineralization interface chemistries (e.g., surfactants, support surfaces); new parental and mutationally altered proteins encoded by the DNA; mechanisms of genetic mutation; spatial proximity of the DNA to the encoded proteins; shape, chemistry, and activities of the polymeric mineral-templating scaffold; chemistry of the mineralization precursors; and additive components of interest (e.g., other mineral-binding polypeptides or aptamers). This simple emulsion-based system thus presents a multitude of ways to begin investigating mineralization via DNA-driven genetic algorithms, which have operated over millions of years of evolution to produce nature’s biomineral fabrication systems. The close genetic relationship of the silicateins with the cathepsin family of digestive proteases and the larger superfamily of hydrolases shows that nature has evolutionarily recruited enzymes with other functions to control biomineralization. Recently, members of this enzyme superfamily (42, 43), as well as other enzyme families (45, 46) that do not normally catalyze mineral synthesis, have been shown to exhibit catalytic mineralization activity in vitro, suggesting that a wide variety of enzymes may be suitable candidates for directed evolution toward new, specific mineral-synthesis activities. The broad biotechnological capacity to evolve and reengineer enzymes suggests exciting possibilities for further mimicking this natural evolutionary process, and directing enzyme evolution toward the development of new solid-state materials. The diverse collection of enzymes in the natural world and their range of unique chemical activities could allow, in principle, a correspondingly diverse range of inorganic materials to be synthesized by this general strategy. As genetic screening pressures are developed to include functional material performance for targeted engineering applications, this approach will begin to allow the same DNA-based evolutionary processes that have created seashells and skeletons to be harnessed to advance human technologies.
Materials and Methods
Preparation of Gene Templates for in Vitro Expression.
The starting recombinant T. aurantia silicatein genes used for all methods encoded the mature 218-aa (silicatein α) or 217-aa (silicatein β) polypeptides (29). Parent or shuffled genes were cloned into T7 in vitro expression vector pIVEX 2.6 (Roche) with NotI and SacI sites. Linear templates that included T7 promoter and terminator regions and the ligated gene were then amplified from this plasmid by PCR using dual biotin-labeled forward primer LMB21-Long and reverse primer pIVB1-Long (20). All primer sequences are provided in ref. 20 or in Table S1, and ligation and PCR conditions, as well as conditions for confirming the presence of recombinant silicatein α on the beads by flow cytometry, are provided in SI Text S1.
Silicatein Gene Library Creation.
Silicatein α and silicatein β genes were PCR-amplified with primers to produce silicatein α genes with one (“bottom”) DNA strand that was 5′-phosphorylated and silicatein β genes that were similarly labeled but in an inverse manner, with the “top” strand 5′-phosphorylated (Fig. S4). Digesting these PCR products with λ-exonuclease (NEB; λ-exonuclease requires 5′-phosphorylation of its DNA substrate for processivity) yielded ssDNA of each parent gene, an intact top strand of silicatein α and an intact bottom strand of silicatein β. The ssDNA products were purified from dsDNA using gel electrophoresis (ssDNA migrates faster than dsDNA; Fig. S4). Each ssDNA sample was then digested with DNase I at 15 °C for 5 min, and 10- to 80-nt digestion products were excised and purified from a 10% (wt/vol) PAGE gel. A primerless gene reconstitution reaction was conducted, followed by a gene recovery PCR using a forward primer for silicatein β and a reverse primer for silicatein α. Amplified products showed a single band at the expected length, and control reactions with mixed primer sets confirmed the presence at least one β-to-α crossover point per shuffled gene (Fig. S4).
In Vitro Protein Expression.
In vitro compartmentalization for protein expression was conducted exactly as described previously (20, 21), except the in vitro expression mixture was supplemented with 40 μg/mL BSA in this study. For control samples subjected to in vitro expression, methionine was withheld from the in vitro expression mixture. After incubation for protein expression, the emulsion was broken and beads were washed as described previously (20), except the carbonate-zinc buffer was omitted in this study. Instead, after washing the beads with PBS with 0.1% Tween 20 (PBS/T) and 8 mg/mL heparin, the beads were further washed twice more with 10 mM Tris⋅HCl, pH 7.4, and resuspended in 55 μL of the same.
