Abstract
Objective
This study investigated the utility of advanced computational techniques to large scale-genome based data to identify novel genes that govern murine pancreatic development.
Methods
An expression dataset for mouse pancreatic development was complemented with “Hanalyzer” (high-throughput data analyzer) to identify and prioritize novel genes. Quantitative-real time polymerase chain reaction, in situ hybridization and immunohistochemistry was used to validate selected genes.
Results
Four new genes whose roles in the development of murine pancreas have not previously been established were identified; Cystathionine beta synthase (Cbs), Meis homeobox 1, Growth factor independent 1 and Aldehyde dehydrogenase 18 family, member A1. Their temporal expression during development was documented. Cbs was localized in the cytoplasm of the tip cells of the epithelial chords of the undifferentiated progenitor cells at E12.5 and was co expressed with the Pancreatic and duodenal homeobox 1 and Pancreas specific transcription factor, 1a positive cells. In the adult pancreas, Cbs was localized primarily within the acinar compartment.
Conclusions
In silico analysis of high-throughput microarray data in combination with background knowledge about genes provides an additional reliable method of identifying novel genes. To our knowledge, the expression and localization of Cbs has not been previously documented during mouse pancreatic development.
Keywords: Pancreas, Development, Microarray, Bioinformatics, Hanalyzer
The molecular mechanism of pancreatic morphogenesis and differentiation is a fundamental enigma during pancreas development. Early pancreatic organogenesis shares a common pool of pancreatic progenitor cells that originate from the foregut endoderm1, 2. The dynamic signaling interactions between surrounding mesoderm, in an autocrine and paracrine manner leads to the branching morphogenesis of the pancreatic epithelium3. The key transcription factors like Pdx1, Hlxb9, Ptf1a, Nkx6-1, and Nkx2-2, Nkx6-2, Sox 9 and many more unidentified such factors mark the pancreatic progenitor cells in the undifferentiated epithelium. These known and unknown entities allow an orchestrated series of complex differentiation and proliferation steps leading to the parallel differentiation of endocrine and exocrine (acinar and ductal) compartments4. The critical events governing the differentiation of endocrine cells occur when Neurog3 is transiently expressed in a subset of cells derived from the pancreatic epithelium from E9.5 to E18.55, 6. These Neurog3 expressing cells are identifiable by lineage tracing as endocrine progenitor cells7, 8. We have previously shown that endocrine differentiation in the human fetal pancreas also takes place in the islands of epithelial tissue but within a much larger volume of mesenchymal tissue9. In humans, endocrine progenitor marker, NGN3-positive cells also persist from 9–23 weeks of gestation9, 10. In mice, unlike humans where secondary transition has not been documented9, the differentiation of the acinar compartment is synchronized with that of endocrine differentiation during the secondary transition11 and is governed by the shift and segregation in expression of Ptf1a from the central to the expanding tips of the pancreatic epithelium3. Unfortunately, the data regarding ductal development is relatively sparse12–14.
Tremendous progress has been made in understanding the molecular dynamics of some of these known factors mentioned above and the identification and validation of novel entities that drive the differentiation process may further enhance our ability to generate therapeutic beta cells from ES and or iPS cells15–17. To document the transcriptome of the developing pancreas in the mouse and highlight the qualitative and quantitative features of global gene expression that contributes to the specification, growth and differentiation of the major endocrine and exocrine (acinar and ductal) cell types, we performed microarray analysis at E12.5, E13.5, E15.5 and E18.5 on murine embryonic pancreas. In order to expedite the discovery of novel factors that govern pancreatic differentiation, we have synergized the differential analysis of significant genes with an advanced computational approach called the Hanalyzer18, 19. The Hanalyzer extracts information from external resources, either by parsing structured data or by using biomedical language processing to glean information from unstructured data, and tracks knowledge provenance about genes.
The use of the tool was critical in the identification of four novel genes whose roles have not been extensively characterized in the murine pancreatic developmental context; Cystathionine beta synthase (Cbs), Growth Factor Independent 1 (Gfi1), Meis homeobox 1 (Meis 1) and Aldehyde dehydrogenase 18 family member a1 (Aldh18a1). In the current study, we validated the temporal expression of Cbs, Gfi1, Meis1 and Aldh18a1 and further documented the localization of Cbs in embryonic and adult pancreas. We identified Cbs as one of the previously unknown cellular markers expressed in early pancreatic epithelium that may contribute to the acinar differentiation in the murine pancreas.
MATERIALS AND METHODS
Mice
CD1 mice were procured from Charles River (Wilmington, MA, USA). All animal procedures were covered by current animal protocols approved by the IUCAC committee of the University of Colorado Denver animal care facility. Embryonic pancreases were harvested from time-mated mouse intercrosses CD1 females and males and stored in RNAlater (Ambion, Austin, TX USA) at 4°C. Adult pancreatic islets were prepared from 6-month-old female CD1 mice by collagenase digestion, isolated by Ficoll/Histopaque gradients and hand picking (6 mice/1000 islets/preparation). Pancreases were pooled from all early embryonic stages (E12.5–E18.5). Mouse embryo numbers assigned at E12.5/E13.5 =11–12, E15.5 = 4, E18.5=2.
