Summary
Silicon microchips with thin electron transparent silicon nitride windows provide a sample support that accommodates both light-, and electron microscopy of whole eukaryotic cells in vacuum or liquid, with minimum sample preparation steps. The windows are robust enough that cellular samples can be cultured directly onto them, with no addition of a supporting film, and no need to embed or section the sample, as is typically required in electron microscopy. By combining two microchips, a microfluidic chamber can be constructed for the imaging of samples in liquid in the electron microscope. We provide microchip design specifications, a fabrication outline, instructions on how to prepare them for biological samples, and examples of images obtained using different light-, and electron microscopy modalities. The use of these microchips is particularly advantageous for correlative light-, and electron microscopy.
Keywords: Microchips, sample preparation, TEM, STEM, Correlated Light and Electron Microscopy, liquid STEM
Introduction
Transmission electron microscopy (TEM) has traditionally been used to study the ultrastructure of cells via the preparation of conventional thin sections, where a cellular sample is fixed, stained, embedded in plastic resin, and sliced (Bozzola & Russell, 1999). Cells can also be prepared into amorphous ice via high-pressure freezing and cryo-sectioning (Barcena & Koster, 2009, Stahlberg & Walz, 2008), although the involved protocols are rather complex. For many questions in cell biology it is useful to study samples containing whole cells with both light microscopy and TEM, which can be accomplished by growing cells directly on 3 mm support grids with carbon or formvar thin films (Sartori et al., 2007a). However, these thin films are typically difficult to handle with cell culture procedures. In recent years, semiconductor-manufacturing processes have resulted in a new type of support film for TEM: thin silicon nitride (SiN) membranes (Ciarlo, 2002, Williamson et al., 2003). These films are highly homogeneous in thickness, extremely robust, and are biocompatible, meaning that cells can be cultured directly on them. In addition, SiN windows can be used as separation windows for vacuum chambers, such that cells in liquid can be studied with electron microscopy (EM) (de Jonge et al., 2009, Nishiyama et al., 2010). The use of microchips as support for cellular samples (Dukes et al., 2010a) is especially beneficial for correlative light microscopy (LM) and EM, wherein lower-resolution images are obtained via LM, and EM is used to “zoom in” on a section of interest with nanometer resolution (Gaietta et al., 2002).
Here, we describe the design and fabrication of the microchips, and provide details of how to prepare biological samples to be imaged on them, including specific labeling with nanoparticles, and drying procedures for use with TEM. We provide examples of images of whole eukaryotic cells, obtained using LM, TEM, scanning TEM (STEM), and liquid STEM. In all examples, the cells were prepared directly on the microchips. We also discuss correlative approaches. This paper serves as a detailed guide for those interested in using silicon microchips with SiN windows as support for cellular samples.
Microchips
Design and testing
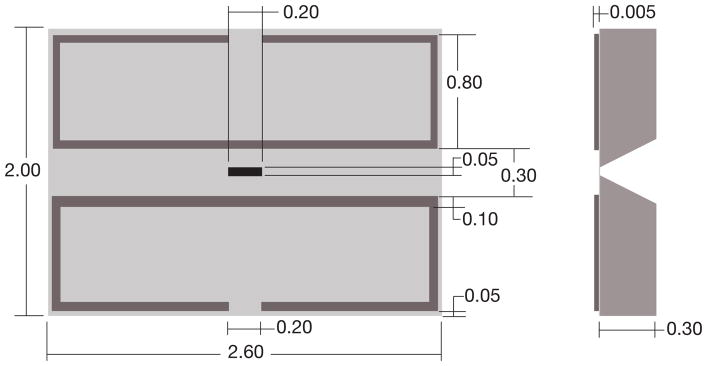
The microchips (manufactured by Protochips Inc, NC) were made of silicon with dimensions of 2.00 mm in height, 2.60 mm in length, and 0.30 mm in thickness, with a silicon nitride (SiN) window in the middle (Ring & de Jonge, 2010), see Fig. 1. SiN is a commonly used material in micro electro mechanical systems (MEMS) fabrication. To determine the optimum SiN window thickness, we deposited gold nanoparticles on the top and bottom of a 50 nm thick test window and imaged the window at high magnification in an aberration corrected 200 kV STEM (JEOL 2200 FS) in a previous study (Ramachandra et al., 2011). Gold fringes with a spacing of 0.2 nm were visible for particles both at the bottom and at the top, thus we concluded that the 50 nm thick window had a negligible effect on the electron beam. A 100 nm-thick test window was also imaged, but the image of the gold layer at the bottom of the window appeared blurred. This is attributed to elastic scattering of the electron beam in the SiN material. With TEM, the resolution would be slightly decreased due to inelastic scattering. Using standard equations, (Reimer & Kohl, 2008) it was calculated that the resolution in TEM would be limited to maximal 0.5 nm for 50 nm-thick SiN, which is more than sufficient for many biological applications. However, when higher resolution is required, for example, for single particle tomography, then thinner SiN windows, or carbon windows should be considered.
Fig. 1.
The design of the spacer microchip. Both the top view and the side-view cross-section are shown. All dimensions are in mm. The microchip had a width of 2.00 mm, a length of 2.60 mm, and a thickness of 0.30 mm. The tolerances were ± 10 μm. The black area in the middle indicates the 50 nm thick silicon nitride (SiN) window. A patterned spacer is shown in dark grey, providing a flow channel over the SiN window.
Several sizes of the SiN window were tested. Windows spanning several hundreds of micrometers allow a large field, but are prone to rupture during cell culture processing steps. The rigidity of the windows is of particular importance for liquid electron microscopy, where two windows are placed in the vacuum of the electron microscope, and enclose a liquid at atmospheric pressure. The windows have to withstand this pressure difference, and each window should not bulge by more than ~1 μm for liquid STEM imaging, as the bulging could render the sample too thick to image. Furthermore, if the thickness throughout the sample varied due to bulging, then the image contrast would vary correspondingly, preventing high-resolution imaging. To optimize window strength and field of view, we tested rectangular windows, since the degree of bulging is mainly determined by the short dimension (Creemer et al., 2010). Two windows with dimensions of 50×200 μm2 and 50 nm thickness were used to enclose a liquid at atmospheric pressure in the vacuum of the electron microscope. The windows were found to bulge by about a micrometer each, which was deemed acceptable. However, windows with dimensions of 70×200 μm2 were found to bulge by about 2 μm each, which was considered too large to allow for high resolution imaging (Ring & de Jonge, 2010).
