Abstract
Progressive telomere shortening from cell division (replicative aging) provides a barrier for human tumor progression. This program is not conserved in laboratory mice, which have longer telomeres and constitutive telomerase. Wild species that do / do not use replicative aging have been reported, but the evolution of different phenotypes and a conceptual framework for understanding their uses of telomeres is lacking. We examined telomeres / telomerase in cultured cells from > 60 mammalian species to place different uses of telomeres in a broad mammalian context. Phylogeny-based statistical analysis reconstructed ancestral states. Our analysis suggested that the ancestral mammalian phenotype included short telomeres (< 20 kb, as we now see in humans) and repressed telomerase. We argue that the repressed telomerase was a response to a higher mutation load brought on by the evolution of homeothermy. With telomerase repressed, we then see the evolution of replicative aging. Telomere length inversely correlated with lifespan, while telomerase expression co-evolved with body size. Multiple independent times smaller, shorter-lived species changed to having longer telomeres and expressing telomerase. Trade-offs involving reducing the energetic / cellular costs of specific oxidative protection mechanisms (needed to protect < 20 kb telomeres in the absence of telomerase) could explain this abandonment of replicative aging. These observations provide a conceptual framework for understanding different uses of telomeres in mammals, support a role for human-like telomeres in allowing longer lifespans to evolve, demonstrate the need to include telomere length in the analysis of comparative studies of oxidative protection in the biology of aging, and identify which mammals can be used as appropriate model organisms for the study of the role of telomeres in human cancer and aging.
Keywords: evolution of telomeres, immortalization, telomerase, replicative aging, senescence
Introduction
The ends of human chromosomes (telomeres) consist of many kilobases of the repeating sequence TTAGGG, ending in approximately 30–600 nucleotides of single-stranded G-rich repeats (Moyzis et al., 1988; Makarov et al., 1997; Chai et al., 2005). The combination of incomplete replication and processing events shortens telomeres with each cell division. This shortening is prevented in the germline and certain stem cells (Lee et al., 1998) by the presence of telomerase. Telomerase is a ribonucleoprotein in which the RNA component hTR (hTERC) (Feng et al., 1995) contains a C-rich sequence that serves as a template for the reverse transcriptase activity of the protein catalytic subunit hTERT (Meyerson et al., 1997; Nakamura et al., 1997). Telomerase reverse transcribes this template to add GGTTAG to the end of the chromosome, where it pauses, repositions the template, then adds additional repeats (Greider & Blackburn, 1989; Blackburn, 1992). Telomerase activity is repressed in most human somatic tissues during development (Wright et al., 1996), after which subsequent cell divisions produce telomere shortening. When telomeres become short enough to produce inadequate telomeric end protection, they generate a DNA damage signal, which causes the growth arrest known as replicative aging in normal cells (Shay & Wright, 2005) and crisis in cancer cells.
It is now widely accepted that replicative aging serves at least partly as a brake against malignancy. Cancer cells require multiple mutations to become malignant (Armitage & Doll, 1954; Luebeck & Moolgavkar, 2002). Each mutation probably uses 20–40 divisions before achieving a population size sufficient for another mutation to occur, so premalignant cells usually come up against the barrier of replicative senescence before accumulating enough mutated pathways to become invasive. One critical step in oncogenesis involves the upregulation or reactivation of telomerase to overcome this limit, and approximately 85–90% of all tumor biopsies are telomerase positive (Shay & Bacchetti, 1997).
There is now substantial evidence that laboratory mice, which have 25–150 kb telomeres compared to 15–20 kb germline human telomeres, do not use replicative aging to ‘count’ cell divisions (Sherr & DePinho, 2000; Wright & Shay, 2000). A comparative study of rodents concluded there was an inverse correlation between levels of telomerase expression and body size (Gorbunova & Seluanov, 2009) but no correlation between telomere length and lifespan. We have shown that primates and Indian muntjac deer have short human-like telomeres and use replicative aging, while lagomorphs have very long telomeres and do not use telomeres to limit the number of cell divisions (Steinert et al., 2002; Zou et al., 2002; Forsyth et al., 2005). To provide a more comprehensive analysis of the phylogenetic distribution of the use of telomere shortening as a tumor-protection mechanism and the role of replicative aging, we undertook a systematic survey of telomere biology covering the mammalian evolutionary tree. A phylogenic-based statistical analysis allowed us to reconstruct the ancestral placental mammalian phenotype as having short humanlike telomeres and repressed telomerase, define multiple transitions to having long telomeres and not repressing telomerase, and provide evidence for one trade-off that could provide compensatory advantages to abandoning replicative aging. Although replicative aging is a key tumor-protection mechanism in humans and many other species, these results put the role of telomeres in cancer and aging in a much broader comparative context needed to appreciate the different functions of telomeres in mammalian biology.
