Abstract
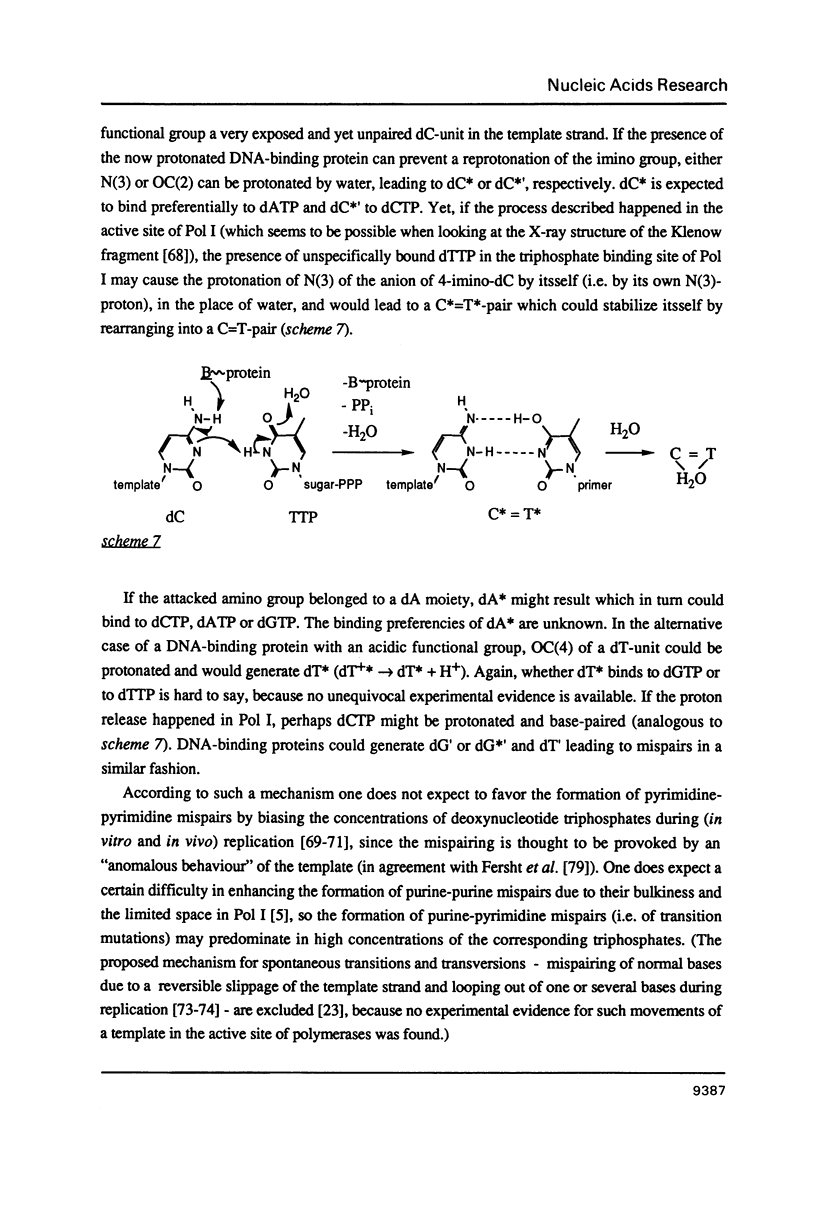
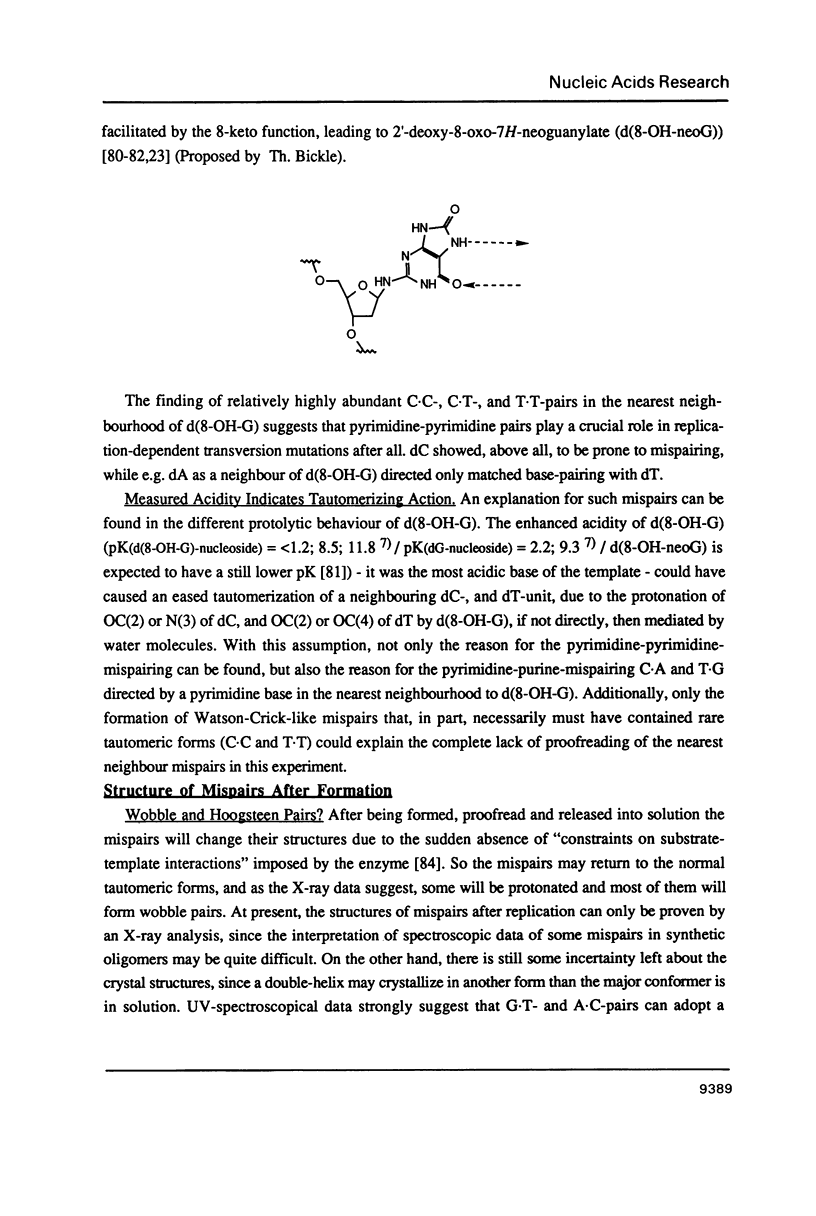
The formation of pyridine-pyrimidine- and pyrimidine-pyrimidine base pairs after in vitro DNA replication with the large fragment of Escherichia coli DNA polymerase I indicates that Watson-Crick-like base pairing between pyrimidine bases can occur in the enzyme due to the presence of the rare tautomers of deoxycytidylate and thymidylate in the template strand. The implications to mispair formation in DNA, such as the difference between the structures of the mispairs during and after replication, are discussed and the possible action of mutagenic DNA protonating and deprotonating agents in vivo is considered.
Full text
PDF















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-ela F., Koh D., Tinoco I., Jr, Martin F. H. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res. 1985 Jul 11;13(13):4811–4824. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks G. R., Brown D. M., Streeter D. G., Grossman L. Mutagenic anologues of cytosine: RNA polymerase template and substrate studies. J Mol Biol. 1971 Sep 28;60(3):425–439. doi: 10.1016/0022-2836(71)90179-3. [DOI] [PubMed] [Google Scholar]
- Bateman J. F., Chan D., Walker I. D., Rogers J. G., Cole W. G. Lethal perinatal osteogenesis imperfecta due to the substitution of arginine for glycine at residue 391 of the alpha 1(I) chain of type I collagen. J Biol Chem. 1987 May 25;262(15):7021–7027. [PubMed] [Google Scholar]
- Bird A. P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980 Apr 11;8(7):1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch A., Dutschman G., Currie B. L., Robins R. K., Robins M. J. Preparation and biological activity of various 3-deazapyrimidines and related nucleosides. J Med Chem. 1973 Mar;16(3):294–297. doi: 10.1021/jm00261a032. [DOI] [PubMed] [Google Scholar]
- Boosalis M. S., Petruska J., Goodman M. F. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J Biol Chem. 1987 Oct 25;262(30):14689–14696. [PubMed] [Google Scholar]
- Bos J. L., Toksoz D., Marshall C. J., Verlaan-de Vries M., Veeneman G. H., van der Eb A. J., van Boom J. H., Janssen J. W., Steenvoorden A. C. Amino-acid substitutions at codon 13 of the N-ras oncogene in human acute myeloid leukaemia. 1985 Jun 27-Jul 3Nature. 315(6022):726–730. doi: 10.1038/315726a0. [DOI] [PubMed] [Google Scholar]
- Both G. W., Shi C. H., Kilbourne E. D. Hemagglutinin of swine influenza virus: a single amino acid change pleiotropically affects viral antigenicity and replication. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6996–7000. doi: 10.1073/pnas.80.22.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker R. J., Wilson T. H. Isolation and nucleotide sequencing of lactose carrier mutants that transport maltose. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3959–3963. doi: 10.1073/pnas.82.12.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. M., Hewlins M. J., Schell P. The tautomeric state of N(4)-hydroxy- and of N(4)-amino-cytosine derivatives. J Chem Soc Perkin 1. 1968;15:1925–1929. doi: 10.1039/j39680001925. [DOI] [PubMed] [Google Scholar]
