Abstract
Objective
Nevirapine is widely prescribed for HIV-1 infection. We characterized relationships between nevirapine-associated cutaneous and hepatic adverse events and genetic variants among HIV-infected adults.
Design
We retrospectively identified cases and controls. Cases experienced symptomatic nevirapine-associated severe (grade III/IV) cutaneous and/or hepatic adverse events within 8 weeks of initiating nevirapine. Controls did not experience adverse events during more than 18 weeks of nevirapine therapy.
Methods
Cases and controls were matched 1 : 2 on baseline CD4 T-cell count, sex, and race. Individuals with 150 or less CD4 T cells/μl at baseline were excluded. We characterized 123 human leukocyte antigen (HLA) alleles and 2744 single-nucleotide polymorphisms in major histocompatibility complex (MHC) and drug metabolism and transport genes.
Results
We studied 276 evaluable cases (175 cutaneous adverse events, 101 hepatic adverse events) and 587 controls. Cutaneous adverse events were associated with CYP2B6 516G→T (OR 1.66, all), HLA-Cw*04 (OR 2.51, all), and HLA-B*35 (OR 3.47, Asians; 5.65, Thais). Risk for cutaneous adverse events was particularly high among Blacks with CYP2B6 516TT and HLA-Cw*04 (OR 18.90) and Asians with HLA-B*35 and HLA-Cw*04 (OR 18.34). Hepatic adverse events were associated with HLA-DRB*01 (OR 3.02, Whites), but not CYP2B6 genotypes. Associations differed by population, at least in part reflecting allele frequencies.
Conclusion
Among patients with at least 150 CD4 T cells/μl, polymorphisms in drug metabolism and immune response pathways were associated with greater likelihood of risk for nevirapine-related adverse events. Results suggest fundamentally different mechanisms of adverse events: cutaneous, most likely MHC class I-mediated, influenced by nevirapine CYP2B6 metabolism; hepatic, most likely MHC class II-mediated and unaffected by such metabolism. These risk variants are insensitive for routine clinical screening.
Keywords: CYP2B6, HIV, human leukocyte antigen, nevirapine, pharmacogenomics, rash, toxicogenomics
Introduction
Nevirapine is widely prescribed for type 1 HIV (HIV-1) infection. Although generally well tolerated and effective, some individuals who receive multiple doses of nevirapine experience severe cutaneous and/or hepatic adverse events during the initial weeks of therapy, especially when initiated at higher CD4 T-cell counts [1]. It is therefore recommended that nevirapine therapy not be initiated in antiretroviral-naive women and men with greater than 250 and 400 CD4 T cells/μl, respectively [2].
Human genetic variants may affect risk for nevirapine-associated adverse events, but results of prior genetic association studies have been inconsistent [3]. A study from Australia implicated HLA-DRB1*0101 as a risk for hepatic adverse events [4], whereas studies from Sardinia and Japan implicated HLA-Cw*08 [5,6]. Studies from South Africa and Mozambique implicated a polymorphism in ABCB1 (which encodes the multidrug efflux pump P-glycoprotein) with hepatic adverse events [7,8] but not CYP2B6 516G→T [7], which is known to increase nevirapine plasma exposure [9–15]. Studies from Thailand implicated HLA-Cw*0401 and HLA-B*3505 as being associated with nevirapine-associated cutaneous adverse events [16,17]. Such inconsistencies may reflect different adverse event phenotypes, patient ancestries, and/or genotyping strategies. False discovery is also possible even with seemingly well-designed studies [18].
To identify genetic variants associated with severe cutaneous rash and/or hepatic adverse events during the initial 8 weeks (the period of highest risk) of nevirapine-containing regimens we recruited cases and controls (matched on sex, race, and CD4 T-cell counts), and genotyped both human leukocyte antigen (HLA) and non-HLA genes among cohorts of African, Asian, and European descent (hereafter referred to as Black, Asian, and White, respectively).
Materials and methods
Study population
This was a retrospective, case-controlled study with prospective DNA collection. Cases and controls were at least 18 years of age, HIV-1-infected, and had previously initiated nevirapine-containing therapy. Cases had experienced one or more of the following cutaneous or hepatic adverse events within the first 8 weeks of initiating nevirapine: severe cutaneous toxicity [grade III or IV categorized by National Institute of Allergy and Infectious Disease (NIAID) Division of AIDS criteria] [19]; symptomatic grade ≥3 hepatic transaminase elevation [alanine transaminase (ALT) or aspartate aminotransferase (AST) >5× upper limit of normal (ULN)]; or acute hepatic failure.
Potential cases and controls were excluded for: 150 or less CD4 T cells/μl at last available date within 6 months before initiating nevirapine; acute viral hepatitis; use of immunomodulatory medications within the first 8 weeks of nevirapine therapy; no hepatic transaminase data within 6 months prior to initiating nevirapine; or previous participation in the 2NN long-term follow-up study. Potential cases were also excluded for: hepatotoxicity or rash that the investigators judged unrelated to nevirapine; initiation of abacavir or trimethoprim/sulfamethoxazole within 2 weeks prior to or within 8 weeks after initiating nevirapine; and ALT or AST values greater than 5 × ULN (grade >3) prior to initiating nevirapine. Potential controls were excluded for: development of grade ≥ 1 rash within 18 weeks of initiating nevirapine or any cutaneous condition potentially attributable to nevirapine; ALT or AST values greater than 2.5× ULN within 18 weeks of starting nevirapine; any hepatobiliary adverse event possibly due to nevirapine; or any systemic reaction (e.g. flu-like symptoms, arthralgia, myalgia, or conjunctivitis) attributable to nevirapine during the first 18 weeks of treatment.
Cases and controls were matched 1:2, respectively, on CD4 T-cell count within 50 cells/μl, sex, and self-reported race. Site personnel were asked to enrol three potential matched controls per case. Final matching was done centrally. Controls that did not match were excluded. The study was approved by the Institutional Review Board at each site, and all participants provided written informed consent. The study was registered at ClinicalTrials.gov (NCT00310843).
Study design
The study involved a screening visit and a subsequent entry and blood sampling visit. A total of 1536 patients were enrolled, of whom 647 failed screening based on eligibility criteria or case-control matching requirements.