Biomimetic Mineralization Reactions with Flow Sorting.
For reactions conducted with the Flc-Si4 polypeptide, 2 μL of 1 mg/mL Flc-Si4 peptide (Sigma–Genosys) (33) (in 10 mM Hepes buffer, pH 7.5) was added to the aqueous phase (giving 57 μL of total H2O) of the emulsion. For silica-targeting reactions, resuspended beads were first reemulsified, after which TEOS (Sigma) was added to 100 mM in the emulsion with stirring by a small magnetic stir bar immersed in the emulsion at 100 rpm for 3 min. For titania-targeting reactions, TiBALDH was added to 100 mM in the water phase immediately before reemulsification. Other than replacement of the aqueous phase (with Tris⋅HCl-buffered water, pH 7.4, used in place of the in vitro expression mixture), all samples were reemulsified with a procedure identical to the emulsification for protein expression (20). Mineralization and control reactions with silicatein α and Flc-Si4 were incubated for 2 h at 16 °C (mimicking the temperature of the sponge’s natural marine environment). Shuffled library screening reactions from which DNA was amplified were incubated for 12 h at 22 °C. Flc-Si4 probe was not added to TiBALDH reactions, nor was it added to the final emulsified TEOS reactions conducted for shuffled library screening. Reacted beads were recovered from the emulsions and washed by centrifugation as described previously (20). After the final wash, the beads were resuspended in 100 μL of PBS/T. Each sample was diluted 20-fold in PBS/T and analyzed with a Becton Dickinson FACSAria cell sorter at a rate of 10,000 events per second; fluorescence analysis was conducted with a 488-nm argon ion excitation laser, and emission was measured through 530 ± 15-nm and 576 ± 13-nm band-pass filters. The forward scatter detector is a photodiode with a 488 ± 10-nm bandpass filter, and the side scatter detector is a photomultiplier also with a 488 ± 10-nm bandpass filter. Sorting gates were set as shown in Figs. 2 and 3 and Fig. S6.
TEM Analysis of Sorted Beads.
TEM analysis was conducted on either a JEM-1230 TEM (JEOL) operated at 80 kV with digital images recorded on an Advantage CCD camera system (AMT) or on an FEI Tecnai G2 Sphera electron microscope with EDS at an operating voltage of 200 kV. For TEM analysis of composite structures mineralized within silicatein α biomimetic vesicles, 1,000 events were sorted through each applied fluorescence subgate (Fig. 2C), washed with 2 mL of ultrapure H2O through a 0.45-μm vacuum filtration unit (AcroPrep GH Polypro filter plates; Pall), and resuspended in 100 μL ultrapure H2O, 20 μL of which was dropped onto a copper-formvar TEM grid and dried at 37 °C overnight before imaging. For selected area diffraction analysis on large mineral structures, the pattern was obtained from near the outer edge of the structure, where the electron beam could penetrate the thickness of the material. For TEM analysis of reacted presorted beads from silicatein library and control biomimetic vesicles, after recovery from emulsions, one-fifth of the bead solution was washed several times in ethanol by centrifugation and evaporated onto a copper-formvar TEM grid for imaging; the remainder of the bead solution was used for flow-sorting analysis.
Amplification, Sequencing, and Homology Modeling of Selected Genes.
After gated subpopulations of the reacted library-displaying beads were flow-sorted, postsorting physical agitation was then conducted as described in Results, with additional details provided in SI Text. Once agitated, beads were filtered (SI Text); they were resuspended in a final volume of 31 μL of 1× Pfu buffer (Stratagene) and transferred to a 50-μL PCR. The PCR included primers T7recF and T7recR (0.2 μM each), 0.2 mM deoxynucleotide triphosphates, and 2.5 units of PfuTurbo (Stratagene). DNA was susceptible to shearing during the sonication step, causing smearing and low-molecular-weight bands in gels of the recovered gene products (Fig. 3B). The PCR cycling schedule is provided in SI Text S6.