The dimensions were chosen such that at least one entire eukaryotic cell could be viewed within the window. It is not uncommon for typical surface adhering eukaryotic cells to spread out to a length of 50 μm (Satulovsky et al., 2008). Thus we aimed for a window with a width that was no smaller than 50 μm. The length of 200 μm facilitated the imaging of multiple cells, which is desirable for biological experiments. Because the width of the window is the limiting dimension in terms of resistance to bulging and breaking, the length can be altered to fit the user’s needs. Additionally, for experiments that involve smaller objects, the width can be made smaller.
For use in liquid EM, two microchips have to be positioned on top of each other. In this application it is crucial to ensure that the windows of both microchips are aligned as closely as possible, in order to maintain a sufficiently large field of view for the transmitted electron beam. A precise overlap of the windows was obtained by 1) manufacturing microchips with precisely diced sides edges, 2) defining the SiN windows in the center of each microchip, and 3) aligning the microchips in a slot in the specimen holder at their sides. All dimensions of the microchips were manufactured within ±10 μm precision. An additional benefit was that the precision-made vertical sides of the microchips allowed for a much easier handling, compared to commercially available microchips. Certain commercially available microchips have tapered sides, which seem to be difficult to secure with tweezers without damaging the sample at the top surface. If a small field of view is acceptable, then a different solution to the alignment problem is to use one of the standard chips, and one chip where the window is oriented perpendicular to that of the standard model. We have successfully used windows with widths as small as 20 μm. Because these windows bulge less than 1 μm each, they can be used for TEM imaging of liquid specimens, which requires thinner samples and has stricter maximum bulging restrictions for the windows than liquid STEM.
Design of spacer layer
To prevent compression of the sample when the microchips are assembled into the microfluidic device, a spacer is needed between the microchips. Two different types of spacers were used. Microspheres were placed between the microchips, with the advantage being that the gap between the microchips was customizable by choosing microspheres of the desired size. (de Jonge et al., 2009). The microspheres were positioned at the four corners of one microchip (not at the window, otherwise rupture is likely to occur), which created a fluid path from the input tubing, through the microchips and surrounding area of the holder, and out through the output tubing. This method accommodated fluid flow, but did not allow for control of the fluid path.
In order to precisely control the rate at which fluid injected into the system reaches the sample, we created a system with a defined fluid path (Ring & de Jonge, 2010). This consisted of a microchip with a patterned spacer layer (Fig. 1) paired with a flat base microchip, providing a straight liquid flow channel with a width of 300 μm over the long side of the microchip. The spacer wall width was 100 μm and its thickness was 6 μm.
Cells were typically grown on base microchips, which have no spacer and provide a flat surface over the whole microchip, allowing the cells to spread and cover the most of this surface. During assembly of the microfluidic chamber the spacer microchip was pressed, with the spacer layer facing down, onto the base microchip containing cells inside the slot in the specimen holder. We patterned a void into the spacer to provide an area for excess cellular material. The wall around the void included an opening at the side where excess cellular material could flow out. Furthermore, the spacer did not extend to the end of the microchips, because in that case, the dicing process (see below) led to detachment of the spacer. The specific design of the spacer layer can be varied by changing the photolithography mask, and the thickness can also be changed, if desired.
Fabrication
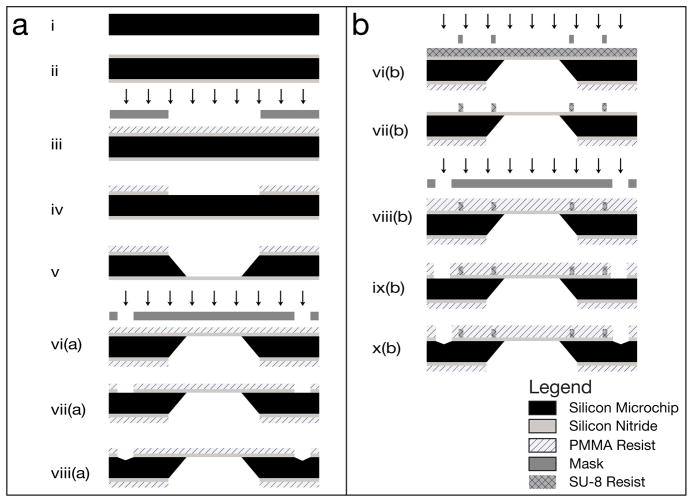
The microchip fabrication procedure was based on standard methods (Grant et al., 2004); the main difference being the highly specified side tolerances, both for the smoothness of the sides, and also for the location of the window with respect to the sides. A schematic of the fabrication of the microchips is shown in Fig. 2 for the base microchip, while Fig. 2b describes the spacer microchip.
Fig. 2.
Schematic of the workflow of the fabrication of (a) the base microchips and (b) the spacer microchips. The arrows indicate lithographic exposure. Drawings not to scale.
A 300 μm thick, double side polished, n-type Si(100) wafer was cleaned using the two step, standard cleaning procedure used in the semiconductor industry, which removes all organic and metal particles, and strips the microchips of their native oxide layer (Step i) (Grant et al., 2004). A low-stress silicon nitride layer was then deposited on both sides of the wafer via low-pressure chemical vapor deposition at 810 °C (Step ii). The thickness of the SiN layer was confirmed using ellipsometry. A layer of resist was applied, and the window was then defined by photolithography using a mask made of soda lime glass (Step iii). The silicon nitride layer was isotropically dry-etched (Step iv), which resulted in a second mask, allowing for an anisotropic KOH etch of the silicon (Step v). This defined the “front” and “back” sides of the microchips – the front side containing the silicon nitride window.