- Brown T., Hunter W. N., Kneale G., Kennard O. Molecular structure of the G.A base pair in DNA and its implications for the mechanism of transversion mutations. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2402–2406. doi: 10.1073/pnas.83.8.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T., Kennard O., Kneale G., Rabinovich D. High-resolution structure of a DNA helix containing mismatched base pairs. Nature. 1985 Jun 13;315(6020):604–606. doi: 10.1038/315604a0. [DOI] [PubMed] [Google Scholar]
- Charczuk R., Tamm C., Suri B., Bickle T. A. An unusual base pairing between pyrimidine and pyridine nucleotides. Nucleic Acids Res. 1986 Dec 9;14(23):9530–9530. doi: 10.1093/nar/14.23.9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyaya R., Ikuta S., Grzeskowiak K., Dickerson R. E. X-ray structure of a DNA hairpin molecule. Nature. 1988 Jul 14;334(6178):175–179. doi: 10.1038/334175a0. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P., Poltev V. I. Alteration of the DNA double helix conformation upon incorporation of mispairs as revealed by energy computations and pathways of point mutations. Nucleic Acids Res. 1985 Jan 11;13(1):141–154. doi: 10.1093/nar/13.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys J. P., Méjean V., Gasc A. M., Sicard A. M. Mismatch repair in Streptococcus pneumoniae: relationship between base mismatches and transformation efficiencies. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5956–5960. doi: 10.1073/pnas.80.19.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro-Stone M., Topal M. D., Kaufman D. G. DNA in proximity to the site of replication is preferentially alkylated in S phase 10T1/2 cells treated with N-methyl-N-nitroso-urea. Carcinogenesis. 1982;3(10):1119–1127. doi: 10.1093/carcin/3.10.1119. [DOI] [PubMed] [Google Scholar]
- Cornelis A. G., Haasnoot J. H., den Hartog J. F., de Rooij M., van Boom J. H., Cornelis A. Local destabilisation of a DNA double helix by a T--T wobble pair. Nature. 1979 Sep 20;281(5728):235–236. doi: 10.1038/281235a0. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Dalbadie-McFarland G., Neitzel J. J., Richards J. H. Active-site mutants of beta-lactamase: use of an inactive double mutant to study requirements for catalysis. Biochemistry. 1986 Jan 28;25(2):332–338. doi: 10.1021/bi00350a008. [DOI] [PubMed] [Google Scholar]
- Dohet C., Wagner R., Radman M. Repair of defined single base-pair mismatches in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):503–505. doi: 10.1073/pnas.82.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W., Baltz R. H. The biochemistry of mutagenesis. Annu Rev Biochem. 1976;45:11–37. doi: 10.1146/annurev.bi.45.070176.000303. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Driggers P. H., Beattie K. L. Effect of pH on the base-mispairing properties of 5-bromouracil during DNA synthesis. Biochemistry. 1988 Mar 8;27(5):1729–1735. doi: 10.1021/bi00405a052. [DOI] [PubMed] [Google Scholar]
- Duncan B. K., Miller J. H. Mutagenic deamination of cytosine residues in DNA. Nature. 1980 Oct 9;287(5782):560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- Early T. A., Olmsted J., 3rd, Kearns D. R., Lezius A. G. Base pairing structure in the poly d(G-T) double helix: wobble base pairs. Nucleic Acids Res. 1978 Jun;5(6):1955–1970. doi: 10.1093/nar/5.