Results
Phylogenetic distribution of telomere phenotypes
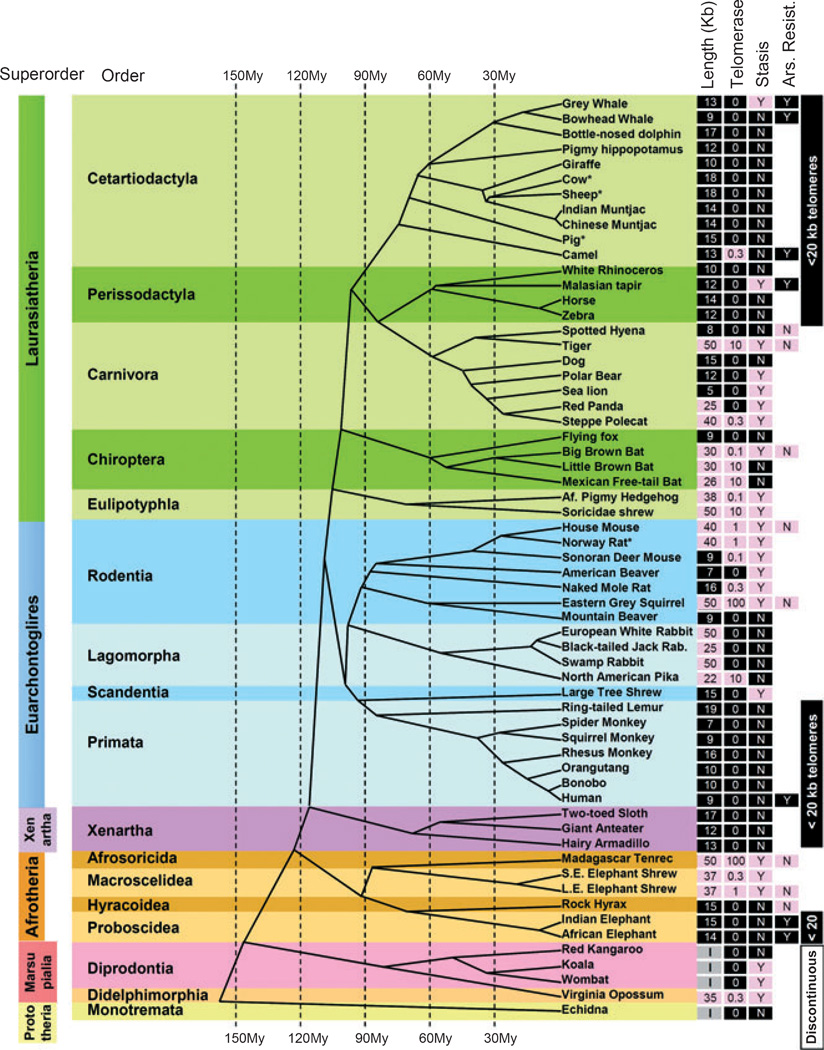
Telomerase expression in cultured fibroblasts from adult donors was used to determine the strength of telomerase repression in somatic mesenchymal cells. Telomere length and telomerase expression from 43 species as well as an additional 18 previously studied species is arrayed on a mammalian phylogeny in Fig. 1 (see Table S1 and Fig. S1, Supporting Information for detailed data for each species). Cells cultured under suboptimal conditions (e.g., lack of a micronutrient, atmospheric rather than physiologic oxygen, other unknown factors) frequently exhibit a growth arrest called stasis (stress or aberrant signaling induced senescence) (Drayton & Peters, 2002) that is independent of telomere shortening. The presence of an early stasis (within 15 doublings) and resistance to the oxidative damage agent sodium arsenite also provided additional phenotypes.
Fig. 1.
Evolutionary distribution of telomere length, telomerase activity and stasis (stress or aberrant signaling induced senescence) in the mammalian evolutionary tree. Telomere length, telomerase activity, and stasis induction run in evolutionary clades. Telomere length was measured by terminal restriction fragment (TRF) analysis: Values < 20 kb are shaded black, while values > 20 kb are shaded pink. Nonplacental mammalian orders such as Marsupials and Monotremes show the presence of restriction enzyme recognition sites intercalated between the telomeric (TTAGGG)n sequences and thus are labeled as being of indeterminate size (I). Telomerase activity was detected with TRAP: Values are expressed as a%of the activity in the reference lung adenocarcinoma tumor line H1299, and shaded black if absent and pink if any activity was detected. Stasis: (N) with black shading = cells grew beyond 15 doublings, (Y) with pink shading = cells growth arrested in culture before population doubling 15. Based on these characteristics, some taxonomic orders could be described as exhibiting uniformly human-like telomere phenotypes or having discontinuous telomeres. Arsenite resistance: (Y) with black shading = LD90 > 20 mm sodium arsenite after 4 h exposure, (N) with pink shading = LD90 < 5 mm (see Fig. 5). Published data on Primates (Steinert et al., 2002), Lagomorphs (Forsyth et al., 2005), and Muntjacs (Zou et al., 2002) are from our laboratory, and thus are directly comparable. Data from other laboratories are indicated by (*) [cow (Lanza et al., 2000), sheep (Cui et al., 2002), pig (Fradiani et al., 2004; Oh et al., 2007), rat (Mathon et al., 2001)]. Scientific names and specific data for each species (scientific name, growth curves, TRF gels, telomerase assays, mass, lifespan) are provided in Table S1 and Fig. S1 (Supporting Information). The telomere lengths for giraffe, rhinoceros, and anteater are adjusted for a large digestion-resistant subtelomeric region using the rate of disappearance of the telomeric signal with increased cell doublings (see Fig. S1, Supporting Information). Cladogram adapted from Bininda-Emonds et al. (2007, 2008).