The edges of the microchips were defined with a tolerance of ±10 μm with respect to the window position. Grooves were etched in the silicon for guidance of the saw-blade. The location of the grooves was patterned into the silicon nitride on the front side of the wafer using CHF3 reactive ion beam etching (Step vii(a)). The silicon was then anisotropically etched to create the groove (Step viii(a)). Before the final step, a resist coating was applied to protect the microchips from damage and from collecting debris. Then, the microchips were placed with the backside down onto double-sided adhesive tape, and were separated by dicing along the grooves with a precision saw.
For the microchips with the spacer layer, the front side of the microchip was coated in a layer of SU-8 photoresist, and the spacer pattern was defined by photolithography (Step vi(b)). The grooves were then established using the same methods as for the base microchips (Steps vii(b)–x(b)), and the microchips were coated with a protective resist layer, and then diced using the same method as the base microchips.
Cleaning
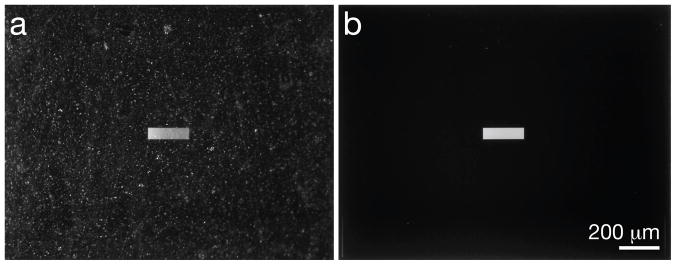
To allow for high resolution imaging, the SiN window surfaces must be free of any contamination, such as dust particles or material fragments. Both types of contamination are easily collected during manufacturing (especially during dicing), and also during exposure outside of a clean room environment. To keep the microchips clean until they were ready to be used, a resist coating was applied on the silicon nitride during the fabrication process. Once the microchips were ready to be used, the coating was removed by washing for 2 minutes in acetone followed by a 2 minute wash in ethanol, taking care that the acetone did not dry during the transfer of the microchips between the two liquids. About 150 mL of each fluid was sufficient to clean up to 12 microchips at a time, and HPLC grade liquids were always used. Fig. 3a shows a window covered in the resist coating and debris from manufacturing, as compared to a clean window, shown in Fig. 3b.
Fig. 3.
Photographs showing the difference between coated and clean microchips. (a) A microchip with a protective resist coat, covered in debris is shown. (b) The same microchip after washing with acetone, ethanol, and water.
The windows were hydrophobic after stripping the resist, but were made hydrophilic for use with biological samples by plasma cleaning for several minutes, usually 3–7. The hydrophilicity lasted for about a day. After plasma cleaning, the microchips were washed with H2O (also HPLC grade) to remove any remainders of debris and salt. Next, the microchips were coated with poly-L-lysine to extend the duration of their hydrophilicity and to promote cell adhesion. This coating was accomplished by immersing the microchips in a solution of 0.01% poly-L-lysine for five minutes at room temperature. Excess poly-L-lysine was removed by soaking the microchips three times briefly in fresh volumes of HPLC grade water. The microchips were inspected under an optical microscope after each cleaning step.
If polystyrene microspheres were used for a spacer layer, they needed to be applied on hydrophobic microchips, thus before plasma cleaning. 0.2 μl droplets containing the beads in aqueous solution were placed at the corners of a microchip. If the droplets were applied after plasma cleaning, the entire microchip surface was wetted, and the microspheres were not confined to the corners. There should be ~5–20 beads in each droplet, depending on the size of microspheres used.
Biological sample preparation
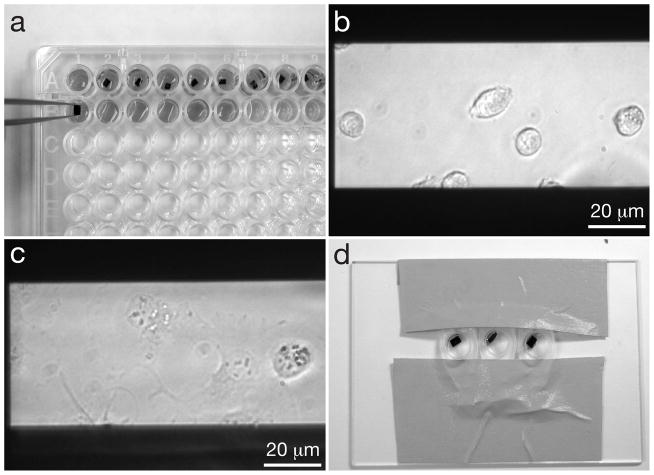
After the microchips had been prepared for cell culture, they were transferred to a ninety-six well cell culture plate, as shown in Fig. 4a, for biological experiments, including cell seeding, labeling, fixation, staining and dehydration.
Fig. 4.
Seeding cells onto microchips and labeling the cells. (a) Microchips in a 96 well plate, where they will be seeded with cells, and the cells will be fixed. Note the orientation of the tweezers gripping microchips by their sides and not their top surfaces. Tweezers with a teflon coated flat tips are recommended. (b) COS7 cells on windows after ~5 minutes of incubation. (c) COS7 cells that have adhered to a window, after ~1 hour of incubation. (d) Microchips in labeling device are inclined against a drop of labeling solution to reduce the amount of non-specifically bound label on the sample.
Cell seeding
Each step of the cell seeding process was accomplished by transferring the microchips to a new well of the 96 well plate. The microchips were transferred between wells using Teflon coated tweezers with flat tips, to minimize the incidence of microchips adhering to the tweezers, and to prevent damage to the edges of the microchips. They were held such that the microchips remained upright (flat-side up) at all times, and the tweezers did not come into contact with the surface of the microchip, except at the edges, as shown in Fig. 4a. If a microchip was placed into a well with a solution and then immediately (within ~10 seconds) placed into a new well, that was termed “rinsing” whereas if a microchip was placed into a well and left for a longer amount of time, that was termed “immersing” or “incubating”.
