Abstract
Background
Studies in the past have shown that perforin expression is up-regulated during acute renal rejection, which provided hopes for a non-invasive and reliable diagnostic method to identify acute rejection. However, a systematic assessment of the value of perforin as a diagnostic marker of acute renal rejection has not been performed. We conducted this meta-analysis to document the diagnostic performance of perforin mRNA detection and to identify potential variables that may affect the performance.
Methodology/Principal Findings
Relevant materials that reported the diagnostic performance of perforin mRNA detection in acute renal rejection patients were extracted from electronic databases. After careful evaluation of the studies included in this analysis, the numbers of true positive, true negative, false positive and false negative cases of acute renal rejection identified by perforin mRNA detection were gathered from each data set. The publication year, sample origin, mRNA quantification method and housekeeping gene were also extracted as potential confounding variables. Fourteen studies with a total of 501 renal transplant subjects were included in this meta-analysis. The overall performance of perforin mRNA detection was: pooled sensitivity, 0.83 (95% confidence interval: 0.78 to 0.88); pooled specificity, 0.86 (95% confidence interval: 0.82 to 0.90); diagnostic odds ratio, 28.79 (95% confidence interval: 16.26 to 50.97); and area under the summary receiver operating characteristic curves value, 0.9107±0.0174. The univariate analysis of potential variables showed some changes in the diagnostic performance, but none of the differences reached statistical significance.
Conclusions/Significance
Despite inter-study variability, the test performance of perforin mRNA detected by polymerase chain reaction was consistent under circumstances of methodological changes and demonstrated both sensitivity and specificity in detecting acute renal rejection. These results suggest a great diagnostic potential for perforin mRNA detection as a reliable marker of acute rejection in renal allograft recipients.
Introduction
Renal transplantation has been the treatment of choice for patients with end-stage renal disease (ESRD) for decades. However, although novel and powerful immunosuppressive drugs have been developed, acute rejection (AR) remains a major cause of allograft dysfunction and allograft failure [1], [2]. Even a single episode of AR can be a strong predictor of graft failure [3].
Currently, the diagnosis of AR is established based on histological evaluation of allograft biopsy samples. However, biopsy is an invasive procedure that may cause biopsy-associated complications such as perirenal hematoma, hematuria and infection [4], [5], which restrict its application for serial surveillance testing. In addition, sampling error and the variability of the pathological changes of AR make it difficult to make definitive diagnoses based on renal biopsy in many cases [6]. Other methods such as ultrasonography and serum creatinine measurements can be indicative of ARbut cannot reach a conclusive diagnosis [7], [8]. Therefore, developing a reliable, specific and non-invasive diagnostic method for identifying ARwould be of great help to improve clinical practice in renal transplantation.
Since allograft infiltration by T lymphocytes is a distinctive feature of rejection, analyzing the expression of specific genes involved in T cell activation provides a new option for AR diagnosis. Among the numerous cell subsets that infiltrate the graft site, cytotoxic T lymphocytes (CTL) are one of the major effector cells during the AR response. Lipman et al. [9] and Suthanthiran et al. [10] revealed a significant increase in transcription of the gene encoding perforin, one of the predominant effector molecules of CTLs [11], in allograft biopsy samples from AR patients using polymerase chain reaction (PCR) techniques. Since that time, many studies had been conducted to validate this approach for AR diagnosis in the clinic, and the samples collected for analysis have been expanded from allograft biopsy samples to less invasive peripheral blood leukocytes (PBL) and urine samples.
Although increased levels of perforin mRNA were a common finding during AR in a series of studies [12]–[14], controversy still exists regarding the clinical utility of this test due to the single study design of the previous work and the variable laboratory methodology used to perform the test among the different studies. Herein, we performed a meta-analysis to document the diagnostic performance of perforin mRNA detection in the identification of AR and try to determine its clinical utility by seeking the potential variables that may affect the performance of this test. These data provide important insights that inform clinical physicians regarding the diagnosis of AR in renal transplantation.
Materials and Methods
Study Protocol
This analysis was conducted in accordance with a predetermined protocol following the recommendations of Deeks et al. [15]. The data collection and reporting were in accordance with Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement (Table S1).
Search Strategy
Relevant materials in the scientific literature were searched in electronic databases including MEDLINE, EMBASE and the Cochrane Database of Systematic Reviews prior to December 1st, 2011, without date or language limitations. The following combinations of key words were used to search for related studies: “perforin” AND (“renal transplant” OR “renal transplantation” OR “kidney transplant” OR “kidney transplantation”) AND “rejection.” The electronic searching was supplemented by checking reference lists from the identified articles for additional original reports.
Inclusion and Exclusion Criteria
Studies were included if they fulfilled the following criteria: (1) Two comparison groups of patients were necessary for every study: AR group and non-rejection group. (2) Patient samples were diagnosed as AR or non-rejection based on the histological evidence according to Banff classification. (3) Quantitative detection of perforin mRNA expression level was accomplished by PCR techniques. (4) The mRNA detection was conducted either at the same time as biopsy pathological evaluation or immediately after with the samples being frozen for preservation during the evaluation. (5) The expression level of perforin was compared to the chosen housekeeping genes which were expressed at a constant level in samples from different patient groups. (6) A specific cutoff value was set to interpret the perforin mRNA results as positive or negative for AR (for those studies which defined the results as “detectable” or “undetectable,” “detectable” results were regarded as positive and vice versa).
The following types of studies were excluded from this meta-analysis: (1) Works designated as conference abstracts, letters, case reports, editorials or reviews. (2) Studies only involving pediatric patients.
Assessment of Study Quality
The quality of each study’s methodology was assessed using the 14-item Quality Assessment of Diagnostic Accuracy Studies (QUADAS) list [16]. Each question was assigned with a response of yes, no, or unclear when evaluating each of the included studies. Since the assessment of quality related strongly to the reporting of results, a well conducted study could score poorly if the methods and results were not reported in sufficient detail. Therefore, we did not report the assessment in scores but in descriptive forms only.