RNA Extraction and Microarray
Total RNA was extracted from tissues with TRIzol reagent® (Invitrogen, Carlsbad, CA, USA), purified by RNeasy columns (Qiagen, Valencia, CA, USA) and analyzed for integrity (Agilent Bioanalyzer 2100, Santa Clara CA, USA)9. Biotin-labeled cRNA was synthesized by the Affymetrix protocol (Affymetrix, Santa Clara, CA) and hybridized to MOE 430–2 microarrays (n=3 chip/embryonic age). Microarray data was analyzed by GeneSpring (Silicon Genetics, Redwood City, CA) and Partek (St. Louis, MI, USA). GC Robust Multi-array Average (GC RMA) was used for normalization. The complete microarray data set will be available publicly on GeneSpeed20, 21
Statistical Analysis
After initial quality control, total number of genes retained in the data set was 38959. ANOVA was applied to the data set and p values were corrected by Benjamini and Hochberg's procedure. P<0.05 and False detection rate (FDR) of 0.1 were considered significant (Supplementary Table 1). The gene lists generated from differential ANOVA analysis are available as supplementary data (Supplementary Tables 2, 3 and 4).
Biomedical Discovery Acceleration by Hanalyzer (High-Throughput Analyzer)
The approach is based on the complementing two weighted networks 1) knowledge network comprising of a large portion of existing knowledge of gene products and their interactions and 2) data network provided by the above microarray analysis of mouse pancreatic development. The detailed description of the Hanalyzer protocol18, 19 and its use and validation in murine cranio-facial development has been previously published18.
Briefly, after combining and integrating varied sources of knowledge between a pair of genes, a score was assigned to attest the reliability of the gene interactions from murine centric databases and literature. Subsequently, this updatable source of knowledge was amalgamated with the parallel data network constructed from our microarray experiment, which was then visualized in Cytoscape22–24. 2,688 significant gene entities with p values less than 0.000816494 (post ANOVA and FDR correction) from the pancreas development microarray data set was used and the average expression for each gene for each time point (2,287 genes) was calculated. Probes/genes were then mapped to their MGI identifiers. In instances where two MGI ID's correlated with multiple correlation values, the largest (absolute value) correlation reported between the pair when duplicates were present was considered. The network produced comprises of the 1000 highest scoring edges as asserted by the Average Metric and the 1000 highest scoring edges as asserted by the Hanisch Metric18. In this instance, this produces a graph consisting of 709 unique nodes (genes) and 1655 edges (data not shown) that were visualized in Cytoscape.
Validation of Microarray Data
Quantitative RTPCR (Q-RTPCR) was performed on cDNA (5 ng) derived from total RNA by iScript cDNA synthesis kits (Bio-Rad Laboratories, Hercules, CA). Assays used FAM dye labeled Taqman MGB probes and an ABI 7000 PCR instrument (Applied Biosystems, Foster City, CA) and were normalized to glyceraldehyde-3-phosphate-dehydrogenase (gapdh) as previously published9. Data was collected in triplicate, Cycle Threshold (CT) values were normalized to control group/calibrator.
Validation of Cbs mRNA by in situ Hybridization
In situ hybridization was performed with digoxigenin-labeled anti-sense riboprobes for Cbs. 3–4 mice embryos were assigned for in situ hybridization. Briefly, the slides with frozen sections were thawed to room temperature and incubated with hybridization buffer containing probe at 1ng/μl, overnight in a humidified chamber at 70°C. The colorimetric in situ reaction was developed with BM Purple (Roche, South San Francisco, CA, USA), over 3 days at room temperature. The slides were finally mounted with aqueous mounting media and counterstained with nuclear fast red.
Immunohistochemistry
3–4 Mouse pancreata were fixed in 4% paraformaldehyde and embedded in Optimum Cutting Temperature (OCT) (Tissue Tek, Sakura Finetek, Inc, Torrance, CA USA) for immunofluorescence microscopy. The sections (6μm) were blocked in Tyramide System Amplification (TSA) buffer (Zymed, Carlsbad, CA, USA) and incubated overnight with guinea pig anti insulin (Dako, Carpinteria, CA, USA) (1:100), mouse anti glucagon (Sigma, St Louis, MO, USA) (1:100), goat anti Pdx1 (1:10,000) (gift, Chris Wright, PhD, Vanderbilt) and rabbit Ptf1a (1:5000) (Developmental Studies Hybridoma Bank, Iowa City, Iowa, USA). Species-specific secondary antibodies conjugated to Alexa and cyanine fluorophores (Jackson Immunoresearch Laboratories, West Grove, PA, USA) were applied for 60 min and sections mounted in glycerol-based media. Images were acquired with an Olympus 1×70 microscope equipped with Photometrics Quantix cooled CCD camera (KAF 1400 chip). Cbs was localized by performing immunoperoxidase staining by mouse monoclonal antibody (1:1000) (gift, Jan Kraus, PhD, University of Colorado Denver) with horseradish peroxidase (HRP) labeled secondary antibody followed by Diaminobenzidine (DAB) oxidation and visualized by bright field microscopy. Immmnofluorescence staining was undertaken after DAB staining in the same sections that were not counterstained or dehydrated.