Genotyping
Genomic DNA was isolated from peripheral blood mononuclear cells using Invitrogen iPrep (Invitrogen Corporation, Carlsbad, California, USA). Single-nucleotide polymorphisms (SNPs) were assayed in drug absorption, disposition, metabolism, and elimination (ADME) and major histocompatibility complex (MHC) genes (including 10-kb upstream and downstream regions), based on dbSNP version 128 and Genome NCBI build version 36 (SNP list available upon request). Some MHC-region SNPs were selected from Illumina MHC Mapping Panel and MHC Exon-Centric Panel (Illumina Inc., San Diego, California, USA). Assay was finalized based on primer designability and validation score. Genotyping was performed using Illumina BeadArray technology (Illumina Inc.): success rate 88.0–93.4% for SNPs, 99.4–100% for patient samples, reproducibility 100% between duplicates. We excluded participants with disagreement between sex indicated by genotype and clinical records.
HLA-A, HLA-B, HLA-C, and HLA-DPB, HLA-DQB, and HLA-DRB typing of all samples was performed with LIFECODES HLA DNA typing kits using Luminex xMAP technology and a reverse SSO protocol (Tepnel Lifecodes Corporation, Stamford, Connecticut, USA). DNA amplification and hybridization conditions followed manufacturer's protocol.
Statistical analysis
HLA alleles and SNPs were analyzed for association with adverse events in the total population and in each race group separately. Validity of self-reported race was supported by principal component analysis of SNP data with two principal components defining three clusters. Concordance with self-reported race was greater than 96%.
Fisher's exact test was used to analyze association of HLA alleles with adverse events in the three race groups and P values were corrected for multiple testing by dividing uncorrected P values by number of alleles with greater than 1% frequency. All alleles with homogeneous odds ratios (ORs) (Breslow-Day test P value >0.01) among the three race groups were tested for association with adverse events in the total population by Cochran-Mantel-Haenszel test.
After filtering SNPs with call rates below 80%, minor allele frequencies less than 1%, or for controls not in Hardy–Weinberg equilibrium (P < 0.001), retained SNPs were tested for association with adverse events by allelic test, genotypic test, Cochran-Armitage trend test, and tests for dominant-effect and recessive-effect models, as implemented in PLINK (version 1.06) [20,21]. All tests were adjusted for multiple testing. Linkage disequilibrium was based on r2 values.
Clinical variables were tested for association with adverse events by conditional logistic regression. HLA alleles and CYP2B6 SNPs associated with cutaneous adverse events, clinical variables, and their two-way product terms were used as explanatory variables in multiple logistic regression models. For CYP2B6 SNPs, an additive effect was assumed, with 0, 1, or 2 encoding the number of risk alleles. Forward and backward elimination were used to select variables associated with cutaneous adverse events.
Results
Patient characteristics
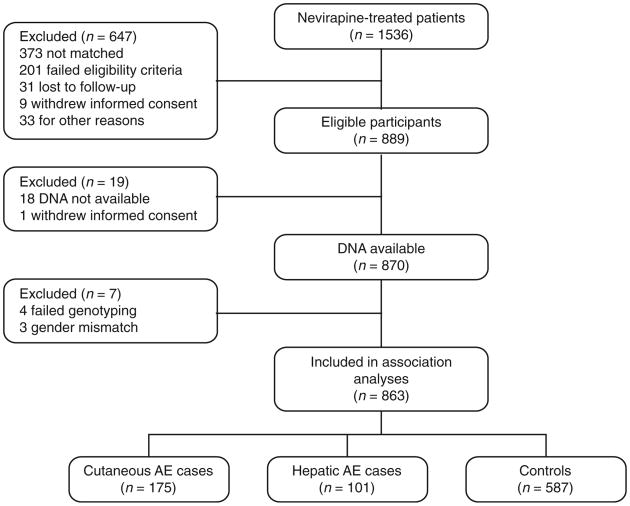
The study enrolled 889 participants from 2006 to 2008 at 76 sites in 11 countries (Argentina, Australia, Canada, France, Germany, Netherlands, Spain, Taiwan, Thailand, UK, and US), including 276 evaluable cases (175 with cutaneous adverse events, 101 with hepatic adverse events) and 587 evaluable controls (Fig. 1). Self-reported race (Asian, African, or European descent and herein referred to as Asian, Black, or White) agreed with the SNP data. Among Asians, 74.3% were Thai and among Whites, 31% self-identified as Hispanic/Latino. Demographic variables and baseline characteristics are shown in Table 1. Baseline CD4 T-cell counts were greater than 250 and 400 cells/ml in 69.7 and 48.7% of women and men, respectively; 71.8% of all participants had prior antiretroviral exposure. No demographic or clinical variable was significantly associated with adverse events.
Fig. 1.
Disposition of study volunteers. AE, adverse events.
Table 1.
Baseline characteristics of study participants.
| Black | Asian | White | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||
| Hepatic (n=14) | Rash (n=27) | Control (n=77) | Hepatic (n=30) | Rash (n=71) | Control (n=233) | Hepatic (n=57) | Rash (n=77) | Control (n=277) | Hepatic (n=101) | Rash (n=175) | Control (n=587) | |