Publication bias was tested using funnel plots and the Egger test by Stata statistical software (STATA) version 11.0 [17]. An asymmetric distribution of data points in the funnel plot and a quantified result of P<0.05 in the Egger test indicated the presence of potential publication bias [18].
Data Extraction
The following data were extracted from each eligible study: year of publication, sample origin, mRNA quantification method, housekeeping gene, and the number of true positive (TP), false positive (FP), false negative (FN) and true negative (TN) cases of AR identified by perforin mRNA levels. All subjects who displayed biopsy results with any degree of AR defined by Banff classification were assigned to the rejection group, regardless of cellular or humoral rejection. The subjects with biopsies showing no evidence of any types of rejection, including normal tissues and tissues with non-rejection pathological changes, were assigned to the non-rejection group. The selected articles were assessed by two reviewers (YS and XL), independently. Disagreements were resolved by consultation with a third reviewer (ZG).
Statistical Analysis
Data were analyzed using Meta-Disc Version 1.4 [19] and STATA version 11.0. The test performance of perforin mRNA detection for the identification of ARwas measured by the following indicators: sensitivity, specificity and diagnostic odds ratio (DOR). Sensitivity was represented by the proportion of AR cases that were correctly identified by the positive results of perforin mRNA levels. Specificity was represented by the proportion of non-rejection cases that were correctly identified by the negative results of perforin mRNA levels [20]. As different cutoff values were used in each study, there was the potential for a threshold effect which would affect the conclusions of this analysis. Therefore, it was more reliable to define the summary of test performance using DOR than simply pooling sensitivity and specificity together across the studies. DOR was an independent indicator ranging from 0 to infinity, which represented how much greater the odds of having AR were for patient with a positive perforin mRNA result than for patient with a negative perforin mRNA result. The higher the DOR, the better the discriminatory ability of the test was [21].
The summary receiver operating characteristic (SROC) curve was plotted based on the combination of sensitivity and specificity, and the area under the curve (AUC) value was then calculated as a global measurement of test performance. The closer the AUC was to 1, the better the test performance [22].
Because of potential heterogeneity between studies, effect sizes were pooled by random-effects models of DerSimonian and Laird in Meta Disc [23]. Empty cells were handled using a 0.5 continuity correction.
Heterogeneity
The χ2 test was used to examine heterogeneity in pooling sensitivity and specificity. The Cochran Q test was used to examine heterogeneity in pooling DOR. Heterogeneity was considered to be statistically significant when P<0.05 in these qualitative tests. We also conducted the I 2 test in every pooling analysis to quantitatively estimate the proportion of total variation across studies that was attributable to heterogeneity rather than chance. The I 2 value would range from 0 to 100%, with a value over 50% indicating significant heterogeneity.
The existence of a threshold effect would manifest as a curvilinear shape in the SROC curves. In addition, we used a Spearman correlation analysis to confirm the absence or presence of a threshold effect by looking for an inverse relationship between sensitivity and specificity. A value of P<0.05 would indicate a significant threshold effect was present [24].
Sensitivity Analysis
To determine whether any single study was incurring undue weight in the analysis, we systematically removed one set of study data and checked the pooled results for the remaining studies to see if they changed significantly. The sensitivity analysis was conducted for every study.
Univariate Analysis
To identify the sources of potential heterogeneity that influenced the results of this analysis, a univariate analysis was conducted. Based on the literature review, the following factors were chosen as potential variables that may have influenced the test performance: year of publication, sample type, mRNA quantification method and housekeeping gene selection. Data sets were stratified based on these factors and the test performance would be compared between subgroups using the DOR values and the AUC of the SROC curves as the major parameters. The comparison was conducted using random-effects models in STATA. A value of P<0.05 in the comparison of DOR indicated a significant change in the test performance due to the covariate.
Results
Literature Search and Characteristics of the Included Studies
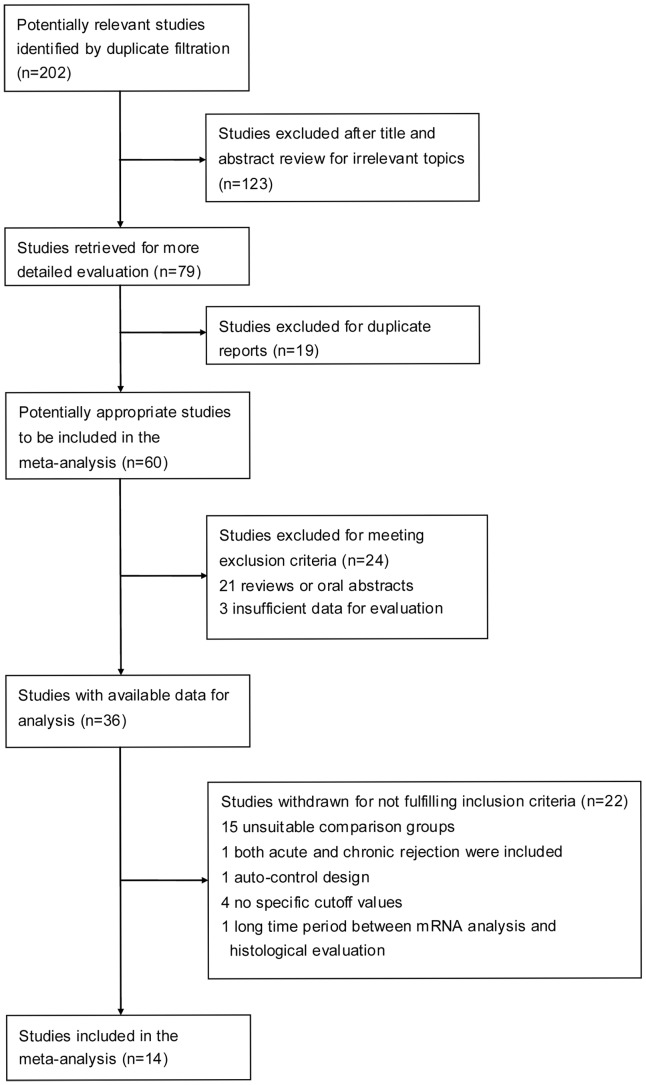
After the primary search of the electronic databases for published work on the subject, 202 studies were identified. Of these studies, 123 were excluded after further review of the title and abstract for irrelevant topics, and an additional 19 were excluded for duplication of the reports, which left 60 studies undergoing full text review. The detailed process of this literature search is shown in Figure 1.