RESULTS
Global Gene Expression Analysis and Validation by Quantitative PCR
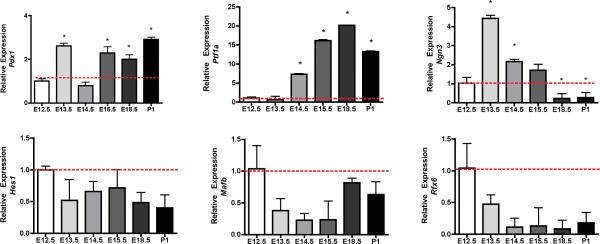
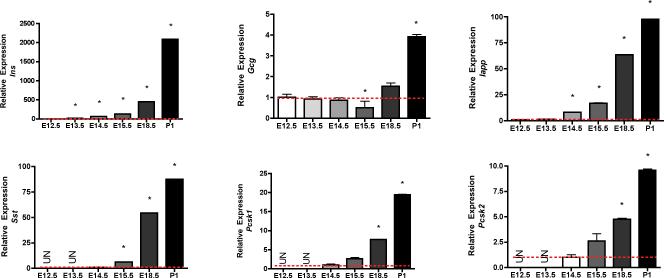
Principal Component Analysis of the microarray data obtained from 18 murine pancreas samples of E12.5 – E18.5 weeks of gestational age was defined by four cluster sets corresponding to E12.5, E13.5, E15.5 and E18.5 weeks. Overlap between the E12.5 and E13.5 clusters (Supplementary Fig. 1) suggested that although organ development proceeded through gradual growth during early development window and shared several common complementary factors. Our microarray data was consistent with the previously published gene expression data of key transcription factors that determine early pancreatic development1, 4. Pdx1 is expressed in the dorsal foregut in areas delineated to differentiate into the pancreas as early as E9.5. Pdx1 deficiency in both man and mouse leads to complete pancreatic agenesis [20, 21]. Pdx1 was upregulated at E13.5 vs. 12.5 (> 2 fold, p value 0.9) and E15.5 Vs. 12.5 (> 2 fold, p value 0.0002) (Supplementary Table 2 and 3). Ptf1a, initially a multipotent pancreatic progenitor marker4, 25, that later functions as a major transcriptional regulator of acinar cell development25, 26 (>3.8-fold, p value 0.003, E13.5 vs E12.5 and >16-fold, p value 0.00000012, E15.5 vs. E12.5 (Supplementary Table 3) was also upregulated. The temporal expression of 1) selected transcription factors: Pdx1, Ptf1a, Neurog3, Rfx6, Hes 1, Sox 9 and Mafb (Figure 1) and 2) selected endocrine markers: Ins, Gcg, Sst, Iapp, Pcsk1 and Pcsk2 (Figure 2) was undertaken in parallel by quantitative Taqman Q-RTPCR to validate the robustness of our microarray data.
FIGURE 1.
Temporal expression of Pdx1, Ptf1a, Neurog3, Hes1, Sox9 and Rfx6 mRNA by quantitative PCR. Q-RTPCR was performed on cDNA synthesized from fetal pancreas and adult CD1 islets using a 5' nuclease assay and FAM ® dye labeled Taqman MGB probes with two PCR primers. Endogenous gapdh was used for normalization. Data (mean ± SE) is expressed relative to a control sample (E12.5). For statistical analysis one-way analysis of variance (ANOVA), probability value (p < 0.05) was considered significant, followed by Student's T- test (p<0.05).
FIGURE 2.
Expression of mature endocrine markers; Ins, Gcg, Sst, Iapp, Pcsk1 and Pcsk2 mRNA by quantitative PCR. Q-RTPCR was performed on cDNA synthesized from fetal pancreas and adult CD1 islets using a 5' nuclease assay and FAM ® dye labeled Taqman MGB probes with two PCR primers. Endogenous gapdh was used for normalization. Data (mean ± SE) is expressed relative to a control sample (E12.5). For statistical analysis one-way analysis of variance (ANOVA), probability value (p < 0.05) was considered significant, followed by Student's T- test (p<0.05).
Identification of Cystathionine Beta Synthase (Cbs) as a Novel Marker of Acinar Differentiation through Hanalyzer Approach
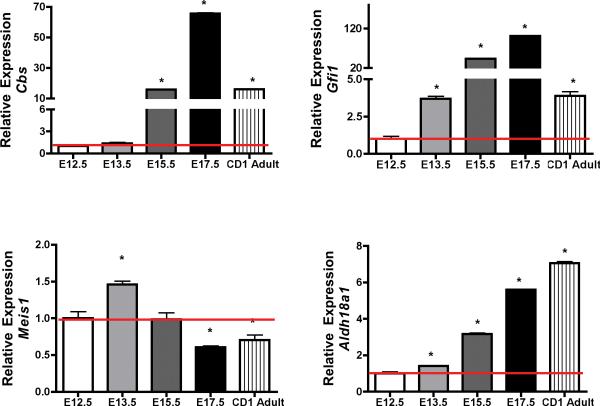
We identified a network of 33 genes associated with early regulation of transcription during pancreatic development (Supplementary Table 5). We selected Cbs, Gfi1, Meis1, and Ald18a1 for their relatively high correlation through expression and lack of strong background knowledge or links in the area of development of the murine pancreas. Their expression in the temporal embryonic series from E12.5 to E17.5 and adult CD1 mouse islets (Figure 3) was validated by Q-RTPCR. Among these selected genes, Cbs, a key enzyme involved in the trans-sulfuration pathway, catalyzes the condensation of serine and homocysteine to cystathionine and had the strongest link to diabetes and metabolic syndrome. Deficiency of CBS activity in human results in elevated levels of homocysteine as well as methionine in plasma and urine and decreased levels of cystathionine and cysteine27–30. Homocysteine metabolism is also perturbed in diabetic patients31. Cbs gene expression was significantly increased at E15.5 to E17.5 (p value <0.05) and adult islets compared to E12.5 pancreas. However, the gene expression was relatively lower in the adult islets as compared to embryonic stage E17.5 (p value =0.01).
FIGURE 3.