Products were electrophoresed on a 1% agarose gel, and DNA from the 0.80-kb product band was excised and purified (gel purification; Qiagen) (Fig. 3B). The purified DNA was used as a template in a PCR with primers BFpET and ARpET to amplify the recovered silicatein genes for cloning into vector pET151-DTOPO (Invitrogen). Cloning and transfection into E. coli top 10 cells were conducted according to the manufacturer’s instructions. After plating on LB-ampicillin–selective media, individual colonies were randomly chosen for culture, DNA purification (Miniprep; Qiagen) and sequencing (University of California DNA sequencing facility, Davis, CA).
Homology models (Fig. 4B and Fig. S7) were generated by SWISS-MODEL (47) and visualized using BALLView v1.3 (www.ball-project.org) (48). Cathepsin L [Protein Data Bank (PDB) ID code 3HHA, Chain A] was selected as the best template for homology modeling of silicatein α through the Automated module on the SWISS-MODEL site, which computationally screens a database of templates. Once identified, this same structural template was used for modeling the silicatein X1 and silicatein XT structures, using the Alignment module on the SWISS-MODEL site.
Bacterial Expression, Purification, and Refolding of Silicatein.
To investigate the mineralizing phenotypes of the identified full-length silicatein variants, genes encoding silicatein X1, silicatein XT, and parent silicatein α were inserted into the pET151-DTOPO plasmid for inducible expression, placing them under control of the T7 promoter and lacO operator; the plasmid also encodes a His6 tag N-terminal to the recombinant protein. Proteins were overexpressed in and purified from E. coli BL21AI cells, as described in SI Text S7. Recombinant silicatein was found expressed as inclusion bodies (Fig. S7), and thus was resolubilized, and then purified and refolded on a 5-mL Ni HisTrap HP column (GE Healthcare) by gradual removal of denaturing buffer using an Äkta purifier (Amersham Biosciences) FPLC system (details are provided in SI Text S7). After eluting the refolded protein from the column, the buffer was exchanged to 20 mM Tris⋅HCl (pH 8.5), 10 mM NaCl, and 0.1% glycerol using a HiPrep 26/10 desalting column (GE Healthcare) with the same Äkta FPLC system. Protein concentration was determined by absorbance at 280 nm using a nanodrop spectrophotometer, with extinction coefficients determined by the respective amino acid sequence of each expressed protein using the Protparam tool on the ExPASy Bioinformatics Resource Portal (http://web.expasy.org/protparam/).
Mineralization Activity of Recombinant Silicateins.
To test for mineralization, 50 μg/mL silicatein was used in 1.5-mL reactions with 100 mM TEOS or TiBALDH, or in 5-mL reactions with 1 M TEOS, in 100 mM Tris⋅HCl (pH 7.5) for 12 h at 22 °C. Denatured protein was obtained by heating in a water bath at 85 °C for 30 min. TEOS reactions that were subject to light physical agitation while incubating were mixed on a Tomy MT-360 microtube mixer at half-full speed. TEOS has low miscibility in water; thus, agitation during incubation constantly refreshed the reaction interface between the two solutions. TiBALDH is completely miscible in water. At the end of the agitated incubation period, before precipitate recovery by centrifugation, no insoluble product was observable by eye from the heat-denatured silicatein reactions. The insoluble product from the silicatein α reactions formed a pellet at the bottom of the test tube. The insoluble product from the silicatein X1 and XT reactions, although clearly visible by eye, remained as a dispersed suspension in solution (a qualitative indication that dispersed particles had been synthesized).
Higher order self-assembled structures from silicatein α and silicatein X1 were observed from reaction with 100 mM TEOS after a static incubation period of 12 h at room temperature, followed by static incubation for 1 wk at 4 °C (Fig. 5B and Fig. S8). To stop the mineralization reactions and recover precipitated products, reaction mixtures were centrifuged at 20,800 × g for 10 min at 4 °C and the supernatants were then discarded. For the TEOS reactions, 1.5 mL of ethanol was added to each reaction before this initial centrifugation to increase the miscibility of TEOS and create a single-phase solution. After the first centrifugation, precipitates were washed five times with 1.5 mL of 70% ethanol by centrifugation and decanting, and an aliquot of the resuspended products was then placed on a TEM copper-formvar grid and dried at 37 °C overnight before analysis with a Tecnai G2 Sphera electron microscope at an operating voltage of 200 kV. High-resolution synchrotron powder diffraction data were collected using beamline 11-BM at the Advanced Photon Source, Argonne National Laboratory, using an average wavelength of 0.13702 Å (44). A mixture of National Institute of Standards and Technology standard reference materials, Si (SRM 640c), and Al2O3 (SRM 676) was used to calibrate the instrument, with the Si lattice constant calibrating the wavelength for each detector.