Figure 1. Flow chart describing the literature search conducted for this meta-analysis.
After careful review, 14 studies with a total of 501 subjects were included in this meta-analysis. In 2 studies [25], [26], perforin expression was detected in both graft biopsy and PBL samples. In another study [27], perforin expression was detected in both PBL and urine samples. We decided to retrieve each group as an independent data set for a total of 17 data sets included in this analysis. The characteristics of each included study are shown in Table 1.
Table 1. Study characteristics of each included study.
| Reference number | Author | Publication year | Sample origin | Messenger RNA quantification method | Housekeeping gene | Number of subjects | Test results | |||
| TP | FP | FN | TN | |||||||
| 9 | Lipman et al. | 1994 | graft biopsy | competitive RT-PCR | GAPDH | 26 | 9 | 2 | 1 | 14 |
| 25 | Netto et al. | 2002 | graft biopsy | RT-PCR | β-actin | 29 | 4 | 1 | 3 | 21 |
| PBL | RT-PCR | β-actin | 29 | 6 | 0 | 1 | 22 | |||
| 26 | Vasconcellos et al. | 1998 | graft biopsy | competitive RT-PCR | GAPDH | 31 | 11 | 2 | 0 | 18 |
| PBL | competitive RT-PCR | GAPDH | 31 | 9 | 3 | 2 | 17 | |||
| 27 | Dias et al. | 2008 | PBL | RT-PCR | cyclophilin | 48 | 20 | 7 | 0 | 21 |
| urine | RT-PCR | cyclophilin | 50 | 20 | 4 | 0 | 26 | |||
| 28 | Sabek et al. | 2002 | PBL | RT-PCR | 18s rRNA | 27 | 5 | 5 | 3 | 14 |
| 29 | Lipman et al. | 1998 | graft biopsy | competitive RT-PCR | GAPDH | 21 | 6 | 0 | 5 | 10 |
| 30 | Strehlau et al. | 1997 | graft biopsy | competitive RT-PCR | GAPDH | 27 | 12 | 1 | 3 | 11 |
| 31 | Li et al. | 2001 | urine | RT-PCR | cyclophilin | 44 | 20 | 3 | 4 | 17 |
| 32 | Øzbay et al. | 2009 | urine | real-time quantitative RT-PCR | cyclophilin | 41 | 21 | 4 | 3 | 13 |
| 33 | Galante et al. | 2006 | urine | real-time quantitative RT-PCR | cyclophilin | 24 | 11 | 1 | 2 | 10 |
| 34 | Shin et al. | 2005 | PBL | competitive RT-PCR | β-actin | 15 | 5 | 1 | 2 | 7 |
| 35 | Simon et al. | 2003 | PBL | real-time quantitative RT-PCR | 18s rRNA | 16 | 4 | 0 | 1 | 11 |
| 36 | Dugr’e et al. | 2000 | PBL | RT-PCR | β-actin | 21 | 4 | 1 | 4 | 12 |
| 37 | Dias et al. | 2004 | graft biopsy | RT-PCR | GAPDH | 21 | 10 | 5 | 1 | 5 |
Abbreviations: TP, true positive; FP, false positive; FN, false negative; TN, true negative; PBL, peripheral blood leukocyte; RT-PCR, reverse transcription polymerase chain reaction; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Study Quality
We used the QUADAS list of questions to review the test quality of the included studies. Most of the studies satisfied a majority of the items on the QUADAS list. The most common missing items in the studies included in this analysis were reports of intermediate results and withdrawn cases. In addition, some of the studies failed to mention the blinded interpretations between the PCR results and the histological evaluation (Table S2).
The Egger test revealed the possibility of significant publication bias among the included reports (P = 0.008). The funnel plot in Figure S1 also presented a certain degree of asymmetry, indicating the potential for publication bias among the studies included in this analysis.
