Abstract
A series of compounds that target reactive metal-chelates to the HIV-1 Rev Response Element (RRE) mRNA have been synthesized. Dissociation constants and chemical reactivity toward HIV RRE RNA have been determined and evaluated in terms of reduction potential, coordination unsaturation, and overall charge associated with the metal-chelate-rev complex. Ethylenediaminetetraacetic acid (EDTA), nitrilotriacetic acid (NTA), diethylenetriaminepentaacetic acid (DTPA), and 1,4,7,10-tetraazacyclo-dodecane-1,4,7,10-tetraacetic acid (DOTA) were linked to a lysine sidechain of a Rev-derived peptide by either EDC/NHS or isothiocyanate coupling. The resulting chelate-Rev (EDTA-Rev, DTPA-Rev, NTA-Rev, and DOTA-Rev) conjugates were used to form coordination complexes with Fe2+, Co2+, Ni2+, and Cu2+ such that the arginine-rich Rev peptide could mediate localization of the metal-chelates to the Rev peptide’s high-affinity mRNA binding partner, RRE stem loop IIB. Metal complexes of the extended peptides GGH-Rev and KGHK-Rev, which also contain N-terminal peptidic chelators (ATCUN motifs), were studied for comparison. A fluorescence titration assay revealed high affinity RRE RNA binding by all 22 metal-chelate-Rev species, with KD values ranging from ~ 0.2 – 16 nM, indicating little to no loss of RNA affinity due to the coupling of the metal-chelates to the Rev peptide. Dissociation constants for binding at a previously unobserved low-affinity site are also reported. Rates of RNA modification by each metal-chelate-Rev species were determined and varied from ~ 0.28 – 4.9 nM/min, but were optimal for Cu2+-NTA-Rev. Metal-chelate reduction potentials were determined and varied from −228 to +1111 mV vs. NHE under similar solution conditions, allowing direct comparison of reactivity with redox thermodynamics. Optimal activity was observed when the reduction potential for the metal center was poised between that of the two principal coreagents for metal-promoted formation of reactive oxygen species: E°ascorbate/ascorbyl radical = −66 mV; E°H2O2/hydroxyl radical = 380 mV. Given the variety of oxidative activities of these metal-complexes and their high-affinity binding to the targeted RRE mRNA following coupling to the Rev peptide, this class of metal-chelate-Rev derivatives constitutes a promising step toward development of multiple-turnover reagents for selective eradication of HIV-1 Rev response element mRNA.
Keywords: Artificial nuclease, metallodrug, HIV, AIDS, Rev Response Element, RRE, Rev, RNA, oxidation, reactive oxygen species, catalysis, redox chemistry
INTRODUCTION
There is a pressing need for reagents that selectively target genetic information (both DNA and RNA) that underlies disease conditions. For example, cancer and HIV affect a large fraction of the population and could each be therapeutically addressed if it were possible to specifically and precisely alter genes or gene products in a host cell. Current gene therapy techniques tend to suffer from a lack of selectivity and subsequent side effects; especially the initiation of new cancers.1,2 A further general limitation is the requirement for stoichiometric saturation of drug targets. Drugs that alter biological targets irreversibly and with multiple turnover have the potential to perform at far lower concentrations than their reversible counterparts, since catalytic drugs that promote irreversible inactivation of a target do not need to saturate the target in order to operate effectively.3 Lower dosage concentrations should result in fewer side-effects and higher efficacy. The metal-chelate-Rev compounds described herein were designed with the goal of progressing toward a catalytic metallodrug that is selective toward a specific gene product with irreversible modification. In this case HIV-1 Rev response element (RRE) mRNA was the transcribed gene product as a result of its critical role in HIV replication, and potential as an anti-HIV therapeutic target.4,5
In this paper we report the synthesis and characterization of several families of metal-chelate-Rev compounds based on combinations of transition metals (Fe2+, Co2+, Ni2+, and Cu2+) and selected chelators, including DOTA, DTPA, EDTA, NTA, tripeptide GGH, and tetrapeptide KGHK, each of which has been coupled to the RNA targeting Rev peptide that promotes high-affinity binding to the RRE stem loop IIB mRNA. These compounds each contained a metal-chelate attached to the N-terminal residue of the Rev peptide and allow a careful comparison of the influence of reduction potential, coordination unsaturation, and overall charge associated with the metal chelate on the reactivity profile for the target RNA. Metal-chelates were used because of their ability to effect irreversible oxidation through a variety of mechanisms under biologically relevant conditions, while the Rev peptide promotes selective binding to RRE RNA.5-8 The metal-chelate complexes are thereby brought in close proximity to the RRE RNA by the Rev peptide and elicit a variety of oxidative chemistries toward the RNA.9-11 To better understand reactivity patterns, the dissociation constants of all of the RNA/metallopeptide pairs were determined. It was found that attachment of metal-chelates to the Rev peptide did not severely diminish its binding to the RRE, although some minor influences were observed, and followed expected trends. Rates of reaction were then determined for each metal-chelate-Rev species when complexed to RRE RNA. These studies build on our prior investigations of Cu-GGH-Rev, which was shown to selectively cleave the RRE RNA both in vitro and in vivo.5,10,11 However, incorporation of distinct metal-chelates is shown to greatly enhance reactivity, and reveals evidence for optimization through control of reduction potential and coordination unsaturation that will allow more precise tuning of catalytic metallodrug reactivity to physiological optimums.
MATERIALS AND METHODS
Chemicals and Reagents
Custom RNAs were purchased from Dharmacon and included AP-RRE RNA, with sequence 5′-GGUCUGGGCGCAGCGCAAGCUGACGG(2-AP)ACAGGCC-3′, and Fl-RRE RNA, with sequence 5′-Fluorescein-UUGGUCUGGGCGCAGCGCAAGCUGACGGUACAGGCC-3′. The N-terminal acetylated and C-terminal amidated peptide (K-Rev) with sequence Ac-KTRQARRNRRRRWRERQR-NH2 used for coupling reactions was purchased from Neo Peptide. The peptides GGH-Rev with sequence GGHTRQARRNRRRRWRERQR-NH2 and KGHK-Rev with sequence KGHKTRQARRNRRRRWRERQR-NH2 were purchased from Genemed Synthesis Inc. RNA and peptide species were immediately aliquoted into single-use containers and stored at −20 °C to minimize freeze-thaw cycles. The bifunctional compounds NHS-DOTA and p-SCN-Bn-DTPA were purchased from Macrocyclics and stored at −20 °C. N-hydroxysuccinimide (NHS) was purchased from GenScript, and 1-ethyl-3-[3-dimethyl aminopropyl] carbodiimide hydrochloride (EDC) was purchased from Pierce and stored at −20 °C. Ethylenediaminetetraacetic acid (EDTA) was purchased from Aldrich. Nitrilotriacetic acid (NTA) and diethylenetriaminepentaacetic acid (DTPA) were purchased from Sigma. The tripeptide GGH-OH (GGH) and tetrapeptide KGHK-NH2 (KGHK) were obtained from Bachem, and 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) was purchased from Macrocyclics. The Fe(II) sulfate heptahydrate, Co(II) chloride hexahydrate, Ni(II) tetrahydrate, and Cu(II) chloride dihydrate salts were purchased from ACROS, J.T.Baker, Aldrich, and J. T. Baker, respectively. Sodium chloride and sodium hydroxide were purchased from Fisher, and HEPES was purchased from Sigma.
Synthesis and Characterization
Compound DOTA-Rev was prepared by making a solution containing 3.8 mM NHS-DOTA (10x stock made in DMSO) and 757 μM K-Rev in a buffer containing 100 mM NaHCO3, pH 8.0. The reaction proceeded for 2 h at RT in the dark, followed by RP-HPLC purification. Elution conditions were: 15 – 35% B, 0 – 50 min; 35 – 100% B, 50 – 55 min; 100% B, 55 – 60 min where mobile phase A = H2O, 0.1% TFA; B = acetonitrile, 0.1% TFA. The HPLC fraction for product DOTA-Rev was collected, lyophilized, resuspended in water, and MALDI-TOF analysis provided the expected mass 2992.3 amu, with no evidence of uncoupled K-Rev reactant. DOTA-Rev concentration was quantified via UV/Vis (240 nm) titration with a solution of known concentration of nickel(II) acetate.