| Age, years (SD) | 40.6 (9.1) | 37.1 (8.4) | 38.4 (7.3) | 38 (9.2) | 38.3 (8.6) | 39.5 (9.2) | 47.6 (11.5) | 45 (9.4) | 46.9 (10) | 43.8 (11.3) | 41.1 (9.6) | 42.8 (10.0) |
| Women [n(%)] | 12 (85.7) | 22 (81.5) | 48 (62.3) | 19 (63.3) | 34 (47.9) | 113 (48.5) | 12 (21.1) | 22 (28.6) | 64 (23.1) | 43 (42.6) | 78 (44.6) | 225 (38.3) |
| CD4 T cellsper μ1 [n (SD)] | 401 (276) | 322 (120) | 366 (175) | 385 (265) | 415 (232) | 412 (247) | 549 (287) | 437 (186) | 471 (227) | 480 (287) | 410 (201) | 434 (232) |
| Women (>250cells/μl) [n (%)] | 6 (50.0) | 16 (72.7) | 36 (75.0) | 9 (47.4) | 24 (70.6) | 74 (65.5) | 8 (66.7) | 19 (86.4) | 49 (76.6) | 23 (53.5) | 59 (75.6) | 159 (70.7) |
| Men (>400 cells/μl) [n (%)] | 1 (50.0) | 1 (20.0) | 8 (27.6) | 6 (54.5) | 16 (43.2) | 50 (41.7) | 29 (64.4) | 26 (47.3) | 115 (54.0) | 36 (62.1) | 43 (44.3) | 173 (47.8) |
| HIV, log10 copies/ml (SD) | 3.8 (1.4) | 3.0 (1.4) | 3.2 (1.5) | 2.9 (1.6) | 2.8 (1.4) | 3.1 (1.6) | 3.2 (1.5) | 3.5 (1.2) | 2.9 (1.5) | 3.2 (1.4) | 3.2 (1.4) | 3.0 (1.5) |
| Body weight [kg (SD)] | 72 (22) | 72 (16) | 76 (19) | 55 (9) | 57(11) | 57(10) | 73 (13) | 66 (12) | 73 (14) | 67 (16) | 63 (13) | 67(15) |
| HCV-positive [n (%)] | 0 (0.0) | 2 (7.4) | 2 (2.6) | 1 (3.3) | 0 (0.0) | 3(1.3) | 1 (1.8) | 9 (11.7) | 19 (6.9) | 2 (2.0) | 11 (6.3) | 24 (4.1) |
| HBV-positive [n (%)] | 1 (7.1) | 2 (7.4)) | 3 (3.9) | 0 (0.0) | 1 (1.4) | 4(1.7) | 4 (7.0) | 8 (10.4) | 21 (7.6) | 5 (5.0) | 11 (6.3) | 28 (4.8) |
| Treatment-native [n(%)] | 8 (57.1) | 7 (25.9) | 24 (31.2) | 16 (53.3) | 13 (18.3) | 98 (42.1) | 15 (26.3) | 14 (18.2) | 48 (17.3) | 39 (38.6) | 34 (19.4) | 170 (29.0) |
| ALT, fold ULN [average(SD)] | 0.7 (0.4) | 0.5 (0.3) | 0.6 (0.4) | 0.6 (0.4) | 0.7 (0.7) | 0.7 (0.7) | 0.7 (0.5) | 0.8 (0.8) | 0.7 (0.5) | 0.7 (0.5) | 0.8 (0.7) | 0.7 (0.6) |
| AST, fold ULN [average(SD)] | 0.7 (0.2) | 0.7 (0.6) | 0.8 (0.3) | 0.7 (0.4) | 0.8 (0.8) | 0.8 (0.3) | 0.7 (0.3) | 0.8 (0.6) | 0.8 (0.4) | 0.7 (0.3) | 0.8 (0.6) | 0.8 (0.6) |
ALT, alanine aminotransferase; AST, aspartate aminotransferase; HBV, hepatitis B; HCV, hepatitisC; ULN, upper limit of normal; SD, standard error.
Associations with ADME gene and major histocompatibility complex polymorphisms
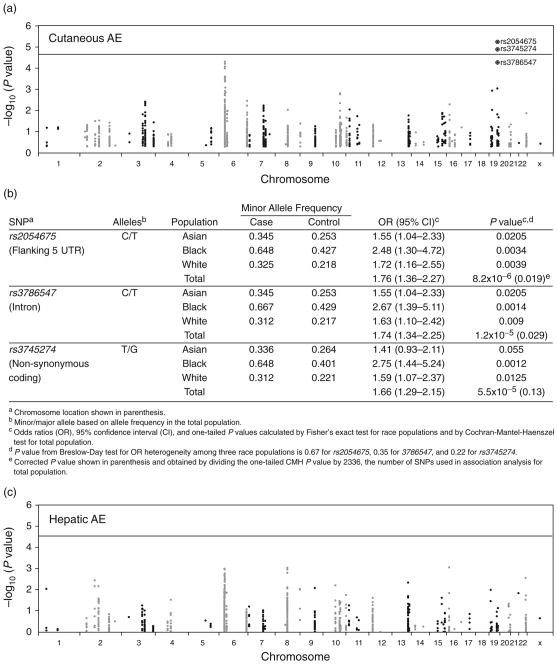
A total of 2744 SNPs across ADME and MHC genes (see supplemental material, http://links.lww.com/QAD/A139) were assessed for associations with adverse events in each race group and among all participants. In analyses controlled for population stratification among all participants, two linked SNPs in CYP2B6 [rs2054675 and rs3786547 (r2 = 0.982)] were significantly associated with cutaneous adverse events after correcting for multiple comparisons (Fig. 2a). Both were in linkage disequilibrium with rs3745274 (CYP2B6 516G→T; r2 > 0.90) (Fig. 2b), which is known to predict delayed plasma nevirapine clearance [9–15].
Fig. 2.
Associations between single-nucleotide polymorphisms and adverse events. Panel (a) is a Manhattan plot of the results for cutaneous adverse events (AEs) compared with controls. Panel (b) shows CYP2B6 SNPs associated with cutaneous AEs. Panel (c) is a Manhattan plot of the results for hepatic AEs compared with controls. In panels (a) and (b), the −log10 P values were plotted against chromosome positions. Lines indicate a Bonferroni corrected P value of 0.05. Panel (a) represents 2336 SNPs. Panel (c) represents 2058 SNPs. The three SNPs labeled are in CYP2B6.
For CYP2B6 516G→T, increasing T-allele count (0, 1, 2) was significantly associated with progressively increased risk of cutaneous adverse events in Blacks and Whites, with a weak trend in Asians (P=0.0025, P=0.021, and P=0.12, respectively, Cochran-Armitage trend test). Among Asians, the association was stronger for rs2054675 and rs3786547 (both P = 0.041). The association with 516G→T was particularly strong in Blacks, in whom the OR for 516TT homozygosity relative to 516GG homozygosity was 5.92 [95% confidence interval (CI) 1.53–26.8]. No SNP was significantly associated with hepatic adverse events after correcting for multiple comparisons (Fig. 2c).
There were possible associations between cutaneous adverse events and MHC polymorphisms (HLA-B, HLA-C, CCHCR1, TCF19), although none withstood multiple comparisons (Fig. 2a). We found no statistically significant association between ABCB1 3435C→T and nevirapine-associated cutaneous or hepatic adverse events, although the OR for hepatic adverse events among Blacks was 0.50 (95% CI 0.14–1.76; P= 0.27).