Overall Diagnostic Performance of Perforin Expression
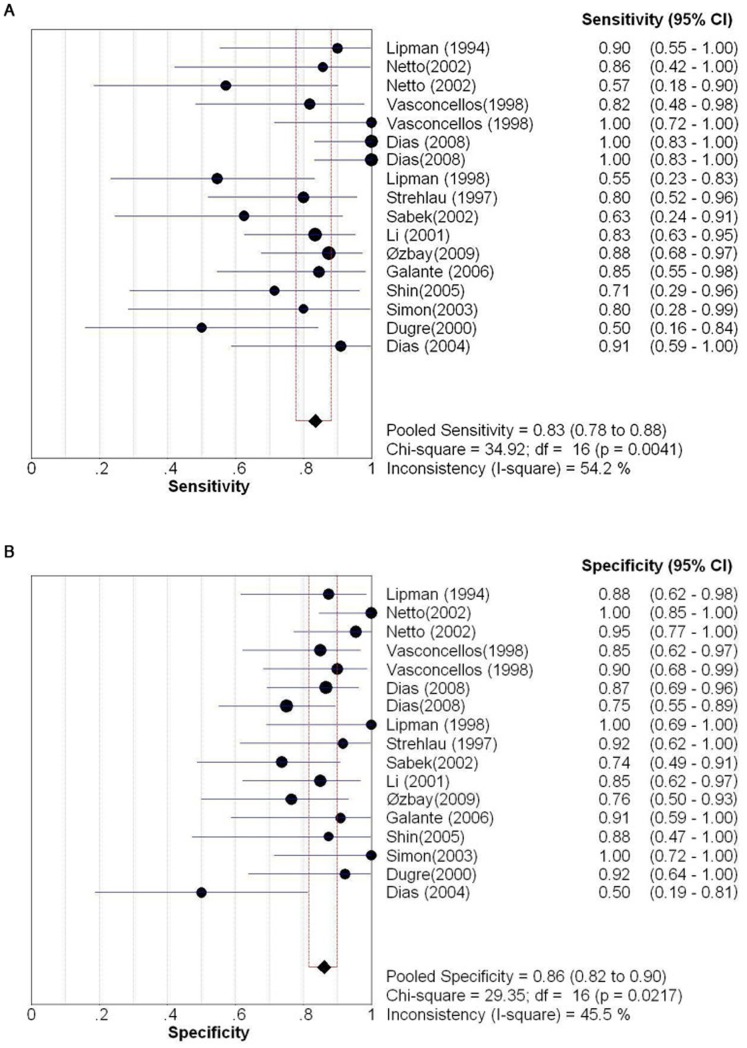
Figure 2 shows the overall diagnostic measurements of perforin expression in detecting AR. The summary sensitivity was 0.83 [95% confidence interval (CI): 0.78 to 0.88], with individual sensitivities ranging from 0.50 to 1.00. The summary specificity was 0.86 (95% CI: 0.82 to 0.90), with individual specificities ranging from 0.50 to 1.00. Both pooled estimations showed significant heterogeneity (Sensitivity: P = 0.0041, χ 2 = 34.92, I 2 = 54.2%; specificity: P = 0.022, χ 2 = 29.35, I 2 = 45.5%).
Figure 2. Sensitivity and specificity of perforin mRNA detection for the diagnosis of AR.
(A) Pooled sensitivity. (B) Pooled specificity. Effect sizes were pooled by random-effects models. The point estimates from each study are shown as solid squares. The pooled estimates are shown as a solid diamond. Error bars represent 95% CIs.
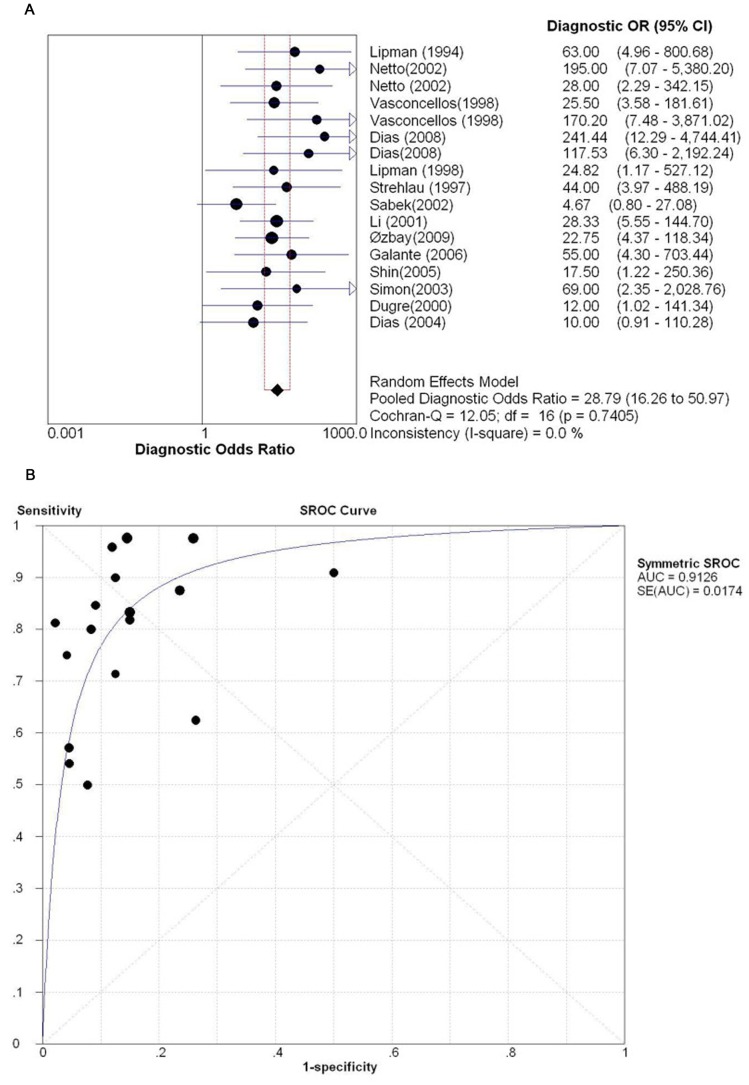
The pooled DOR and the SROC curves based on summary sensitivity and specificity across all data sets are shown in Figure 3. The pooled DOR was 28.79 (95% CI: 16.26 to 50.97), with individual DORs ranging from 4.67 to 241.44. The results of DOR showed consistency accross the included reports, without noticeable heterogeneity (P = 0.74, Cochran-Q = 12.05, I 2 = 0.0%). The point size in the SROC curve represented the proportional study weight. Most data gathered near the top left corner where sensitivity and specificity were both the highest. The AUC value was 0.9107±0.0174.
Figure 3. Overall DOR and SROC curves for all data sets describing the diagnostic performance of perforin mRNA detection in identifying AR.
(A) Overall DOR. (B) The SROC curves for all data sets. Effect sizes were pooled by random-effects models. The pooled DOR is shown as a solid diamond. Each square in the SROC curve represents one study. Sample size is indicated by the size of the square.
Although we did not notice a curvilinear shape distribution of the data in the SROC curve, the Spearman correlation analysis revealed a significant result (P = 0.032), suggesting the potential presence of a threshold effect.