Compounds EDTA-Rev and NTA-Rev were made separately by first preparing a solution of 38 mM EDTA (or NTA), 38 mM NHS, 15.6 mM EDC in a buffer containing 20 mM HEPES, 100 mM NaCl, pH 5.7 and allowed to react for 1 min. After 1 min, K-Rev was added to a final concentration of 757 μM. The reaction proceeded for 2 - 4 h at RT in the dark, followed by RP-HPLC purification. Elution conditions were: 15 – 20% B, 0 – 50 min; 20 – 100% B, 50 – 55 min; 100% B, 55 – 60 min where mobile phase A = H2O, 0.1% TFA; B = acetonitrile, 0.1% TFA. Due to poor separation and <100% coupling, the collected HPLC fraction containing a mixture of the coupled and unreacted K-Rev was lyophilized, resuspended in water and recycled through the above procedure with additional EDTA (or NTA) to increase yield, and RP-HPLC purification was repeated. The lyophilized HPLC fraction was resuspended in water, and the EDTA-Rev and NTA-Rev concentrations were determined by UV/Vis titrations with solutions of known concentrations of CuCl2 (at 270 nm) and FeSO4 (at 250 nm), respectively. The purity of the coupled K-Rev product (also a measure of the extent of coupling) was determined by comparison of the concentration of coupled EDTA (or NTA) determined by metal ion titration, and the concentration of the internal Trp residue of EDTA-Rev/K-Rev (or NTA-Rev/K-Rev) independently determined by absorbance at 280 nm. Coupling efficiency was found to be 83% (EDTA-Rev) and 137% (NTA-Rev)—it remains unclear why the apparent extent is >100%, and it may be a data fitting artifact. MALDI analysis provided the expected masses of 2880.7 and 2779.7 amu for EDTA-Rev and NTA-Rev, respectively, as well as the remaining uncoupled K-Rev (2606.9 amu) reactant.
Compound DTPA-Rev was prepared from a solution containing 3.8 mM p-SCN-Bn-DTPA (10x stock made in DMSO) and 757 μM K-Rev in a buffer containing 100 mM NaHCO3, pH 9.0. The reaction proceeded for 2 h at RT in the dark, followed by RP-HPLC purification using the same elution conditions as used for DOTA-Rev. The HPLC fraction for DTPA-Rev was collected, lyophilized, resuspended in water, and MALDI-TOF analysis provided the expected mass of 3146.4 amu for DTPA-Rev as well as a much smaller amount of residual K-Rev at 2606.9 amu. Both DTPA-Rev and K-Rev products were easily separable via HPLC, and so the observed K-Rev at 2606.9 amu may be artifactual from the MALDI analysis. The DTPA-Rev concentration was quantified by UV/Vis (290 nm) titration with a solution of known concentration of CuCl2.
Divalent iron, cobalt, nickel, and copper complexes of DOTA-Rev, DTPA-Rev, EDTA-Rev, NTA-Rev, GGH-Rev, and KGHK-Rev were prepared by mixing the respective metal salts with the chelate-Rev species in a 1:1.2 ratio (to ensure essentially all metal was chelated) and mixing at RT for 30 min prior to each measurement. Metal-chelate complex formation was verified by UV/Vis titration. Fe2+ complexes with chelators GGH-Rev and KGHK-Rev were not used in later experiments due to insufficient complex formation. Experimentally determined extinction coefficients for all metal-chelate and metal-chelate-Rev species, as well as stability constants for metal-chelates, are summarized in the supplementary information.
Determination of Dissociation Constants
Dissociation constants (KD) were determined by fluorimetry on a Varian Cary Eclipse fluorescence spectrophotometer with excitation at 310 nm (SW = 10 nm) and emission at 371 nm (SW = 10 nm). A 660 μL volume of 100 nM AP-RRE RNA was first heated to 90 °C for 5 min and then allowed to cool to, but not below, the titration temperature (37 °C). A 650 μL volume of this sample was added to the pre-thermostatted cuvette. After 10 min of temperature equilibration a solution of 5 μM M-chelate-Rev complex was titrated into the solution. After each addition, 100 μL of the solution was mixed by gentle pipeting to ensure complete mixing. The fluorescence change associated with the change of environment of the aminopurine label was monitored following peptide binding. Titration response curves were dilution and background corrected and normalized to initial intensity values. Curves were fit to a biphasic equation (1) for two independent binding sites where Fobs is the total observed fluorescence intensity, P0 is the total concentration of added M-chelate-Rev, F0, F1, and F2 are fitting parameters corresponding to the intensities of the unbound, singly-bound, and doubly-bound AP-RNA, respectively, R0 is the initial concentration of AP-RNA, and KD1 and KD2 are dissociation constants for binding at the high- and low-affinity sites, respectively. All binding experiments were performed in a binding buffer composed of 20 mM HEPES, 100 mM NaCl, pH 7.4.
| (1) |
PAGE Analysis of RRE RNA Cleavage Kinetics
Reactions of 100 nM metal-chelate-Rev (1:1.2 M:chelate ratio), 1 mM H2O2, 1 mM ascorbate, and 100 nM 5′ fluorescein end-labeled RRE stem IIB RNA (Fl-RNA) were conducted at 37 °C in separate tubes, each containing 20 μL total reaction volume – one tube corresponding to one gel lane. A buffer consisting of 20 mM HEPES, 100 mM NaCl, pH 7.4 was used in all experiments. Fl-RNA was heated to 90 °C and allowed to cool to, but not below 37 °C prior to reaction, and the Fl-RNA was immediately added to each pre-incubated tube. Reactants were added to pre-incubated tubes at time-points corresponding to the beginning of each reaction, which were performed in a dark incubator. Reaction start times were staggered and all reactions were quenched at the same time by addition of a quenching/gel-loading buffer solution (80 μL/tube) consisting of standard 1x tris-EDTA-acetic acid buffer, 8M urea, 20 mM HEPES, 100 mM NaCl. The tubes were then heated to 90 °C for 5 min and a 5 μL aliquot from each tube was then loaded into its corresponding lane of a 10% polyacrylamide 8 M urea gel, resulting in 1 pmole Fl-RNA per lane. Gel electrophoresis was performed at 250 V for 1.5 h. Gels were analyzed on a GE Typhoon variable mode imager using 488 nm excitation and a 526 nm single pass emission filter. Bands were quantified with the program ImageQuant. Loss of initial Fl-RNA reactant bands were fit to a first-order reaction equation using the software Origin, and observed rate constants and initial rates were determined. Control reactions with respective metal-chelates lacking the Rev domain were conducted in the same manner.
Reaction Kinetics Monitored by Real-Time Fluorimetry
Reactions of 100 nM metal-chelate-Rev (1:1.2 M:chelate ratio), 1 mM H2O2, 1 mM ascorbate, and 100 nM AP-RNA were conducted at 37 °C in a fluorescence cuvette in a Varian Cary Eclipse fluorescence spectrophotometer. Reactions were monitored using excitation at 310 nm (SW = 10 nm) and emission at 371 nm (SW = 10 nm). Prior to reaction the AP-RNA was pre-heated to 90 °C for 5 min and then added to the pre-warmed 37 °C cuvette. The metal-chelate-Rev complexes were added after 5 min, and reactions were initiated by the addition of H2O2 and ascorbate after an additional 10 min. Each resulting fluorescence trace was fit to a first-order reaction equation using the software Origin, and observed rate constants and initial rates were determined; the magnitude of fluorescence change that corresponded to complete reaction was established by averaging the magnitudes of fluorescence change from many trials known to react to completion. All concentrations and buffer were identical to those used in the PAGE analysis assay with Fl-RNA. Control reactions with respective metal-chelates lacking the Rev domain were conducted in the same manner.