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eritja R., Horowitz D. M., Walker P. A., Ziehler-Martin J. P., Boosalis M. S., Goodman M. F., Itakura K., Kaplan B. E. Synthesis and properties of oligonucleotides containing 2'-deoxynebularine and 2'-deoxyxanthosine. Nucleic Acids Res. 1986 Oct 24;14(20):8135–8153. doi: 10.1093/nar/14.20.8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eritja R., Kaplan B. E., Mhaskar D., Sowers L. C., Petruska J., Goodman M. F. Synthesis and properties of defined DNA oligomers containing base mispairs involving 2-aminopurine. Nucleic Acids Res. 1986 Jul 25;14(14):5869–5884. doi: 10.1093/nar/14.14.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrin L. J., Mildvan A. S. NMR studies of conformations and interactions of substrates and ribonucleotide templates bound to the large fragment of DNA polymerase I. Biochemistry. 1986 Sep 9;25(18):5131–5145. doi: 10.1021/bi00366a023. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W. DNA polymerase accuracy and spontaneous mutation rates: frequencies of purine.purine, purine.pyrimidine, and pyrimidine.pyrimidine mismatches during DNA replication. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4251–4255. doi: 10.1073/pnas.78.7.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W. Fidelity of replication of bacteriophage phi X174 DNA in vitro and in vivo. J Mol Biol. 1983 Apr 25;165(4):633–654. doi: 10.1016/s0022-2836(83)80271-x. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W., Tsui W. C. Kinetic basis of spontaneous mutation. Misinsertion frequencies, proofreading specificities and cost of proofreading by DNA polymerases of Escherichia coli. J Mol Biol. 1982 Mar 25;156(1):37–51. doi: 10.1016/0022-2836(82)90457-0. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Shi J. P., Tsui W. C. Kinetics of base misinsertion by DNA polymerase I of Escherichia coli. J Mol Biol. 1983 Apr 25;165(4):655–667. doi: 10.1016/s0022-2836(83)80272-1. [DOI] [PubMed] [Google Scholar]
- Fowler R. G., Schaaper R. M., Glickman B. W. Characterization of mutational specificity within the lacI gene for a mutD5 mutator strain of Escherichia coli defective in 3'----5' exonuclease (proofreading) activity. J Bacteriol. 1986 Jul;167(1):130–137. doi: 10.1128/jb.167.1.130-137.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojobori T., Li W. H., Graur D. Patterns of nucleotide substitution in pseudogenes and functional genes. J Mol Evol. 1982;18(5):360–369. doi: 10.1007/BF01733904. [DOI] [PubMed] [Google Scholar]
- Gray D. M., Cui T., Ratliff R. L. Circular dichroism measurements show that C.C+ base pairs can coexist with A.T base pairs between antiparallel strands of an oligodeoxynucleotide double-helix. Nucleic Acids Res. 1984 Oct 11;12(19):7565–7580. doi: 10.1093/nar/12.19.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEMMENS W. F. NEO-GUANYLIC ACID PRODUCED BY THE ACTION OF ACID ON RIBONUCLEIC ACID. Biochim Biophys Acta. 1964 Oct 16;91:332–334. doi: 10.1016/0926-6550(64)90260-9. [DOI] [PubMed] [Google Scholar]
- HEMMENS W. F. neo-Guanylic acid, a new nucleotide from ribonucleic acid. Biochim Biophys Acta. 1963 Feb 26;68:284–292. doi: 10.1016/0006-3002(63)90144-6. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., den Hartog J. H., de Rooij J. F., van Boom J. H., Altona C. Loopstructures in synthetic oligodeoxynucleotides. Nucleic Acids Res. 1980 Jan 11;8(1):169–181. doi: 10.1093/nar/8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall Z. W., Lehman I. R. An in vitro transversion by a mutationally altered T4-induced DNA polymerase. J Mol Biol. 1968 Sep 28;36(3):321–333. doi: 10.1016/0022-2836(68)90158-7. [DOI] [PubMed] [Google Scholar]