Sensitivity Analysis
We systematically removed one data set at a time and recalculated the DOR and AUC values for the remaining studies. The largest change occurred when removing the data set from Sabek et al. [28], which changed the pooled DOR from 28.79 to 35.68 (+23.9%), and the corresponding change in AUC value was from 0.9107 to 0.9228 (+1.33%). The second largest change occurred when removing the urine subgroup from Dias et al. [27], which changed the pooled DOR from 28.79 to 26.54 (−7.81%) and the corresponding AUC value from 0.9107 to 0.9058 (−0.54%). These results indicated that no single data set carried enough weight to significantly influence the pooled test performance reported for the ability of perforin mRNA detection to identify cases of acute renal rejection.
Univariate Analysis
Publication year
Based on the year of publication of the studies included in this analysis, we divided the data sets into two subgroups: those reported prior to the year 2000 and those reported after the year 2000 (including studies published in 2000). This time point was chosen because significant progress was made in the PCR technology and experimental methodology at the beginning of the 21st century, which may have had an effect on the perforin mRNA detection performance. We noticed a remarkable difference in the amount of publications in each subgroup. Only 4 reports were published prior to 2000 [9], [26], [29], [30], one of them contained 2 data sets which made it 5 data sets in this subgroup. The remaining 10 reports (12 data sets) were published after 2000 [25], [27], [28], [31]–[37]. The DOR of studies before the year 2000 was 43.52 while the DOR of studies after 2000 was 24.90. The difference was not statistically significant (P = 0.59).
Sample origin
The studies were stratified according to the 3 types of samples: allograft biopsy tissue, PBL and urine. The biopsy subgroup contained 6 data sets [9], [25], [26], [29], [30], [37], the PBL subgroup contained 7 data sets [25]–[27], [28], [34]–[36], and the urine subgroup contained 4 data sets [27], [31]–[33]. The DORs were 35.11, 21.32, 36.76 for biopsy group, PBL group and urine group, respectively. However, the difference in DORs did not reach a level of statistical significance (P = 0.77).
Messenger RNA quantification method
There were 3 different PCR techniques used in the included studies to quantify perforin mRNA: 8 data sets used reverse transcriptase PCR (RT-PCR) [25], [27], [28], [31], [36], [37], 6 data sets used competitive RT-PCR [9], [26], [29], [30], [34], and 3 data sets used real-time quantitative RT-PCR [32], [33], [35]. The DOR was 25.23 for RT-PCR, 37.94 for competitive RT-PCR and 33.35 for real-time quantitative RT-PCR. The difference between the three techniques was not statistically significant (P = 0.89).
Housekeeping gene
Four different housekeeping genes were used as the standard expression in the included studies to measure the relative expression level of perforin: cyclophilin for 4 data sets [27], [31]–[33], β-actin for 5 [25], [27], [34], [36], 18s rRNA for 2 [28], [35], and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for 6 [9], [26], [29], [30], [37]. The DOR was 36.76 for cyclophilin group, 34.38 for β-actin group, 12.03 for 18s rRNA group and 33.45 for GAPDH group. However, the result was not statistically significant either (P = 0.95).
Table 2 summarizes the results of univariate analysis.
Table 2. Univariate analysis of potential variables influencing the test performance of perforin during AR.
| Variables | Subgroups | Number of independent data sets | Sensitivity (95% CI) | Specificity (95% CI) | DOR (95% CI) | AUC | |
| Publication year | before 2000 | 5 | 0.81 (0.69–0.90) | 0.90 (0.81–0.95) | 43.52 (14.19–133.46) | 0.9406 | |
| after 2000 | 12 | 0.84 (0.78–0.90) | 0.85 (0.79–0.89) | 24.90 (12.82–48.37) | 0.9028 | ||
| Sample origin | graft biopsy | 6 | 0.80 (0.68–0.89) | 0.88 (0.79–0.94) | 35.11 (12.02–102.56) | 0.9210 | |
| PBL | 7 | 0.80 (0.69–0.89) | 0.86 (0.78–0.92) | 21.32 (7.97–57.09) | 0.9028 | ||
| urine | 4 | 0.89 (0.80–0.95) | 0.85 (0.75–0.92) | 36.76 (13.59–99.39) | 0.9158 | ||
| PCR techniques | RT-PCR | 8 | 0.85 (0.76–0.91) | 0.84 (0.78–0.89) | 25.23 (9.59–66.37) | 0.9056 | |
| competitive RT-PCR | 6 | 0.80 (0.68–0.89) | 0.90 (0.81–0.95) | 37.94 (13.51–106.56) | 0.9370 | ||
| real-time quantitative RT-PCR | 3 | 0.86 (0.71–0.95) | 0.87 (0.73–0.96) | 33.35 (9.26–120.09) | 0.9196 | ||
| Housekeeping gene | cyclophilin | 4 | 0.89 (0.80–0.95) | 0.85 (0.75–0.92) | 36.76 (13.59–99.39) | 0.9158 | |
| β-actin | 5 | 0.80 (0.66–0.90) | 0.89 (0.81–0.95) | 34.38 (10.16–116.35) | 0.9372 | ||
| 18s rRNA | 2 | 0.69 (0.39–0.91) | 0.83 (0.65–0.94) | 12.03 (0.96–151.24) | unavailable a | ||
| GAPDH | 6 | 0.83 (0.72–0.91) | 0.85 (0.76–0.92) | 33.45 (12.12–92.34) | 0.9194 | ||
| All | 17 | 0.83 (0.78–0.88) | 0.86 (0.82–0.90) | 28.79 (16.26–50.97) | 0.9107 | ||
Three independent data points are required at least to draw an SROC curve.
