Knowledge that microbes lead fascinating chemical lives is as old as microbiology itself. Observations in the late 19th century by Burdon-Sanderson (1) that were elaborated on by Pasteur and Joubert (2) gave rise to the idea that compounds secreted by some microbes could have remarkable effects on the lives of others. The accretion of such knowledge continued steadily through the early years of the 20th century until the quantum leap emanating from Fleming’s seminal discovery of penicillin (3). Chain and Florey’s subsequent development of penicillin as a therapeutic agent ushered in not only the golden era of antibiotics but an entire pharmaceutical industry based largely on small-molecule natural products. Since those days, the chemical lives of microbes have been studied largely through labor-intensive and time-consuming approaches that addressed one molecule at a time. Although this time-honored approach has yielded much new knowledge, the rate of discovery slowed down dramatically in recent decades. Perhaps all there was to discover about small-molecule natural products had been discovered. The advent of genomics put a halt to that idea; genomes revealed a great potential diversity of secreted microbial products waiting to be found (4). However, progress in realizing that potential has been slow (5). In PNAS, Watrous et al. offer a brand new way of viewing secreted microbial products that holds great promise in providing the next quantum leap in understanding the fascinating world of microbial chemical ecology (6).
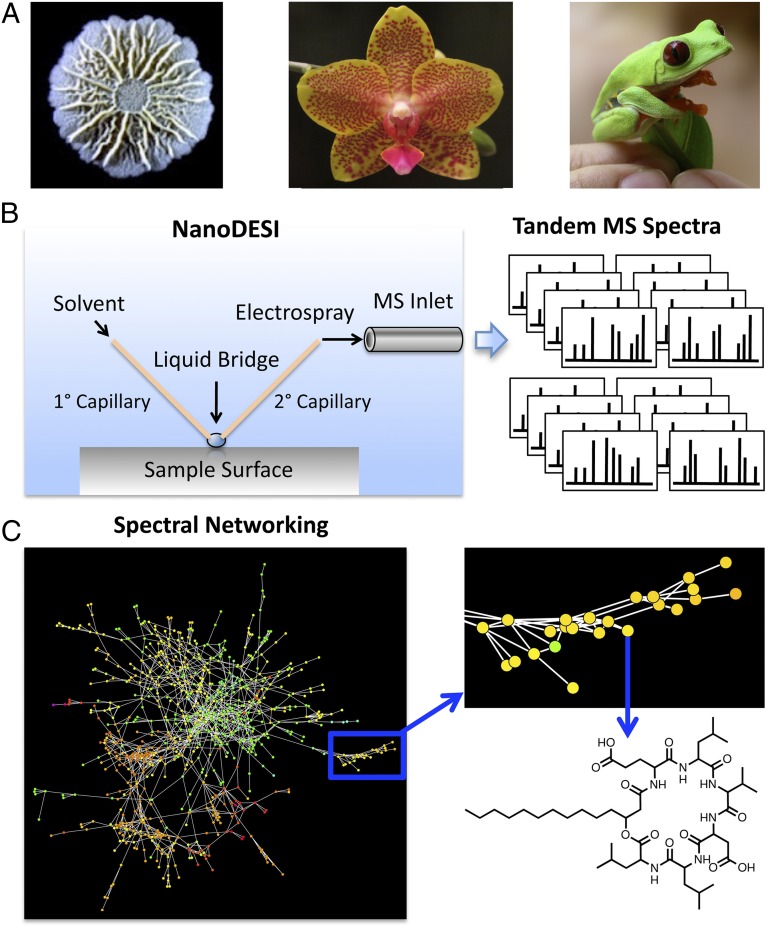
The new approach applies rapid and highly sensitive MS to directly identify and characterize molecules secreted by bacterial colonies (6), thus circumventing many shortcomings of conventional MS approaches (reviewed recently in ref. 7). In theory, this approach could be applied to any accessible surface colonized by microbes, abiotic or biotic. The microbial communities living on the surfaces of plants and animals might some day be queried with this methodology, providing us with heretofore unimaginable views of the richness of microbial chemistry (Fig. 1A).
Fig. 1.
NanoDESI MS workflow. (A) NanoDESI allows for direct sampling of almost any surface (biotic or otherwise), including bacterial colonies, plants, and animals. (B) Solvent is pumped onto and then off of the sample surface and subsequently electrosprayed into the mass spectrometer. Ions of individual parent mass are fragmented yielding hundred or thousands of tandem mass spectra. (C) Parent masses represented as nodes are built into networks based on MS2 spectral similarity, allowing for analysis of individual compounds within the context of the entire chemical profile.
Watrous et al. (6) integrate two methodological advances, namely nanospray desorption electrospray ionization (nanoDESI) MS and spectral network construction, into this exciting new platform for metabolomic profiling and natural products discovery. In the nanoDESI workflow (Fig. 1B), solvent is pumped through a primary capillary directly onto the sample surface, where molecules of interest are desorbed. A secondary capillary with one end positioned near the surface draws up the sample analyte, which is subsequently electrosprayed into the mass spectrometer inlet. This setup offers several compelling advantages. The approximately microliter-sized solvent bridge between the two capillaries allows for direct sampling with relatively high resolution. When the analyte has been aspirated into the mass spectrometer, the hundreds or thousands of different ions from the sample are fragmented, and their characteristic fragmentation spectra (i.e., MS2) are obtained. The overall ease and comprehensive nature of the sampling, combined with the high sensitivity of MS2 analysis, allows researchers an unprecedented view of the compounds present. When Watrous et al. (6) sampled living bacterial colonies of several species in this way, they found an astonishing diversity of molecules, surpassing by far our current views of bacterial secondary metabolism.