Supplementary Material
Acknowledgments
We thank the expert beamline staff at 11-BM at the Advanced Photon Source at Argonne National Laboratory; the laboratory of P. Daugherty in the Department of Chemical Engineering at the University of California, Santa Barbara, for assistance with flow sorting; J. Fong, J. Kunkel, J. León, G. Mantalas, J. Stuart, and A. Ibish for experimental assistance; past and present members of D.E.M.’s laboratory for helpful discussions; and F. Meldrum for careful reading of the manuscript. This work was supported by US Department of Energy Grant DEFG03-02ER46006. L.A.B. was partially supported by University of California Systemwide Biotechnology Research and Education Program Graduate Research and Education in Adaptive Bio-Technology Training Grant 2007-19. J.R.N. was partially supported by a National Science Foundation Graduate Research Fellowship. Use of the Advanced Photon Source at the Argonne National Laboratory was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 10142 (volume 109, number 26).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116958109/-/DCSupplemental.
References
- 1.Smith JV. Biochemical evolution. I. Polymerization On internal, organophilic silica surfaces of dealuminated zeolites and feldspars. Proc Natl Acad Sci USA. 1998;95:3370–3375. doi: 10.1073/pnas.95.7.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohmoto H, Kakegawa T, Lowe DR. 3.4-Billion-year-old biogenic pyrites from Barberton, South Africa: Sulfur isotope evidence. Science. 1993;262:555–557. doi: 10.1126/science.11539502. [DOI] [PubMed] [Google Scholar]
- 3.Brutchey RL, Morse DE. Silicatein and the translation of its molecular mechanism of biosilicification into low temperature nanomaterial synthesis. Chem Rev. 2008;108:4915–4934. doi: 10.1021/cr078256b. [DOI] [PubMed] [Google Scholar]
- 4.Mayer G. Rigid biological systems as models for synthetic composites. Science. 2005;310:1144–1147. doi: 10.1126/science.1116994. [DOI] [PubMed] [Google Scholar]
- 5.Meldrum FC, Heywood BR, Mann S. Magnetoferritin: In vitro synthesis of a novel magnetic protein. Science. 1992;257:522–523. doi: 10.1126/science.1636086. [DOI] [PubMed] [Google Scholar]
- 6.Sarikaya M, Tamerler C, Jen AKY, Schulten K, Baneyx F. Molecular biomimetics: Nanotechnology through biology. Nat Mater. 2003;2:577–585. doi: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- 7.Weiner S, Addadi L. Design strategies in mineralized biological materials. J Mater Chem. 1997;7:689–702. [Google Scholar]
- 8.Whaley SR, English DS, Hu EL, Barbara PF, Belcher AM. Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly. Nature. 2000;405:665–668. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]
- 9.Xiang XD, et al. A combinatorial approach to materials discovery. Science. 1995;268:1738–1740. doi: 10.1126/science.268.5218.1738. [DOI] [PubMed] [Google Scholar]
- 10.Brown S. Engineered iron oxide-adhesion mutants of the Escherichia coli phage lambda receptor. Proc Natl Acad Sci USA. 1992;89:8651–8655. doi: 10.1073/pnas.89.18.8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee YJ, et al. Fabricating genetically engineered high-power lithium-ion batteries using multiple virus genes. Science. 2009;324:1051–1055. doi: 10.1126/science.1171541. [DOI] [PubMed] [Google Scholar]
- 12.Rothberg JM, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 13.Mann S. Biomineralization: Principles and Concepts in Bioinorganic Materials Chemistry. UK: Oxford Univ Press; 2001. [Google Scholar]
