Abstract
The pharmacologically important tropane alkaloids have a scattered distribution among angiosperm families, like many other groups of secondary metabolites. To determine whether tropane alkaloids have evolved repeatedly in different lineages or arise from an ancestral pathway that has been lost in most lines, we investigated the tropinone-reduction step of their biosynthesis. In species of the Solanaceae, which produce compounds such as atropine and scopolamine, this reaction is known to be catalyzed by enzymes of the short-chain dehydrogenase/reductase family. However, in Erythroxylum coca (Erythroxylaceae), which accumulates cocaine and other tropane alkaloids, no proteins of the short-chain dehydrogenase/reductase family were found that could catalyze this reaction. Instead, purification of E. coca tropinone-reduction activity and cloning of the corresponding gene revealed that a protein of the aldo-keto reductase family carries out this reaction in E. coca. This protein, designated methylecgonone reductase, converts methylecgonone to methylecgonine, the penultimate step in cocaine biosynthesis. The protein has highest sequence similarity to other aldo-keto reductases, such as chalcone reductase, an enzyme of flavonoid biosynthesis, and codeinone reductase, an enzyme of morphine alkaloid biosynthesis. Methylecgonone reductase reduces methylecgonone (2-carbomethoxy-3-tropinone) stereospecifically to 2-carbomethoxy-3β-tropine (methylecgonine), and has its highest activity, protein level, and gene transcript level in young, expanding leaves of E. coca. This enzyme is not found at all in root tissues, which are the site of tropane alkaloid biosynthesis in the Solanaceae. This evidence supports the theory that the ability to produce tropane alkaloids has arisen more than once during the evolution of the angiosperms.
Keywords: convergent evolution, pseudotropine, immunoprecipitation, immunolocalization
Tropane alkaloids consist of over 200 known compounds with a tropane ring in their structures, such as the anticholinergic drugs atropine and scopolamine and the stimulant cocaine (1). Like other plant secondary metabolites, tropane alkaloids have a scattered distribution in the angiosperms, being reported from seven families, including the Proteaceae, Convolvulaceae, Brassicaceae, Euphorbiaceae, Rhizophoraceae, Solanaceae, and Erythroxylaceae (2), many of which are taxonomically distant from one another. For example, the Erythroxylaceae and Solanaceae are members of completely different clades, and their last common ancestor is believed to have lived ∼120 million y ago (3). The uneven distribution of tropane alkaloids in the angiosperms and the evolutionary distances between those families producing them suggest two alternative hypotheses: Either the ability to produce tropane alkaloids has arisen independently in multiple plant lineages, or the tropane alkaloid biosynthetic pathway was present in an ancestor basal to much of the angiosperms and was subsequently lost in most lineages. Determining which hypothesis is correct may help explain why many other groups of secondary metabolites also have scattered distributions among the angiosperms.
Studies of the biosynthesis of tropane alkaloids have been predominantly performed with members of the Solanaceae and to a lesser extent with cultivated species of the Erythroxylaceae. The majority of these studies used in vivo feeding of radiolabeled precursors (4–6) to elucidate the outlines of a general tropane alkaloid biosynthetic pathway (7, 8). Biosynthesis is initiated from the polyamine putrescine, which is derived from the amino acids ornithine or arginine (Fig. S1). Putrescine becomes N-methylated via the action of putrescine methyltransferase in what is generally considered to be the first committed step in tropane alkaloid production (9). This compound is then oxidized to yield 4-(methyl-1-amino)butanal, which under normal physiological conditions is thought to spontaneously rearrange into the five-membered-ring compound N-methyl-Δ1-pyrrolinium. The formation of the second ring in tropane alkaloid biosynthesis is a subject of debate. One hypothesis is that N-methyl-Δ1-pyrrolinium condenses with acetoacetate and the ring closure occurs following oxidation and another round of aldol condensation (10). Another hypothesis is that the N-methyl-Δ1-pyrrolinium acts as a starter unit for two rounds of polyketide-type extension with malonyl-CoA (11). In both schemes, the resulting second ring contains a keto function at the 3 position. Reduction at this position is required for subsequent ester formation.
In members of the Solanaceae, reduction of the keto group in the tropane ring is catalyzed by enzymes known as tropinone reductases (TRs) (12). These enzymes belong to the short-chain dehydrogenase/reductase (SDR) enzyme family, which are NAD(P)(H)-dependent monomeric oxidoreductases with low sequence identities and a catalytic Asn-Ser-Tyr-Lys tetrad (13). There are two separate tropinone reductases (TRI and TRII) in the Solanaceae, and they lead to a significant bifurcation of tropane alkaloid biosynthesis in this family. TRI converts the 3-keto function exclusively to a product with a 3α-configuration, producing a tropine (3α-tropanol), which is the precursor to a variety of esterified tropane alkaloids, such as atropine. On the other hand, TRII produces exclusively an alcohol with a 3β-configuration, referred to as pseudotropine (3β-tropanol), which is then converted to various nonesterified tropane alkaloids called calystegines. The occurrence of these two separate TRs in the Solanaceae has been attributed to a gene-duplication event (14). TRs are characteristic of tropane alkaloid biosynthesis, and are even proposed to be limited to tropane alkaloid-producing plants (15). However, it is unclear whether all tropane alkaloid biosynthesis relies on TRs for catalysis. Besides the TRs of the short-chain dehydrogenase/reductase family, there are several other major groups of plant proteins that contain enzymes that could supply reductase activity for tropane alkaloid biosynthesis, including the medium-chain dehydrogenase/reductases, the aldehyde dehydrogenases, and the aldo-keto reductases.