Abbreviations: DOR, diagnostic odds ratio; CI, confidence interval; AUC, area under the curve of the SROC curve; PBL, peripheral blood leukocyte; PCR, polymerase chain reaction; RT-PCR, reverse transcription polymerase chain reaction; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Discussion
Since the middle of the 20th century, great success has been made in renal transplantation with the progress in surgical techniques, expanded organ sources, organ preservation techniques, novel immunosuppressants and management of complications. However, transplant patients are still facing many challenges, among which AR draws the greatest attention. Despite the fact that histological evaluation for AR has been well defined in guidelines such as Banff criteria [38] and Cooperative Clinical Trials in Transplantation (CCTT) criteria [39], novel and less invasive methods are still required to improve the diagnostic evaluation of AR. The effector molecules of CTLs such as perforin, granzyme B, Fas and Fas ligand are potential diagnostic markers for AR, especially when they can be detected in samples such as PBLs and urine that do not require invasive procedures to obtain. The major objectives of conducting this meta-analysis were to explore the diagnostic performance of perforin mRNA expression in AR and to determine its clinical utility. To our knowledge, this is the first pooled estimation of the diagnostic performance of perforin mRNA detection for the evaluation of AR in renal transplant recipients.
In this meta-analysis, we included 14 relevant studies with a total of 501 subjects. Although results were not consistent across the different studies, the overall diagnostic performance of detecting perforin mRNA in kidney transplant patients showed pooled sensitivity and specificity of 0.83 (95% CI: 0.78 to 0.88) and 0.86 (95% CI: 0.82 to 0.90), respectively. The pooled DOR and AUC of the SROC curves for all data sets were 28.79 (95% CI: 16.26 to 50.97) and 0.9107±0.0174, respectively. These results represented a good diagnostic efficacy for perforin mRNA detection in identifying AR, regardless of the sample origin and methodology variation. Furthermore, to investigate potential variables that might have influenced the diagnostic performance, we conducted a univariate analysis trying to provide clues for methodology standardization. In this analysis, none of the chosen factors appeared to have a significant effect on the diagnostic performance. This lack of variation from the chosen factors may be due to the small sample sizes of the included data sets since this diagnostic method had not been widely used in transplant centers. In addition, the perforin gene sequences used in the included reports were not uniform, which could be another potentail source of variation that may have influenced test performance. However, the limited number of data sets using each perforin sequence restricted us from categorizing the studies into subgroups for the univariate analysis.
In several clinical studies during the 1990s [9], [10], [30], cytotoxic gene expression was found to be up-regulated in allografts during AR. However, these discoveries had limited impact as diagnostic tests that supplemented the histological diagnosis of AR at that time. More recently, given the fact that lymphocytes would infiltrate the kidneys during AR and present in urine sediment cells, Li et al. [31] explored the utilization of perforin mRNA detection in urine cells as a non-invasive diagnostic marker of AR. Subsequent studies conducted in other centers confirmed the feasibility of this approach [32], [33]. Although in our analysis, the diagnostic performance of urine sample didn’t stand out particularly, the result was still encouraging since it brought hope for a non-invasive method for the diagnosis of AR which was as reliable as biopsy sample.
Debates about the application of urine perforin detection mainly focus on the differential diagnosis between rejection and other complications such as delayed graft function (DGF) and urinary tract infection (UTI). In the study conducted by Yannaraki et al. [14], an increase in perforin mRNA was found in both the AR group and the UTI group. Their experience suggested a significant overlap of perforin mRNA levels in different clinical conditions, which made it difficult to establish a threshold value for differential diagnosis. Øzbay et al. [32] reported similar results when trying to differentiate AR from bacteriuria. This may be explained by the similar cytolytic response of the activated T lymphocytes during both rejection and infection. In three of the 4 included studies in the urine subgroup of this meta-analysis [31]–[33], the non-rejection samples were composed of stable grafts only, while in the other study [27], the non-rejection samples contained chronic allograft nephropathy, toxic tubulopathy, nonspecific changes, acute tubular necrosis and renal-vein thrombosis. The non-rejection controls in other included studies also contained samples with multiple other types of kidney dysfunction other than graft rejection, which did not allow us to carry out a meta-analysis of the differential diagnostic performance of perforin. Therefore, we could not conclude that a high perforin expression level would definitely point to the diagnosis of AR, which would require supplemental laboratory tests to rule out other complications.
Although urine is the ideal choice for a non-invasive procedure, there are some potential limitations to this approach. Most importantly, the test depends on urine production. This limits the utilization in patients under anuric conditions which can appear during AR, acute tubular necrosis (ATN), DGF, as well as other conditions. In the study conducted by Dias et al. [27], nearly 20% of the patients were unable to provide sufficient urine samples for analysis. Given these circumstances, the evaluation of perforin mRNA levels in PBLs and allografts are important alternatives. An increase in perforin mRNA can help clinical decision-making for early enhanced immunosuppression intervention before histological evidence of substantial damage develops, and a decrease in perforin mRNA levels may provide an indication of response to therapy.
There are several limitations in this meta-analysis. First, qualities of the included studies were not uniform. The essential demographical data like age and gender distributions were missing in some studies, which might be a potential heterogeneity source in the analysis. Also, the specific cut-off values for the mRNA level were not provided in most of the studies. In addition, only 14 studies met the inclusion criteria in this analysis. The small sample size limited the generalization of the results and did not allow us to test the differential diagnostic performance of perforin mRNA detection. All these limitations provide room for future evaluation.
In conclusion, the test performance of perforin mRNA detected by PCR techniques was impressive and consistent under circumstances of methodological changes. The test in urine stood out as a potential novel and non-invasive method for the reliable diagnosis of AR, or at least as an indicator that a biopsy is warranted. Prospective studies with larger sample sizes would reinforce the findings revealed in the current meta-analysis and may be able to reveal how perforin mRNA detection would help to differentiate between diagnoses that are clinically similar to AR, providing more conclusive evidence for its clinical utility in the evaluation of renal transplant recipients.
Supporting Information
Funnel plot for the assessment of potential publication bias. The funnel graphs plot the log of the DOR against the standard error (SE) of the log of the DOR. Each solid circle represents each study in the meta-analysis. Asymmetry of the circle distribution between two sides indicates potential publication bias.