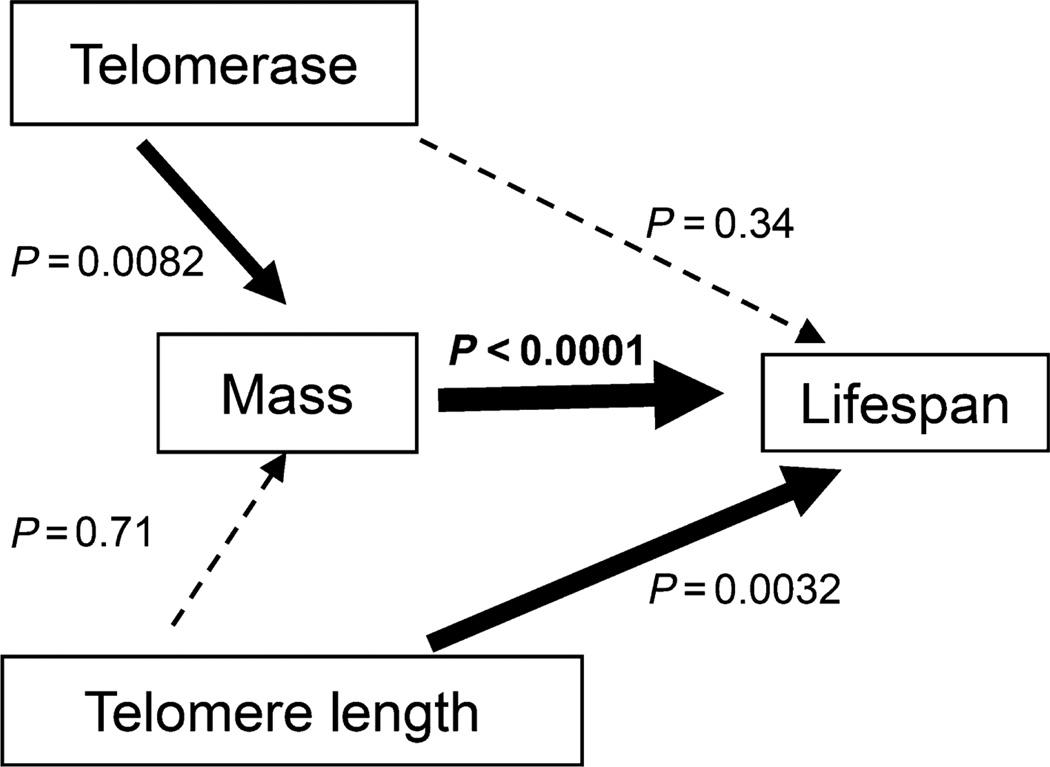
We examined telomerase expression, telomere length, body size and lifespan using regression models which account for the shared ancestry as implied by the phylogeny within the phylogenetic generalized least squares (PGLS) framework (Pagel, 1999). Telomerase expression significantly co-evolved with body size (P = 0.0082), but showed no independent effects on lifespan (P = 0.34). Telomere length significantly co-evolved with lifespan (P = 0.0032) over and above what we would expect from body size alone, but there was no independent association of telomere length with size (P = 0.71) (Fig. 2).
Fig. 2.
Relationship of telomeres and telomerase to mass and lifespan. Dark arrows indicate significance while dotted arrows show lack of significance. Phylogenetic generalized least squares framework (PGLS) analysis demonstrates that telomerase expression inversely co-evolves with increased mass, while telomere length inversely correlates with increased lifespan.
Ancestral placental mammals had human-like telomeres and repressed telomerase
The ancestral telomere length at the base of the placental mammals was reconstructed using maximum likelihood models that yield the most likely value at the root of a given phylogenetic tree under the evolutionary model of Brownian motion, along with the parameter ‘lambda’, which measures the strength of the phylogenetic signal (Freckleton et al., 2002; Pagel et al., 2004; Organ et al., 2009). The ancestral state was determined to be 18.6 kb with a lambda equal to 1, indicating a very strong phylogenetic signal. A Markov-transition approach (Pagel et al., 2004) was used to estimate the ancestral state for the expression of telomerase. The probability that ancestral placental mammals repressed telomerase was calculated to be high (0.998) compared to the probability that it was expressed (0.002). These results remained qualitatively the same even if each order was analyzed separately (Table S2, Supporting Information). The transition rate from expressed to repressed was estimated to be close to zero, meaning that only transitions from repressed to expressed occurred.