The microchips were first immersed in ~150 μL of cell media per well. Confluent cells were detached from a culture flask using a cell stripper solution such as trypsin, and then re-suspended in cell media. We typically re-suspended cells grown in a 25 cm3 culture flask in 5 mL of media, which we counted to be ~(1–3)×105 cells/mL for COS7 cells, as determined using a hemocytometer. A droplet of the cell suspension was added to each well containing a microchip and media. After ~1 minute the microchips were inspected with an inverted light microscope to verify that cells had begun to adhere to the windows. A microchip should have no more than ~1 adhered cell per 50 × 50 μm2 area. The presence of more cells than this inhibits them from flattening against the window. If after ~5 minutes, the desired amount of cells had not begun to adhere to the window, we added another droplet of the cell suspension. If too many cells were present, we typically discarded the microchips, although the amount of cells could sometimes be sufficiently reduced by transferring the chip into a new well with fresh media, which washed away cells that had not yet tightly adhered to the window surface.
Once the desired amount of cells had begun adhering to the window, as seen in Fig. 4b, the microchips were immersed in a new well with media and incubated for at least 4 hours at 37°C in 5% CO2. For certain experiments, the cells were incubated in serum-free media for 2–12 hours prior to labeling, as this enhances the cells’ ability to uptake certain protein-coupled labels. An example of cells adhered to the surface is shown in Fig. 4c.
Cell labelling
Just as fluorescent tags can be used to image specific proteins in LM, nanoparticles made of high atomic number materials, such as gold, can be used to image specific proteins in EM. For example, it has been shown that epidermal growth factor (EGF) coupled to either quantum dots or gold nanoparticles could be used to label the EGF receptor (Dukes et al., 2010b, de Jonge et al., 2009).
To label the cells with protein-coupled labels, the following method was used, based on live cell labeling protocols developed by others (Ibaraki et al., 1996, Driskell et al., 2007). The microchips with adhered cells were first rinsed with Tyrode’s buffer containing 1–3% Bovine Serum Albumen (BSA) to reduce non-specific label binding. The chips were then placed in the labeling solution. Incubating the microchips in an inclined position, with the cells facing downwards, was found to reduce unspecific binding with certain labels, such as larger diameter gold nanoparticles, probably by reducing the effect of gravity on the labels. The rim of the 96 well plate wells are too high to allow for the maneuvering of the microchips into such a position, so we made simple labeling devices using lids of 0.2 mL microcentrifuge tubes that were clipped from the tubes and fixed with tape onto a large glass microscope slide, as shown in Fig. 4d. For the labeling, a droplet of gold nanoparticle label solution was deposited at the edge of such a plastic well, and a microchip was then placed onto this droplet. For long incubation times (longer than 5 minutes) these labeling devices were placed into a humid environment, such as a plastic box containing damp paper towels, to prevent evaporation of the labeling solution. The duration of incubation was defined by the protein-label pair used, and the purpose of the experiment. For example, to examine membrane bound EGF with gold nanoparticles (EGF-Au) before internalization occurs (Lidke et al., 2004b), the microchips were incubated for 5 – 10 minutes at room temperature (de Jonge et al., 2009). After incubation with the label, the microchips were rinsed with Tyrode’s buffer containing BSA to remove any unbound label, and further processed for fixation.
Cell fixation and staining
Fixation of biological samples can help to protect them from degradation during sample preparation and imaging. Fixation of cells attached to SiN windows for EM is analogous to the fixation of fluorescence microscopy samples. However, instead of the typical LM fixatives (i.e. paraformaldehyde, formalin), glutaraldehyde is used, as it is a stronger EM fixative (Hopwood, 1969).
For fixation, the microchips were immersed in a 4% glutaraldehyde solution in Phosphate Buffer Saline (PBS) at room temperature for at least 20 minutes. After this step, the cells were rinsed three times in PBS to remove excess glutaraldehyde, followed by three five-minute immersions in 10% PBS in water to remove excess salt. If the cells were to be air dried, an additional soaking step in pure water was performed.
In some cases it was required to post-fix and stain the samples with osmium tetroxide, which aids fixation by increasing lipid stability, and also increases contrast during EM (Gluaert & Lewis, 1998). In that case, the microchips were first rinsed with cacodylate buffer (0.1M, pH 7.4), and then immersed in a osmium tetroxide solution 1% in cacodylate buffer for 1 hour at 37°C with 5% CO2. We later found that a much lower osmium tetroxide concentration of 0.001% provided sufficient contrast and sample stability; and also that the required time could be lowerd to 30 min, even at room temperature. Because the sample consists of a monolayer of thin cells only, presumably the fixation conditions are relaxed as compared to tissue samples. The microchips were then rinsed for 5 minutes each, first 3 times in cacodylate buffer, and then 3 times in water times to remove salts. The samples were stored in the refrigerator overnight before use.
Sample drying
For standard EM imaging, the sample must first be dried before it can be imaged in the vacuum of the electron microscope. Any water left in a sample will evaporate upon exposure to vacuum, resulting in structural damage and instability of the sample (Bozzola & Russell, 1999).
The sample can be air dried in principle (after removal of salts via soaking in pure water),but the high surface tension of water typically causes cellular damage that can be seen in high-resolution imaging. Preferably, the water is replaced by ethanol, which causes less damage upon drying as it has a smaller surface tension than water. A graded ethanol series was used for dehydration. The samples were immersed in each of the following grades of ethanol for 5 minutes: 20%, 30%, 50%, 70%, 95%; and then immersed 3 times in 100% ethanol for 10 minutes each. EM-grade ethanol, with a very low concentration of water, was used. Samples were then air dried for 30 min, and used for imaging. We have also used hexamethildisilazane (HMDS) (Braet et al., 1997) to replace the ethanol for a further reduction of the surface tension, and thus to reduce the damaging occurring during drying. Just after ethanol dehydration, microchips were immersed for 5 minutes in an ethanol/HMDS (1:2) mixture, then 2 times in pure HMDS, and allowed to air dry for 30 minutes. An example of the results obtained using TEM with this method is shown in Fig. 6b.