(TIF)
PRISMA 2009 check list.
(DOC)
Quality assessment of the included articles. Abbreviation: QUADAS, Quality Assessment of Diagnostic Accuracy Studies.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by the National High Technology Research and Development Program of China (863 Program) (2012AA021008), the Key Clinical Project from the Ministry of Health (2010159), the National Natural Science Foundation of China (30972951, 81102244, 81102245, and 81170448), the Special Fund for science research by Ministry of Health (201002004), the Research Fund for the Doctoral Program of Higher Education of China by Ministry of Education (20100171110063 and 20110171120077), the Science and Technology Planning Key Clinical Project of Guangdong Province (2011A030400005), and Project by Division of Medical Service Management of Ministry of Health(2010). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Reference
- 1.Matas AJ, Gillingham KJ, Payne WD, Najarian JS. The impact of an acute rejection episode on long-term renal allograft survival (t1/2). Transplantation. 1994;57:857–859. doi: 10.1097/00007890-199403270-00015. [DOI] [PubMed] [Google Scholar]
- 2.Matas AJ. Acute rejection is a major risk factor for chronic rejection. Transplant Proc. 1998;30:1766–1768. doi: 10.1016/s0041-1345(98)00422-9. [DOI] [PubMed] [Google Scholar]
- 3.Hariharan S, Johnson CP, Bresnahan BA, Taranto SE, McIntosh MJ, et al. Improved graft survival after renal transplantation in the United States, 1988 to 1996. N Engl J Med. 2000;342:605–612. doi: 10.1056/NEJM200003023420901. [DOI] [PubMed] [Google Scholar]
- 4.Huraib S, Goldberg H, Katz A, Cardella CJ, deVeber GA, et al. Percutaneous needle biopsy of the transplanted kidney: technique and complications. Am J Kidney Dis. 1989;14:13–17. doi: 10.1016/s0272-6386(89)80087-3. [DOI] [PubMed] [Google Scholar]
- 5.Beckingham IJ, Nicholson ML, Bell PR. Analysis of factors associated with complications following renal transplant needle core biopsy. Br J Urol. 1994;73:13–15. doi: 10.1111/j.1464-410x.1994.tb07449.x. [DOI] [PubMed] [Google Scholar]
- 6.Rush D. Can protocol biopsy better inform our choices in renal transplantation? Transplant Proc. 2009;41(6 Suppl.):S6–S8. doi: 10.1016/j.transproceed.2009.06.092. [DOI] [PubMed] [Google Scholar]
- 7.Perella RR, Duerinckx AJ, Tessler FN, Danovitch GM, Wilkinson A, et al. Evaluation of renal transplant dysfunction by duplex Doppler sonography: a prospective study and review of the literature. Am J Kidney Dis. 1990;15:544–550. doi: 10.1016/s0272-6386(12)80524-5. [DOI] [PubMed] [Google Scholar]
- 8.Schold JD, Kaplan B. The elephant in the room: failings of current clinical end points in kidney transplantation. Am J Transplant. 2010;10(5):1163–1166. doi: 10.1111/j.1600-6143.2010.03104.x. [DOI] [PubMed] [Google Scholar]
- 9.Lipman ML, Stevens AC, Strom TB. Heightened intragraft CTL gene expression in acutely rejecting renal allografts. J Immunol. 1994;152(10):5120–5127. [PubMed] [Google Scholar]
- 10.Suthanthiran M, Manikkam MD. Clinical application of molecular biology: a study of allograft rejection with polymerase chain reaction. Am J Med Sci. 1997;313(5):264–267. doi: 10.1097/00000441-199705000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Lichtenheld MG, Olsen KJ, Lu P, Lowrey DM, Hameed A, et al. Structure and function of human perforin. Nature. 1988;335:448–451. doi: 10.1038/335448a0. [DOI] [PubMed] [Google Scholar]
- 12.Muthukumar T, Dadhania D, Ding R, Snopkowski C, Naqvi R, et al. Messenger RNA for foxp3 in the urine of renal-allograft recipients. N Engl J Med. 2005;353:2342–51. doi: 10.1056/NEJMoa051907. [DOI] [PubMed] [Google Scholar]
- 13.Nickel P, Lacha J, Ode-Hakim S, Sawitzki B, Babel N, et al. Cytotoxic effector molecule gene expression in acute renal allograft rejection: correlation with clinical outcome; histopathology and function of the allograft. Transplantation. 2001;72(6):1158–1161. doi: 10.1097/00007890-200109270-00031. [DOI] [PubMed] [Google Scholar]
- 14.Yannaraki M, Rebibou JM, Ducloux D, Saas P, Duperrier A, et al. Urinary cytotoxic molecular markers for a noninvasive diagnosis in acute renal transplant rejection. Transpl Int. 2006;19(9):759–768. doi: 10.1111/j.1432-2277.2006.00351.x. [DOI] [PubMed] [Google Scholar]
- 15.Deeks J. Egger M, Davey Smith G, Altman D, editors. Systematic reviews of evaluations of diagnostic and screening tests. 2001. Systematic Reviews in Health Care: Meta-analysis in context. London: BMJ Publishing Group.
- 16.Whiting P, Rutjes AW, Reitsma JB, Bossuyt PM, Kleijnen J. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol. 2003;3:25. doi: 10.1186/1471-2288-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stata Corporation website. Available: http://www.stata.com/stata11/. Accessed: 2012 May 30.