Identification of 4 novel genes during murine pancreatic development. Expression of Cbs, Gfi1, Meis1, and Aldh18a1 mRNA by quantitative PCR. Q-RTPCR was performed on cDNA synthesized from fetal pancreas and adult CD1 islets using a 5' nuclease assay and FAM ® dye labeled Taqman MGB probes with two PCR primers. Endogenous GAPDH was used for normalization. Data (mean ± SE) is expressed relative to a control sample (E12.5). For statistical analysis one-way analysis of variance (ANOVA), probability value (p < 0.05) was considered significant, followed by Student's T- test (p<0.05).
Growth factor independent-1 Gfi-1 is a zinc finger transcriptional repressor displaying multiple and essential roles in controlling hematopoietic stem cell biology, myeloid and lymphoid differentiation and lymphocytic functions32–34. However, its role in pancreatic biology has not been established. Gfi-1 expression was significantly increased in the embryonic pancreas from E13.5 to E17.5 and adult islets (p value <0.05) compared to E12.5. The expression in the adult islets was relatively lower than later embryonic time points (E15.5 (p value 0.001) and E17.5 (p value 0.001) (Figure 3). Myeloid ecotropic viral integration site-1 (Meis-1) belongs to the family of three amino acid loop extension (TALE) homeodomain proteins. Our data shows that Meis1 expression peaks at E13.5 and declined subsequently from E15.5. The relative expression of Meis1 was significantly lower in the early embryonic tissue than at E17.5 and adult islets (Figure 3).
The expression of Aldh18a1 was also validated by Q-RTPCR. The relative expression of Aldh18a1 increased temporally from E13.5 to E17.5 and unlike other genes that were profiled, Aldh18a1 expression was highest in the adult CD1 islets (Figure 3). Although ALDH18A1 has been previously described in relation to Down's syndrome and an autosomal recessive neurocutaneous syndrome in humans35, 36, the expression of Aldh18a1 has not been previously documented in the context of murine pancreatic development.
Validation and Localization of Cbs Expression by in situ Hybridization and Immunohistochemistry
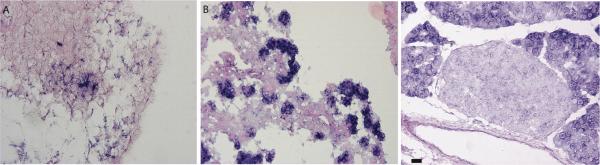
The localization of Cbs mRNA temporally in the developing murine pancreas was validated by in situ hybridization using anti-sense probes. Cbs mRNA was localized as early as E12.5 in the undifferentiated pancreatic epithelial cords. Between E13.5 to E15.5 Cbs mRNA was localized in the cytoplasm of developing clusters of neo-acinar cells at the tips of the epithelial chords. In the adult CD1 pancreas, Cbs mRNA was present ubiquitously, although the expression was higher in the acinar cells than endocrine cells. The ductal cells were devoid of Cbs mRNA expression (Figure 4).
FIGURE 4.
In situ localization of Cbs mRNA using anti sense probes in mouse embryos at various gestation ages A) E12.5, B) E15.5 C) CD1 adult pancreas. The slides were counterstained with nuclear fast red. Scale bar =15 μm.
Localization of Cbs at E12.5 by immunoperoxidase method showed cytoplasmic localization in the early pancreatic buds, which are known to contain undifferentiated multipotent pancreatic progenitor cells4, 37. Co-localization studies undertaken with Pdx1 and β-catenin at E12.5 (Figure 5) showed that Cbs was localized in the cytoplasm of the branching epithelium of the developing pancreas. The cells were marked by Pdx1, expressed as nuclear protein and β-catenin expression was noted in the cell membranes. The cells expressing higher intensity of Cbs were localized at the tips of the branching epithelium. These cells exhibited relatively decreased levels of Pdx1 and β-catenin, indicating greater degree of differentiation within the epithelial cell clusters. It has been previously shown that during early secondary transition, endocrine cells upregulate PDX1 and differentiating exocrine cells show relatively lower PDX138. Localization of Cbs in other embryonic ages (E13.5, E17.5, P0 and Adult) with Pdx1, β-catenin and insulin is shown in Supplementary Figure 2. The tip cells were also positive for Ptf1a, suggesting their commitment to acinar lineage at E12.5 (Figure 6). Cbs could not be detected in the cells expressing strong neurog3 nuclear signals (data not shown). At time points beyond E15.5 of pancreatic development, where much of the acinar cell neogenesis has occurred and the cells committed to the acinar fate are undergoing rapid mitotic expansion, Cbs was localized predominantly in the amylase positive cells at E18.5 (Figure 7). The intensity of Cbs staining, in the endocrine compartment was however much decreased as compared to the acinar cells in the adult pancreatic tissue (Supplementary Figure 3, panel P) as documented previously with in situ hybridizations. As expected, the ductal cells did not show any Cbs expression during any stage of pancreatic development.
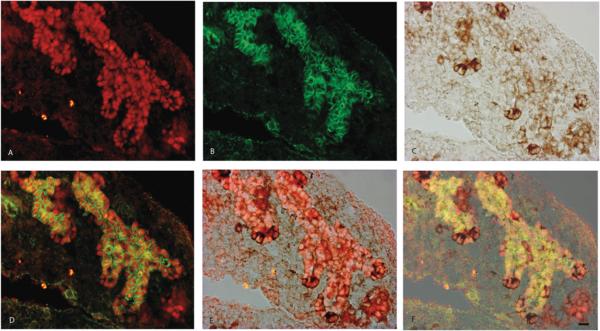
FIGURE 5.