Fig. 6.
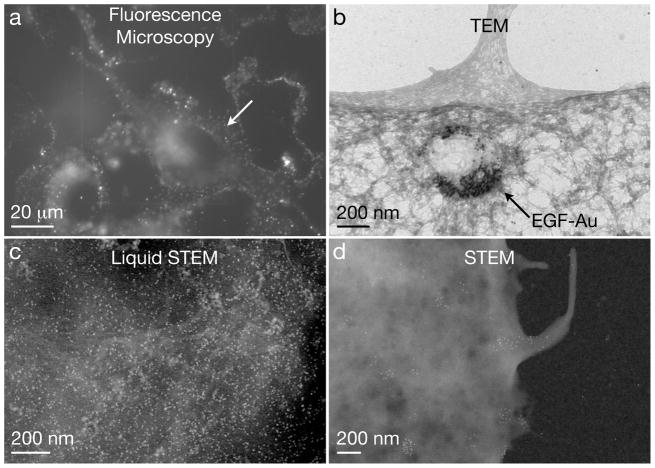
Examples of micrographs of cellular samples on microchips obtained with different microscopy modalities. (a) Fluorescence image showing epidermal growth factor (EGF)-quantum dot (QD) labeled COS7 fibroblast cells in saline water. Image with permission from (Dukes et al., 2010a). (b) TEM image of EGF-Au nanoparticles in a vesicle in a fixed and dried COS7 cell. (c) Liquid STEM image recorded at the location of the arrow in (a). Individual quantum dots are visible on a background of biological material. (d) Dry STEM image of EGF-QD labels on the edge of a COS7 cell.
An alternate way to dry samples with minimal damage to the cellular structure is critical point drying. Critical point drying is the process of drying a sample by raising the temperature and pressure such that the sample transitions from the liquid phase to the gas phase above the critical point. This eliminates damage deriving from the surface tension effects that occur if the sample crosses the phase boundary (Bozzola & Russell, 1999), and thus prevents collapse of the sample during dehydration. For critical point drying the microchips were placed in the chamber of the critical point drier (CPD) after the graded ethanol dehydration, while remaining immersed in 100% ethanol. Liquid CO2 was flown through the chamber of the CPD for one minute, followed by a five-minute soak, a cycle that was repeated until the fluid exchange was complete. The chamber was then heated under pressurized conditions to the critical point of CO2 (31.1 °C, 1072 psi) (NIST, 2008), and once this point was reached, the pressure was reduced over a period of 45 minutes. After any drying method that was used, samples were always stored under desiccated conditions until imaging.
Comparison to current techniques
The sample preparation process using SiN windows shares some similarities to current TEM preparation procedures (Bozzola & Russell, 1999, Glauert & Lewis, 1998), but also offers several advantages. Most conventional cell preparations involve fixation, post-staining, and drying of the samples. The cells are typically embedded in resin, such that they can be sliced into thin sections using an ultramicrotome. The embedded sections are transferred to metal or carbon grids, typically by floating the sample on water above a grid, and then draining that water. The inclusion of fluorescent tags can be used for correlative fluorescence microscopy and TEM of thin sections (Grabenbauer et al., 2005, Gaietta et al., 2002).
Whole-mount cell samples can be prepared on grids coated with a thin film such as carbon (Hosoi et al., 1981). Cells can be cultured, fixed, stained, and dried, for example, with critical-point drying (Bozzola & Russell, 1999). Alternatively, wet samples can be rapidly frozen for cryo TEM. This type of sample preparation is often used in correlative microscopy (van Rijnsoever et al., 2008, Sartori et al., 2007b).
The silicon microchips are compatible with both LM and EM and provide the most robust sample support. All steps in the preparation process are compatible with current biological protocols and equipment, e.g., the microchips fit in the well of a standard 96 well plate, and live cells on the microchips can be imaged with light microscopy. When the size and the thickness of the SiN windows are chosen correctly, they do dot break, or wrinkle, athough the window area is limited. The cells are cultured, labeled, fixed, stained, dried, and imaged directly on the microchip. In order to visualize labels, the samples can be easily air-dried, or critical-point drying can be used. We have shown that the microchips can be used to image fixed cells in a liquid environment (de Jonge et al., 2009), which is not possible when using grids. We have not tested if cryo samples can be prepared, but this should be possible in principle.
Microscopy
The cells prepared on microchips accommodate several different imaging modalities. The cells can be imaged with LM (dry or wet), either placed in a standard dish, or in the liquid (S)TEM specimen holder. Dry microchips can be examined with both STEM and TEM. Eukaryotic cells in the microfluidic chamber can be imaged with liquid STEM. Fig. 5 summarizes the possible imaging modalities, of which examples are provided in the following section.
Fig. 5.
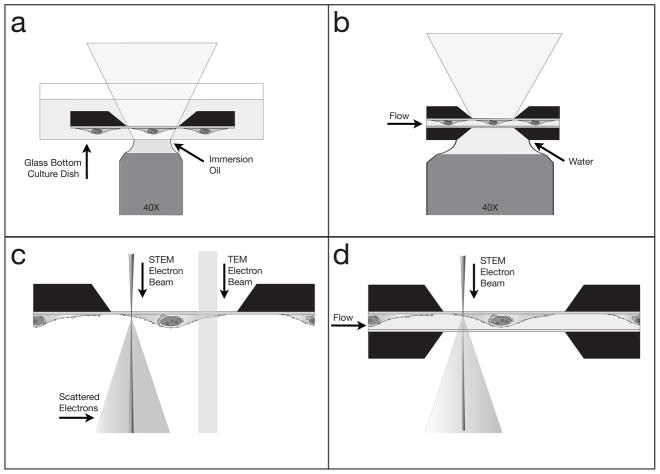
Schematics showing different ways microchips can be used for imaging. Images not to scale. (a) The microchip can be imaged in liquid in a standard dish with light microscopy (LM). (b) Two microchips enclosing a sample in liquid can be imaged with LM. (c) A dry sample on a microchip can be imaged with transmission electron microscopy (TEM), or the scanning TEM (STEM). (d) Two microchips enclosing a cell in liquid can be imaged with STEM; thin samples can be imaged in liquid TEM as well. Drawings not to scale.