- Hare D., Shapiro L., Patel D. J. Wobble dG X dT pairing in right-handed DNA: solution conformation of the d(C-G-T-G-A-A-T-T-C-G-C-G) duplex deduced from distance geometry analysis of nuclear Overhauser effect spectra. Biochemistry. 1986 Nov 18;25(23):7445–7456. doi: 10.1021/bi00371a029. [DOI] [PubMed] [Google Scholar]
- Hillebrand G. G., Beattie K. L. Influence of template primary and secondary structure on the rate and fidelity of DNA synthesis. J Biol Chem. 1985 Mar 10;260(5):3116–3125. [PubMed] [Google Scholar]
- Hillebrand G. G., McCluskey A. H., Abbott K. A., Revich G. G., Beattie K. L. Misincorporation during DNA synthesis, analyzed by gel electrophoresis. Nucleic Acids Res. 1984 Apr 11;12(7):3155–3171. doi: 10.1093/nar/12.7.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P. S., Frederick C. A., Quigley G. J., van der Marel G. A., van Boom J. H., Wang A. H., Rich A. G.T wobble base-pairing in Z-DNA at 1.0 A atomic resolution: the crystal structure of d(CGCGTG). EMBO J. 1985 Dec 16;4(13A):3617–3623. doi: 10.1002/j.1460-2075.1985.tb04125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter W. N., Brown T., Anand N. N., Kennard O. Structure of an adenine-cytosine base pair in DNA and its implications for mismatch repair. Nature. 1986 Apr 10;320(6062):552–555. doi: 10.1038/320552a0. [DOI] [PubMed] [Google Scholar]
- Hunter W. N., Brown T., Kennard O. Structural features and hydration of a dodecamer duplex containing two C.A mispairs. Nucleic Acids Res. 1987 Aug 25;15(16):6589–6606. doi: 10.1093/nar/15.16.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter W. N., Brown T., Kennard O. Structural features and hydration of d(C-G-C-G-A-A-T-T-A-G-C-G); a double helix containing two G.A mispairs. J Biomol Struct Dyn. 1986 Oct;4(2):173–191. doi: 10.1080/07391102.1986.10506338. [DOI] [PubMed] [Google Scholar]
- Hunter W. N., Brown T., Kneale G., Anand N. N., Rabinovich D., Kennard O. The structure of guanosine-thymidine mismatches in B-DNA at 2.5-A resolution. J Biol Chem. 1987 Jul 25;262(21):9962–9970. doi: 10.2210/pdb113d/pdb. [DOI] [PubMed] [Google Scholar]
- Hunter W. N., Kneale G., Brown T., Rabinovich D., Kennard O. Refined crystal structure of an octanucleotide duplex with G . T mismatched base-pairs. J Mol Biol. 1986 Aug 20;190(4):605–618. doi: 10.1016/0022-2836(86)90246-9. [DOI] [PubMed] [Google Scholar]
- Hübscher U. DNA polymerases in prokaryotes and eukaryotes: mode of action and biological implications. Experientia. 1983 Jan 15;39(1):1–25. doi: 10.1007/BF01960616. [DOI] [PubMed] [Google Scholar]
- Jukes T. H. Silent nucleotide substitutions and the molecular evolutionary clock. Science. 1980 Nov 28;210(4473):973–978. doi: 10.1126/science.7434017. [DOI] [PubMed] [Google Scholar]
- Jukes T. H. Transitions, transversions, and the molecular evolutionary clock. J Mol Evol. 1987;26(1-2):87–98. doi: 10.1007/BF02111284. [DOI] [PubMed] [Google Scholar]