To determine whether tropane alkaloid formation in distant angiosperm lineages has a common evolutionary origin, we investigated the tropinone-reduction step in Erythroxylum coca (Erythroxylaceae), a species that accumulates cocaine and other tropane alkaloids in abundance (16) and is taxonomically very remote from the Solanaceae. For the tropane alkaloids of E. coca, tropinone reduction involves the conversion of 2β-carbomethoxy-3-tropinone (methylecgonone) to 2β-carbomethoxy-3β-tropine (methylecgonine). Interestingly, tropinone reductases from the Solanaceae have not been shown to have significant methylecgonone reducing activity (15, 17), suggesting the existence of a different type of catalyst in the Erythroxylaceae.
In this study, we report the biochemical and molecular characterization of the tropinone-reduction step of tropane alkaloid biosynthesis in E. coca, which converts methylecgonone to methylecgonine, the penultimate step in cocaine biosynthesis. Our initial approach was based on the assumption that this reduction step is carried out by a TR-like member of the SDR family of reductases. However, homology-based cloning and heterologous expression of TR-like sequences from organs involved in tropane alkaloid biosynthesis failed to yield an enzyme with the requisite activity. We therefore purified the tropinone reductase activity from E. coca leaves, isolated the corresponding gene, and found it to encode a member of the aldo-keto reductase protein family. The result demonstrates that a major enzyme of tropane alkaloid biosynthesis has been independently recruited in the Solanaceae and the Erythroxylaceae, and suggests that the entire pathway has evolved more than once in the evolution of the angiosperms.
Results
Homology-Based Attempt to Isolate a Methylecgonone Reductase from E. coca Was Not Successful.
The tropane alkaloid biosynthetic pathway in the Erythroxylaceae is thought to be similar to that in the Solanaceae (7, 18). Therefore, enzymes such as the tropinone reductases identified in the Solanaceae that reduce tropinone to tropines should exist as homologs in the Erythroxylaceae with the ability to reduce methylecgonone (2-carbomethoxy-3-tropinone) to methylecgonine (2-carbomethoxy-3β-tropine) (Fig. 1 and Fig. S1). Starting with this hypothesis, known TR protein sequences (GenBank accession nos. AAA33281, AAA33282, ACG34080, CAC34420, CAD20555, BAA85844, AAB09776, and CAB52307) were blasted individually with “tblastn” against an E. coca EST library (19). Three sequences were identified that encoded for polypeptides ranging between 258 and 275 amino acids (GenBank accession nos. JQ015102, JQ015103, and JQ015104). The full-length ORFs of these tropinone reductase-like genes were cloned and expressed in Escherichia coli, and the crude supernatants were tested for TR activity. To verify the appropriateness of the assay conditions, Solanum tuberosum tropinone reductase II (GenBank accession no. CAB52307) was used as a positive control. The expressed empty pET-28a vector served as a negative control. Tropinone reductase activity was detected in the positive control and the crude plant extract, but not for any of the heterologously expressed tropinone reductase-like enzymes from E. coca.
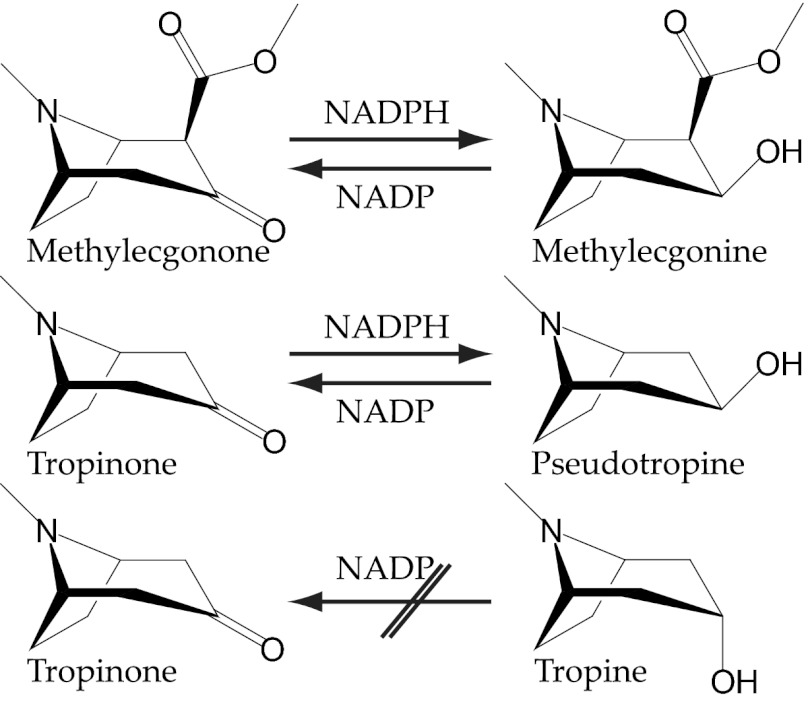
Fig. 1.
MecgoR catalyzes the reduction of methylecgonone (2-carbomethoxy-3-tropinone) to methylecgonine (2-carbomethoxy-3β-tropine) and tropinone to 3β-tropine (pseudotropine). The oxidation in the reverse direction is possible, but stereoselective: The 3β-isomers are converted, but not the 3α-isomers.
Methylecgonone Reductase Activity Was Detected in Various Above-Ground Organs of E. coca.