Associations with HLA alleles
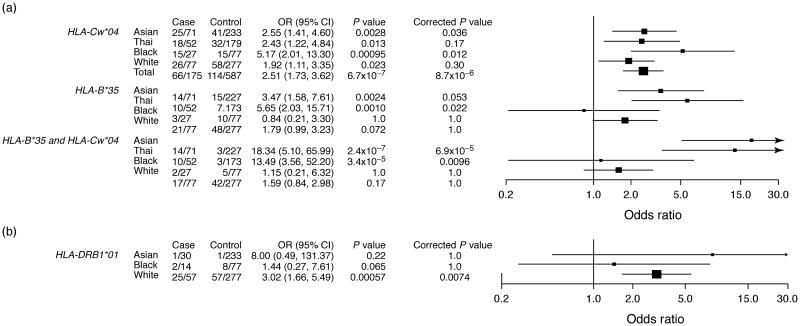
A total of 863 patients were HLA typed. HLA-Cw*04 was significantly associated with cutaneous adverse events among all study participants, most notably among Blacks and Asians (Fig. 3a). HLA-B*35 was strongly associated with cutaneous adverse events in Asian and Thai participants, but not in non-Thai Asians (mainly Taiwanese); there was a weak association with HLA-B*35 among Whites that did not withstand multiple testing correction. In analyses that excluded individuals with concomitant cutaneous and hepatic adverse events, OR for cutaneous adverse events increased from 5.65 to 7.31 for HLA-B*35 in Thai participants and from 2.51 to 2.98 for HLA-Cw*04 among all participants (data not shown). Among Asians, but not Blacks or Whites, the OR for cutaneous adverse events was markedly increased among those carrying both HLA-Cw*04 and HLA-B*35 (OR 18.34, 95% CI 5.10–65.99). Among Whites, a weak association between cutaneous adverse events and the relatively infrequent HLA-Cw*15 was no longer apparent when considering patients with only isolated cutaneous adverse events.
Fig. 3.
Associations between HLA alleles and nevirapine-related adverse events. Panel (a) shows cutaneous adverse events (AEs). Panel (b) shows hepatic AEs. Odds ratios (OR) and two-tailed P value were calculated by Fisher's exact test in race groups for all HLA alleles, and by Cochran-Mantel-Haenszel (CMH)test in the total population for HLA-Cw*04. All Thai participants were also included in the Asian group and were excluded from the CMH test. P values were corrected by Bonferroni procedure and corrected P value = Pvalue/n, where n is 22 for HLA-B*35, 13 for HLA-Cw*04, 13 for HLA-DRB1*01, and 286 for HLA-B35-Cw*04. Cases and controls are presented as numbers of participants (n) with the indicated genotypes divided by the numbers of cases or controls. Height of a square is proportional to the sample size. Horizontal lines represent 95% confidence intervals (CIs).
HLA-DRB1*01 was significantly associated with hepatic adverse events in Whites (Fig. 3b). This HLA allele was infrequent among Blacks and rare among Asians. In analyses that excluded individuals with coincident cutaneous and hepatic adverse events, OR for hepatic adverse events among Whites increased from 3.02 to 3.63 for HLA-DRB1*01. Similarly, HLA-DQB1*05 was significantly associated with hepatic adverse events among Whites only, despite its high frequency among Asians and Blacks but not when considering isolated hepatic adverse events (data not shown). We found no association between HLA-Cw*08 (present in 7% of participants) and cutaneous or hepatic adverse events.
Associations with CYP2B6 and HLA combinations
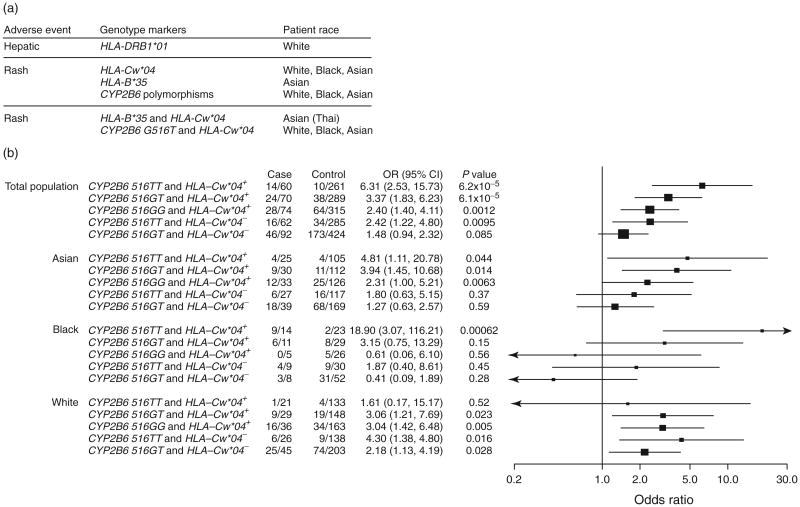
Major associations with genetic polymorphism in CYB2B6 and HLA are summarized in Fig. 4a. Relationships between HLA-Cw*04 and CYP2B6 516G→T and effects of clinical variables, including age, sex, CD4 T-cell count, race, plasma HIV-1 RNA, and antiretroviral history, were examined. By logistic regression analysis that adjusted for HLA-Cw*04 and CYP2B6 516G→T, the only clinical variable associated with cutaneous adverse events was prior antiretroviral status (OR 0.57 for antiretroviral experienced, 95% CI 0.37–0.87; P= 0.0097; data not shown), particularly history of abacavir use in Whites (P= 0.010; data not shown), which was independent of HLA-Cw*04 and CYP2B6 516G→T. History of use for stavudine were not associated with cutaneous or hepatic adverse events. Patients carrying both CYP2B6 516TT and HLA-Cw*04 were at highest risk in the total population (OR 6.31, 95% CI 2.53–15.73), and in Blacks and Asians analyzed separately (Fig. 4b).
Fig. 4.
Association between polymorphisms and related adverse events. Panel (a) shows major association between genetic polymorphisms and nevirapine-related adverse events (AEs). CYP2B6 polymorphisms include rs2054675, rs3786547, and rs3745274. Panel (b) shows associations between CYP2B6 516G→T, HLA-Cw*04, and cutaneous AEs. The lowest risk genotype (CYP2B6 516GG and lacking HLA-Cw*04) was the reference for calculating odds ratios (OR) and two-tailed P values by Fisher's exact test for race groups and by Cochran-Mantel-Haenszel test for total population. A continuity correction was made to calculate OR by adding one to all cells of a contingency table if a cell harbours zero. Cases and controls are presented as numbers of participants with the indicated genotypes divided by the numbers of cases or controls. Height of a square is proportional to sample size. Horizontal lines represent 95% confidence intervals (CIs).
Among Asians, the three-way combination of HLA-Cw*04, HLA-B*35, and CYP2B6 516G→T was too infrequent to evaluate. Our matching of cases and controls on CD4 T-cell count prevented direct assessment of its relationship with adverse events. In post hoc analyses stratified by CD4 T-cell count, the latter did not contribute to adverse event risk in the presence of risk HLA alleles or SNPs.
Concomitant hepatic and cutaneous adverse events
Among the 276 evaluable cases, 74 were known to have had concomitant hepatic and cutaneous adverse events. We repeated the above analyses, limited to this subgroup of cases. After correcting for multiple comparisons, none of the above HLA alleles or SNP associations was significantly associated in this subgroup of cases, and no additional significant associations were identified (data not shown).