Immunohistological localization of Pdx1 (A), B-catenin (B), Cbs (C) in E12.5 mouse embryos. Pdx1 and B-catenin mark the branching epithelial chords (D). Cbs is localized in the Pdx1 positive tip cells of the epithelial chords (E). The cells expressing higher intensity of Cbs have decreased Pdx1 and B-catenin intensity marking the degree of differentiation.
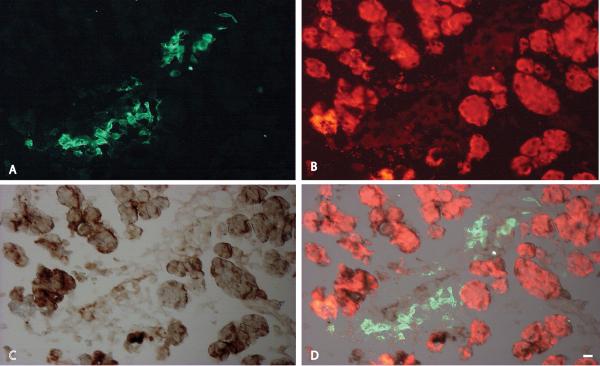
FIGURE 6.
Immunohistochemical localization of Ptf1A (A) and Cbs (B) in the E14.5 mouse pancreas. Ptf1a marks the nuclei of differentiating epithelium and Cbs is expressed in the cytoplasm tips of the branching epithelium (C).
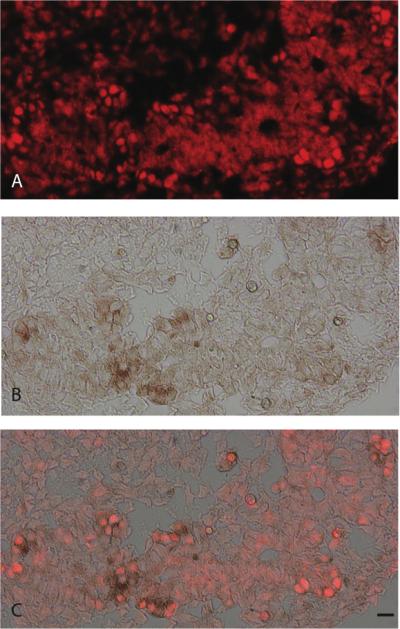
FIGURE 7.
Immunohistochemical localization of Insulin (A) Cbs (B) and amylase (C) in E18.5 mouse pancreas. Cbs is co-expressed with mature acinar cells (D). Scale bar = 20μm.
DISCUSSION
The purpose of this study was to elucidate the transcriptome of murine pancreatic development and assess the role of advanced computational tools to identify novel genes that govern pancreatic differentiation. High throughput microarray technology and other global genomics applications are powerful tools that allow investigations at the transcriptome levels39. Current methods for/of microarray analysis results in gene prioritization based on the production of ranked gene lists or the enrichment or clustering of annotation or functional terms. These methods can be of limited use, particularly when dealing with poorly annotated/studied genes and processes40. A more integrated approach to microarray analyses would be to employ protein-interaction networks to examine gene products in dynamic functional groups. Unfortunately, such networks are sparsely populated in mice and human for whom experimental protein interaction data is limited compared to the yeast.
In this study, we have used a newly developed scientific discovery tool, Hanalyzer18, 19 to facilitate knowledge-based analysis of the experimental data, this approach used 3 broad classes of algorithms“3 R's” 1) Reading methods to extract information by parsing structured data or the use of biomedical language processing to glean information regarding murine pancreatic development from unstructured data. 2) Reasoning methods to enrich pancreatic development related knowledge gathered from reading by noting genes sharing similar ontology terms or database entries. Reasoning method also integrates all background knowledge in a network that encompassed all possible relationships between pairs of genes and calculated a combined reliability score for the association. 3) Reporting methods to combine pancreatic development related knowledge network with congruent network constructed from our microarray data derived from E12.5 to E18.5 stages of murine development and helps to visualize the combined network in Cytoscape®.
Apart from the unstructured gene lists generated by ANOVA (Supplementary Table 2, 3 and 4), we identified four genes in the pancreas (Cbs, Meis1, Gfi1 and Aldh18a1) (Figure 3) that were supported by highest correlated expression but with fewer sources of shared characterization, which therefore provided an unique opportunity to search for truly novel genes regulating early pancreatic development machinery. The bioinformatics analysis was validated by our ability to document the temporal expression profiles of these genes from E12.5–E18.5 in the developing mouse embryos and adult islets. It has been previously shown that Meis1 is essential for embryonic development since Meis1 knockout mice die during embryogenesis at E14.541 due to hematopoietic and vasculature defects. Recently, it has been shown that Meis1 is localized within the nuclei of ductal cells of the murine pancreas, and can regulate Keratin 19 (Krt19) through Pdx142. The role of Gfi1, is far clearer in the development of the murine gut epithelium43 than the pancreas. Deletion of the Gfi1 results in an increased enteroendocrine cell population at the expense of Paneth and goblet cells44, due to cellular reprogramming of secretory cells toward a Neurog3+ enteroendocrine cell phenotype43 suggesting that Gfi1 may be upstream of Neurog3. A similar interaction in the pancreas can also be envisioned since Neurog3 is a key mediator of endocrine development in the pancreas.