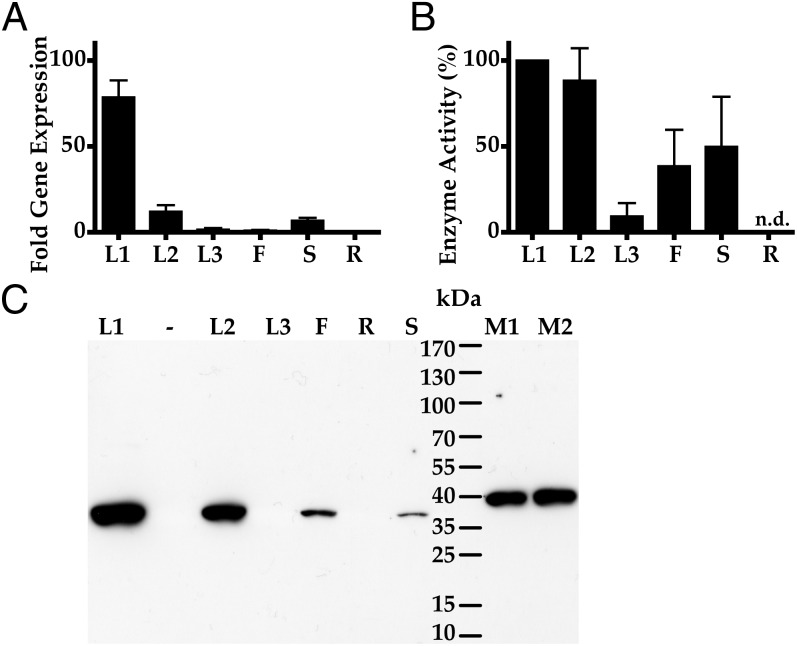
Although none of the proteins encoded by tropinone reductase-like genes from E. coca exhibited any detectable TR activity, it was possible to detect this activity in crude plant extracts. Assays of desalted plant extracts from various organs showed the ability to reduce 2-carbomethoxy-3-tropinone (methylecgonone) to 2-carbomethoxy-3β-tropine (methylecgonine), the expected reaction in the biosynthesis of cocaine. Methylecgonine was initially identified by comparison of chromatographic retention time and mass spectral fragmentation patterns with those reported in the literature for an authentic standard and later confirmed by 1H NMR comparison (see below). The highest methylecgonone reductase (MecgoR) activity was found in the youngest leaves (stage 1; Fig. S2) at 249.4 pkat (mg protein)−1 (Fig. 2B), and the next-highest activity in stage 2 leaves. Stem and flower extracts exhibited less than 20% of the activity of the youngest leaves. No methylecgonone reducing activity was detected in either the stage 3 (mature) leaves or the roots.
Fig. 2.
Comparison of methylecgonone reductase gene expression, enzyme activity, and protein levels in different E. coca organs and development stages: leaf stage 1 (L1), leaf stage 2 (L2), leaf stage 3 (L3), flowers (F), stems (S), and roots (R). n.d., not detected. (A) Relative transcript levels of MecgoR in 4-mo-old plants normalized to internal reference genes. Transcript levels in the flowers were set to a value of 1. Values displayed are means ± SD of three technical replicates from each of three biological replicates. DNA blot analysis and other information (Fig. S7) suggest that MecgoR is a single-copy gene. (B) Enzyme activity. Depicted are the means of activity obtained from desalted protein extracts from three replicates of each organ and developmental stage. Highest activity was detected in stage 1 leaves at 249.4 pkat (mg protein)−1. The reaction was run in the reducing direction, converting methylecgonone to methylecgonine. (C) Protein levels were determined by immunoblotting. Samples consisting of 50 μg of protein extracted from each organ as well as 250 ng (M1) and 500 ng (M2) 9× His-tagged MecgoR (40.4 kDa) were run on SDS/PAGE and gels were blotted onto filters. The filters were first probed with anti-MecgoR antibodies, followed by incubation with secondary antibodies conjugated to horseradish peroxidase. Polyclonal antibodies are specific for MecgoR protein (Fig. S8). Bands were visualized with chemiluminescence.
Localization of MecgoR activity is consistent with the previously reported localization of alkaloid (cocaine and cinnamoyl cocaine) content in E. coca plants (20). No alkaloids were detected in roots, with only low cocaine and cinnamoyl cocaine levels found in the stem. The highest alkaloid levels (cinnamoyl cocaine) were present in the early leaf developmental stages, the same stages that show high MecgoR activity in the present study.
MecgoR Activity Was Purified from E. coca and the Corresponding Gene Was Isolated.
Because we were able to readily detect MecgoR activity in young leaves, this activity was purified from young leaf extracts (Table S1). After ammonium sulfate fractionation and purification of the 40–80% ammonium sulfate pellet via hydrophobic-interaction chromatography (phenyl Sepharose), a >100-fold purification was achieved. Purity was increased over fourfold further by dye-interaction chromatography on a column with the Cibacron blue F3G-A dye, which has been previously shown to bind proteins that use nucleotide cofactors (21), such as the NADPH used in the reduction of methylecgonone. Additional purification was achieved by anion-exchange chromatography, although there was a drop in the specific activity, most likely due to the interference of high salt concentrations in the enzyme assay. The fraction containing the highest activity was subsequently subjected to gel filtration to determine a mass of 36.37 kDa for the active protein.
A preparative 2D gel of the active fraction from the anion-exchange column revealed over 30 spots following staining with colloidal Coomassie blue. All visually identified proteins were then eluted for de novo sequencing via protein mass spectrometry, and the resulting peptide fragment sequences corresponding to the isolated proteins were blasted against the E. coca EST library (22). Three full-length sequences coding for polypeptides of 253–364 amino acid residues were isolated and cloned into the expression vector pH9GW for heterologous expression in E. coli (GenBank accession nos. GU562618, JQ804916, and JQ804917) (Fig. S3). One polypeptide consisting of 327 amino acid residues exhibited reducing activity with both tropinone and methylecgonone, and was designated MecgoR.
Immunoprecipitation Confirmed That the Protein Encoded by the MecgoR Gene Is Responsible for the Corresponding Activity in Planta.
Polyclonal antibodies were produced against the heterologously expressed MecgoR protein. These reduced the methylecgonone reductase activity of the young leaf extract to 6% of that of a control extract without antibodies. Preimmune serum reduced enzyme activity to 60% of the control. The immunoprecipitated proteins were separated on a protein gel and sequencing identified MecgoR within the precipitate formed by the anti-MecgoR antibodies, but this protein was not detected in the precipitate formed by the preimmune serum.
MecgoR Is Not Related to the TRs of the Solanaceae, but Is a Member of the Aldo-Keto Reductase Protein Family.