Discussion
The present study characterizes associations between genetic variants and symptomatic hepatic and cutaneous reactions among HIV-1 patients who had initiated nevirapine with relatively high CD4 T-cell counts (>150 cells/μl). We convincingly demonstrated that polymorphisms in CYP2B6 and multiple HLA loci were associated with a higher likelihood of risk for nevirapine-related adverse events. We also showed that genetic predictors of cutaneous versus hepatic adverse events differ, and that relationships between genetic variants and adverse events vary by race.
This is the first study that CYP2B6 variants affect risk for nevirapine-associated adverse events. Major metabolic pathways for nevirapine involve hydroxylation by CYP2B6 and CYP3A4 [22,23]. In addition, nevirapine induces CYP2B6 and CYP3A4 expression over several weeks, increasing its own clearance [23]. Previous studies associated CYP2B6 516G→T with increased plasma nevirapine concentrations [9–15], but separate analyses found no relationship between plasma nevirapine concentrations and adverse events [24]. It is possible that previous analyses missed a true association with plasma nevirapine concentrations. Alternatively, a metabolite, rather than parent compound, may mediate toxicity. With loss of CYP2B6 function, more nevirapine may be shunted through pathways that generate minor metabolites. Consistent with this hypothesis, a rodent model implicated a quinine methide of nevirapine formed in the skin following sulfation of the 12-hydroxy metabolite [25]. Among 129 SNPs in 11 sulfotransferase and sulfatase genes assayed herein, none were significantly associated with adverse events. We found no association between CYP2B6 variants and hepatic adverse events, consistent with limited previous data [7,8] and with a rat model [26].
Cutaneous adverse events were associated with MHC class I alleles (HLA-B*35 and HLA-Cw*04) and hepatic adverse events with MHC class II alleles (HLA-DRB1*01 and possibly HLA-DQB1*05). These findings suggest distinct mechanisms with CD8 T cells (i.e. MHC class I) possibly more important for cutaneous adverse events, and CD4 T cells (i.e. MHC class II) more important for hepatic adverse events. The increased ORs observed when concomitant hepatitis and rash cases were excluded support this concept. Without more direct evidence, however, this hypothesis is tentative, as HLA associations do not prove causation. We suspect that associations with other polymorphisms in the MHC locus reflect linkage with risk HLA alleles.
Our results support the association between HLA-B*3505, HLA-Cw*0401, and nevirapine-associated cutaneous adverse events previously reported among Thai populations [16,17]. In analyses comprising 147 cases and 185 controls, Chantarangsu et al. [17] identified an association between HLA-B*3505 and nevirapine-associated cutaneous adverse events (OR 18.96); also predictive were HLA-Cw*0401 (OR 5.36) and the HLA-Cw*0401-HLA-B*3501 haplotype (OR 12.12). Similarly, Likanonsakul et al. [16] reported an association between HLA-Cw*04 and cutaneous adverse events among 39 cases and 60 controls. Our data confirmed strong associations between HLA-B*35, HLA-Cw*04, and cutaneous adverse events among Asians, particularly in Thai participants, and extended the HLA-Cw*04 association to Black and White populations. The study results also showed that among HLA-Cw*04 carriers, cutaneous adverse events were increased with concomitant CYP2B6 516G→T. In contrast, this study did not find significant associations with HLA-B*35 in Blacks and Whites, perhaps reflecting its infrequency in these populations [27].
The results of our study also support the association between HLA-DRB1*0101 and nevirapine-associated adverse events reported in the Western Australian HIV Cohort [4]. That study included 26 cases (25 Whites) who experienced ALT elevation, fever, and/or rash with nevirapine and 209 controls. HLA-DRB1*0101 was significantly associated with hepatic/systemic reactions (OR 4.8), but not with isolated cutaneous adverse events. The association was only apparent in patients with at least 25% CD4 T cells [4]. The current study confirmed the association between HLA-DRB1*01 and hepatic adverse events among Whites. We did not find any significant association with HLA-DRB1*01 in Blacks and Asians, perhaps because HLA-DRB1*01 is infrequent in these populations [27]. Results also indicated an association between HLA-DQB*05 and hepatic adverse events in Whites, likely reflecting linkage between HLA-DRB1*01 and HLA-DQB1*05 [27]. However, among individuals with isolated hepatic adverse events the association with HLA-DQB*05 was no longer significant, whereas the HLA-DRB1*01 association increased.
This analysis did not replicate findings from studies in Sardinia and Japan that suggested an association between HLA-Cw*08 and nevirapine-associated hypersensitivity [5,6]. Two previous studies involving individuals of African descent suggested that ABCB1 3435C→T was associated with decreased nevirapine-associated hepatic adverse events [5,6]. Whereas we found no statistically significant association between ABCB1 3435C→T and nevirapine-associated adverse events, the ORs for ABCB1 3435C→T and hepatic adverse events among Blacks was consistent with previous studies. There was no such association in Asians or Whites despite increased T-allele frequencies.
Our study had limitations. Matching on CD4 T-cell count, sex, and race allowed more efficient discovery of genetic associations, but prevented assessment of these variables independently. Excluding individuals with mild or equivocal adverse events prevented us from assessing whether the same genetic variants predict less severe events. The retrospective design excluded individuals who died from any cause and limited the types of data we could reliably collect. The associations identified cannot be generalized to individuals with less than 150 CD4 T cells/μl, an exclusion criterion in our study. Although we performed extensive genotyping focused on ADME genes and MHC region, important HLA and non-HLA polymorphisms may have been missed. High-resolution HLA typing would likely increase ORs and specificities for at least some associations identified, but would likely not increase sensitivity.
Clinically useful markers for serious adverse events must be highly sensitive. Unfortunately, the genetic variants identified herein lacked sensitivity, and therefore have little or no clinical utility for patients initiating nevirapine-containing regimens. Among Asians with cutaneous adverse events, 80% were HLA-B* 35-negative; among all participants with cutaneous adverse events, 62% were HLA-Cw* 04-negative; and among Whites with hepatic adverse events, 56% were HLA-DRB1*01-negative. Among all participants with any adverse event, 48% had neither an HLA risk allele (HLA-Cw*04, -B*35, -DRB1*01) nor homozygosity for CYP2B6 516TT This suggests additional, not yet identified risk factors. It is almost certain that some adverse events in the present study were unrelated to nevirapine, further limiting our ability to assess how effectively genetic testing would prevent true nevirapine reactions.