Square Wave Voltammetry
Solutions of 1 mM metal chelates were formed in 20 mM HEPES, 100 mM NaCl, pH 7.4 under anaerobic conditions at RT. M-chelate ratios used ranged from 1:1.5 to 1:2, depending on the chelator used. Buffer, reagents, and the electrochemical cell were degassed and argon purged before complex formation for each experiment. Reduction potentials for each metal complex were determined via square wave voltammetry (SWV) using an EG&G Princeton Applied Research potentiostat/galvanostat, glassy carbon working electrode, Ag/AgCl reference electrode, and platinum auxiliary electrode. Instrument settings: Int. = 10 mV, Freq. = 100 Hz, I = 10−4 A. Potentials vs. Ag/AgCl were converted to potentials vs. NHE.
Molecular Modeling
RRE RNA–metallopeptide interactions were modeled using Spartan software. Molecular mechanics energy minimization was performed using the Merck molecular force field MMFFaq, in which structural energy minimization occurred without solvent considerations. Model geometries were constrained to the average NMR structure of the RNA-Rev peptide complex (PDB ID: 1ETF) and M-chelate X-Ray crystal structures, while the N-terminal lysine (threonine in GGH-Rev and KGHK-Rev) and lysine-chelate linker geometries were allowed to vary during energy minimization. Structures were accepted once 10 successive rounds of energy minimization lowered the energy by no more than 0.01 kcal/mol. Modeling parameters: equilibrium geometry at ground state, subject to frozen atoms and symmetry, multiplicity = singlet. For each complex, the total charge used during energy minimization corresponded to that expected at pH 7.4. Charge assignments were made as follows: 1− or 2− for each internal or terminal phosphate, respectively; 1+ for each lysine or arginine sidechain; 1− for each carboxylic acid functional group; 1− for each metal-chelating amide functional group (for GGH and KGHK); and 2+ for each transition metal cation.
RESULTS
Synthesis and Characterization
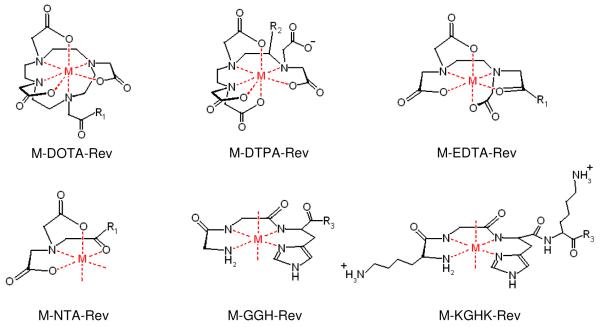
Chelate-Rev conjugates, including DOTA-Rev, DTPA-Rev, EDTA-Rev, and NTA-Rev were synthesized by coupling the respective chelators with the N-terminal lysine sidechain of the Rev-derivative peptide, K-Rev. Syntheses of DOTA-Rev and DTPA-Rev were achieved by direct reaction of K-Rev with the bifunctional chelators NHS-DOTA (at pH 8.0) and p-SCN-Bn-DTPA (at pH 9.0), which contained an amine-reactive NHS-ester and isothiocyanate, respectively. For EDTA-Rev and NTA-Rev, EDTA and NTA were first separately reacted at pH 5.7 with EDC/NHS to form activated NHS-esters of EDTA/NTA, and then a limiting concentration (to minimize coupling of more than one K-Rev to each chelator) of K-Rev was added to each, yielding the products EDTA-Rev and NTA-Rev. The chelate-Rev products were separated from reactants by RP-HPLC. The compounds DOTA-Rev and DTPA-Rev were easily separated from reactants. However elution times for EDTA-Rev and NTA-Rev overlapped with that of K-Rev, and a repeat reaction proved necessary to achieve quantitative product formation. HPLC purified products were further characterized by MALDI-TOF mass spectrometry, and the expected masses for DOTA-Rev, DTPA-Rev, EDTA-Rev, and NTA-Rev were observed: 2992.3 amu, 3146.4 amu, 2880.7 amu, 2779.7 amu, respectively. Complex formation between each metal ion (Fe2+, Co2+, Ni2+, and Cu2+) and each chelator-Rev species, as well as each chelator lacking Rev, was monitored by UV/Vis spectroscopy, and metal complex extinction coefficients were determined (supplementary information). As expected, formation of complexes between Fe2+ and GGH/KGHK was not observed under the experimental conditions used, and so combinations of Fe2+ with GGH-Rev/KGHK-Rev were not used in later experiments. Absorption spectra for each of the metal-chelate-Rev species closely matched the characteristic absorption spectra for each of the respective metal-chelates obtained without the attached Rev peptide, although the extinction coefficients for the charge transfer bands of Fe-DOTA, Ni-EDTA, and Co-NTA were each observed to increase significantly once coupled to K-Rev. The metal-chelate-Rev species used in this study are summarized in Figure 1.
Figure 1.
Summary of the metal-chelate complexes and their modes of attachment to the Rev peptide. M = Fe2+, Co2+, Ni2+, Cu2+; R1 = N(H)-K-Rev; R2 = p-Bn-N(H)C(S)-K-Rev; R3 = N(H)-Rev.
RRE RNA Binding
Dissociation constants for complex formation between the RRE stem loop IIB RNA and each of the metal-chelate-Rev species were determined by titration. This assay used a version of the RNA with an internal 2-aminopurine fluorescent probe (AP-RNA), where Rev binding at the high-affinity RRE RNA site resulted in a conformational change that increased the quantum yield of 2-aminopurine fluorescence, as previously described.12 Analysis of the binding curves by fitting to equation (1) generally revealed one high affinity binding site (KD1 ~ 1 nM), as expected, and one low affinity binding site (KD2 ~ 1000 nM), which has not been reported previously. The KD1 values for binding of M-chelate-Rev complexes to the high affinity RNA site varied from 0.2 nM to 16 nM, depending on the complex, while KD2 values for low affinity binding at a second RNA site varied from 148 nM to 2550 nM. For comparison, dissociation constants of the canonical Rev peptide to the RNA were 1.60 nM and 770 nM for the high- and low-affinity sites, respectively. In general there was an approximate 10-fold variation of dissociation constants in either direction from that of the Rev peptide upon attachment of M-chelates. However, high affinity binding was retained for each compound. Dissociation constants of each M-chelate-Rev species for the high-affinity and low-affinity RNA sites are summarized in Table 1, and titration response curves are provided in the supplementary section.
Table 1.
Dissociation constants for binding at the high-affinity (KD1) and low-affinity (KD2) RRE RNA sites by M-Chelate-Rev species determined by AP-RNA fluorescence titration experiments.