Nonplacental mammals have discontinuous telomeres
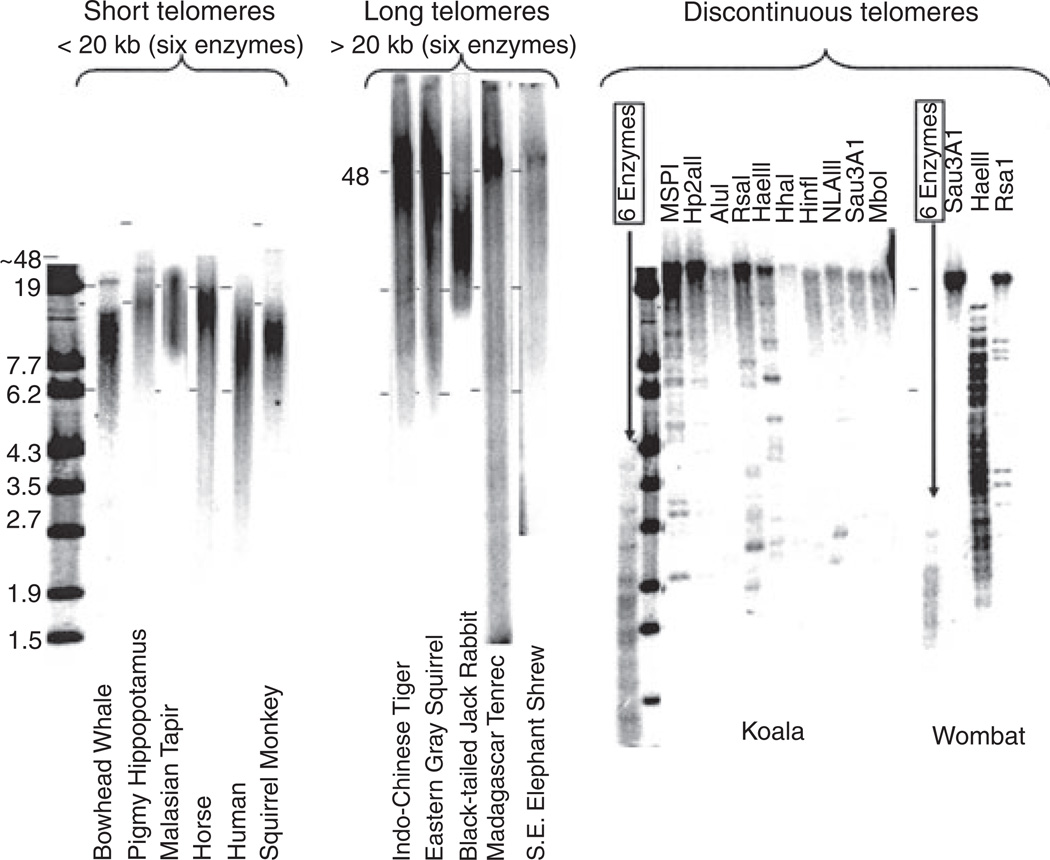
The nonplacental mammals are generally considered to express the ancestral mammalian state, but we were unable to unambiguously determine their telomere length phenotype. The sequence TTAGGG lacks restriction sites. After digesting genomic DNA with a mixture of four-base recognition restriction enzymes to remove sequence-diverse DNA from the internal (centromeric) side of the telomere, the size of the telomeres was determined on agarose gels. The nonplacental mammals contained discontinuous telomeres in which kilobase long stretches of telomeric repeats were interrupted by DNA containing restriction sites. Figure 3 compares telomeres from species from orders with uniformly short telomeres, long telomeres from orders with variable telomere lengths, and two marsupials. The wombat and koala telomeres are < 2 kb when digested with a mixture of six enzymes, but exhibit completely different patterns when digested with individual enzymes. We thus could not assign a telomere length to the nonplacental species.
Fig. 3.
Nonplacental mammals have discontinuous telomeres. Telomere restriction fragment analysis (TRF) involves digestion of genomic DNA with a mixture of six 4-base restriction enzymes (HaeIII, AluI, HinfI, MspI, RsaI, and CfoI) and yields telomeres of a variety of sizes, examples of which are shown. Nonplacental mammalian repeats are interrupted by DNA containing restriction sites, so their size depends on which enzyme is used. Dashes corresponding to the 19- and 6.2-kb size markers were digitally fixed to the image for each species, and then the size of each image adjusted so that all the marks aligned to allow a direct visual comparison between the different species. The DNA from species with very long telomeres was analyzed on FIGE gels.
Oxidative resistance and telomere trade-offs
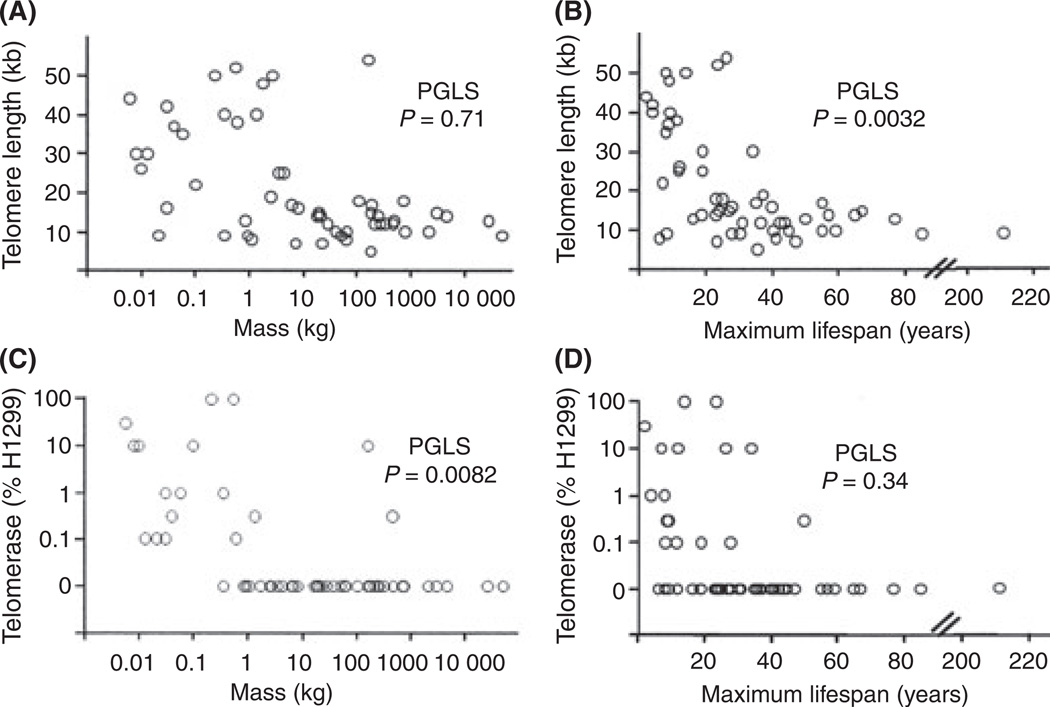
The relationships among the variables analyzed in the regressions are shown in Fig. 4, independent of their phylogenetic connections. Most of the diversity in telomere size and telomerase activity occurs in species weighing < 1 kg. Our phylogenetic analysis suggested that the ancestral mammalian phenotype had short telomeres and repressed telomerase as the initial adaptation to homeothermy (see Discussion), and that the acquisition of long telomeres and not repressing telomerase represents secondary changes that provided adaptive advantage to species that