Light microscopy
For LM imaging, the microchip with the cellular sample can simply be inverted onto a glass bottom culture dish containing saline water, and imaged in an inverted LM, following typical LM protocols, as shown in Fig. 5a. A fluorescence image of quantum dot (QD) labeled epidermal growth factor (EGF) receptors on COS7 cells in 10% PBS in water buffer is shown in Fig. 6a (Dukes et al., 2010b). The sample was incubated with labeling solution for 5 minutes, and then fixed with glutaraldehyde, such that the E GF-QDs remained on the cell membrane (Lidke et al., 2004a, de Jonge et al., 2009). The cellular surface is covered with bright spots. Possibly, activated EGF receptors at the cell membrane were in the process of forming clusters and coated pits at the onset of endocytosis. Additional fluorescence is visible in the nuclear region, presumably autofluorescence of the fixative. The image in Fig. 5a was recorded using a widefield microscope (TE300, Nikon), with a 40X oil immersion objective, equipped with a farred bandpass exitation filter (hq615/40 X) and a far-red emission filter set (hq710/100 m). The exposure time was 5 seconds. To reduce the autofluorescence of the glutaraldehyde fixative the samples where illuminated with the light source of the microscope for several minutes. The brightness and contrast of all images were optimized for contrast and brightness using ImageJ software (NIH).
The sample can also be assembled in a microfluidic chamber in the tip of the liquid (S)TEM holder, and the holder can be positioned on a microscope stage to study the sample with LM, as illustrated in Fig. 6b. The best microscopy results were obtained with a water immersion objective (data not shown). The space between the lens and the lower window was filled with water. Water immersion objectives allow working distances of several mm for a large numerical aperture (NA) values (typically NA = 1). A dry objective (with a lower NA than the water immersion objective) was also used, leaving an air gap between the lens and the window. It turned out to be impractical to use an oil immersion objective, because it was difficult to 1) ensure that the gap was entirely filled with oil, and 2) remove the oil from the microchips and specimen holder for subsequent liquid STEM.
Transmission electron microscopy
Dried samples can be viewed with standard TEM and STEM. The imaging setup for TEM, as well as for (dry) STEM imaging is shown schematically in Fig. 6c. Fig. 7b shows an example of TEM of an EGF-Au labeled COS7 cell. The sample was incubated with the labeling solution for 10 minutes, and then with Tyrode’s Buffer for 10 minutes more. After fixation, it was post-fixed with osmium tetroxide, and dried using a graded ethanol series/HMDS drying method. The EGF coupled nanoparticles are visible, concentrated in a large vesicle located near a filipodium, consistent with the idea that filipodia may be found in proximity of receptors like the EGF receptor (Lidke et al., 2005). This image was taken on a TEM (CM12, Philips) operating at 120 kV equipped with a 2k × 2k CCD camera. The microchip was placed in the TEM specimen holder with the cells facing downward; thus the SiN window did not interfere with the electron beam between the specimen and the projection system.
Fig. 7.
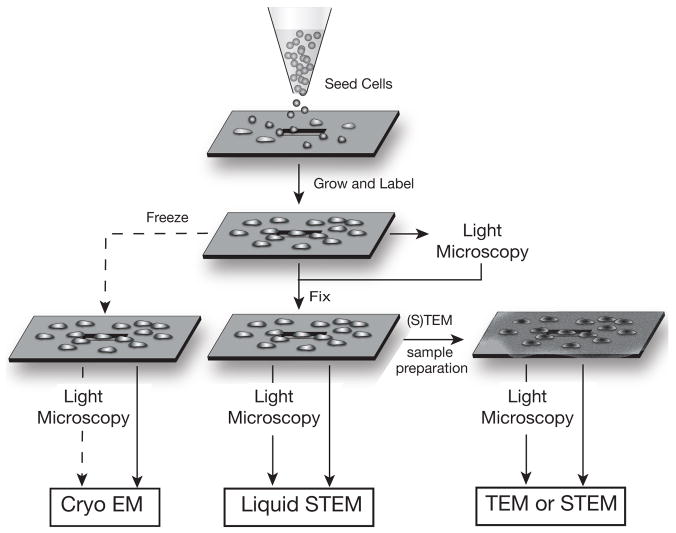
Diagram illustrating the versatility of the silicon microchip support for whole mount cell samples for imaging with different modalities. Cells are seeded onto a clean poly-L-lysine coated microchip, and grown and labeled (if needed) directly on the support. Live cells can be imaged with LM. The cells can then be fixed, or frozen (freezing was not yet tested and this branch in the schematics is, therefore, indicated with a dashed line). Fixed cells can undergo additional sample preparation steps for EM processing such as staining, dehydrating and evaporative coating, as needed. This sample preparation system permits multiple samples to be prepared under the same conditions, and efficiently processed for cryo EM, liquid STEM, TEM, and dry STEM. Thin biological samples can also be imaging with liquid TEM. The cells can be imaged at different stages with LM enabling correlative imaging studies to be performed. Drawings not to scale.