| Complex | KD1 (nM) | KD2 (nM) |
|---|---|---|
| [DOTA]3−–Rev | 0.2 ± 0.2 | 500 ± 30 |
| [Fe-DOTA]1−–Rev | 0.36 ± 0.08 | 1120 ± 20 |
| [Co-DOTA]1−–Rev | 2.1 ± 0.2 | 2550 ± 40 |
| [Ni-DOTA]1−–Rev | 2.3 ± 0.2 | 1180 ± 20 |
| [Cu-DOTA]1−–Rev | 0.4 ± 0.1 | 488 ± 9 |
| [EDTA]3−–Rev | 0.8 ± 0.4 | 1150 ± 30 |
| [Fe-EDTA]1−–Rev | 5.6 ± 0.7 | 630 ± 10 |
| [Co-EDTA]1−–Rev | 0.5 ± 0.3 | 560 ± 10 |
| [Ni-EDTA]1−–Rev | 0.8 ± 0.3 | 520 ± 10 |
| [Cu-EDTA]1−–Rev | 1.9 ± 0.3 | 1160 ± 20 |
| [DTPA]4−–Rev | 23 ± 1 | 980 ± 20 |
| [Fe-DTPA]2−–Rev | 0.7 ± 0.8 | 430 ± 20 |
| [Co-DTPA]2−–Rev | 13.7 ± 0.7 | 1330 ± 20 |
| [Ni-DTPA]2−–Rev | 4.2 ± 0.3 | 1840 ± 20 |
| [Cu-DTPA]2−–Rev | 16 ± 2 | 1570 ± 80 |
| [NTA]2−–Rev | 25.3 ± 0.8 | 910 ± 10 |
| [Fe-NTA]0–Rev | 7.0 ± 0.4 | 950 ± 10 |
| [Co-NTA]0–Rev | 9.4 ± 0.4 | 1210 ± 10 |
| [Ni-NTA]0–Rev | 2.3 ± 0.1 | 1160 ± 10 |
| [Cu-NTA]0–Rev | 3.5 ± 0.3 | 857 ± 9 |
| [GGH]1+–Rev | 2.1 ± 0.2 | 924 ± 9 |
| [Co-GGH]0–Rev | 3.8 ± 0.6 | 360 ± 10 |
| [Ni-GGH]0–Rev | 1.3 ± 0.3 | 458 ± 8 |
| [Cu-GGH]0–Rev | 4.6 ± 0.5 | 363 ± 9 |
| [KGHK]3+–Rev | 77 ± 1 | 1020 ± 10 |
| [Co-KGHK]2+–Rev | 1.4 ± 0.4 | 300 ± 6 |
| [Ni-KGHK]2+–Rev | 3.2 ± 0.4 | 173 ± 3 |
| [Cu-KGHK]2+–Rev | 1.3 ± 0.5 | 148 ± 5 |
| Rev peptide | 1.6 ± 0.3 | 770 ± 20 |
Structural Modeling
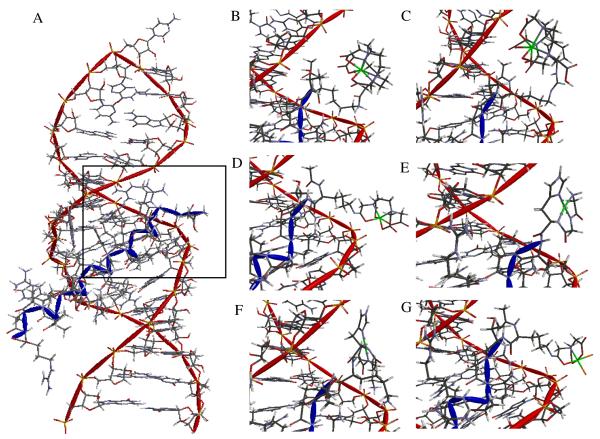
Complex formation between the RRE stem loop IIB RNA and M-chelate-Rev species was structurally modeled to visually assess the structural alignment of the reactive metal centers with the RNA (Figure 2). Model geometries were constrained to the average NMR structure of the RRE-Rev peptide complex and M-chelate X-Ray crystal structures of Fe-DOTA, Fe-DTPA, Fe-EDTA, Fe-NTA, Cu-GGH (for both M-GGH and M-KGHK), but the chelate and linker geometries N-terminal to the point of attachment to the Rev peptide were allowed to vary, and molecular mechanics energy minimization was performed.13-18 The M-NTA- and M-EDTA-Rev derivative metal centers each demonstrated a preference for very close association (< 5 Å) with the RNA backbone, while the M-DOTA-Rev, M-DTPA-Rev, M-KGHK-Rev, and M-GGH-Rev metal centers were held more distally, relative to the RNA backbone. Only M-KGHK-Rev made specific chelate-RNA contacts, which were made through the two additional lysine-phosphate salt bridges, although these contacts constrained the M-KGHK center to a poor alignment of the metal center with respect to the RNA backbone. The M-NTA-Rev metal center demonstrated a very close association of its vacant metal-coordination sites relative to the RNA backbone, and this proximity was expected to increase reactivity with the RNA, as was observed (Table 2).
Figure 2.
High-affinity site binding of RRE stem loop IIB RNA by (A) Rev peptide (NMR structure). Inset: Energy minimized structural models of high-affinity site binding by M-chelate-Rev peptides (B) Fe-DOTA-Rev, (C) Fe-DTPA-Rev, (D) Fe-EDTA-Rev, (E) Cu-GGH-Rev, (F) Cu-KGHK-Rev, and (G) Fe-NTA-Rev.
Table 2.
Initial rates of Fl-RNA cleavage and AP-RNA modification and metal-chelate reduction potentials.
| Complex | Fl-RNA Cleavage Ratea (nM/min) |
AP-RNA Modificationa Rate (nM/min) |
Red. Potential vs. NHE of M- Chelate (mV) |
Redox Couple |
|---|---|---|---|---|
| Cu-DOTA-Rev | 0.8 ± 0.1 | 2.8 ± 0.1 | 180 | 2+/1+ |
| Ni-DOTA-Rev | 0.009 ± 0.008 | 0.323 ± 0.004 | −35 | 2+/1+ |
| Co-DOTA-Rev | 0.063 ± 0.005 | 0.332 ± 0.008 | 142 | 3+/2+ |
| Fe-DOTA-Rev | 0.026 ± 0.006 | 0.367 ± 0.009 | 396 | 3+/2+ |
| Cu-DTPA-Rev | 0.496 ± 0.008 | 1.25 ± 0.04 | 148 | 2+/1+ |
| Ni-DTPA-Rev | 0.054 ± 0.008 | 0.169 ± 0.002 | None Obs. | None Obs. |
| Co-DTPA-Rev | 0.063 ± 0.007 | 0.66 ± 0.01 | 1111 | 3+/2+ |
| Fe-DTPA-Rev | 0.16 ± 0.04 | 0.248 ± 0.004 | 280 | 3+/2+ |
| Cu-EDTA-Rev | 0.14 ± 0.03 | 0.81 ± 0.01 | 47 | 2+/1+ |
| Ni-EDTA-Rev | 0.18 ± 0.05 | 0.524 ± 0.003 | 172 | 2+/1+ |
| Co-EDTA-Rev | 0.17 ± 0.03 | 0.328 ± 0.003 | 146 | 3+/2+ |
| Fe-EDTA-Rev | < 0.02 | 0.109 ± 0.001 | 391 | 3+/2+ |
| Cu-GGH-Rev | 0.045 ± 0.005 | 0.4 ± 0.3 | 1038 | 3+/2+ |
| Ni-GGH-Rev | 0.026 ± 0.005 | 0.58 ± 0.01 | 1000 | 3+/2+ |
| Co-GGH-Rev | 0.003 ± 0.007 | 0.20 ± 0.01 | −119 | 3+/2+ |
| Cu-KGHK-Rev | 0.060 ± 0.005 | 0.511 ± 0.003 | 1058 | 3+/2+ |
| Ni-KGHK-Rev | 0.043 ± 0.003 | 0.227 ± 0.006 | 1055 | 3+/2+ |
| Co-KGHK-Rev | 0.031 ± 0.003 | 0.257 ± 0.009 | −228 | 3+/2+ |
| Cu-NTA-Rev | 2.1 ± 0.2 | 4.9 ± 0.2 | 215 | 2+/1+ |
| Ni-NTA-Rev | 0.067 ± 0.005 | 0.78 ± 0.01 | 176 | 2+/1+ |
| Co-NTA-Rev | 0.069 ± 0.003 | 0.99 ± 0.02 | 274 | 3+/2+ |
| Fe-NTA-Rev | 0.031 ± 0.006 | 0.170 ± 0.001 | 464 | 3+/2+ |
| Background | 0.015 ± 0.005 | 0.28 ± 0.03 |
|
|
Rates above the limit of detection are shown in bold.