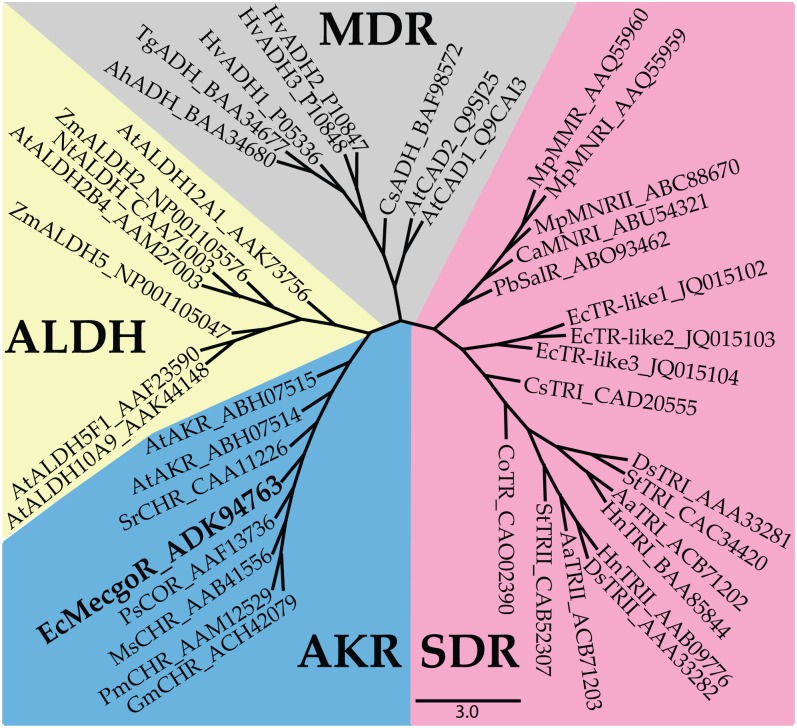
Amino acid analysis of MecgoR revealed that the enzyme is most similar to three proteins from the aldo-keto reductase (AKR) enzyme family: chalcone reductase from Sesbania rostrata, codeinone reductase from Papaver somniferum, and deoxymugineic acid synthase from Zea mays. An alignment with these three sequences revealed that MecgoR shares 48%, 50%, and 58% identity at the amino acid level with deoxymugineic acid synthase, codeinone reductase, and chalcone reductase, respectively. MecgoR and other proteins of the AKR family are distinct from the tropinone reductases of the Solanaceae, which are assigned to the short-chain dehydrogenase/reductase family. The overall identity of MecgoR with any TRI or TRII is less than 10% at the amino acid level. MecgoR is also not closely related to members of the two other large protein families containing reductases, the medium-chain dehydrogenases/reductases and the aldehyde dehydrogenases (Fig. 3).
Fig. 3.
Phylogenetic relationships of reductases of plant secondary metabolism. Selected reductases of four reductase superfamilies were aligned using the CLUSTAL X program with standard settings for protein alignment. The phylogenetic tree was built by the Bayesian method using the MRBAYES program. The four reductase superfamilies shown are ALDH (aldehyde dehydrogenases), SDR (short-chain dehydrogenases/reductases), MDR (medium-chain dehydrogenases/reductases), and AKR (aldo-keto reductases). MecgoR, which belongs to the AKR family, is depicted in bold. The tropinone reductases carrying out similar reactions in the biosynthesis of tropane alkaloids in the Solanaceae belong to the SDR family. The scale bar represents 3.0 amino acid substitutions per site. Please refer to Table S2 for an explanation of the abbreviated names.
MecgoR Shows Considerable Specificity for Substrate, Cofactor, and Other Properties.
To obtain enough pure protein for biochemical characterization, the coding region of MecgoR was cloned into a vector that introduced an N-terminal strep tag and was used for heterologous expression in the yeast Pichia pastoris. The recombinant protein increased in size from 36.9 kDa to 39.8 kDa with the N-terminal strep tag, and was purified to homogeneity following StrepTrap HP and HiTrap Blue affinity chromatography. The pH optimum for the reduction of methylecgonone to methylecgonine was determined to be 6.8, and 9.8 for the reverse reaction. When preincubated at 4, 25, 37, 50, and 65 °C for 30 min, MecgoR activity was reduced over 30% at 25 °C and over 70% at 37 °C compared with the 4 °C sample and was undetectable at higher temperatures. MecgoR activity was not stimulated by the addition of monovalent or divalent metal cations. Cu2+ and Zn2+ had a strong inhibitory effect on enzyme performance when tested at concentrations of 5 mM, reducing activity to 8% and 58%. However, both Co2+ and Fe2+ only inhibited MecgoR by less than 30%.
Of all of the compounds tested as substrates besides methylecgonone, only 6-hydroxytropinone, tropinone, and nortropinone were reduced to their corresponding alcohols with 45%, 36%, and 6%, respectively, of the activity with methylecgonone as substrate (Fig. S4). No activity was detected for 8-thiabicyclo[3.2.1]octan-3-one (TBON), cyclohexanone, cyclooctanone, and N-methyl-4-piperidone. The Michaelis–Menten constant (Km) for methylecgonone and NADPH were 69.6 ± 14.1 μM and 5.1 ± 0.9 μM, respectively. For the reverse reaction in the oxidative direction, Km values of 151.2 ± 16.8 μM and 29.6 ± 5.3 μM were determined for methylecgonine and NADP, respectively (Table 1). Substrate inhibition was also observed, and the inhibitory constant (Ki) was 1,136.0 ± 257.9 μM for methylecgonone, 2,480.0 ± 1,149.0 μM for NADPH, 2,673.0 ± 484.6 μM for methylecgonine, and 506.3 ± 97.3 μM for NADP. NADH can successfully substitute for NADPH in the reduction of methylecgonone, but the activity is decreased to 14% of the value with NADPH. No activity was detected in the absence of a nucleotide cofactor.