- 14.Gugliotti LA, Feldheim DL, Eaton BE. RNA-mediated metal-metal bond formation in the synthesis of hexagonal palladium nanoparticles. Science. 2004;304:850–852. doi: 10.1126/science.1095678. [DOI] [PubMed] [Google Scholar]
- 15.Aizenberg J, et al. Skeleton of Euplectella sp.: Structural hierarchy from the nanoscale to the macroscale. Science. 2005;309:275–278. doi: 10.1126/science.1112255. [DOI] [PubMed] [Google Scholar]
- 16.Mann S, Frankel RB, Blakemore RP. Structure, morphology and crystal-growth of bacterial magnetite. Nature. 1984;310:405–407. [Google Scholar]
- 17.Sundar VC, Yablon AD, Grazul JL, Ilan M, Aizenberg J. Fibre-optical features of a glass sponge. Nature. 2003;424:899–900. doi: 10.1038/424899a. [DOI] [PubMed] [Google Scholar]
- 18.Arnold FH, Georgiou G, editors. Directed Evolution Library Creation: Methods and Protocols. Totowa, NJ: Humana Press; 2003. [Google Scholar]
- 19.Bershtein S, Tawfik DS. Advances in laboratory evolution of enzymes. Curr Opin Chem Biol. 2008;12(2):151–158. doi: 10.1016/j.cbpa.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths AD, Tawfik DS. Directed evolution of an extremely fast phosphotriesterase by in vitro compartmentalization. EMBO J. 2003;22(1):24–35. doi: 10.1093/emboj/cdg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller OJ, et al. Directed evolution by in vitro compartmentalization. Nat Methods. 2006;3:561–570. doi: 10.1038/nmeth897. [DOI] [PubMed] [Google Scholar]
- 22.Tawfik DS, Griffiths AD. Man-made cell-like compartments for molecular evolution. Nat Biotechnol. 1998;16:652–656. doi: 10.1038/nbt0798-652. [DOI] [PubMed] [Google Scholar]
- 23.Cha JN, et al. Silicatein filaments and subunits from a marine sponge direct the polymerization of silica and silicones in vitro. Proc Natl Acad Sci USA. 1999;96:361–365. doi: 10.1073/pnas.96.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curnow P, et al. Enzymatic synthesis of layered titanium phosphates at low temperature and neutral pH by cell-surface display of silicatein-alpha. J Am Chem Soc. 2005;127:15749–15755. doi: 10.1021/ja054307f. [DOI] [PubMed] [Google Scholar]
- 25.Kisailus D, Truong Q, Amemiya Y, Weaver JC, Morse DE. Self-assembled bifunctional surface mimics an enzymatic and templating protein for the synthesis of a metal oxide semiconductor. Proc Natl Acad Sci USA. 2006;103:5652–5657. doi: 10.1073/pnas.0508488103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mugnaioli E, et al. Crystalline nanorods as possible templates for the synthesis of amorphous biosilica during spicule formation in Demospongiae. Chembiochem. 2009;10:683–689. doi: 10.1002/cbic.200800623. [DOI] [PubMed] [Google Scholar]
- 27.Murr MM, Morse DE. Fractal intermediates in the self-assembly of silicatein filaments. Proc Natl Acad Sci USA. 2005;102:11657–11662. doi: 10.1073/pnas.0503968102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schröder HC, et al. Apposition of silica lamellae during growth of spicules in the demosponge Suberites domuncula: Biological/biochemical studies and chemical/biomimetical confirmation. J Struct Biol. 2007;159:325–334. doi: 10.1016/j.jsb.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Shimizu K, Cha J, Stucky GD, Morse DE. Silicatein alpha: Cathepsin L-like protein in sponge biosilica. Proc Natl Acad Sci USA. 1998;95:6234–6238. doi: 10.1073/pnas.95.11.6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sumerel JL, et al. Biocatalytically templated synthesis of titanium dioxide. Chem Mater. 2003;15:4804–4809. [Google Scholar]