RRE RNA Modification Kinetics
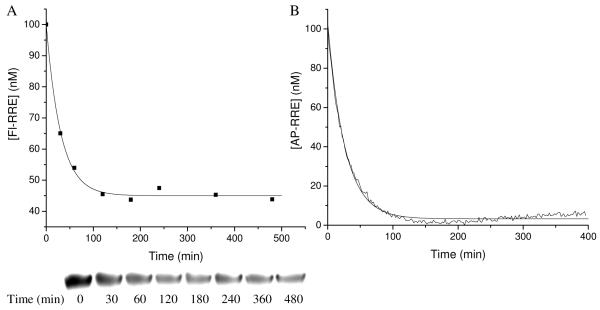
Initial rates of modification of the RRE RNA promoted by each M-chelate-Rev species were determined by two independent kinetic assays. The rate of RNA cleavage mediated by each M-chelate-Rev species was determined by following the disappearance of full-length end-labeled RRE (Fl-RNA) over time as monitored either by polyacrylamide gel electrophoresis (PAGE), or by the change in the intrinsic fluorescence of internally-labeled RRE (AP-RNA) that results from the conformational change of AP-RNA following reaction with each M-chelate-Rev species. These two assays provided an independent assessment of the reaction (Figure 3). Each reaction was run in the presence of physiologically available coreactants, H2O2, ascorbate and ambient oxygen, and initial rates are summarized in Table 2.
Figure 3.
Time-dependent modification of RRE RNA by Cu-DOTA-Rev. (A) Disappearance of full-length Fl-RNA monitored by PAGE analysis. (B) Conformational change of AP-RNA following reaction monitored by real-time fluorescence.
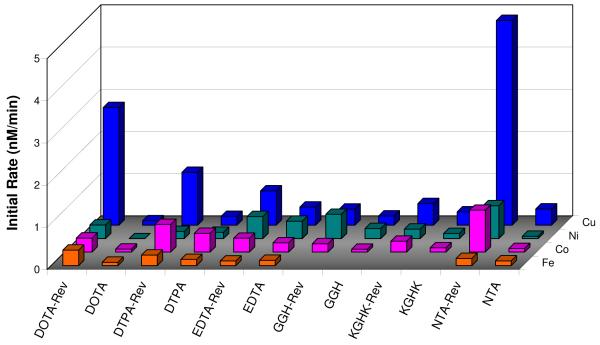
The highest rate of RRE RNA modification was seen with Cu-NTA-Rev, and in general, the most reactive M-chelate-Rev species were Cu-chelate-Rev complexes, including Cu-DOTA-Rev, Cu-DTPA-Rev, and Cu-EDTA-Rev. Surprisingly, the Fe-chelate-Rev complexes were among the slowest in reactivity, and intermediate rates were observed with Ni- and Co-chelate-Rev species. Overall, a remarkable range of activity was observed for each metal, and activity was highly dependent on the chelating ligand used.
Control assays were run with the isolated M-chelate domains, and as expected, RNA degradation by M-chelate-Rev species was faster than observed for the respective M-chelates lacking Rev, but to varying extents. This was especially apparent for DOTA-Rev and NTA-Rev complexes, and for most Cu-chelate-Rev species. These results demonstrate the positive effect of Rev targeting of M-chelates toward the RRE RNA (Figure 4).
Figure 4.
Rate of RRE RNA modification by M-chelate-Rev vs. M-chelate complexes. Enhanced rates were observed for M-chelates attached to Rev for 100% of the species with above-background rates. The background rate with no complex was 0.28 ± 0.03 nM/min.
To compare the catalytic efficiency of RNA degradation promoted by M-chelate-Rev complexes relative to other catalysts, second-order rate constants for RNA degradation reactions were established by use of equation (2), where kobs is the observed first order rate constant for RNA degradation (after subtraction of the observed background rate constant obtained in the absence of complex), and [M] is the concentration of M-chelate-Rev (10−7 M). Table 3 compares the second-order rate constants obtained from the study described here with those of several other catalysts known to cleave nucleic acids or nucleic acid analogues. The RNA degradation reactions promoted by several M-chelate-Rev complexes demonstrate significant activity enhancement (over six orders of magnitude) relative to other artificial nucleases.
| (2) |
Table 3.
Comparison of second-order rate constants for nucleic acid cleavage promoted both by several M-chelate-Rev complexes, and by other artificial catalysts.
| Complex | Substrate | k2 (M−1 min−1) | Coreagents | Conditions | Ref. |
|---|---|---|---|---|---|
| [Cu-NTA-Rev]9+/8+ | RRE RNA | 460000 | ascorbate + H2O2 | pH 7.4, 37 °C | a |
| [Cu-DOTA-Rev]8+/7+ | RRE RNA | 250000 | ascorbate + H2O2 | pH 7.4, 37 °C | a |
| [Cu-KGHK-Rev]12+/11+ | RRE RNA | 23000 | ascorbate + H2O2 | H 7.4, 37 °C | a |
| [Cu-GGH-Rev]10+/9+ | RRE RNA | 12000 | ascorbate + H2O2 | pH 7.4, 37 °C | a |
| [Cu-GGH-Rev]10+/9+ | RRE RNA | 700 | ascorbate | pH 7.4, 37 °C | 10 |
| [Cu-KGHK]2+/1+ | tRNAPhe | 6.3 | ascorbate | pH 7.4, 25 °C | 19 |
| [Cu-KGHK]2+/1+ | dsDNA | 5580 | ascorbate | pH 7.4, 37 °C | 6 |
| [Cu-GGH]0/1− | dsDNA | 2340 | ascorbate | pH 7.4, 37 °C | 6 |
| [Cu-neamine]4+ | dsDNA | 7690b | none | pH 7.3, 37 °C | 20 |
| [Co-cyclen](4-5)+ | dsDNA | 13-90b | none | pH 7.0, 37 °C | 21 |
| Eu3+ | dsDNA | 108b | none | pH 7.0, 37 °C | 22 |
| [Rh-(phi)2bpy’-peptide(Zn)]0 | dsDNA | 300 | none | pH 6.0, 37 °C | 23 |
| [Co-tamen]3+ | dsDNA | 3 | none | pH 7.6, 37 °C | 24 |
| [Ce2-HXTA]3+ | ssDNA | 840 | none | pH 8.0, 37 °C | 25 |
| [Ce2-HXTA]3+ | dsDNA | 78 | none | pH 8.0, 37 °C | 25 |
| (Zn)2-bis(TACN)-linker | HPNP | 0.348-15 | none | pH 7.6, 25 °C | 26 |
| (Zn)-TACN | HPNP | 0.126 | none | pH 7.6, 25 °C | 26 |
| (Zn)2(calix[4]arene) | HPNP | 78.8 | none | pH 7, 25 °C | 27 |
| LuCl3 | ApA | 38 | none | pH 7.2, 30 °C | 28 |
This study.
Enzyme-like activity is expressed as kcat/KM. dsDNA = double stranded DNA; ssDNA = single stranded DNA; HPNP = 2-hydroxypropyl p-nitrophenyl phosphate; ApA = adenylyl(3′-5′)adenosine.
Metal-Chelate Reduction Potentials
Reduction potentials for the metal-chelates were determined by square-wave voltammetry under anaerobic conditions and using similar solution buffer conditions as used in the RNA degradation experiments. The reduction potential for each metal cofactor was significantly modulated by the chelator used. As expected, reduction potentials were generally shifted toward more negative values for complexes with more negatively charged chelators and/or complexes with greater ligand field splitting. Observed potentials for Cu2+/1+ ranged from +47 mV (Cu-EDTA) to +215 mV (Cu-NTA), while the complexes Cu-GGH and Cu-KGHK provided the expected Cu3+/2+ potentials ~1000 mV.29 The Fe3+/2+ reduction potentials ranged from +280 mV (Fe-DTPA) to +464 (Fe-NTA), while potentials for Co3+/2+ ranged from −228 mV (Co-KGHK) to +1111 mV (Co-DTPA). Observed potentials for Ni2+/1+ ranged from −35 mV (Ni-DOTA) to +176 (Ni-NTA), while the complexes Ni-GGH and Ni-KGHK provided the expected Ni3+/2+ potentials ~1000 mV.29 Table 2 summarizes the M-chelate reduction potentials for all of the metal-chelates studied. These are expected to be similar to potentials in the Rev conjugates, although this was not tested as a result of the relatively small amounts of Rev-coupled material in hand.