either remained small or which evolved from larger precursors [such as may have occurred in bats (Simmons et al., 2008)]. These smaller species came to have longer telomeres / express telomerase at least 5–7 independent times in the evolutionary tree. These multiple convergent changes suggest trade-offs between the benefits of tumor suppression and the costs of other processes, and two potential candidates are the costs of limiting regeneration and the costs of oxidative damage protection of telomeres (see Discussion).
Fig. 4.
Telomere length, telomerase, lifespan, and body mass distributions. Telomere length (A and B) or telomerase (C and D) vs. log body mass (A and C) or lifespan (B and D). The probability of a significant relationship as an independent variable is indicated by the phylogenetic generalized least squares (PGLS) analysis, which takes into account phylogenetic relationships (which are not indicated in the figure). Telomerase is significantly inversely related to mass while telomere length is inversely correlated with lifespan. Most of the variability occurs within smaller shorter-lived species.
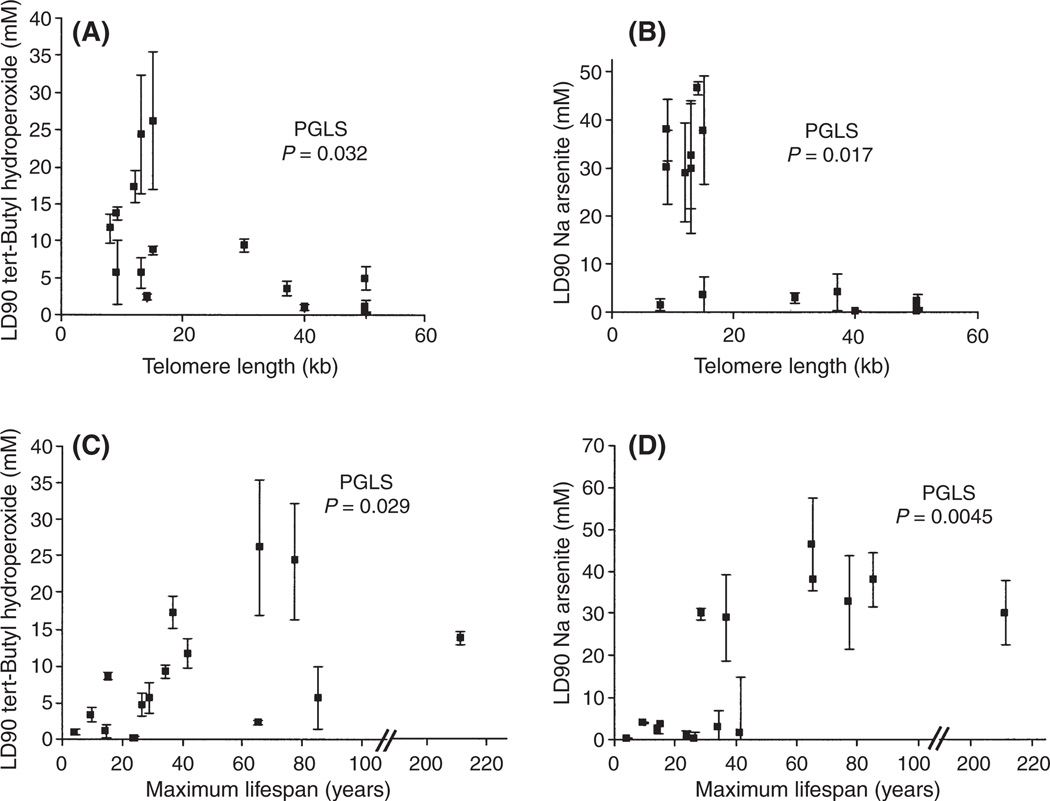
The working hypothesis that a trade-off could involve oxidation damage to telomeres was examined by determining the sensitivity of 15 species to two types of oxidative stress, tert-butyl hydroperoxide and sodium arsenite (Fig. 5). The relationship of drug sensitivity to telomere length independent of size or lifespan was significant by maximum likelihood in a PGLS framework analysis for both agents (tert-butyl hydroperoxide P = 0.032, Fig. 5A; sodium arsenite P = 0.017, Fig. 5B). Many studies have found an association between lifespan or body weight and resistance to oxidative stress in cultured cells [e.g., (Kapahi et al., 1999; Harper et al., 2007)]. This relationship also held with either lifespan (Fig. 5C,D) or mass (Fig. S2, Supporting Information: tert-butyl hydroperoxide, PGLS body size P = 0.016, sodium arsenite, PGLS body size P = 0.0045) as independent variables. The response to sodium arsenite was more dramatic than for tert-butyl hydroperoxide. Species appeared to fall into two groups differing by 6-fold in their sensitivity. Sodium arsenite resistance was independent of the ability of the cells to grow well under standard culture conditions, because two species with short telomeres exhibiting stasis (grey whales and Malaysian tapirs, Table S3, Supporting Information) still exhibited the resistant phenotype.