Table 1.
Kinetic constants for E. coca MecgoR reaction measured in forward (reductive) and reverse (oxidative) directions
| Km (μM) | kcat (s−1) | kcat/Km (M−1s−1) | Ki (μM) | |
| Methylecgonone | 69.6 ± 14.1 | 0.240 ± 0.023 | 3,443.7 | 1,136.0 ± 257.9 |
| NADPH | 5.1 ± 0.9 | 0.121 ± 0.005 | 24,052.1 | 2,480.0 ± 1,149.0 |
| Methylecgonine | 151.2 ± 16.8 | 5.538 ± 0.329 | 36,627.0 | 2,673.0 ± 484.6 |
| NADP | 29.6 ± 5.3 | 5.238 ± 0.418 | 176,704.8 | 506.3 ± 97.3 |
The stereochemistry of the MecgoR reaction was first studied with tropinone as substrate. The configuration of the product was determined by comparing its 1H NMR spectrum to those of tropine (3α-tropanol) and pseudotropine (3β-tropanol) (Fig. S5). The product of the reduction of tropinone was found to be pseudotropine, without any trace of tropine detected. To confirm this, the reverse assay was performed with both tropine and pseudotropine as substrates, but only pseudotropine was converted to tropinone (Fig. 1). When the native substrate methylecgonone was used, the 1H NMR spectrum of the product was compared with that of a methylecgonine standard (Fig. S5). Although 3α-methylecgonine was not available for comparison, the spectrum of the product of the reduction of methylecgonone was found to match that of the commercial methylecgonine standard, including the expected pattern of signals for the H-3 position. Thus, the reduction of methylecgonone to methylecgonine is stereospecific, with the product C3-alcohol having exclusively a β-configuration.
MecgoR Gene Expression Was Highest in the Youngest Leaf Stage.
To compare the MecgoR enzymatic activity obtained from crude plant extracts of various organs and developmental stages with the expression of the MecgoR gene, quantitative real-time PCR was performed with transcript levels normalized to those in the flowers, the organ exhibiting the lowest detectable amounts of transcript (Fig. 2A). MecgoR expression was highest in stage 1 leaves, with an expression of 78-fold relative to the flowers. Transcript levels dropped to a value of 11-fold for stage 2 leaves, followed by 1.5-fold for stage 3 leaves. The stems had average transcript levels of 6.7-fold compared with flowers. MecgoR transcripts were not detected in the roots at a level above that in reactions with no-RNA controls.
MecgoR Protein Levels Were also Highest in Young Leaves.
The polyclonal antibodies raised against MecgoR were used to detect protein in different plant organs and developmental stages. Equal amounts of extracted protein from different organs were separated by a protein gel, transferred, and subjected to immunoblotting. MecgoR was detected as a single band with a size of ∼37 kDa in stage 1 and 2 leaves, flowers, and stems, in decreasing order of abundance. No MecgoR protein was detected in either the stage 3 leaves or in the roots.
The heterologously expressed His-tagged MecgoR was used as a control on the gel and appears slightly bigger due to the addition of 31 amino acids from the tag (Fig. 2C). The co-occurrence of the highest amount of MecgoR protein with the highest amount of gene transcript and the highest enzyme activity in stage 1 leaves suggests that MecgoR activity is controlled at the transcriptional level.
MecgoR Is Localized in the Leaf Mesophyll.
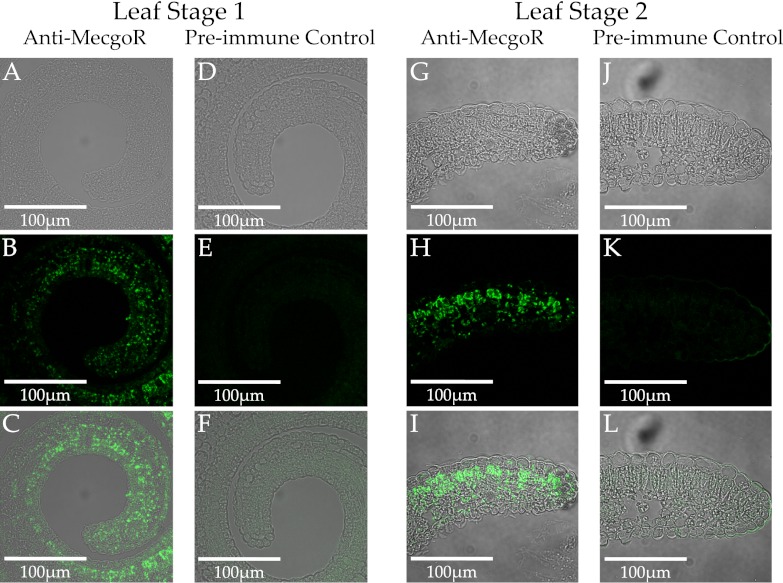
Based on the immunoblot results of various organs, young leaves and flowers were used for subsequent tissue-level immunolocalization experiments. Cross-sections of young leaves and longitudinal sections of flowers are depicted in Fig. 4 and Fig. S6, respectively. Preimmune serum was used as a negative control, and the fluorescence signal obtained from these samples was considered unspecific. Whereas there was only little fluorescence detected in the preimmune images of all three analyzed organs, a strong signal was detected after the application of anti-MecgoR antibodies. The overlay of transmitted light and fluorescent images localized the detected signal to the palisade and spongy mesophyll tissue of leaves and sepals. No MecgoR signal was detected in the upper or lower epidermis tissue or cuticle of these organs, and none in any tissue in the stems, roots, and mature leaves (Fig. 4).
Fig. 4.