DISCUSSION
RRE RNA Binding Affinity
Analysis of the dissociation constants for binding of M-chelate-Rev species to the RRE RNA reveals that all M-chelate-Rev species bind the RNA with high affinity. However several trends were observed. Binding at the high affinity site (KD1), which is known to be primarily governed by specific recognition events such as salt bridge formation and hydrogen bonding networks, appeared to be additionally and modestly affected by a combination of electrostatic and steric factors, as well as metal coordination number.13 At the low affinity site (KD2), there was a clear correlation between electrostatics and binding affinity. Figure 2 illustrates energy minimized structural models of complexes for M-chelate-Rev species bound to the high affinity RNA site.
Attachment of bulkier chelate groups to the Rev peptide appeared to diminish affinity slightly at the high-affinity site, as can be seen in the case of metal-free DTPA-Rev and KGHK-Rev, the most sterically hindered of the chelate-Rev species. The addition of steric bulk at the high-affinity RNA binding site is expected to perturb some of the specific Rev-RNA contacts typically responsible for high-affinity binding. Metal-free DTPA-Rev and KGHK-Rev binding to the high-affinity RNA site was 10- to 400-fold weaker than the other chelate-Rev compounds, in spite of the additional Lys residues for the latter.
The metal coordination environment of the complexes also appeared to affect the RRE RNA affinity at both sites. Comparison of KGHK-Rev with M-KGHK-Rev complexes reveals an ~30-fold increase in binding affinity upon metal chelation, which most likely reflects either direct coordination of metal ion to RRE RNA functional groups, or reorganization of the chelator into a less sterically hindered conformation following metal chelation. A similar result was observed for NTA-Rev and DTPA-Rev. Comparison of Fe-DTPA-Rev, in which the expected coordination number of iron is 7, with other M-DTPA-Rev species, in which the expected coordination number is 6, reveals the 7-coordinate species has a 6- to 30-fold higher affinity for the RRE RNA at the high-affinity site, relative to the other M-DTPA-Rev species.15,30 For DTPA-Rev, a lower coordination number is equated with higher number of uncoordinated carboxylates with subsequent repulsion of the RNA backbone through steric and/or electrostatic factors.
Electrostatics appeared to affect the dissociation constants modestly at the high-affinity site and strongly at the low-affinity site, such that more positively-charged M-chelate-Rev species generally had higher affinities toward the negatively-charged RRE RNA than did less positive M-chelate-Rev species. This is consistent with binding being dominated by the overall electrostatic interactions rather than by specific recognition events. Comparison of the M-KGHK-Rev species (+11 overall charge in the divalent metal state) with M-DTPA-Rev species (+6 overall charge), reveals that non-specific binding at the low affinity site (KD2) was greatly enhanced for the more positively-charged species. At the high-affinity site, electrostatics only modestly affected the dissociation constant (KD1), most likely because binding at the high-affinity site is governed primarily by specific Rev-RNA contacts. In fact, in the case of M-KGHK-Rev, it appears that two new contacts with the RRE RNA were introduced, because the two lysine residues of KGHK can form additional salt bridge contacts with RNA backbone phosphates, and adding to the Rev-RRE RNA network of salt bridges and hydrogen bonds. Comparison of KD1 values between M-KGHK-Rev and M-GGH-Rev indeed reveals overall tighter binding of M-KGHK-Rev to RRE RNA. Comparison of KD2 values between M-KGHK-Rev and M-GGH-Rev similarly reveal tighter non-specific binding of M-KGHK-Rev to the low-affinity RNA site, although this interaction is likely due to addition of non-specific electrostatic attraction.
The Rev peptide binds with high selectivity to RRE RNA. The observed range for the low-affinity KD2 values obtained in this study is strikingly similar to previously reported KD values for binding of Rev peptide to yeast tRNA, which is more than three orders of magnitude weaker than binding to the high-affinity RRE RNA site (KD ~ 1 nM for RRE RNA vs. KD ~ 2.4 μM for yeast tRNA).5 One would expect the binding affinity at the low-affinity RNA site to be similar to the binding affinity toward other RNAs, because the factors that govern binding at the low-affinity RNA site (primarily electrostatic) would also increase non-selective binding to other RNAs in an in-vivo context. Therefore the best candidates for selective binding to the RRE RNA are most likely those M-chelate-Rev complexes which bind very tightly at the high affinity site but very weakly at the low affinity site.
RRE RNA Modification Kinetics
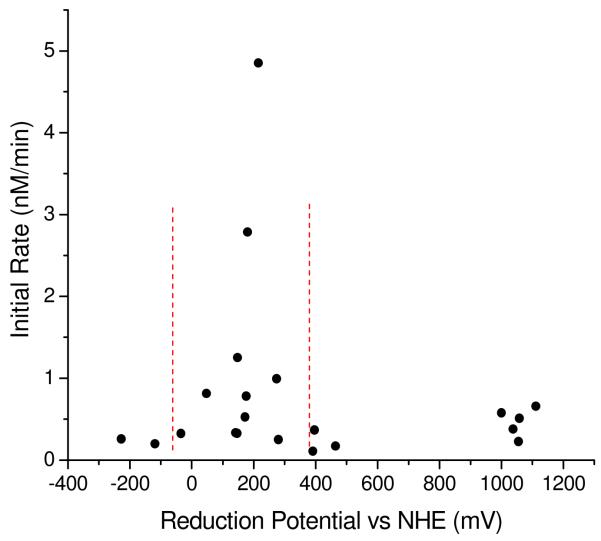
Degradation of the RRE RNA by M-chelate-Rev compounds was observed for all of the compounds tested, although considerable variation in reactivity was found. The ability of the M-chelates to react with the RRE RNA was facilitated by two major factors: (1) localization to the RRE RNA by the attached Rev peptide and (2) reduction potentials that are close to the potentials for relevant redox half reactions of reactive oxygen species. Many of the M-chelates used in this study are known to undergo oxidative reactions that produce reactive oxygen species. Localization of reactive M-chelates to the RRE RNA is expected to increase the exposure of the RNA to these reactive intermediates. Indeed, increased rates of RNA modification by M-chelate-Rev complexes were generally observed (Figure 4). This ability of M-chelates to generate reactive intermediates is directly related to their ability to switch between oxidation states, since the reaction of M-chelates with the biological coreactants H2O2, O2, and ascorbate occur via electron transfer. Those M-chelate-Rev species with the highest rates of RNA modification contained M-chelates whose reduction potentials fall in the range of 0 – 300 mV (Table 2). Given that the reported reduction potentials at neutral pH for O2/superoxide, superoxide/H2O2, H2O2/HO•, and ascorbyl radical/ascorbate are −330 mV, 890 mV, 380 mV, and −66 mV,31,32 respectively, multiple turnover single-electron oxidation/reduction by H2O2/ascorbate is thermodynamically favored for M-chelates with reduction potentials lying between 380 mV and −66 mV. Indeed, the most reactive M-chelate-Rev species fell in this range (Figure 5). M-chelate mediated reduction of oxygen is also possible, although direct superoxide generation is less thermodynamically favored (E° = −330 mV). A few M-chelate-Rev complexes with reduction potentials > 1000 mV also showed modest activity (Co-DTPA-Rev, Cu-GGH-Rev, Ni-GGH-Rev, Cu-KGHK-Rev, Ni-KGHK-Rev), and they most likely operate via an alternate mechanism. The reduction potentials for these complexes are such that the oxidation of substrate is strongly thermodynamically favored, but the requirement for prior generation of the oxidized metal is not favored, which could account for the slow rates observed by these complexes.