As with many high-throughput techniques, nanoDESI tandem mass spectral analysis generates data at a rate that drastically outpaces our current capacity for interpretation. Watrous et al. (6) present an ingenious way to overcome this obstacle. They have developed a data analysis approach that allows for visualization of compounds (as represented by nodes of discrete parent masses) in the context of networks based on chemical similarity (Fig. 1C). The resulting molecule networks, which serve as “chemical fingerprints,” can be considered at multiple levels—from system-wide to structural characterization of individual compounds.
In their study, Watrous et al. (6) provide examples of analyses that function at both levels. By sampling compounds from a single colony of Bacillus subtilis at four times, they were able to observe temporal activation and suppression of discrete sections of B. subtilis secondary metabolism. In some cases, these changes involved known compounds. Interestingly, they found that many of these known compounds were produced as multiple structural variants, indicative of unexpected biosynthetic plasticity over time. They also demonstrate that nanoDESI can be useful for detection and characterization of compounds that lay beyond the reach of more traditional biochemical approaches. A recent metagenomic study of the sugar beet rhizosphere found that certain Pseudomonads play a role in defending the plant from fungal infections (8). This protection was dependent on genes for production of a putative lipopeptide dubbed thanamycin. However, chemical detection and characterization of thanamycin had remained elusive. Through live-colony nanoDESI analysis of WT producer and nonproducing mutant strains of the bacteria, Watrous et al. were able to detect thanamycin and provide a partial chemical structure for this new molecule (6).
The stunning complexity of the initial chemical datasets presented by Watrous et al. call to mind other important conceptual shifts in (micro)biological thinking that have also been brought about by important advances in technology. An apt comparison might be the implementation of cloning of environmental DNA to examine microbial diversity found in wide-ranging ecosystems. After a short time of
NanoDESI MS combined with spectral networking may usher in a new era of discovery in the context of human therapeutics.
applying that technique, the community of investigators realized that the vast majority of microbial species had yet to be cultivated. Similarly, with the advent of genome sequencing came the realization that, while hundreds of genes had been assigned a function, thousands—if not millions—of genes of unknown function reside in microbial genomes. The inherent complexity of both cases limited the utility of purely reductionist approaches. This prompted the development of new paradigms and lexicons to facilitate the study of these new frontiers at a systems level. In the case of environmental diversity surveys, this led to development of algorithms for routinely building phylogenetic trees from sequence data. High-throughput genome sequencing has likewise given rise to the fields of comparative and functional genomics. An analogous change in worldview is likely to occur in the field of microbial secreted metabolites with the advent of nanoDESI.
Although some of the compounds observed in the datasets of Watrous et al. (6) are commonplace adducts of known compounds, many more are likely to be completely new compounds and/or important new structural variants of known compounds. In either case, the data imply that the chemical landscape inhabited and manipulated by bacteria is vastly more complex and sophisticated than previously thought. Because almost any sample type can be directly probed by using nanoDESI, the door is open to explore the chemical networks that underlie microbial interactions in myriad environments. For example, what chemical ecology underlies the microbiome associated with human skin? What signals potentiate relationships between hosts and symbionts? How do soil bacteria, long known to make a variety of antibiotics, deploy their arsenals in the presence of competitors? Beyond the characterization of chemical networks associated with a given environment or population, one can also see that correlating metabolic output with genome-wide transcriptomics might offer yet another dimension for systems-level analysis.
Natural products research has enjoyed a recent renaissance prompted by bioinformatic analyses of biosynthetic gene clusters. However, the discovery of new, therapeutically useful compounds has remained a challenge as a result of the laborious nature of structure determination and the high occurrence of compound rediscovery. Beyond facilitating systems-level analysis, nanoDESI also holds great promise in addressing this challenge. The ease of this technique will allow for metabolic profiling of sample types heretofore unexplored. Perhaps even more importantly, the spectral networks segregate compounds into distinct clusters of structural relatedness. This makes it possible to seed the networks with MS2 spectra of known compounds, and also opens the door to querying entire datasets against databases of defined MS2 spectra. Both cases may provide information to guide researchers to compounds of novel structure and activity. As such, nanoDESI MS combined with spectral networking may usher in a new era of discovery in the context of human therapeutics.
Acknowledgments
Research on interspecies interactions in our lab is supported by the National Institutes of Health (NIH) Grant GM82137 (to R.K.). M.F.T. is a NIH Postdoctoral Fellow (Grant 5F32GM089044-02).
Footnotes
References
- 1.Burdon-Sanderson J. 13th Report of the Medical Officer of the Privy Council [John Simon], with Appendix, 1870. London: Her Majesty’s Stationery Office; 1871. Appendix No. 5—Further report of researches concerning the intimate pathology of contagion. The origin and distribution of microzymes (bacteria) in water, and the circumstances which determine their existence in the tissue and liquids of the living body; pp. 56–66. [Google Scholar]
- 2.Pasteur L, Joubert JF. Charbon et septicemie. C R Soc Biol Paris. 1877;85:101–115. [Google Scholar]
- 3.Fleming A. On the antibacterial action of cultures of a Penicillium, with special reference to their use in the isolation of B. influenzæ. Br J Exp Pathol. 1929;10:226–236. [Google Scholar]
- 4.Walsh CT, Fischbach MA. Natural products version 2.0: Connecting genes to molecules. J Am Chem Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baltz RH. Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol. 2008;8:557–563. doi: 10.1016/j.coph.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Watrous J, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA. 2012;109:E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watrous JD, Dorrestein PC. Imaging mass spectrometry in microbiology. Nat Rev Microbiol. 2011;9:683–694. doi: 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendes R, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]