Immunolabeling of MecgoR in cross-sections of E. coca leaf stage 1 (A–F) and leaf stage 2 (G–L). The samples were labeled with either anti-MecgoR antibodies (A–C and G–I) or preimmune serum (D–F and J–L). (Top) Transmitted light (A, D, G, and J). (Middle) Fluorescent images (B, E, H, and K). (Bottom) Overlays of transmitted light and fluorescent images (C, F, I, and L). Single sections were probed with primary antibody (anti-MecgoR or preimmune serum) and secondary antibody (anti-rabbit conjugated to horseradish peroxidase assayed with chemiluminescent substrate). Fluorescence excitation was at 543 nm and detection used a BP 585–615 filter.
Discussion
Tropane alkaloids, like other major categories of plant secondary metabolites, are scattered among the major clades of angiosperms. They are present in four major lineages of dicotyledons: the peripheral eudicots (Proteaceae), the malvid (Brassicaceae) and fabid (Erythroxylaceae, Rhizophoraceae, Euphorbiaceae) clusters of the rosid lineage, and the lamid cluster of the asterid lineage (Solanaceae, Convolvulaceae) (23). To determine whether this distribution is a result of independent evolution or a shared ancestral angiopserm pathway that was selectively lost in most lineages, we compared a key step of tropane biosynthesis between the Solanaceae and the Erythroxylaceae. The reduction of tropinone to tropine or pseudotropine in Solanaceae species is catalyzed by the well-described tropinone reductase enzymes of the SDR family (24). Screening of an E. coca young leaf EST database yielded three distinct TR-like genes; however, the corresponding proteins exhibited no reducing activity either with the substrate tropinone, typical of the Solanaceae, or the substrate methylecgonone, typical of the Erythroxylaceae. The methylecgonone reductase of E. coca, identified by purification of the enzymatic activity from an extract of young leaves and sequencing of the resulting protein, proved to be a member not of the short-chain dehydrogenase/reductase family but rather of the aldo-keto reductase family.
Confirmation of MecgoR’s role in tropane alkaloid biosynthesis came from immunoprecipitation experiments. In addition, the stereospecificity of the expressed protein (all products had a β-configuration at C-3) perfectly matched the C-3–configuration of the tropane alkaloids reported from E. coca (25). Both 3α- and 3β-tropane alkaloids are known in nature. In the Solanaceae, one group of TRs forms 3α-configured products and the other group forms products with a 3β-configuration (12), with several 3α- and 3β-configured products becoming esterified (1).
E. coca MecgoR belongs to the AKR protein family, a large group of enzymes not restricted to plants but also described from mammals, amphibians, yeast, protozoa, and bacteria (26). Previously characterized AKR proteins are very diverse in their biological roles, ranging from participating in reduction reactions in carbohydrate and steroid metabolism to various detoxification reactions (27). To date, all plant AKR enzymes described belong to one group (family 4) that shares a minimum of 40% amino acid identity (28). This group includes other enzymes of alkaloid metabolism (codeinone reductase), an enzyme of flavonoid biosynthesis (chalcone reductase), and a protein catalyzing a step in siderophore formation in graminaceous plants (deoxymugineic acid synthase). There are at least 21 members of the AKR family represented in the whole genome of Arabidopsis thaliana, although very few of the encoded enzymes have been characterized (29). All AKRs characterized so far share a common α/β-barrel motif that uses either NADH or NADPH as cofactors (26), and activity is mediated through the conserved catalytic AKR tetrad His, Lys, Tyr, and Asp.
The fact that species of the Solanaceae and Erythroxylaceae (E. coca) have recruited enzymes from two completely different protein groups to carry out the same reaction in tropane alkaloid biosynthesis strongly suggests that other steps of this pathway have also evolved independently in the two plant families. The distinct location of tropane alkaloid formation in the two families also supports this hypothesis. In the Solanaceae, tropane alkaloids are produced in the roots and transported to the leaves and reproductive organs. All biosynthetic enzymes, including the TRs, have been localized in root tissue (30, 31). However, in the Erythroxylaceae, tropane alkaloids are found in shoots. Moreover, we showed that the site of biosynthesis of these compounds in E. coca is the young leaves, based on the incorporation of 13CO2 in these compounds (20). In the present paper, MecgoR enzyme activity was found to be highest in young leaf tissue (Fig. 2), with lower levels in stems and flowers and no activity in roots. A similar pattern was found for the accumulation of MecgoR protein and transcript (Fig. 2). Within the leaf, the majority of MecgoR protein was localized to the palisade parenchyma and spongy mesophyll (Fig. 4).
Many other types of plant secondary metabolites have scattered distributions in the plant kingdom, and for these an independent origin of biosynthesis in different lineages is also likely. For example, the pyrrolizidine alkaloids are found in such diverse families as the Orchidaceae, Fabaceae, Boraginaceae, and Asteraceae. Analysis of the first biosynthetic step, homospermidine synthase, indicated that this enzyme has been recruited from a primary metabolic enzyme, deoxyhypusine synthase, at least four times over the course of evolution (32). Besides alkaloids, groups such as the cardenolides, a type of glycosylated steroid found in the Liliaceae, Ranunculaceae, Brassicaceae, and Apocynaceae, among other families, have also been proposed to have originated polyphyletically (33). Thus, the great diversity of secondary metabolites in plants is likely a product of the continual gain (and loss) of biosynthetic pathways throughout evolution.
The independent recruitment of novel enzymes into biochemical pathways often takes advantage of their broad substrate specificity (34), which becomes narrower under the influence of natural selection. E. coca MecgoR is different from the tropinone reductases of the Solanaceae in a variety of properties. Whereas MecgoR can use the Solanaceae TRII substrate tropinone, the TRII enzymes have no activity with the MecgoR substrate methylecgonone (15, 17). In addition, MecgoR is capable of catalyzing the reverse reaction whereas the TRII enzymes are not (35–37), although the reverse reaction has an optimal pH of 9.8 and is thus unlikely to be physiologically relevant for an enzyme that is thought to be localized in the cytosol. Other differences between the Solanaceae TRII enzymes and E. coca MecgoR are that the TRIIs are not able to substitute NADPH with NADH, as we have shown for MecgoR (35–37). Furthermore, the relative activity data presented in Fig. 4 suggest that MecgoR contains an active site that is optimized for the recognition of the carbomethoxy group of methylecgonone. In addition, we have determined that MecgoR can be inhibited by its substrate, which has not been reported for any of the previously characterized TRs or closely related members of the AKR family. Thus, MecgoR has not only been recruited to tropinone reduction separately from the tropinone reductases of the Solanaceae but has a number of very different biochemical characteristics.