Scanning transmission electron microscopy
One of the main advantages of these microchips is the ability to assemble cellular samples in buffer solution into a microfluidic chamber placed in a specimen holder for liquid STEM (de Jonge et al., 2009, Dukes et al., 2010b). The specimen holder accommodates liquid flow in the specimen region (Ring & de Jonge, 2010), such that the microfluidic chamber is entirely filled with liquid even when placed in the vacuum of the electron microscope, and this system opens the possibility of live-cell experiments. We have previously demonstrated nanometer resolution on gold nanoparticles and QDs used to tag EGF on whole fixed eukaryotic cells (COS7) in their native liquid state. A schematic of the imaging setup is shown in Fig. 5d.
Liquid STEM is particularly useful in correlation with LM. Cells can be maintained in liquid in the microfluidic chamber and imaged with LM to provide overview images of the cells. Cellular regions can then be imaged at high resolution with liquid STEM, zooming-in on regions of interest with nanoscale resolution. If bi-modal labels are used, LM can reveal the locations of regions of tagged proteins with respect to the cellular structure, while liquid STEM provides information of the distribution of the labels at the level of individual proteins. We have for example, imaged the same cell of which the fluorescence image is shown in Fig. 6a, with liquid STEM. Fig. 6c reveals the locations of individual QDs on a background of signal from the biological material (the resolution obtained on the cellular material is much lower than that obtained on the labels, because the contrast mechanism of STEM depends on the atomic number(s) of the material(s) in the sample). The QDs are densely packed, which suggest that the EGF receptor had begun to concentrate together at the cellular surface, which is a step preceding the formation of endocytotic pits (Schreiber et al., 1983). Thus, where we observed a bright spot in the LM image, we found a high density of QDs in the liquid STEM image. We did not find QDs in regions outside of the cell. Fig. 6c was recorded with a 200 kV STEM (CM200, FEI), a pixel-dwell time of 20 μs, 1024×1024 pixels, a probe current of 0.68 nA, an electron probe semi-angle of 11 mrad, an annular dark field detector semi-angle of 70 mrad, a pixel size of 1.6 nm, and a magnification of 80,000. The noise in the mage was reduced by using a convolution filter (in ImageJ) with a kernel of (1, 1, 1; 1, 3, 1; 1, 1, 1). In Fig. 6a and c we assembled the microfluidic chamber after LM, but we have also successfully tested LM and subsequent liquid STEM of cells already in the microfluidic chamber. In that case, the amount of time needed for the transfer between the two microscopy modalities can be as short as a minute, mainly set by the vacuum pumping time, and this time determines the temporal correlation. Shorter times can be obtained using integrated LM and (S)TEM (Agronskaia et al., 2008). For samples thinner than about 1 μm liquid samples can also be viewed with TEM (Klein et al., 2011).
The microchips can also be used for imaging dried samples in the STEM, as shown schematically in Fig. 5c. An example of an image of a dried STEM sample is shown in Fig. 6d. The sample was prepared in the same way as that shown in Fig. 6a and c, (i.e., used EGF-QDs), and then dehydrated using a graded ethanol series, and air-dried. Individual EGF-QDs are visible along the edge of a dried COS7 cell. In this case, the cellular ultrastructure is visible with a much better contrast and resolution than in the case of liquid STEM, since the contrast is now obtained against a background of the SiN window only, instead of against a column of liquid as in liquid STEM. Fig. 5d was recorded at the same microscope settings as Fig. 5c, but with a magnification of 48,000 and a pixel size of 2.7 nm. The microchip was placed in a modified single-tilt specimen holder that had a 0.2 mm deeper sample slot. The microchip was positioned with the cell side facing upwards, such that the STEM probe was unperturbed by the SiN window, and the sample was imaged at the highest possible resolution.
An important benefit of using SiN membranes for EM imaging is their versatility. Typically, biological sample preparation techniques are chosen based on the type of imaging modality desired, and the possibility for using different or additional modalities may be limited based on the type of support chosen. However, this is not a problem when microchips supports are used. As illustrated in Fig. 7, starting with the same basic cell culture steps, essentially any desired imaging modality may be used. Cells are grown and labeled directly on the microchips, and may be viewed in the light microscope at any point after this. While sample freezing and cryo-EM have not yet been demonstrated, we expect that this should be possible. Alternatively, the samples could be fixed and then imaged using both LM and STEM for samples in liquid, with no further preparation steps required. If imaging the samples in liquid is not desired, the samples could be prepared using any other standard (S)TEM sample preparation procedure, and be imaged in both light and electron microscopes. The silicon microchips are compatible with all current biological protocols and equipment. Thus, they can be easily incorporated into current practices, streamlining laboratory procedures.
Conclusions
Silicon microchips with thin electron transparent SiN windows provide robust supports for both light and electron microscopy of whole eukaryotic cells in vacuum, or liquid. These microchip supports are suitable for a range of imaging modalities including LM, TEM, and dry, and liquid STEM, without imposing the need for different preparation protocols, except for the drying step, if desired. This minimizes sample processing. When using liquid STEM, the sample preparation is similar to standard light microscopy sample preparation using fixation. Most importantly, the presented microchip sample support can be used for experimental series where samples are studied with different microscopy modalities in correlative approaches.
Acknowledgments
We are grateful to Protochips Inc. (NC) provided the silicon microchips and specimen holder. STEM images were recorded at the SHaRE User Facility, which is sponsored by the Division of Scientific User Facilities, Office of Basic Energy Sciences, U.S. Department of Energy. TEM images and fluorescence data were recorded at the VUMC Cell Imaging Shared Resource. This work was supported by Vanderbilt University Medical Center, and by NIH grant R01GM081801.