Cbs was chosen for further investigation because of its strong links indirectly to human disease and diabetes. CBS is a key enzyme in the trans-sulfuration pathway that catalyzes the irreversible, serine dependant conversion of homocysteine to cystathionine. The synthesis of cystathionine is the first reaction in the irreversible pathway for the catabolism of homocysteine via sequential conversion to cysteine and sulfate in the liver, kidney, small intestine and pancreas45. It is now known that homocysteine metabolism is altered in diabetes and plasma homocysteine levels which may contribute to their increased cardiovascular risk31. In vitro, regulation of Cbs in rat hepatoma cell lines have shown increased Cbs enzyme activity after glucocorticoid exposure and decreased activity after insulin treatment46. The above results implicate that insulin could be an important regulator of Cbs gene and activity or altered levels of insulin in diabetes may influence local homocysteine metabolism and thus further contribute to islet dysfunction by alterations in glutathione and H2S levels47, 48.
In this manuscript, we have documented by temporal gene expression, in situ (Figure 4) and histochemical analysis that Cbs is an early marker expressed in the cytoplasm of the undifferentiated pancreatic epithelium (Figures 5, 6 and 7). Co-expression of Pdx1 and Ptf1a positive cells marks the undifferentiated cells of the early pancreatic epithelium as early as E9.5 to E12.54. This is followed temporally (E12.5 to E14.5) by a phase that is characterized by expansion of the branching network of the epithelial chords37. We documented the expression of Cbs in the epithelial cords at E12.5 with cells expressing Pdx1 and β-catenin. Since the cells expressing strongest Cbs were localized in the distal tips of the epithelial cords, we further localized its expression with Ptf1a. The commitment to the acinar cell type, in the distal tips of the epithelial cords is also marked by the expression of cMyc, carboxypeptidase A and Ptf1a expression37. It is therefore not surprising that the temporal gene expression patterns of Ptf1a (Figure 3) and Cbs (Figure 4) are similar. Ptf1a being the nuclear factor and Cbs expressed as a cytoplasmic marker of similar differentiation pathways. Due to technical constrains, Cbs, Pdx1 and Ptf1a could not be co-localized together, however from the pattern of expression of Cbs, it can be speculated that alterations in the level of Pdx1 or Ptf1a could regulate Cbs expression and activity thereby, modulating the development of the acinar compartment in mouse. The supporting evidence comes from microarray profiling of Pdx+/− mice that show decreased Cbs mRNA expression as compared to wild type49, further adding to the complexity of the transcriptional network governing the differentiation of the pancreas. Moreover, we can only speculate about the enzymatic activity of Cbs at this early stage of development and whether homocysteine metabolism or H2S as a gaseous byproduct of this pathway has any potential role in development.
Given the complexity of the transcriptional network governing the development of the pancreas, the proximity of the cells expressing these 4 embryonic makers and how these markers interact among themselves and other well known players still remains open to speculation. Since our studies in human pancreatic development still rely heavily on extrapolated data from mouse genomic and transgenic study, we believe it would be useful to apply advanced bioinformatics tools to identify novel genes, which of course would have to be thoroughly validated using wet laboratory techniques. Such queries can be further refined to include background knowledge specifically related to β-cell development from the Neurog3 knockout background9.
The algorithms used in the Hanalyzer protocol are uniquely adaptable to any newer high throughput data like microRNA or high throughput sequencing. Our global microarray dataset will be available publicly on GeneSpeed20, 21 and EPConDB50 databases and will provide an important tool for further scientific endeavors.
In conclusion, apart from generating robust and useful microarray dataset related to temporal development of murine pancreas, the current study also implemented the use of an advanced computational tool to analyze large-scale gene expression dataset relevant to mouse pancreatic development. The use of this tool was critical in identifying 4 new embryonic markers that have limited characterization in terms of pancreatic development and validated the expression of Cbs in the early undifferentiated epithelial buds and later in the developing acinar cells.
Supplementary Material
ACKNOWLEDGEMENTS
S.A.S obtained grant funding, designed study and wrote manuscript.
C.E.L performed individual experiments and calculated data.
K.J and J.D.D performed microarray experiments.
H.T, A. K-F and L.H provided bioinformatics support.
JAW assisted with time-mated breeding and edited manuscript.
J.C.H provided scientific input and edited manuscript.
The authors are grateful to Jan Kraus, PhD from University of Colorado Denver for providing Cbs monoclonal antibody. Dr. Chris Wright, PhD is thanked for the Pdx1 antibody.
DERC (P30 DK057516 to J.C.H) molecular core is thanked for the personnel and equipment used to conduct gene validations.