In summary, we have shown that the reduction of the 3-keto function of tropane alkaloids in E. coca is catalyzed by MecgoR, which belongs to a class of enzymes that is very different from that of the Solanaceae enzymes catalyzing a similar reaction. This finding provides a strong indication that tropane alkaloid biosynthesis has evolved more than once in different plant lineages. There are several other lines of evidence suggesting that the pathway of tropane alkaloid biosynthesis in the Erythroxylaceae has an independent origin from tropane alkaloid formation in the Solanaceae and other plant families. For example, the Erythroxylaceae tropane alkaloids possess a carbomethoxy function on the tropane ring that is not found in tropane alkaloids of the Solanaceae. In addition, tropane alkaloids with 3β-aromatic ester functions, which are dominant in the Erythroxylaceae, are rarely found in the Solanaceae. There are also prominent spatial and temporal differences in tropane alkaloid biosynthesis between the Erythroxylaceae and the Solanaceae. To gain further insight into the evolution of tropane alkaloids in the Erythroxylaceae, focus on the early steps of the pathway is required.
Materials and Methods
A crude enzyme extract from young expanding E. coca leaves was purified over several chromatographic steps using an ÄKTApurifier preparative protein liquid chromatography system (GE Healthcare). Fractions were assayed for tropinone reducing activity, and both substrate and products were analyzed by LC-MS. A high-activity fraction from the final ion-exchange column was subjected to 2D gel electrophoresis, and selected spots were picked for de novo peptide sequence identification. Peptide sequences were used to screen a young leaf E. coca EST library. For biochemical characterization, heterologous expression was performed in P. pastoris KM71. The sequence of MecgoR was registered at GenBank (GenBank accession no. GU562618) as well as on the AKR website (http://www.med.upenn.edu/akr; accession no. AKR4B10). Polyclonal rabbit antibodies, produced from purified recombinant MecgoR protein, were used for immunoprecipitation and immunolocalization. Details of plant material, reagents, protein purification/sequencing, LC-MS, GC-MS, NMR, localization experiments, quantitative RT-PCR, phylogenetic analysis, and all other methods used in this study are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Birgit Dräger for providing a plasmid containing the tropinone reductase II from S. tuberosum as an enzyme assay control and the chemical TBON; Katrin Luck, Dr. Katharina Schramm, Dr. Daniel Giddings Vassão, Dr. Michael Phillips, Dr. Stefan Bartram, and Dr. Bettina Hause for technical assistance; and Dr. Alexander Muck and Dr. Natalie Wielsch for sequencing of the purified plant protein and the immunoprecipitated protein, respectively. We also thank the greenhouse team of the Max Planck Institute for Chemical Ecology for nursing and taking care of the plants.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GU562618, JQ015102–JQ015104, JQ804916, and JQ804917).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200473109/-/DCSupplemental.
References
- 1.Lounasmaa M, Tamminen T. The tropane alkaloids. In: Cordell GA, editor. The Alkaloids. Vol 44. New York: Academic; 1993. pp. 1–114. [Google Scholar]
- 2.Griffin WJ, Lin GD. Chemotaxonomy and geographical distribution of tropane alkaloids. Phytochemistry. 2000;53:623–637. doi: 10.1016/s0031-9422(99)00475-6. [DOI] [PubMed] [Google Scholar]
- 3.Magallón S, Castillo A. Angiosperm diversification through time. Am J Bot. 2009;96:349–365. doi: 10.3732/ajb.0800060. [DOI] [PubMed] [Google Scholar]
- 4.Leete E, Marion L, Spenser ID. The biogenesis of alkaloids. 12. The mode of formation of the tropine base of hyoscyamine. Can J Chem/Rev Can Chim. 1954;32:1116–1123. [Google Scholar]
- 5.Ahmad A, Leete E. Biosynthesis of tropine moiety of hyoscyamine from δ-N-methylornithine. Phytochemistry. 1970;9:2345–2347. [Google Scholar]
- 6.Leete E. Biosynthesis of cocaine and cuscohygrine in Erythroxylon coca. J Chem Soc Chem Commun. 1980:1170–1171. [Google Scholar]
- 7.Leete E. Recent developments in the biosynthesis of the tropane alkaloids. Planta Med. 1990;56:339–352. doi: 10.1055/s-2006-960979. [DOI] [PubMed] [Google Scholar]
- 8.Leete E, Bjorklund JA, Couladis MM, Kim SH. Late intermediates in the biosynthesis of cocaine: 4-(1-Methyl-2-pyrrolidinyl)-3-oxobutanoate and methyl ecgonine. J Am Chem Soc. 1991;113:9286–9292. [Google Scholar]
- 9.Hashimoto T, Yamada Y. Alkaloid biogenesis: Molecular aspects. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:257–285. [Google Scholar]
- 10.Humphrey AJ, O’Hagan D. Tropane alkaloid biosynthesis. A century old problem unresolved. Nat Prod Rep. 2001;18:494–502. doi: 10.1039/b001713m. [DOI] [PubMed] [Google Scholar]
- 11.Leete E, Kim SH. A revision of the generally accepted hypothesis for the biosynthesis of the tropane moiety of cocaine. J Am Chem Soc. 1988;110:2976–2978. [Google Scholar]