Figure 5.
Observed initial rates for RRE RNA modification by M-chelate-Rev and coreactants H2O2/ascorbate were found to be highest for M-chelate complexes with reduction potentials lying between those of ascorbyl radical/ascorbate (−66 mV) and H2O2/hydroxyl radical (380 mV), indicated by the vertical dashed lines.
Possible reactive intermediates generated by the M-chelate-Rev complexes include superoxide radical, hydroxyl radical, and metal-bound oxygen intermediates. However, M-chelates known to efficiently generate diffusible hydroxyl radicals, including Fe-EDTA and Fe-NTA, were found to be among the slowest RNA modifiers when coupled to Rev, suggesting that other reactive oxygen intermediates or metal-associated reactive oxygen species may be more effective in the case of Cu-NTA-Rev, Cu-DOTA-Rev, Cu-DTPA-Rev, and Cu-EDTA-Rev. For M-chelate-Rev targeted degradation of RRE RNA, it is expected that metal-associated reactive oxygen intermediates would more efficiently modify the RNA than freely diffusible reactive oxygen intermediates, due to their prolonged proximity to the RNA in the metal-bound case. The data are seen to be consistent with such a proximity effect.
Structural models of complex formation between RRE RNA and M-chelate-Rev suggest that M-NTA-Rev and M-EDTA-Rev are well suited for RNA modification, because their metal centers prefer close proximity with the RNA backbone (Figure 2). The M-NTA-Rev complexes demonstrate pronounced levels of activity for RNA modification, most likely because they are closely aligned with the RNA backbone and have two cis open metal-coordination sites to which an RNA functional group and reactive oxygen species may simultaneously coordinate and react by a template effect. Indeed, the fastest rate of RNA cleavage was observed with Cu-NTA-Rev, although Cu-DOTA-Rev, Cu-DTPA-Rev, and Cu-EDTA-Rev also demonstrated significant RNA modification activity, despite their lack of open coordination sites.
Previous studies have shown that transient opening of metal-coordination sites allows for coordination of redox coreactants and the observed generation of reactive oxygen species in M-chelate complexes such as Fe-EDTA.7 Such transient opening of metal-coordination sites may underlie the observed RNA modification activity by Cu-DOTA-Rev, Cu-DTPA-Rev, and Cu-EDTA-Rev. The M-KGHK-Rev complexes demonstrated relatively slow rates of RNA modification, which may in part reflect the structural confinement of M-KGHK to a non-ideal alignment with the RNA backbone as a result of salt bridge contacts between the lysines of KGHK and RNA phosphates. In addition, M-KGHK-Rev and M-GGH-Rev complexes contain relatively short chelate-Rev linkers, relative to the other M-chelate-Rev compounds (reflecting different modes of attachment), and this lack of flexibility may also contribute the relatively low RNA modification activity observed for M-KGHK-Rev and M-GGH-Rev. The large positive reduction potential for the Cu3+/2+ redox couple is also likely to mitigate against the effective production of reactive oxygen species, which could be a very favorable factor in the physiological application of such a metal-chelate, and may reflect the innate presence of this amino terminal Cu and Ni binding (ATCUN) sequence in natural peptides such as albumins, histatins, and neuromedin C.14,33-35 Conversely, the relatively high rates of RNA modification observed for Cu-DOTA-Rev, Cu-DTPA-Rev, Cu-EDTA-Rev, and Cu-NTA-Rev may be partially attributable to the flexibility of the tethered M-chelates, which would allow transient formation of states of ideal alignment with the RNA backbone. These elevated levels of activity will also be mollified by the lower available physiological concentrations of peroxide. In that regard, and keeping in mind the observation that copper and nickel derivatives of ATCUN peptides are known to promote reactive oxygen species formation at a slower rate than other M-chelates, most likely as a result of the distinct redox pairs involved (Cu3+/2+ and Ni3+/2+), a measure of sluggishness in promoting formation of ROS could be beneficial in a therapeutic setting. That is, intrinsic reactivity is not necessarily the only, or even the most important factor in assessing catalytic metallodrug candidates.
Comparison of observed rates between the two kinetics assays provided information concerning the different stages of RNA modification. Initial rates observed for RNA modification that is monitored internally by use of AP-RNA were generally faster than the observed rates for RNA cleavage monitored by Fl-RNA, even though the reaction conditions were similar. This fact suggests the existence of oxidative reactions that cause chemical and structural changes to the RRE RNA but do not immediately result in cleavage. That is, the two assays provide distinct, but complimentary information on the various stages of RNA modification. Scission events represent only a fraction of all of the modifications arising from oxidative damage to RNA, and so general oxidative modifications to the RNA are expected to accumulate faster than the subset of reactions that result in immediate cleavage. Reactions monitored by real-time fluorimetry through changes in AP emission are generally observed to be faster than those determined from gel assays since the general oxidative modifications of the RNA that are monitored by the former arise more quickly than the fraction of these that result in immediate strand scission (monitored by Fl-RNA gels). Indeed there is much literature precedent for oxidative modifications of nucleic acids that do not result in immediate strand scission at neutral pH and 37°C, including base oxidation, non-scissile ribose oxidation, or racemization of stereocenters.6,36,37 There are many possible pathways to oxidative RNA modification, however, only a small number of these are likely to result in cleavage. Many of these oxidative modifications would most likely inactivate the RRE RNA in an in vivo context without the need for strand scission, because these modifications would cause the mRNA to be improperly recognized and/or processed by the ribosome, while cells lack a general RNA repair mechanism.38 Previous studies have indeed demonstrated that significant translation errors, reduced translation, and translated protein truncation and degradation arise as a result of non-scissile oxidation of full-length mRNA.39,40 Therefore, if the overall in vivo rate of RRE RNA inactivation by such M-chelate-Rev species were to sufficiently exceed the rates of RRE RNA transcription and translation, the available pool of cytosolic HIV-1 mRNA would be knocked down, and HIV viral particle production would be inhibited.
CONCLUSIONS
For drugs to function with multiple turnover capability in an in-vivo setting, they must meet several functional criteria. These include selective binding to the desired target; irreversible inactivation of the target; rapid dissociation from the modified target; and facile regeneration of the active state such that the cycle may be repeated. The first two prerequisites for multiple-turnover, targeting and irreversible inactivation of the target, were the primary focus of this study. Relatively high-affinity binding was indeed observed between all of the M-chelate-Rev species and the targeted RRE RNA. Additionally, the selectivity of binding of each M-chelate-Rev species to the RRE RNA vs. other RNAs can be predicted by comparison of the KD1 to KD2 values, since KD1 values were governed primarily by specific (most likely hydrogen bonding) interactions, whereas KD2 values were governed primarily by non-specific electrostatic interactions.13,41 Since the majority of metal-chelates used in this study are known to produce highly-reactive oxidative intermediates in the presence of O2 or H2O2, and since oxidation of mRNA is an irreversible process with no known in-vivo repair pathway, these M-chelate-Rev compounds are ideal candidates for irreversible inactivation of the RRE RNA. Indeed, it was observed that the majority of the M-chelate-Rev compounds tested were able to mediate RNA degradation. Following oxidative modification of the RNA, it is likely that the affinities of the M-chelate-Rev species toward the modified RNA will be significantly reduced as a result of the loss of specific RNA-Rev contacts upon reaction, which would favor dissociation of the M-chelate-Rev species from inactivated RRE RNA. Finally, regeneration of the reduced M-chelate-Rev species in-vivo would be mediated by biological reducing agents such as ascorbic acid (vitamin C) or glutathione, both present at mM concentrations in-vivo, as has indeed been shown for many of the respective metal-chelates used in this study. Accordingly, many of the M-chelate-Rev species reported and studied herein have desirable properties for catalytic multiple-turnover inactivation of RRE RNA. Even if multiple turnover function proved problematic, the irreversible nature of such oxidative mRNA modification would be a remarkable improvement over the majority of current drugs, which operate reversibly and thus require higher concentrations in order to attain effectiveness. Further studies are necessary to elucidate the complex oxidative chemistry and RNA modification/cleavage chemistry elicited by these compounds.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health [HL093446 and AA016712]. Jeff Joyner was supported by an NIH Chemistry/Biology Interface training grant (T32 GM08512). MALDI-TOF results were obtained through The Ohio State University Campus Chemical Instrumentation Center.