- 18.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zamora J, Abraira V, Muriel A, Khan KS, Coomarasamy A. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol. 2006;6:31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altman DG, Bland JM. Diagnostic tests 1: sensitivity and specificity. BMJ. 1994;308:1552. doi: 10.1136/bmj.308.6943.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol. 2003;56:1129–1135. doi: 10.1016/s0895-4356(03)00177-x. [DOI] [PubMed] [Google Scholar]
- 22.Moses LE, Shapiro D, Littenberg B. Combining independent studies of a diagnostic test into a summary ROC curve: data-analytic approaches and some additional considerations. Stat Med. 1993;12:1293–1316. doi: 10.1002/sim.4780121403. [DOI] [PubMed] [Google Scholar]
- 23.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: An update. Contemp Clin Trials. 2007;28:105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Devillé WL, Buntinx F, Bouter LM, Montori VM, de Vet HC, et al. Conducting systematic reviews of diagnostic studies: didactic guidelines. BMC Med Res Methodol. 2002;2:9. doi: 10.1186/1471-2288-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Netto MVP, Fonseca BAL, Dantas M, Saber LTS, Castro MCR, et al. Granzyme B, Fas-ligand and perforin expression during acute cellular rejection episodes after kidney transplantation: comparison between blood and renal aspirates. Transplant Proc. 2002;34(2):476–478. doi: 10.1016/s0041-1345(02)02601-5. [DOI] [PubMed] [Google Scholar]
- 26.Vasconcellos L, Lauro M, Asher F, Schachter D, Zheng X. Cytotoxic lymphocyte gene expression in peripheral blood leukocytes correlates with rejecting renal allografts. Transplantation 15. 1998;66(5):562–566. doi: 10.1097/00007890-199809150-00002. [DOI] [PubMed] [Google Scholar]
- 27.Dias ECA, Joelsons G, da Silva DM, Berdichewski RH, Ribeiro AR, et al. Non-invasive diagnosis of acute rejection in kidney transplants with delayed graft function. Kidney Int. 2008;73(7):877–884. doi: 10.1038/sj.ki.5002795. [DOI] [PubMed] [Google Scholar]
- 28.Sabek O, Dorak MT, Kotb M, Gaber A, Gaber L. Quantitative detection of t-cell activation markers by real-time PCR in renal transplant rejection and correlation with histopathologic evaluation. Transplantation 15. 2002;74(5):701–707. doi: 10.1097/00007890-200209150-00019. [DOI] [PubMed] [Google Scholar]
- 29.Lipman ML, Shen Y, Jeffery JR, Gough J, McKenna RM, et al. Immune-activation gene expression in clinically stable renal allograft biopsies: molecular evidence for subclinical rejection. Transplantation 27. 1998;66(12):1673–1681. doi: 10.1097/00007890-199812270-00018. [DOI] [PubMed] [Google Scholar]
- 30.Strehlau J, Pavlakis M, Lipman ML, Shapiro M, Vasconcellos L, et al. Quantitative detection of immune activation transcripts as a diagnostic tool in kidney transplantation. Proc Natl Acad Sci USA 21. 1997;94(2):695–700. doi: 10.1073/pnas.94.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B, Hartono C, Ding R, Sharma VK, Ramaswamy R, et al. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med. 2001;344:947–954. doi: 10.1056/NEJM200103293441301. [DOI] [PubMed] [Google Scholar]
- 32.Øzbay A, Tørring C, Olsen R, Carstens J. Transcriptional profiles in urine during acute rejection, bacteriuria, CMV infection and stable graft function after renal transplantation. Scand J Immunol. 2009;69(4):357–365. doi: 10.1111/j.1365-3083.2009.02226.x. [DOI] [PubMed] [Google Scholar]
- 33.Galante NZ, Câmara NOS, Kallas EG, Salomão R, Pacheco-Silva A, et al. Noninvasive immune monitoring assessed by flow cytometry and real time RT-PCR in urine of renal transplantation recipients. Transpl Immunol. 2006;16(2):73–80. doi: 10.1016/j.trim.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 34.Shin GT, Kim SJ, Lee TS, Oh CK, Kim HS. Gene expression of perforin by peripheral blood lymphocytes as a marker of acute rejection. Nephron Clin Pract. 2005;100(3):c63–70. doi: 10.1159/000085050. [DOI] [PubMed] [Google Scholar]
- 35.Simon T, Opelz G, Wiese M, Ott RC, Susal C. Serial peripheral blood perforin and granzyme B gene expression measurements for prediction of acute rejection in kidney graft recipients. Am J Transplant. 2003;3(9):1121–1127. doi: 10.1034/j.1600-6143.2003.00187.x. [DOI] [PubMed] [Google Scholar]
- 36.Dugr’e FJ, Gaudreau S, Belles-Isles M, Houde I, Roy R. Cytokine and cytotoxic molecule gene expression determined in peripheral blood mononuclear cells in the diagnosis of acute renal rejection. Transplantation 15. 2000;70(7):1074–1080. doi: 10.1097/00007890-200010150-00014. [DOI] [PubMed] [Google Scholar]
- 37.Dias ECA, Veronese FJV, Gonçalves LFS, Manfro RC. Molecular markers in subclinical acute rejection of renal transplants. Clin Transplant. 2004;18:281–287. doi: 10.1111/j.1399-0012.2004.00161.x. [DOI] [PubMed] [Google Scholar]
- 38.Racusen LC, Solez K, Colvin RB. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55:713–723. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- 39.Colvin RB, Cohen AH, Saiontz C, Bonsib S, Buick M, et al. Evaluation of pathologic criteria of acute renal allograft rejection: Reproducibility, sensitivity, and clinical correlation. J Am Soc Nephrol. 1997;8:1930–1941. doi: 10.1681/ASN.V8121930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Funnel plot for the assessment of potential publication bias. The funnel graphs plot the log of the DOR against the standard error (SE) of the log of the DOR. Each solid circle represents each study in the meta-analysis. Asymmetry of the circle distribution between two sides indicates potential publication bias.
(TIF)
PRISMA 2009 check list.
(DOC)
Quality assessment of the included articles. Abbreviation: QUADAS, Quality Assessment of Diagnostic Accuracy Studies.
(DOC)