- Kalnik M. W., Kouchakdjian M., Li B. F., Swann P. F., Patel D. J. Base pair mismatches and carcinogen-modified bases in DNA: an NMR study of A.C and A.O4meT pairing in dodecanucleotide duplexes. Biochemistry. 1988 Jan 12;27(1):100–108. doi: 10.1021/bi00401a017. [DOI] [PubMed] [Google Scholar]
- Kalnik M. W., Kouchakdjian M., Li B. F., Swann P. F., Patel D. J. Base pair mismatches and carcinogen-modified bases in DNA: an NMR study of G.T and G.O4meT pairing in dodecanucleotide duplexes. Biochemistry. 1988 Jan 12;27(1):108–115. doi: 10.1021/bi00401a018. [DOI] [PubMed] [Google Scholar]
- Kan L. S., Chandrasegaran S., Pulford S. M., Miller P. S. Detection of a guanine X adenine base pair in a decadeoxyribonucleotide by proton magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4263–4265. doi: 10.1073/pnas.80.14.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keepers J. W., Schmidt P., James T. L., Kollman P. A. Molecular-mechanical studies of the mismatched base analogs of d(CGCGAATTCGCG)2:d(CGTGAATTCGCG)2, d(CGAGAATTCGCG)2, d(CGCGAATTCACG)2, d(CGCGAATTCTCG)2, and d(CGCAGAATTCGCG).d(CGCGAATTCGCG). Biopolymers. 1984 Dec;23(12):2901–2929. doi: 10.1002/bip.360231214. [DOI] [PubMed] [Google Scholar]
- Kennard O. Structural studies of DNA fragments: the G.T wobble base pair in A, B and Z DNA; the G.A base pair in B-DNA. J Biomol Struct Dyn. 1985 Oct;3(2):205–226. doi: 10.1080/07391102.1985.10508412. [DOI] [PubMed] [Google Scholar]
- Kneale G., Brown T., Kennard O., Rabinovich D. G . T base-pairs in a DNA helix: the crystal structure of d(G-G-G-G-T-C-C-C). J Mol Biol. 1985 Dec 20;186(4):805–814. doi: 10.1016/0022-2836(85)90398-5. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Fratini A. V., Drew H. R., Dickerson R. E. Ordered water structure around a B-DNA dodecamer. A quantitative study. J Mol Biol. 1983 Jan 5;163(1):129–146. doi: 10.1016/0022-2836(83)90033-5. [DOI] [PubMed] [Google Scholar]
- Kornberg A. Enzyme studies of replication of the Escherichia coli chromosome. Adv Exp Med Biol. 1984;179:3–16. doi: 10.1007/978-1-4684-8730-5_1. [DOI] [PubMed] [Google Scholar]
- Kozma S. C., Bogaard M. E., Buser K., Saurer S. M., Bos J. L., Groner B., Hynes N. E. The human c-Kirsten ras gene is activated by a novel mutation in codon 13 in the breast carcinoma cell line MDA-MB231. Nucleic Acids Res. 1987 Aug 11;15(15):5963–5971. doi: 10.1093/nar/15.15.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Kuchino Y., Mori F., Kasai H., Inoue H., Iwai S., Miura K., Ohtsuka E., Nishimura S. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature. 1987 May 7;327(6117):77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Alexander P. S. The base substitution fidelity of eucaryotic DNA polymerases. Mispairing frequencies, site preferences, insertion preferences, and base substitution by dislocation. J Biol Chem. 1986 Jan 5;261(1):160–166. [PubMed] [Google Scholar]
- Lai M. D., Beattie K. L. Influence of DNA sequence on the nature of mispairing during DNA synthesis. Biochemistry. 1988 Mar 8;27(5):1722–1728. doi: 10.1021/bi00405a051. [DOI] [PubMed] [Google Scholar]
- Lin T. S., Cheng J. C., Ishiguro K., Sartorelli A. C. 8-Substituted guanosine and 2'-deoxyguanosine derivatives as potential inducers of the differentiation of Friend erythroleukemia cells. J Med Chem. 1985 Sep;28(9):1194–1198. doi: 10.1021/jm00147a012. [DOI] [PubMed] [Google Scholar]