Fig. 5.
Resistance to tert-butyl hydroperoxide and sodium arsenite. The LD90 of a 4-h treatment with different oxidative damage-inducing agents was examined for 15 different species. Data are shown for telomere length (A and B), maximum lifespan (C and D), tert-butyl hydroperoxide (A and C), and sodium arsenite (B and D). Phylogenetic generalized least squares (PGLS) analysis demonstrates that resistance is significantly associated with telomere length independent of the effects on lifespan or mass. Virtually identical patterns are observed if plotted against body mass (data not shown) instead of lifespan. Table S3 (Supporting Information) gives the actual values for the specific species analyzed. ±SEM of 2–6 titration curves.
Discussion
Telomerase, telomere length, and body size vs. lifespan
One previous study reported that telomerase expression in vivo decreased with increasing mass but did not find a relationship with lifespan for either telomerase activity or telomere length in rodents (Seluanov et al., 2007). We present here a much more extensive data set on diverse mammalian species. The phylogenetic analysis in Fig. 2 demonstrates that telomere length does co-evolve with lifespan. One needs both human-like telomeres (short enough that replication-dependent telomere attrition can produce enough shortening to ‘count’ cell divisions) and repress telomerase to use the full power of replicative aging as a tumor suppression mechanism. The independent association of short telomere lengths with increased lifespan suggests that the full establishment of replicative aging is required to suppress tumor formation over long periods of time in large mammals.
Homeothermy and ancestral mammalian telomeres / telomerase
The maximum likelihood and Markov-transition analysis indicated that ancestral placental mammals had short telomeres and repressed telomerase. Although only limited data are available, a broad range of aquatic poikilothermic species from echinoderms to cartilaginous and bony fishes have short telomeres and express telomerase in many of their tissues (Elmore et al., 2008). The presence of human-like telomeres with repressed telomerase as the ancestral placental mammalian phenotype leads to the hypothesis that one of the early adaptations to becoming homeothermic, with its accompanying increased mutational load (Adelman et al., 1988; Martin & Palumbi, 1993; Rand, 1994), was the repression of telomerase in adult somatic cells, where the consequent progressive telomere shortening could function as a potent tumor-protection mechanism.
There are examples of poikilothermic amphibians, reptiles, and turtles that have heterogeneous telomere lengths ranging up to 50 kb (reviewed in (Gomes et al., 2010). Additional study will be needed, but these may represent other examples of trade-offs and transitions to having very long telomeres resulting from elevated body temperatures as these species exposed themselves to prolonged periods in hot environments. The role of homeothermy in birds awaits better data on the function of micro / macrochromosomal telomeres, replicative aging and telomerase expression in different tissues.
Nonplacental mammals
We were unable to classify telomere length in nonplacental mammals because the TTAGGG telomeric repeats were interrupted by DNA containing restriction endonuclease sites. The nature of the interspersed sequences in the nonplacental mammals and whether they reflect past recombination / insertion events or an ongoing process involved in telomere maintenance remains to be determined. The lack of telomerase expression by koala cells, their growth arrest after only 38 population doublings and their immortalization following the introduction of hTERT (Fig. S1, Supporting Information) suggest that at least one marsupial is using replicative aging and that only the most terminal stretch of uninterrupted telomere repeats is functioning in this process.
Replicative aging and long telomeres / telomerase
Although we believe that most of the species with telomeres > 20 kb do not use replicative aging, additional evidence is required to establish this conclusion. There is a clear tendency for species larger than 1 kg to have short telomeres and repress telomerase (Fig. 4). Most of these may use replicative aging, because they growth-arrested in culture with short telomeres and were immortalized following the introduction of hTERT, often without needing to block cell cycle regulatory genes such as Tp53 (Fig. S1, Supporting Information).
Evolutionary trade-offs and long telomeres / telomerase
There are likely to be trade-offs involving advantages for the expression of telomerase in somatic tissues of small animals. Human diseases involving mutations in telomerase cause premature stem cell depletion and a variety of age-related diseases such as sporadic bone marrow failure, dyskeratosis congenita, and idiopathic pulmonary fibrosis (Garcia et al., 2007). One advantage of not using replicative aging would be an increase in regenerative capacity.