- 12.Dräger B. Tropinone reductases, enzymes at the branch point of tropane alkaloid metabolism. Phytochemistry. 2006;67:327–337. doi: 10.1016/j.phytochem.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Kavanagh KL, Jörnvall H, Persson B, Oppermann U. Medium- and short-chain dehydrogenase/reductase gene and protein families: The SDR superfamily: Functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci. 2008;65:3895–3906. doi: 10.1007/s00018-008-8588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakajima K, Hashimoto T, Yamada Y. Two tropinone reductases with different stereospecificities are short-chain dehydrogenases evolved from a common ancestor. Proc Natl Acad Sci USA. 1993;90:9591–9595. doi: 10.1073/pnas.90.20.9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimoto T, Nakajima K, Ongena G, Yamada Y. 2-Tropinone reductases with distinct stereospecificities from cultured roots of Hyoscyamus niger. Plant Physiol. 1992;100:836–845. doi: 10.1104/pp.100.2.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plowman T, Rivier L. Cocaine and cinnamoylcocaine content of Erythroxylum species. Ann Bot (Lond) 1983;51:641–659. [Google Scholar]
- 17.Couladis MM, Friesen JB, Landgrebe ME, Leete E. Chemistry of the tropane alkaloids and related compounds. 47. Enzymes catalyzing the reduction of tropinone to tropine and ψ-tropine isolated from the roots of Datura innoxia. Phytochemistry. 1991;30:801–805. [Google Scholar]
- 18.Abraham TW, Leete E. New intermediate in the biosynthesis of the tropane alkaloids in Datura innoxia. J Am Chem Soc. 1995;117:8100–8105. [Google Scholar]
- 19.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Docimo T, et al. The first step in the biosynthesis of cocaine in Erythroxylum coca: The characterization of arginine and ornithine decarboxylases. Plant Mol Biol. 2012;78:599–615. doi: 10.1007/s11103-012-9886-1. [DOI] [PubMed] [Google Scholar]
- 21.McGettrick AF, Worrall DM. Dye-ligand affinity chromatography. In: Cutler P, editor. Methods in Molecular Biology: Protein Purification Protocols. Vol 244. Totowa, NJ: Humana; 2004. pp. 151–157. [DOI] [PubMed] [Google Scholar]
- 22.Shevchenko A, et al. Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal Chem. 2001;73:1917–1926. doi: 10.1021/ac0013709. [DOI] [PubMed] [Google Scholar]
- 23.The Angiosperm Phylogeny Group An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 2009;161:105–121. [Google Scholar]
- 24.Brock A, Bieri S, Christen P, Dräger B. Calystegines in wild and cultivated Erythroxylum species. Phytochemistry. 2005;66:1231–1240. doi: 10.1016/j.phytochem.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 25.Johnson EL. Alkaloid content in Erythroxylum coca tissue during reproductive development. Phytochemistry. 1996;42:35–38. [Google Scholar]
- 26.Jez JM, Bennett MJ, Schlegel BP, Lewis M, Penning TM. Comparative anatomy of the aldo-keto reductase superfamily. Biochem J. 1997;326:625–636. doi: 10.1042/bj3260625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penning TM. Introduction and overview of the aldo-keto reductase superfamily. In: Penning TM, Petrash JM, editors. Aldo-Keto Reductases and Toxicant Metabolism. Vol 865. Washington, DC: Am Chem Soc; 2004. pp. 3–20. ACS Symposium Series. [Google Scholar]
- 28.Jez JM, Penning TM. The aldo-keto reductase (AKR) superfamily: An update. Chem Biol Interact. 2001;130–132:499–525. doi: 10.1016/s0009-2797(00)00295-7. [DOI] [PubMed] [Google Scholar]
- 29.Simpson PJ, et al. Characterization of two novel aldo-keto reductases from Arabidopsis: Expression patterns, broad substrate specificity, and an open active-site structure suggest a role in toxicant metabolism following stress. J Mol Biol. 2009;392:465–480. doi: 10.1016/j.jmb.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 30.Kaiser H, et al. Immunolocalisation of two tropinone reductases in potato (Solanum tuberosum L.) root, stolon, and tuber sprouts. Planta. 2006;225:127–137. doi: 10.1007/s00425-006-0335-8. [DOI] [PubMed] [Google Scholar]
- 31.Nakajima K, Hashimoto T. Two tropinone reductases, that catalyze opposite stereospecific reductions in tropane alkaloid biosynthesis, are localized in plant root with different cell-specific patterns. Plant Cell Physiol. 1999;40:1099–1107. doi: 10.1093/oxfordjournals.pcp.a029494. [DOI] [PubMed] [Google Scholar]
- 32.Reimann A, Nurhayati N, Backenköhler A, Ober D. Repeated evolution of the pyrrolizidine alkaloid-mediated defense system in separate angiosperm lineages. Plant Cell. 2004;16:2772–2784. doi: 10.1105/tpc.104.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wink M. Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry. 2003;64:3–19. doi: 10.1016/s0031-9422(03)00300-5. [DOI] [PubMed] [Google Scholar]
- 34.Tawfik DS. Messy biology and the origins of evolutionary innovations. Nat Chem Biol. 2010;6:692–696. doi: 10.1038/nchembio.441. [DOI] [PubMed] [Google Scholar]
- 35.Portsteffen A, Dräger B, Nahrstedt A. The reduction of tropinone in Datura stramonium root cultures by two specific reductases. Phytochemistry. 1994;37:391–400. doi: 10.1016/0031-9422(94)85066-6. [DOI] [PubMed] [Google Scholar]
- 36.Dräger B, Schaal A. Tropinone reduction in Atropa belladonna root cultures. Phytochemistry. 1994;35:1441–1447. [Google Scholar]
- 37.Keiner R, Kaiser H, Nakajima K, Hashimoto T, Dräger B. Molecular cloning, expression and characterization of tropinone reductase II, an enzyme of the SDR family in Solanum tuberosum (L.) Plant Mol Biol. 2002;48:299–308. doi: 10.1023/a:1013315110746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.