- Martin F. H., Castro M. M., Aboul-ela F., Tinoco I., Jr Base pairing involving deoxyinosine: implications for probe design. Nucleic Acids Res. 1985 Dec 20;13(24):8927–8938. doi: 10.1093/nar/13.24.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhaskar D. N., Goodman M. F. On the molecular basis of transition mutations. Frequency of forming 2-aminopurine-cytosine base mispairs in the G X C----A X T mutational pathway by T4 DNA polymerase in vitro. J Biol Chem. 1984 Oct 10;259(19):11713–11717. [PubMed] [Google Scholar]
- Mizrahi V., Henrie R. N., Marlier J. F., Johnson K. A., Benkovic S. J. Rate-limiting steps in the DNA polymerase I reaction pathway. Biochemistry. 1985 Jul 16;24(15):4010–4018. doi: 10.1021/bi00336a031. [DOI] [PubMed] [Google Scholar]
- Muster-Nassal C., Kolodner R. Mismatch correction catalyzed by cell-free extracts of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7618–7622. doi: 10.1073/pnas.83.20.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Deoxyadenosine-deoxycytidine pairing in the d(C-G-C-G-A-A-T-T-C-A-C-G) duplex: conformation and dynamics at and adjacent to the dA X dC mismatch site. Biochemistry. 1984 Jul 3;23(14):3218–3226. doi: 10.1021/bi00309a016. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Deoxyguanosine-deoxyadenosine pairing in the d(C-G-A-G-A-A-T-T-C-G-C-G) duplex: conformation and dynamics at and adjacent to the dG X dA mismatch site. Biochemistry. 1984 Jul 3;23(14):3207–3217. doi: 10.1021/bi00309a015. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Dynamics of DNA duplexes containing internal G.T, G.A, A.C, and T.C pairs: hydrogen exchange at and adjacent to mismatch sites. Fed Proc. 1984 Aug;43(11):2663–2670. [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Dallas J., Itakura K., Breslauer K. J. Structure, dynamics, and energetics of deoxyguanosine . thymidine wobble base pair formation in the self-complementary d(CGTGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):437–444. doi: 10.1021/bi00532a003. [DOI] [PubMed] [Google Scholar]
- Patten J. E., So A. G., Downey K. M. Effect of base-pair stability of nearest-neighbor nucleotides on the fidelity of deoxyribonucleic acid synthesis. Biochemistry. 1984 Apr 10;23(8):1613–1618. doi: 10.1021/bi00303a005. [DOI] [PubMed] [Google Scholar]
- Preston B. D., Singer B., Loeb L. A. Mutagenic potential of O4-methylthymine in vivo determined by an enzymatic approach to site-specific mutagenesis. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8501–8505. doi: 10.1073/pnas.83.22.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privé G. G., Heinemann U., Chandrasegaran S., Kan L. S., Kopka M. L., Dickerson R. E. Helix geometry, hydration, and G.A mismatch in a B-DNA decamer. Science. 1987 Oct 23;238(4826):498–504. doi: 10.1126/science.3310237. [DOI] [PubMed] [Google Scholar]
- Rabinovich D., Haran T., Eisenstein M., Shakked Z. Structures of the mismatched duplex d(GGGTGCCC) and one of its Watson-Crick analogues d(GGGCGCCC). J Mol Biol. 1988 Mar 5;200(1):151–161. doi: 10.1016/0022-2836(88)90340-3. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Genetic recombination: strand transfer and mismatch repair. Annu Rev Biochem. 1978;47:847–880. doi: 10.1146/annurev.bi.47.070178.004215. [DOI] [PubMed] [Google Scholar]