This work was supported by funding from Juvenile Diabetes Research Foundation (1-2008-1021) to SAS. SAS is additionally supported by NIH/NIDDK (K01DK80193). LH is supported by R01LM008111 and AFK is supported by T15LM009451
Abbreviations
- CT
Cycle threshold
- ES
Embryonic Stem Cells
- FDR
False Detection Rate
- iPS
Induced Pluripotent Stem Cells
- OCT
Optimum Cutting Temperature
- PCA
Principal Component Analysis
- Q-RTPCR
Quantitative-Real time polymerase Chain Reaction
- TSA
Tyramide System Amplification
Footnotes
Disclosure: The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Jorgensen MC, Ahnfelt-Ronne J, Hald J, et al. An illustrated review of early pancreas development in the mouse. Endocr Rev. 2007 Oct;28(6):685–705. doi: 10.1210/er.2007-0016. [DOI] [PubMed] [Google Scholar]
- 2.Spagnoli FM. From endoderm to pancreas: a multistep journey. Cell Mol Life Sci. 2007 Sep;64(18):2378–2390. doi: 10.1007/s00018-007-7184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pictet RL, Clark WR, Williams RH, et al. An ultrastructural analysis of the developing embryonic pancreas. Dev Biol. 1972 Dec;29(4):436–467. doi: 10.1016/0012-1606(72)90083-8. [DOI] [PubMed] [Google Scholar]
- 4.Seymour PA, Sander M. Historical perspective: beginnings of the beta-cell: current perspectives in beta-cell development. Diabetes. Feb;60(2):364–376. doi: 10.2337/db10-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gradwohl G, Dierich A, LeMeur M, et al. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000 Feb 15;97(4):1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S, Jensen JN, Seymour PA, et al. Sustained Neurog3 expression in hormone-expressing islet cells is required for endocrine maturation and function. Proc Natl Acad Sci U S A. 2009 Jun 16;106(24):9715–9720. doi: 10.1073/pnas.0904247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002 May;129(10):2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 8.Mellitzer G, Martin M, Sidhoum-Jenny M, et al. Pancreatic islet progenitor cells in neurogenin 3-yellow fluorescent protein knock-add-on mice. Mol Endocrinol. 2004 Nov;18(11):2765–2776. doi: 10.1210/me.2004-0243. [DOI] [PubMed] [Google Scholar]
- 9.Juhl K, Sarkar SA, Wong R, et al. Mouse pancreatic endocrine cell transcriptome defined in the embryonic Ngn3-null mouse. Diabetes. 2008 Oct;57(10):2755–2761. doi: 10.2337/db07-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugiyama T, Rodriguez RT, McLean GW, et al. Conserved markers of fetal pancreatic epithelium permit prospective isolation of islet progenitor cells by FACS. Proc Natl Acad Sci U S A. 2007 Jan 2;104(1):175–180. doi: 10.1073/pnas.0609490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanders TG, Rutter WJ. The developmental regulation of amylolytic and proteolytic enzymes in the embryonic rat pancreas. J Biol Chem. 1974 Jun 10;249(11):3500–3509. [PubMed] [Google Scholar]
- 12.Hick AC, van Eyll JM, Cordi S, et al. Mechanism of primitive duct formation in the pancreas and submandibular glands: a role for SDF-1. BMC Dev Biol. 2009;9:66. doi: 10.1186/1471-213X-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobberup S, Schmerr M, Dang ML, et al. Conditional control of the differentiation competence of pancreatic endocrine and ductal cells by Fgf10. Mech Dev. Apr;127(3–4):220–234. doi: 10.1016/j.mod.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wescott MP, Rovira M, Reichert M, et al. Pancreatic ductal morphogenesis and the Pdx1 homeodomain transcription factor. Mol Biol Cell. 2009 Nov;20(22):4838–4844. doi: 10.1091/mbc.E09-03-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higuchi Y, Shiraki N, Kume S. In vitro models of pancreatic differentiation using embryonic stem or induced pluripotent stem cells. Congenit Anom (Kyoto) Mar;51(1):21–25. doi: 10.1111/j.1741-4520.2010.00307.x. [DOI] [PubMed] [Google Scholar]
- 16.Noguchi H. Production of pancreatic beta-cells from stem cells. Curr Diabetes Rev. May;6(3):184–190. doi: 10.2174/157339910791162934. [DOI] [PubMed] [Google Scholar]
- 17.Zhang D, Jiang W, Liu M, et al. Highly efficient differentiation of human ES cells and iPS cells into mature pancreatic insulin-producing cells. Cell Res. 2009 Apr;19(4):429–438. doi: 10.1038/cr.2009.28. [DOI] [PubMed] [Google Scholar]
- 18.Leach SM, Tipney H, Feng W, et al. Biomedical discovery acceleration, with applications to craniofacial development. PLoS Comput Biol. 2009 Mar;5(3):e1000215. doi: 10.1371/journal.pcbi.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tipney HJ, Leach SM, Feng W, et al. Leveraging existing biological knowledge in the identification of candidate genes for facial dysmorphology. BMC Bioinformatics. 2009;10(Suppl 2):S12. doi: 10.1186/1471-2105-10-S2-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kutchma A, Quayum N, Jensen J. GeneSpeed: protein domain organization of the transcriptome. Nucleic Acids Res. 2007 Jan;35(Database issue):D674–679. doi: 10.1093/nar/gkl990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quayum N, Kutchma A, Sarkar SA, et al. GeneSpeed Beta Cell: an online genomics data repository and analysis resource tailored for the islet cell biologist. Exp Diabetes Res. 2008;2008:312060. doi: 10.1155/2008/312060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol. 696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 23.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003 Nov;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeung N, Cline MS, Kuchinsky A, et al. Exploring biological networks with Cytoscape software. Curr Protoc Bioinformatics. 2008 Sep; doi: 10.1002/0471250953.bi0813s23. Chapter 8:Unit 8 13. [DOI] [PubMed] [Google Scholar]