Footnotes
Supporting Information Available. Titration curves, sample kinetics traces, sample square wave voltammogram, metal-chelate extinction coefficients, and a summary of observed rates for metal-chelate modification of RRE RNA
REFERENCES
- (1).Hacein-Bey-Abina S, Kalle CVS,M, M.P. M, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, Sorenson R, Forster A, Fraser P, Cohen JI, de Saint Basile G, Alexander I, Wintergest U, Frebourg T, Aurias A, Stoppa-Lyonnet D, Romana S, Radford-Weiss I, Gross F, Valensi F, Delabesse E, Macintyre E, Sigaux F, Soulier J, Leiva LE, Wissler M, Prinz C, Rabbitts TH, Deist F. Le, Fischer A, Cavazzana-Calvo M. Science. 2003;302:415. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- (2).Baum C, von Kalle C, Staal F, Li X, Fehse B, Schmidt M, Weerkamp F, Karlsson S, Wagemaker G, Williams D. Molecular Therapy. 2004;9:5. doi: 10.1016/j.ymthe.2003.10.013. [DOI] [PubMed] [Google Scholar]
- (3).Hocharoen L, Cowan JA. Chemistry - A European Journal. 2009;15:8670. doi: 10.1002/chem.200900821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Malim MH, Hauber J, Le S, Maizel J, Cullen B. Nature. 1989;338:254. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- (5).Luedtke NW, Tor Y. Biopolymers. 2003;70:103. doi: 10.1002/bip.10428. [DOI] [PubMed] [Google Scholar]
- (6).Jin Y, Cowan JA. Journal of the American Chemical Society. 2005;Vol. 127:8408. doi: 10.1021/ja0503985. [DOI] [PubMed] [Google Scholar]
- (7).Graf E, Mahoney J, Bryant R, Eaton J. The Journal of Biological Chemistry. 1984;259:3620. [PubMed] [Google Scholar]
- (8).Inoue S, Kawanishi S. Cancer Research. 1987;47:6522. [PubMed] [Google Scholar]
- (9).Dreyer G, Dervan P. PNAS. 1985;82:968. doi: 10.1073/pnas.82.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jin Y, Cowan JA. J Am Chem Soc. 2006;128:410. doi: 10.1021/ja055272m. [DOI] [PubMed] [Google Scholar]
- (11).Jin Y, Cowan JA. J Biol Inorg Chem. 2007;12:637. doi: 10.1007/s00775-007-0221-2. [DOI] [PubMed] [Google Scholar]
- (12).Lacourciere KA, Stivers JT, Marino JP. Biochemistry. 2000;Vol. 39:5630. doi: 10.1021/bi992932p. [DOI] [PubMed] [Google Scholar]
- (13).Battiste J, Mao H, Rao N, Tan R, Muhandiram D, Kay L, Frankel A, Williamson J. Science. 1996;273:1547. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- (14).Camerman N, Camerman A, Sarkar B. Canadian Journal of Chemistry. 1976 [Google Scholar]
- (15).Finnen D, Pinkerton A, Dunham W, Sands R, Funk M. Inorganic Chemistry. 1990;30:3960. [Google Scholar]
- (16).Viola-Villegas N, Doyle R. Coordination Chemistry Reviews. 2009;253:1906. [Google Scholar]
- (17).Mizuta T, Wang J, Miyoshi K. Inorganica Chimica Acta. 1995;230:119. [Google Scholar]
- (18).Walters MA, Vapnyar V, Bolour A, Incarvito C, Rheingold AL. Polyhedron. 2003;22:941. [Google Scholar]
- (19).Bradford S, Kawarasaki Y, Cowan JA. J. Inorg. Biochem. 2009;103:871. doi: 10.1016/j.jinorgbio.2009.03.003. [DOI] [PubMed] [Google Scholar]
- (20).Sreedhara A, Freed JD, Cowan JA. J. Am. Chem. Soc. 2000;122:8814. [Google Scholar]
- (21).Hettich R, Schneider HJ. J. Am. Chem. Soc. 1997;119:5638. [Google Scholar]
- (22).Rammo J, Hettich R, Roigk A, Schneider HJ. Chem. Commun. 1996:105. [Google Scholar]
- (23).Fitzsimons MP, Barton JK. J. Am. Chem. Soc. 1997;119:3379. [Google Scholar]
- (24).Dixon NE, Geue RJ, Lambert JN, Moghaddas S, Pearce DA, Sargeson AM. Chem. Commun. 1996:1287. [Google Scholar]
- (25).Branum ME, Tipton AK, Zhu S, Que LJ. J. Am. Chem. Soc. 2001;123:1898. doi: 10.1021/ja0010103. [DOI] [PubMed] [Google Scholar]
- (26).Iranzo O, Elmer T, Richard J, Morrow J. Inorganic Chemistry. 2003;42:7737. doi: 10.1021/ic030131b. [DOI] [PubMed] [Google Scholar]
- (27).Molenveld P, Kapsabelis S, Engbersen J, Reinhoudt D. J. Am. Chem. Soc. 1997;119:2948. [Google Scholar]
- (28).Matsumura K, Komiyama M. J. Biochem. 1997;122:387. doi: 10.1093/oxfordjournals.jbchem.a021765. [DOI] [PubMed] [Google Scholar]
- (29).Jin Y, Lewis MA, Gokhale NH, Long EC, Cowan JA. Journal of the American Chemical Society. 2007;129:8353. doi: 10.1021/ja0705083. [DOI] [PubMed] [Google Scholar]
- (30).Fomenko V, Polynova T, Porai-Koshits M, Varlamova G, Pechurova N. Zhurnal Strukturnoi Khimii. 1973;14:571. [Google Scholar]
- (31).Borsook H, Keighley G. PNAS. 1933;19:875. doi: 10.1073/pnas.19.9.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wood P. Biochem. J. 1988;253:287. doi: 10.1042/bj2530287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Sankararamakrishnan R, Verma S, Kumar S. PROTEINS: Structure, Function, and Bioinformatics. 2005;58:211. doi: 10.1002/prot.20265. [DOI] [PubMed] [Google Scholar]
- (34).Grogan J, McKnight CJ, Troxler RF, Oppenheim FG. FEBS Letters. 2001;491:76. doi: 10.1016/s0014-5793(01)02157-3. [DOI] [PubMed] [Google Scholar]
- (35).Lau S-J, Sarkar B. J. Biol. Chem. 1971;246:5938. [PubMed] [Google Scholar]
- (36).Pogozelski W, Tullius T. Chemical Reviews. 1998;98:1089. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- (37).Fortini P, Pascucci B, Parlanti E, D’Errico M, Simonelli V, Dogliotti E. Mutation Research. 2003;531:127. doi: 10.1016/j.mrfmmm.2003.07.004. [DOI] [PubMed] [Google Scholar]
- (38).Wurtmann E, Wolin S. Crit Rev Biochem Mol Biol. 2009;44:34. doi: 10.1080/10409230802594043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Tanaka M, Chock B, Stadtman E. PNAS. 2006;104:66. doi: 10.1073/pnas.0609737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Shan X. The Ohio State University; 2005. [Google Scholar]
- (41).Cowan JA, Ohyama T, Wang D, Natarajan K. Nucleic Acids Research. 2000;28:2935. doi: 10.1093/nar/28.15.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.