- Reddy E. P., Reynolds R. K., Santos E., Barbacid M. A point mutation is responsible for the acquisition of transforming properties by the T24 human bladder carcinoma oncogene. Nature. 1982 Nov 11;300(5888):149–152. doi: 10.1038/300149a0. [DOI] [PubMed] [Google Scholar]
- Reyland M. E., Lehman I. R., Loeb L. A. Specificity of proofreading by the 3'----5' exonuclease of the DNA polymerase-primase of Drosophila melanogaster. J Biol Chem. 1988 May 15;263(14):6518–6524. [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Ryan J. P., Duncan M. C., Bankaitis V. A., Bassford P. J., Jr Intragenic reversion mutations that improve export of maltose-binding protein in Escherichia coli malE signal sequence mutants. J Biol Chem. 1986 Mar 5;261(7):3389–3395. [PubMed] [Google Scholar]
- Schaaper R. M., Danforth B. N., Glickman B. W. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J Mol Biol. 1986 May 20;189(2):273–284. doi: 10.1016/0022-2836(86)90509-7. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M., Dunn R. L. Spectra of spontaneous mutations in Escherichia coli strains defective in mismatch correction: the nature of in vivo DNA replication errors. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6220–6224. doi: 10.1073/pnas.84.17.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B., Fraenkel-Conrat H. Messenger and template activities of chemically modified polynucleotides. Biochemistry. 1970 Sep 15;9(19):3694–3701. doi: 10.1021/bi00821a007. [DOI] [PubMed] [Google Scholar]
- Sinha N. K., Haimes M. D. Molecular mechanisms of substitution mutagenesis. An experimental test of the Watson-Crick and topal-fresco models of base mispairings. J Biol Chem. 1981 Oct 25;256(20):10671–10683. [PubMed] [Google Scholar]
- Sinha N. K. Specificity and efficiency of editing of mismatches involved in the formation of base-substitution mutations by the 3'----5' exonuclease activity of phage T4 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Feb;84(4):915–919. doi: 10.1073/pnas.84.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers L. C., Fazakerley G. V., Eritja R., Kaplan B. E., Goodman M. F. Base pairing and mutagenesis: observation of a protonated base pair between 2-aminopurine and cytosine in an oligonucleotide by proton NMR. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5434–5438. doi: 10.1073/pnas.83.15.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers L. C., Fazakerley G. V., Kim H., Dalton L., Goodman M. F. Variation of nonexchangeable proton resonance chemical shifts as a probe of aberrant base pair formation in DNA. Biochemistry. 1986 Jul 15;25(14):3983–3988. doi: 10.1021/bi00362a002. [DOI] [PubMed] [Google Scholar]
- Strazewski P. Nucleosides and nucleotides: Part 28 [1]. 13C-NMR spectra of 2'-deoxycytidine and 3-deaza-2'-deoxycytidine. Nucleic Acids Res. 1988 Jun 10;16(11):5191–5191. doi: 10.1093/nar/16.11.5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S. S., Lahue R. S., Au K. G., Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J Biol Chem. 1988 May 15;263(14):6829–6835. [PubMed] [Google Scholar]