An additional trade-off might involve a need for a higher amount of resources invested in oxidative damage protection in species with short telomeres. Free radicals preferentially damage GGG triplets (Hall et al., 1996; Oikawa & Kawanishi, 1999), present every 6 bp in TTAGGG repeats. Furthermore, a fundamental property of telomeres (suppressing local DNA damage signaling so that the ends of linear chromosomes are not recognized as DNA double-strand breaks) results in oxidative telomeric damage being repaired much more slowly than elsewhere in the genome (Petersen et al., 1998), increasing the probability that single-strand damage gets converted into double-strand breaks and loss of telomeric sequences. Very long telomeres (too long to effectively count cell divisions for physiological purposes) might allow large segments of the telomere to be lost without compromising cell division. Similarly, expressing telomerase would allow repair and elongation of any highly truncated telomeres. Maintaining telomeres short enough to limit cellular proliferative capacity and function as a tumor suppressor mechanism might require an increased amount of resources invested in some oxidative protection mechanisms.
We observed quantitatively different responses for tert-butyl hydroperoxide and sodium arsenite (Fig. 5 and Table S2, Supporting Information). Although the mechanism for this is unknown, it may reflect cellular compartmentalization, different propensities to damage lipid, protein, or DNA (Harper et al., 2007), or the ability of arsenite to react with critical cysteines in some proteins (Flamigni et al., 1989; Kapahi et al., 2000). Oxidative damage has long been hypothesized to be a major determinant of longevity (Harman, 1956). The present result showing dependency on telomere length for both tert-butyl hydroperoxide and sodium arsenite demonstrates that it will be important to consider telomere length as an independent variable in comparing the level of resistance to different oxidative stresses as a function of lifespan.
We observed a minimum of 5–7 independent events in which a transition between short telomeres / repressed telomerase to long telomeres / expression of telomerase occurred. As these are independent events, there is no reason to assume that the molecular / genetic mechanisms underlying these changes are identical.
Conclusion
In summary, we have presented a general survey of telomere biology covering mammalian evolution that provides a conceptual framework for understanding the different uses of telomeres in different species. We show the ancestral mammalian phenotype had human-like telomeres and repressed telomerase, consistent with the hypothesis that the initial adaptation to homeothermy involved the adoption of replicative aging to compensate for the increased mutational load of elevated body temperatures. In addition, we find that telomere length inversely correlates with lifespan, while telomerase expression correlates with mass. The role of replicative aging as a tumor suppression mechanism is well accepted; however, its contribution to lifespan remains controversial. The demonstration that telomere length inversely correlates with lifespan provides support for the interpretation that replicative aging is one of many factors contributing to lifespan in a large number of species. The evidence that oxidative protection mechanisms are lower in species with long telomeres suggests one evolutionary advantage of abandoning replicative aging in favor of long telomeres and not repressing telomerase in smaller mammals. Comparative aging studies of stress resistance / oxidative protection will need to evaluate the changes that function in lifespan determination vs. those reflecting changes in telomere phenotype. These results now allow the role of telomeres in human cancer and aging to be put in the larger context of mammalian telomere biology.
Discussion of additional considerations affecting the interpretation of these results can be found in the Data S1 (Supporting Information).
Materials and methods
Data and phylogeny
Data on size and maximum lifespan were obtained from the AnAGE database (The Animal Ageing & Longevity Database. http://genomics.senescence.info/species/).
We used a pruned version of Bininda-Emonds et al.’s mammal supertree (Bininda-Emonds et al., 2007, 2008), with branch lengths measured by estimated divergence times in millions of years.
Phylogenetic regression analyses
We conducted simultaneous multiple regression analyses (so all P-values reported are independent of all other variables in the model) within a maximum likelihood PGLS framework using the computer program BayesTraits (Pagel et al., 2004). This method explicitly accounts for the shared ancestry as implied by the phylogeny by converting the tree topology and branch lengths into a variance–covariance matrix which is used to weigh the points accordingly. By incorporating λ into the regression model, the PGLS framework offers a more flexible approach relative to other statistical methods, as it can quantify and account for the precise phylogenetic signal in the residuals. This well-established, and widely used, methodology is fully described elsewhere (Pagel, 1999; Freckleton et al., 2002). Because telomere length, mass and lifespan are all bounded by zero, calculations were based on the log of the values.
Phylogenetic ancestral state reconstruction
To estimate the ancestral telomere length, we used a model introduced by Organ et al. (2009). This model also uses PGLS framework. It has been shown to outperform other methods of ancestral state reconstruction as the model of evolution estimated across the whole tree is used to infer the ancestral states (Montgomery et al., 2010). This method is fully described by Organ et al. (2009).
To infer whether the ancestral mammal repressed or expressed telomerase, we used a continuous time Markov model of trait evolution, as implemented in the computer program BayesTraits (Pagel et al., 2004), allowing independent gains and losses in each branch of the phylogenetic tree. This method differs from that used to infer telomere length as it varies discretely rather than continuously. This methodology provides a probability for each state (in this case repressed or expressed) at any given node. This method is fully described by Pagel et al. (2004).
Cell culture
Fibroblasts from the skin, kidney, lung or cornea of various adult mammalian species were grown in a variety of media (detailed for each species in Fig. S1, Supporting Information). All cells were grown at 37°C in 5%CO2 and low oxygen conditions (2–5% O2) except as noted in Fig. S1 (Supporting Information).