References
- Agronskaia AV, Valentijn JA, van Driel LF, Schneijdenberg CT, Humbel BM, van Bergen en Henegouwen PM, Verkleij AJ, Koster AJ, Gerritsen HC. Integrated fluorescence and transmission electron microscopy. J Struct Biol. 2008;164:183–189. doi: 10.1016/j.jsb.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Barcena M, Koster AJ. Electron tomography in life science. Semin Cell Dev Biol. 2009;20:920–930. doi: 10.1016/j.semcdb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzola JJ, Russell LD. Electron Microscopy Principles and Techniques for Biologists. Jones and Barlett Publishers; Boston: 1999. [Google Scholar]
- Braet F, De Zanger R, Wisse E. Drying cells for SEM, AFM and TEM by hexamethyldisilazane: a study on hepatic endothelial cells. J Micros. 1997;186:84–87. doi: 10.1046/j.1365-2818.1997.1940755.x. [DOI] [PubMed] [Google Scholar]
- Ciarlo DR. Silicon nitride thin windows for biomedical microdevices. Biomed Microdev. 2002;4:63–68. [Google Scholar]
- Creemer JF, Helveg S, Hoveling GH, Ullmann S, Molenbroek AM, Sarro PM, Zandbergen HW. A MEMS reactor for atomic-scale microscopy of nanomaterials under industrially relevant conditions. J Microelectromech Syst. 2010:19. [Google Scholar]
- de Jonge N, Peckys DB, Kremers GJ, Piston DW. Electron microscopy of whole cells in liquid with nanometer resolution. Proc Natl Acad Sci. 2009;106:2159–2164. doi: 10.1073/pnas.0809567106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell O, Mironov A, Alayan P, Woodman P. Dynein is required for receptor sorting and the morphogenesis of early endosomes. Nat Cell Biol. 2007;9:113–120. doi: 10.1038/ncb1525. [DOI] [PubMed] [Google Scholar]
- Dukes MJ, Peckys DB, de Jonge N. Correlative fluorescence microscopy and scanning transmission electron microscopy of quantum-dot-labeled proteins in whole cells in liquid. ACS Nano. 2010a;4:4110–4116. doi: 10.1021/nn1010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- Glauert AM, Lewis PR. Biological specimen preparation for transmission electron microscopy. Portland press; London: 1998. [Google Scholar]
- Grabenbauer M, Geerts WJ, Fernadez-Rodriguez J, Hoenger A, Koster AJ, Nilsson T. Correlative microscopy and electron tomography of GFP through photooxidation. Nat Methods. 2005;2:857–862. doi: 10.1038/nmeth806. [DOI] [PubMed] [Google Scholar]
- Grant AW, Hu QH, Kasemo B. Transmission electron microscopy ‘windows’ for nanofabricated structures. Nanotechnol. 2004;15:1175–1181. [Google Scholar]
- Hopwood D. A comparison of the crosslinking abilities of glutaraldehyde, formaldehyde and alpha-hydroxyadipaldehyde with bovine serum albumin and casein. Histochem Cell Biol. 1969;17:151–161. doi: 10.1007/BF00277781. [DOI] [PubMed] [Google Scholar]
- Hosoi J, Hama K, Kosaka T. Whole mount observation of cultured cells by 200 kV ultrahigh resolution TEM. J Electron Microsc. 1981;30:57–62. [PubMed] [Google Scholar]
- Ibaraki N, Lin L, Reddy V. A study of growth factor receptors in human lens epithelial cells and their relationship to fiber differentiation. Exp Eye Res. 1996;63:683–692. doi: 10.1006/exer.1996.0162. [DOI] [PubMed] [Google Scholar]
- Klein KL, Anderson IM, de Jonge N. Transmission electron microscopy with a liquid flow cell. J Microscopy. 2011 doi: 10.1111/j.1365-2818.2010.03484.x. early online. [DOI] [PubMed] [Google Scholar]
- Lidke DS, Lidke KA, Rieger B, Jovin TM, Arndt-Jovin DJ. Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J Cell Biol. 2005;170:619–626. doi: 10.1083/jcb.200503140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, Jares-Erijman EA, Jovin TM. Quantum dot ligands provide new insights into erB/HER receptor-mediated signal transduction. Nat Biotechnol. 2004a;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- Nishiyama H, Suga M, Ogura T, Maruyama Y, Koizumi M, Mio K, Kitamura S, Sato C. Atmospheric scanning electron microscope observes cells and tissues in open medium through silicon nitride film. J Struct Biol. 2010;169:438–449. doi: 10.1016/j.jsb.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Ramachandra R, Demers H, de Jonge N. Atomic-resolution scanning transmission electron microscopy through 50 nm-thick silicon nitride membranes. Appl Phys Lett. 2011 doi: 10.1063/1.3561758. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer L, Kohl H. Transmission electron microscopy: physics of image formation. Springer; New York: 2008. [Google Scholar]
- Ring EA, de Jonge N. Microfluidic system for transmission electron microscopy. Microsc Microanal. 2010;16:622–629. doi: 10.1017/S1431927610093669. [DOI] [PubMed] [Google Scholar]
- Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, Plitzko JM. Correlative microscopy: Bridging the gap between fluorescence light microscopy and cryo-electron tomography. J Struct Biol. 2007b;160:135–145. doi: 10.1016/j.jsb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Satulovsky J, Lui R, Wang Y-l. Exploring the Control Circuit of Cell Migration by Mathematical Modeling. Biophys J. 2008;94:3671–3683. doi: 10.1529/biophysj.107.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber AB, Libermann TA, Lax I, Yarden Y, Schlessinger J. Biological role of epidermal growth factor-receptor clustering - investigation with monoclonal anit-receptor antibodies. J Biol Chem. 1983;258:846–853. [PubMed] [Google Scholar]
- Stahlberg H, Walz T. Molecular electron microscopy: state of the art and current challenges. ACS Chem Biol. 2008;3:268–281. doi: 10.1021/cb800037d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijnsoever C, Oorschot V, Klumperman J. Correlative light-electron microscopy (CLEM) combining live-cell imaging and immunolabeling of ultrathin cryosections. Nat Meth. 2008;5:973–980. doi: 10.1038/nmeth.1263. [DOI] [PubMed] [Google Scholar]
- Williamson MJ, Tromp RM, Vereecken PM, Hull R, Ross FM. Dynamic microscopy of nanoscale cluster growth at the solid-liquid interface. Nat Mater. 2003;2:532–536. doi: 10.1038/nmat944. [DOI] [PubMed] [Google Scholar]