- 31.Wang XH, et al. Evagination of cells controls bio-silica formation and maturation during spicule formation in sponges. PLoS ONE. 2011;6:e20523. doi: 10.1371/journal.pone.0020523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y, Shimizu K, Cha JN, Stucky GD, Morse DE. Efficient catalysis of polysiloxane synthesis by silicatein alpha requires specific hydroxy and imidazole functionalities. Angew Chem Int Ed Engl. 1999;38:780–782. doi: 10.1002/(SICI)1521-3773(19990315)38:6<779::AID-ANIE779>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 33.Naik RR, Brott LL, Clarson SJ, Stone MO. Silica-precipitating peptides isolated from a combinatorial phage display peptide library. J Nanosci Nanotechnol. 2002;2(1):95–100. doi: 10.1166/jnn.2002.074. [DOI] [PubMed] [Google Scholar]
- 34.Mena MA, Treynor TP, Mayo SL, Daugherty PS. Blue fluorescent proteins with enhanced brightness and photostability from a structurally targeted library. Nat Biotechnol. 2006;24:1569–1571. doi: 10.1038/nbt1264. [DOI] [PubMed] [Google Scholar]
- 35.Stephens CJ, Kim YY, Evans SD, Meldrum FC, Christenson HK. Early stages of crystallization of calcium carbonate revealed in picoliter droplets. J Am Chem Soc. 2011;133:5210–5213. doi: 10.1021/ja200309m. [DOI] [PubMed] [Google Scholar]
- 36.Stemmer WPC. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 37.Zha W, Zhu T, Zhao H. Family shuffling with single-stranded DNA. In: Arnold FH, Georgiou G, editors. Directed Evolution Library Creation: Methods and Protocols. Totowa, NJ: Humana Press; 2003. pp. 91–97. [DOI] [PubMed] [Google Scholar]
- 38.Hecky RE, Mopper K, Kilham P, Degens ET. Amino-acid and sugar composition of diatom cell-walls. Marine Biology. 1973;19:323–331. [Google Scholar]
- 39.Tilburey GE, Patwardhan SV, Huang J, Kaplan DL, Perry CC. Are hydroxyl-containing biomolecules important in biosilicification? A model study. J Phys Chem B. 2007;111:4630–4638. doi: 10.1021/jp068539s. [DOI] [PubMed] [Google Scholar]
- 40.Fairhead M, et al. Crystal structure and silica condensing activities of silicatein alpha-cathepsin L chimeras. Chem Commun. 2008:1765–1767. doi: 10.1039/b718264c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murr MM, et al. New pathway for self-assembly and emergent properties. Nano Today. 2009;4(2):116–124. [Google Scholar]
- 42.Frampton M, Vawda A, Fletcher J, Zelisko PM. Enzyme-mediated sol-gel processing of alkoxysilanes. Chem Commun. 2008:5544–5546. doi: 10.1039/b812389f. [DOI] [PubMed] [Google Scholar]
- 43.Smith GP, Baustian KJ, Ackerson CJ, Feldheim DL. Metal oxide formation by serine and cysteine proteases. J Mater Chem. 2009;19:8299–8306. [Google Scholar]
- 44.Wang J, et al. A dedicated powder diffraction beamline at the advanced photon source: Commissioning and early operation results. Rev Sci Instrum. 2008;79 doi: 10.1063/1.2969260. 085105. [DOI] [PubMed] [Google Scholar]
- 45.Abbate V, Bassindale AR, Brandstadt KF, Taylor PG. A large scale enzyme screen in the search for new methods of silicon-oxygen bond formation. J Inorg Biochem. 2011;105:268–275. doi: 10.1016/j.jinorgbio.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Unuma H, Matsushima Y, Kawai T. Enzyme-mediated synthesis of ceramic materials. Journal of the Ceramic Society of Japan. 2011;119:623–630. [Google Scholar]
- 47.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 48.Moll A, Hildebrandt A, Lenhof HP, Kohlbacher O. BALLView: An object-oriented molecular visualization and modeling framework. Journal of Computer-Aided Molecular Design. 2005;19(11):791–800. doi: 10.1007/s10822-005-9027-x. [DOI] [PubMed] [Google Scholar]