Nevirapine is generally well tolerated when initiated in patients who are treatment-naïve and have lower CD4 T-cell counts (<250 cells/μl in women, <400 cells/μl in men) [2]. These cut-offs may reduce nevirapine-related symptomatic hepatotoxicity to approximately 1% [28]. We show that both ADME and HLA variants affect risk for nevirapine-associated adverse events. These genetic variants, however, identified only a minority of adverse event cases. Continued search for additional genetic predictors is warranted, as are studies of underlying pathogenic mechanisms.
Supplementary Material
Acknowledgments
The work was sponsored by Boehringer Ingelheim Pharmaceuticals, Inc. We are grateful to the study volunteers, clinical investigators, and site personnel. We thank Olimpia Disorber for assistance with sample collection and DNA processing; Thorsten Lehr, Vijo Joseph, Iosif Yuabov, and Yirong Wang for program and database design and informatics contributions; George Austen and Pek-Ju Hu for clinical data collection and database; Tom MacGregor for scientific advice and helpful discussion; and Warren Ku, Kerry Blanchard, Holger Gellermann, and Peter Piliero for project management and manuscript review. We thank all members of Non-Clinical Drug Safety and Medical Affairs, Boehringer Ingelheim Pharmaceuticals, for their contributions towards study completion. We also thank UBC-Envision Group, Southport, Connecticut, for editorial and formatting assistance with this manuscript.
Source(s) of support: This work was sponsored by Boehringer Ingelheim Pharmaceuticals, Inc. The sponsor was involved in study design, data collection, analysis, interpretation, and study reporting. Additional support to D.W.H. was through grants AI077505 and AI54999 from National Institute of Allergy and Infectious Diseases, and to K.R. was through a professional researcher-strengthening grant from the National Science and Technology Development Agency (NSTDA), Pathumthani, Thailand.
Disclaimers: J.Y., S.G., S.J., A.M.C., D.H., S.S., and Z.H. are employees of Boehringer Ingelheim Pharmaceuticals, Inc. M.D. is an employee of Boehringer Ingelheim GmbH. K.R. has been involved in a Boehringer Ingelheim GmbH-sponsored study. D.P. has received research grants and/or honoraria for advisories/conferences from pharmaceutical companies that manufacture antiretroviral drugs, including Boehringer Ingelheim. D.W.H. has received research grants from Bristol-Myers Squibb, Boehringer Ingelheim, Merck & Co., and Gilead Sciences, and performs ad hoc consulting to Boehringer Ingelheim.
We thank the following investigators and personnel of the Nevirapine Toxicogenomics Study Team, who enrolled participants at clinical sites: Jorge Benetucci, Gerardo Ortega (Fundación de Ayuda al Inmunodeficiente (FUNDAI) Ciudad de Buenos Aires, Argentina); Pedro Cahn, Carina Cesar (Fundacion Huesped, Buenos Aires, Argentina); Isabel Cassetti, Emiliano Bissio (FUNCEI, Buenos Aires, Argentina); Sergio Lupo (Instituto CAICI, Rosario, Argentina); John Chuah (Gold Coast Sexual Health Clinic, Queensland, Australia); Cassy Workman, Vanessa Rees (Ground Zero Medical Centre, Sydney, Australia); David A Cooper, Rebecca Hickey (St Vincent's Hospital, Sydney, Australia); Jonathan Anderson, Richard Moore (Carlton Clinic, Melbourne, Australia); Jennifer Hoy, Cath Downs (The Alfred Hospital, Melbourne, Australia); Robert Finlayson, Neil Bodsworth (Taylor Square Private Clinic, Sydney, Australia); Beng Eu, Helen Lau (Prahran Market Clinic, Victoria, Australia); Julio Montaner, Marianne Harris (BC Centre for Excellence in HIV/AIDS, at Providence Healthcare and the University of British Columbia, Vancouver, BC, Canada); Sharon Walmsley, Adrianna d'Aquila (University Health Network, University of Toronto, Toronto, Canada); Brian Conway, Harout Tossonian (University of British Columbia, Vancouver, BC, Canada); Philippe Morlat, Isabelle Louis (Hôpital Saint André; University Hospital of Bordeaux, Bordeaux, France); Yazdan Yazdanpanah, Faiza Ajana (Centre Hospitalier de Tourcoing Hospital, Tourcoing, France); Diane Bollens, Pierre-Marie Girard (Hôpital St Antoine, Paris, France); Thierry May (Centre Hospitalier Universitaire de Nancy, Nancy, France); Gilles Pialoux, Laurence Slama, Thomas Lyavanc (Unit of Infectious Diseases Hopital Tenon, APHP, Paris, France); Christophe Piketty, Christel Pluot (Hôpital Européen Georges Pompidou, Paris, France); Laurent Cotte, Patrick Miailhes (Hôtel-Dieu Hospital, Hospices Civils de Lyon, Lyon, France); Veronique Reliquet, François Raffi (University Hospital, Nantes, France); Bao Phung, Patrick Yeni (Hopital Bichat, Paris, France); Anne Simon, Manuela Bonmarchand (Pitié Salpétriére Hospital, Paris, France); Diane Ponscarme, Willy Rozenbaum(UniversityHospital Saint-Louis, Paris, France); Muriel Alvarez, Bruno Marchou (Hôpital Purpan, Cedex, France); Andreas Plettenberg, Albrecht Stoehr (Institut für Interdisziplinäre Medizin an der Asklepios Klinik St. Georg, Hamburg, Germany); Jürgen Rockstroh, Carolynne