- Summers M. F., Byrd R. A., Gallo K. A., Samson C. J., Zon G., Egan W. Nuclear magnetic resonance and circular dichroism studies of a duplex--single-stranded hairpin loop equilibrium for the oligodeoxyribonucleotide sequence d(CGCGATTCGCG). Nucleic Acids Res. 1985 Sep 11;13(17):6375–6386. doi: 10.1093/nar/13.17.6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J. L., Trifonov E. N. Possibility of nonkinked packing of DNA in chromatin. Proc Natl Acad Sci U S A. 1978 Jan;75(1):103–107. doi: 10.1073/pnas.75.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabin C. J., Bradley S. M., Bargmann C. I., Weinberg R. A., Papageorge A. G., Scolnick E. M., Dhar R., Lowy D. R., Chang E. H. Mechanism of activation of a human oncogene. Nature. 1982 Nov 11;300(5888):143–149. doi: 10.1038/300143a0. [DOI] [PubMed] [Google Scholar]
- Tibanyenda N., De Bruin S. H., Haasnoot C. A., van der Marel G. A., van Boom J. H., Hilbers C. W. The effect of single base-pair mismatches on the duplex stability of d(T-A-T-T-A-A-T-A-T-C-A-A-G-T-T-G) . d(C-A-A-C-T-T-G-A-T-A-T-T-A-A-T-A). Eur J Biochem. 1984 Feb 15;139(1):19–27. doi: 10.1111/j.1432-1033.1984.tb07970.x. [DOI] [PubMed] [Google Scholar]
- Toorchen D., Topal M. D. Mechanisms of chemical mutagenesis and carcinogenesis: effects on DNA replication of methylation at the O6-guanine position of dGTP. Carcinogenesis. 1983 Dec;4(12):1591–1597. doi: 10.1093/carcin/4.12.1591. [DOI] [PubMed] [Google Scholar]
- Topal M. D., DiGuiseppi S. R., Sinha N. K. Molecular basis for substitution mutations. Effect of primer terminal and template residues on nucleotide selection by phage T4 DNA polymerase in vitro. J Biol Chem. 1980 Dec 25;255(24):11717–11724. [PubMed] [Google Scholar]
- Topal M. D., Eadie J. S., Conrad M. O6-methylguanine mutation and repair is nonuniform. Selection for DNA most interactive with O6-methylguanine. J Biol Chem. 1986 Jul 25;261(21):9879–9885. [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Complementary base pairing and the origin of substitution mutations. Nature. 1976 Sep 23;263(5575):285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- Topal M. D., Sinha N. K. Products of bacteriophage T4 genes 32 and 45 improve the accuracy of DNA replication in vitro. J Biol Chem. 1983 Oct 25;258(20):12274–12279. [PubMed] [Google Scholar]
- Varmus H. E. The molecular genetics of cellular oncogenes. Annu Rev Genet. 1984;18:553–612. doi: 10.1146/annurev.ge.18.120184.003005. [DOI] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953 May 30;171(4361):964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- Wang M. C., Bloch A. Studies on the mode of action of 3-deazapyrimidines. 1. Metabolism of 3-deazauridine and 3-deazacytidine in microbial and tumor cells. Biochem Pharmacol. 1972 Apr 15;21(8):1063–1073. doi: 10.1016/0006-2952(72)90100-1. [DOI] [PubMed] [Google Scholar]
- Webb T. R., Matteucci M. D. Hybridization triggered cross-linking of deoxyoligonucleotides. Nucleic Acids Res. 1986 Oct 10;14(19):7661–7674. doi: 10.1093/nar/14.19.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werntges H., Steger G., Riesner D., Fritz H. J. Mismatches in DNA double strands: thermodynamic parameters and their correlation to repair efficiencies. Nucleic Acids Res. 1986 May 12;14(9):3773–3790. doi: 10.1093/nar/14.9.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. H., Lusnak K., Fogel S. Mismatch-specific post-meiotic segregation frequency in yeast suggests a heteroduplex recombination intermediate. Nature. 1985 May 23;315(6017):350–352. doi: 10.1038/315350a0. [DOI] [PubMed] [Google Scholar]