- 25.Kawaguchi Y, Cooper B, Gannon M, et al. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002 Sep;32(1):128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 26.Schaffer AE, Freude KK, Nelson SB, et al. Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev Cell. Jun 15;18(6):1022–1029. doi: 10.1016/j.devcel.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraus JP. Komrower Lecture. Molecular basis of phenotype expression in homocystinuria. J Inherit Metab Dis. 1994;17(4):383–390. doi: 10.1007/BF00711354. [DOI] [PubMed] [Google Scholar]
- 28.Kraus JP. Biochemistry and molecular genetics of cystathionine beta-synthase deficiency. Eur J Pediatr. 1998 Apr;157(Suppl 2):S50–53. doi: 10.1007/pl00014304. [DOI] [PubMed] [Google Scholar]
- 29.Kraus JP, Janosik M, Kozich V, et al. Cystathionine beta-synthase mutations in homocystinuria. Hum Mutat. 1999;13(5):362–375. doi: 10.1002/(SICI)1098-1004(1999)13:5<362::AID-HUMU4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 30.Meier M, Oliveriusova J, Kraus JP, et al. Structural insights into mutations of cystathionine beta-synthase. Biochim Biophys Acta. 2003 Apr 11;1647(1–2):206–213. doi: 10.1016/s1570-9639(03)00048-7. [DOI] [PubMed] [Google Scholar]
- 31.Wijekoon EP, Brosnan ME, Brosnan JT. Homocysteine metabolism in diabetes. Biochem Soc Trans. 2007 Nov;35(Pt 5):1175–1179. doi: 10.1042/BST0351175. [DOI] [PubMed] [Google Scholar]
- 32.Phelan JD, Shroyer NF, Cook T, et al. Gfi1-cells and circuits: unraveling transcriptional networks of development and disease. Curr Opin Hematol. Jul;17(4):300–307. doi: 10.1097/MOH.0b013e32833a06f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramirez J, Lukin K, Hagman J. From hematopoietic progenitors to B cells: mechanisms of lineage restriction and commitment. Curr Opin Immunol. Apr;22(2):177–184. doi: 10.1016/j.coi.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Meer LT, Jansen JH, van der Reijden BA. Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia. Nov;24(11):1834–1843. doi: 10.1038/leu.2010.195. [DOI] [PubMed] [Google Scholar]
- 35.Bicknell LS, Pitt J, Aftimos S, et al. A missense mutation in ALDH18A1, encoding Delta1-pyrroline-5-carboxylate synthase (P5CS), causes an autosomal recessive neurocutaneous syndrome. Eur J Hum Genet. 2008 Oct;16(10):1176–1186. doi: 10.1038/ejhg.2008.91. [DOI] [PubMed] [Google Scholar]
- 36.Patel A, Rees SD, Kelly MA, et al. Association of variants within APOE, SORL1, RUNX1, BACE1 and ALDH18A1 with dementia in Alzheimer's disease in subjects with Down syndrome. Neurosci Lett. 7 Jan;487(2):144–148. doi: 10.1016/j.neulet.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Q, Law AC, Rajagopal J, et al. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell. 2007 Jul;13(1):103–114. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Oster A, Jensen J, Serup P, et al. Rat endocrine pancreatic development in relation to two homeobox gene products (Pdx-1 and Nkx 6.1) J Histochem Cytochem. 1998 Jun;46(6):707–715. doi: 10.1177/002215549804600602. [DOI] [PubMed] [Google Scholar]
- 39.Bassett DE, Jr., Eisen MB, Boguski MS. Gene expression informatics--it's all in your mine. Nat Genet. 1999 Jan;21(1 Suppl):51–55. doi: 10.1038/4478. [DOI] [PubMed] [Google Scholar]
- 40.Baumgartner WA, Jr., Cohen KB, Fox LM, et al. Manual curation is not sufficient for annotation of genomic databases. Bioinformatics. 2007 Jul 1;23(13):i41–48. doi: 10.1093/bioinformatics/btm229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hisa T, Spence SE, Rachel RA, et al. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 2004 Jan 28;23(2):450–459. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Burstin J, Reichert M, Wescott MP, et al. The pancreatic and duodenal homeobox protein PDX-1 regulates the ductal specific keratin 19 through the degradation of MEIS1 and DNA binding. PLoS One. 5(8):e12311. doi: 10.1371/journal.pone.0012311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bjerknes M, Cheng H. Cell Lineage metastability in Gfi1-deficient mouse intestinal epithelium. Dev Biol. 1 Sep;345(1):49–63. doi: 10.1016/j.ydbio.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 44.Shroyer NF, Wallis D, Venken KJ, et al. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005 Oct 15;19(20):2412–2417. doi: 10.1101/gad.1353905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finkelstein JD. Pathways and regulation of homocysteine metabolism in mammals. Semin Thromb Hemost. 2000;26(3):219–225. doi: 10.1055/s-2000-8466. [DOI] [PubMed] [Google Scholar]
- 46.Ratnam S, Maclean KN, Jacobs RL, et al. Hormonal regulation of cystathionine beta-synthase expression in liver. J Biol Chem. 2002 Nov 8;277(45):42912–42918. doi: 10.1074/jbc.M206588200. [DOI] [PubMed] [Google Scholar]
- 47.Kaneko Y, Kimura T, Taniguchi S, et al. Glucose-induced production of hydrogen sulfide may protect the pancreatic beta-cells from apoptotic cell death by high glucose. FEBS Lett. 2009 Jan 22;583(2):377–382. doi: 10.1016/j.febslet.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 48.Kaneko Y, Kimura Y, Kimura H, et al. L-cysteine inhibits insulin release from the pancreatic beta-cell: possible involvement of metabolic production of hydrogen sulfide, a novel gasotransmitter. Diabetes. 2006 May;55(5):1391–1397. doi: 10.2337/db05-1082. [DOI] [PubMed] [Google Scholar]
- 49.Oliver-Krasinski JM, Kasner MT, Yang J, et al. The diabetes gene Pdx1 regulates the transcriptional network of pancreatic endocrine progenitor cells in mice. J Clin Invest. 2009 Jul;119(7):1888–1898. doi: 10.1172/JCI37028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazzarelli JM, Brestelli J, Gorski RK, et al. EPConDB: a web resource for gene expression related to pancreatic development, beta-cell function and diabetes. Nucleic Acids Res. 2007 Jan;35(Database issue):D751–755. doi: 10.1093/nar/gkl748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.