Schwarze-Zander (Oberarzt an der Medizinischen Universitätsklinik Innere-Rheuma-Tropen Ambulanz, Bonn-Venusberg, Germany);Norbert H Brockmeyer(German Competence Network for HIV/AIDS, Ruhr-University, Bochum, Germany); Hartwig Klinker, Werner Heinz (Universität Würzburg, Würzburg, Germany); Christian Hoffmann (ICH Mitte, Infektionsmedizinisches Centrum, Hamburg, Germany); Frank Bergmann, Dirk Schuermann (Charité-Universitätsmedizin Berlin, Berlin, Germany); Thomas Lutz, Peter Gute (Infektiologikum Frankfurt, Frankfurt, Germany); Keikawus Arastéh, Jukka Hartikainen, Michael Rittweger (EPIMED c/o Vivantes Auguste-Viktoria-Klinikum, Berlin, Germany); Peter Kern, Georg Härter (University Hospital and Medical Center, Ulm, Germany); Stefan Reuter, Björn Jensen (University Hospital Düsseldorf, Düsseldorf, Germany); Stefan Esser, Robert Jablonka (University Hospital Essen, Essen, Germany); Karin Morbach (MUC Research, Munich, Germany); Eva Jaegel-Guedes (MVZ Karlsplatz – HIV Research and Clinical Care Centre, Munich, Germany); Peter Reiss, Guido van den Berk, Judith Branger (Academic Medical Center, University of Amsterdam, Amsterdam, the Netherlands); Kees Brink-man (Onze Lieve Vrouwe Gasthuis, Amsterdam, the Netherlands); Jordi Puig, Eugénia Negredo (Fundacio de la Lluita contra la SIDA, Hospital Universitàri ‘Germans Trias i Pujol’ Badalona, Catalonia, Spain); Pompeyo Viciana, Luis Lopez-Cortes (Universitary Hospital Virgen del Rocío, Seville, Spain); Ignacio De Los Santos, Jesús Sanz (Hospital Universitario de la Princesa, Instituto de Investigación de la Princesa, Madrid, Spain); Josep Mallolas, Montserrat Laguno (Hospital Clínic-IDIBAPS, Barcelona, Spain); Esteban Ribera, Sara Villar del Saz (Hospital Universitari Vall d'Hebron, Universitat Auton-oma de Barcelona, Barcelona, Spain); Jorge Vergas, M. Jesús Téllez (Hospital Clinico S. Carlos, Madrid, Spain); Juan Carlos López, Matilde Sanchez-Conde (Hospital General Universitario Gregorio Marañon, Madrid, Spain); Pere Domingo (Hospital de la Santa Creu i Sant Pau, Univeristy of Barcelona, Spain); José L. Casado, María Pumares (Ramon y Cajal Hospital, Madrid, Spain); Hsi-Hsun Lin (E-Da Hospital/I-Shou University, Kaohsiung City, Taiwan); Yung-Ching Liu (School of Medicine, Taipei Medical University, Shuang-Ho Hospital, Taipei, Taiwan); Yung-Hsing Wang, Hung-Chin Tsai (Kaohsiung Veterans General Hospital, Kaohsiung, Taiwan); Jen-Hsien Wang (China Medical University Hospital, Taichung, Taiwan); Wing Wai Wong, Bor-Shen Hu (Taipei City Hospital, Taipei Veterans General Hospital, and College of Medicine, National Yang Ming University, Shipai, Taipei, Taiwan); Szu-Min Hsieh, Wang-Huei Sheng (National Taiwan University Hospital and Medical College of National Taiwan University, Taipei, Taiwan); Yu-Hui Lin (Taichung Veteran General Hospital, Taichung, Taiwan); Po-Liang Lu, Tun-Chieh Chen (Kaohsiung Medical University Hospital, Kaohsiung Medical University, Kaohsiung, Taiwan); Anchalee Avihingsanon (Thai Red Cross AIDS Research Center, Chulalongkorn University, Thailand); Thanomsak Anekthananon, Winai Ratanasuwan (Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand); Cheryl Tawana, Simon Limb (Newham University Hospital NHS Trust, London, UK); Francisco Javier Vilar, Yvonne Clowes (North Manchester General Hospital, Manchester, UK); Sarah Barrett, Stephen Taylor (Birmingham Heartlands Hospital, Birmingham, UK); Sris Allan, Justine Barnes (Coventry and Warwickshire Hospital, Coventry, UK); Martin Fisher, Nicky Perry (Brighton and Sussex University Hospitals, Brighton, UK); Anton L.Pozniak, Marta Boffito (SSAT, Chelsea and Hospital NHS, Foundation Trust, London, UK); Tariq Sadiq, Ade Adebiyi (The Courtyard Clinic, St George's Healthcare NHS Trust, London. UK); Charles Davis, Robert R Redfield (University of Maryland, Baltimore, MD, USA); MichaelJ Kozal, Laurie Andrews (Yale School of Medicine, New Haven, CT, USA); Mark Miller, Barbara Johnston (Mount Sinai Medical Center, New York, NY, USA); Karen Savage, Michael Saag (University of Alabama at Birmingham Center for AIDS Research, Birmingham, AL, USA); Howard A Grossman, Daniel E Cohen (Fenway Community Health, Boston, MA, USA); Edgar Turner Overton, Mariea B. Snell, (Washington University School of Medicine, St. Louis, MO, USA); William Burman, Dale Britt (Denver Public Health, Denver, CO, USA); Claudia Martorell, Jacqueline Rivera (The Research Institute, Springfield, MA, USA); Joseph J Eron, Jr., Prema Menezes (University of North Carolina at Chapel Hill, Chapel Hill, NC, USA).
References
- 1.Leith J, Piliero P, Storfer S, Mayers D, Hinzmann R. Appropriate use of nevirapine for long-term therapy. J Infect Dis. 2005;192:545–546. doi: 10.1086/431606. [DOI] [PubMed] [Google Scholar]
- 2.Boehringer-Ingelheim Pharmaceuticals Inc. Viramune( (nevirapine) prescribing information. [Accessed 31 January 2011];2009 http://bidocs.boehringer-ingelheim.com/BIWebAccess/ViewServlet.ser?docBase=re-netnt&folderPath=/Prescribing+Information/PIs/Viramune/Vira-mune.pdf.
- 3.Hoskins JM, Roederer MW, McLeod HL. Nevirapine hypersen-sitivity reaction: will the real HLA please stand up? Curr Opin Mol Ther. 2009;11:226–230. [PubMed] [Google Scholar]
- 4.Martin AM, Nolan D, James I, Cameron P, Keller J, Moore C, et al. Predisposition to nevirapine hypersensitivity associated with HLA-DRB1M0101 and abrogated by low CD4 T-cell counts. AIDS. 2005;19:97–99. doi: 10.1097/00002030-200501030-00014. [DOI] [PubMed] [Google Scholar]
- 5.Gatanaga H, Yazaki H, Tanuma J, Honda M, Genka I, Teruya K, et al. HLA-Cw8 primarily associated with hypersensitivity to nevirapine. AIDS. 2007;21:264–265. doi: 10.1097/QAD.0b013e32801199d9. [DOI] [PubMed] [Google Scholar]
- 6.Littera R, Carcassi C, Masala A, Piano P, Serra P, Ortu F, et al. HLA-dependent hypersensitivity to nevirapine in Sardinian HIV patients. AIDS. 2006;20:1621–1626. doi: 10.1097/01.aids.0000238408.82947.09. [DOI] [PubMed] [Google Scholar]
- 7.Haas DW, Bartlett JA, Andersen JW, Sanne I, Wilkinson GR, Hinkle J, et al. Pharmacogenetics of nevirapine-associated hepatotoxicity: an adult AIDS Clinical Trials Group collaboration. Clin Infect Dis. 2006;43:783–786. doi: 10.1086/507097. [DOI] [PubMed] [Google Scholar]
- 8.Ciccacci C, Borgiani P, Ceffa S, Sirianni E, Marazzi MC, Altan AM, et al. Nevirapine-induced hepatotoxicity and pharmacogenetics: a retrospective study in a population from Mozambique. Pharmacogenomics. 2010;11:23–31. doi: 10.2217/pgs.09.142. [DOI] [PubMed] [Google Scholar]
- 9.Saitoh A, Sarles E, Capparelli E, Aweeka F, Kovacs A, Burchett SK, et al. CYP2B6 genetic variants are associated with nevir-apine pharmacokinetics and clinical response in HIV-1-infected children. AIDS. 2007;21:2191–2199. doi: 10.1097/QAD.0b013e3282ef9695. [DOI] [PubMed] [Google Scholar]
- 10.Chantarangsu S, Cressey TR, Mahasirimongkol S, Capparelli E, Tawon Y, Ngo-Giang-Huong N, et al. Influence of CYP2B6 polymorphisms on the persistence of plasma nevirapine concentrations following a single intra-partum dose for the prevention of mother to child transmission in HIV-infected Thai women. J Antimicrob Chemother. 2009;64:1265–1273. doi: 10.1093/jac/dkp351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahungu T, Smith C, Turner F, Egan D, Youle M, Johnson M, et al. Cytochrome P450 2B6 516G–>T is associated with plasma concentrations of nevirapine at both 200mg twice daily and 400mg once daily in an ethnically diverse population. HIV Med. 2009;10:310–317. doi: 10.1111/j.1468-1293.2008.00689.x. [DOI] [PubMed] [Google Scholar]
- 12.Penzak SR, Kabuye G, Mugyenyi P, Mbamanya F, Natarajan V, Alfaro RM, et al. Cytochrome P450 2B6 (CYP2B6) G516T influences nevirapine plasma concentrations in HIV-infected patients in Uganda. HIV Med. 2007;8:86–91. doi: 10.1111/j.1468-1293.2007.00432.x. [DOI] [PubMed] [Google Scholar]
- 13.Wyen C, Hendra H, Vogel M, Hoffmann C, Knechten H, Brockmeyer NH, et al. Impact of CYP2B6 983T>C polymorphism on nonnucleoside reverse transcriptase inhibitor plasma concentrations inHIV-infected patients. J Antimicrob Chemother. 2008;61:914–918. doi: 10.1093/jac/dkn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rotger M, Colombo S, Furrer H, Bleiber G, Buclin T, Lee BL, et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and ne-virapine in HIV-infected patients. Pharmacogenet Genomics. 2005;15:1–5. doi: 10.1097/01213011-200501000-00001. [DOI] [PubMed] [Google Scholar]
- 15.Chou M, Bertrand J, Segeral O, Verstuyft C, Borand L, Comets E, et al. Population pharmacokinetic-pharmacogenetic study of nevirapine in HIV-infected Cambodian patients. Antimicrob Agents Chemother. 2010;54:4432–4439. doi: 10.1128/AAC.00512-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Likanonsakul S, Rattanatham T, Feangvad S, Uttayamakul S, Prasithsirikul W, Tunthanathip P, et al. HLA-CwM04 allele associated with nevirapine-induced rash in HIV-infected Thai patients. AIDS Res Ther. 2009;6:22. doi: 10.1186/1742-6405-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chantarangsu S, Mushiroda T, Mahasirimongkol S, Kiertibur-anakul S, Sungkanuparph S, Manosuthi W, et al. HLA-BM3505 allele is a strong predictor for nevirapine-induced skin adverse drug reactions in HIV-infected Thai patients. Pharmacogenet Genomics. 2009;19:139–146. doi: 10.1097/FPC.0b013e32831d0faf. [DOI] [PubMed] [Google Scholar]
- 18.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 19.National Institute of Allergy and Infectious Diseases (NIAID) Division of AIDS (DAIDS) Division of AIDS table for grading severity of adult adverse experiences. [Accessed 31 January 2011];1992 http://rsc.tech-res.com/Document/safetyandpharmacovigilance/Table_for_Grading_Severity_of_Adult_AEs_v01a.pdf.
- 20.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. [Accessed 28 January 2011];Am J Human Genet. 2007 81:559–575. doi: 10.1086/519795. http://pngu.mgh.harvard.edu/purcell/plink/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S. PLINK version 1.06. [Accessed 28 January 2011];2009 http://pngu.mgh.harvard.edu/purcell/plink/
- 22.Erickson DA, Mather G, Trager WF, Levy RH, Keirns JJ. Characterization of the in vitro biotransformation of the HIV-1 reverse transcriptase inhibitor nevirapine by human hepatic cytochromes P-450. Drug Metab Dispos. 1999;27:1488–1495. [PubMed] [Google Scholar]
- 23.Riska P, Lamson M, MacGregor T, Sabo J, Hattox S, Pav J, et al. Disposition and biotransformation of the antiretroviral drug nevirapine in humans. [Accessed January 31,2011];Drug Metab Dispos. 1999 27:895–901. [PubMed] [Google Scholar]
- 24.Kappelhoff BS, van Leth F, Robinson PA, MacGregor TR, Baraldi E, Montella F, et al. Are adverse events of nevirapine and efavirenz related to plasma concentrations? Antivir Ther. 2005;10:489–498. [PubMed] [Google Scholar]
- 25.Chen J, Mannargudi BM, Xu L, Uetrecht J. Demonstration of the metabolic pathway responsible for nevirapine-induced skin rash. Chem Res Toxicol. 2008;21:1862–1870. doi: 10.1021/tx800177k. [DOI] [PubMed] [Google Scholar]
- 26.Walubo A, Barr S, Abraham AM. RAT CYP3A and CYP2B1/2 were not associated with nevirapine-induced hepatotoxicity. Methods Find Exp Clin Pharmacol. 2006;28:423–431. doi: 10.1358/mf.2006.28.7.1003580. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. [Accessed 31 January 2011];Nucleic Acid Res. 2011 39:D913–D919. doi: 10.1093/nar/gkq1128. http://www.allelefrequencies.net. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dieterich DT, Robinson PA, Love J, Stern JO. Drug-induced liver injury associated with the use of nonnucleoside reverse-transcriptase inhibitors. Clin Infect Dis. 2004;38(2):S80–S89. doi: 10.1086/381450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.