Telomere length and telomerase
Mean telomere length was determined using terminal restriction fragment (TRF) analysis (Herbert et al., 2003). Genomic DNA was digested with one or a mixture of six enzymes (HaeIII, AluI, HinfI, MspI, RsaI, and CfoI) and resolved on 0.7% agarose gels. Cell telomerase activity was measured using the telomeric repeat amplification protocol (TRAP) (Herbert et al., 2003), and activity expressed with respect to the reference human lung adenocarcinoma line H1299. Telomerase is expressed during embryogenesis in humans and presumably all mammals. The expression of telomerase in cells cultured from adult animals is being interpreted as a reflection of the regulation of telomerase in adult somatic cells that are not stem or transient amplifying cells. Tissue biopsies often show weak telomerase activity when first put in culture that disappears within the first few doublings. Whether this represents the presence of stem cells that are overgrown by fibroblasts / more differentiated cells or simply a downregulation of telomerase is unknown. Telomerase activity was quantitated from the activity sustained for more than a few doublings in culture.
Telomere size varies by 3–4 kb between different human donors in addition to decreasing with donor age. As in most cases cultures from a single animal of undetermined age were analyzed, telomere size was estimated (accurate within a few kb) rather than calculated to emphasize the limits of this analysis (i.e., only > 5 kb differences are significant).
Cytotoxicity assay
Thawed cells were grown for 11 days then plated at 20 000 cells per 100 µL per well in 96-well plates. After a 4 h recovery, 100 µL of RPMI-1640 media containing dilutions of tert-butyl hydroperoxide or sodium arsenite were added for 4 h at 37°C. After three washes with D-PBS, each well was incubated for 4 h at 37°C with 100 µL MTT (Sigma) diluted in RPMI-1640 (with 25 mm Hepes and 1 mm NaPyruvate). To each well, 200 µL of solubilizing solution (10% Triton X-100, 0.1 N HCl, 80% Isopropanol) was added, and the plate was shaken for 10 min. Absorbance at 570 nm was measured using an automated plate reader (700 nm reference wavelength).
Viral transfections
Some cells rapidly entered stasis, a growth arrest because of inadequate growth conditions. To force these cells to proliferate, we overexpressed SV40 Large-T antigen to block cell cycle checkpoints, cyclin-dependent kinase 4 (Cdk4), or human papillomavirus (HPV) E6 / E7. In cases where the cells were nondividing at arrival, they were first infected with adenovirus expressing SV40 LgT antigen to initiate cell division, and then infected with retroviral vectors to produce stable expression. Stable infection with retrovirus expressing SV40 LgT antigen was performed 72 h after adenoviral infection. Stable expression of the human telomerase protein catalytic component (hTERT) used the retroviral vector pBabepurohTERT (Ouellette et al., 1999).
Nondenaturing in-gel hybridization to detect telomere DNA
In some cases, it was difficult to distinguish telomeres from abundant internal telomeric repeats. The true telomeric signal was then determined on native gels by hybridizing ds genomic DNA to a C-rich telomeric probe that would only anneal to the single-stranded G-rich overhang at telomeres.
Fluorescence in situ hybridization analysis
Metaphase spreads were hybridized to 3′-Cy3 or Cy5-conjugated (CCCTAA)3 2′-deoxyoligonucleotide N3′-P5′ phosphoramidate telomeric probes (kindly provided by Geron Corp., Menlo Park, CA, USA) as described by Zou et al. (2002).
Supplementary Material
Acknowledgments
We thank John Wise for providing the Bowhead Whale lung cells with permission from the National Marine Sanctuary Foundation, and Julie Fronczek, Ying S. Zou, Jason Sherrell, Michael Wang, Maeve Hsieh, Gregory Thompson, Donna Meng and Martin Bayer for their excellent technical assistance. Animal Photos provided courtesy of the Zoological Society of San Diego. Supported by the European Union Programs POCI 2010 & FSE and by national funds from the Portuguese Ministry for Science, Technology and Superior Education (N.M.V.G), by the Keck Foundation (W.E.W. & J.W.S.) and the National Institute on Aging AG01228 (W.E.W.). O.A.R., M.L.H., S.J.C., and S.N.A. provided primary cultures of multiple animal species. W.W. (bowhead whale) and N.R.F. (dog) obtained some data on those species. N.M.V.G. obtained the data on all species and performed the oxidative sensitivity analysis. C.V. and M.P. performed the phylogenetic analysis. N.M.V.G., C.V., M.P., J.W.S., and W.E.W. analyzed the data and prepared the manuscript.
Footnotes
Supporting Information
Additional supporting information may be found in the online version of this article:
Data S1 Discussion.
Fig. S1 Data on growth, telomere length and telomerase expression for 43 species.
Fig. S2 Resistance to tert-Butyl hydroperoxide and sodium arsenite.
Table S1 Species, mass and lifespan.
Table S2 PGLS analysis of individual non-placental mammalian orders.
Table S3 Species analyzed for resistance to oxidative stressors.
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Adelman R, Saul RL, Ames BN. Oxidative damage to DNA: relation to species metabolic rate and life span. Proc. Natl Acad. Sci. USA. 1988;85:2706–2708. doi: 10.1073/pnas.85.8.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage P, Doll R. The age distribution of cancer and a multi-stage theory of carcinogenesis. Br. J. Cancer. 1954;8:1–12. doi: 10.1038/bjc.1954.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bininda-Emonds OR, Cardillo M, Jones KE, MacPhee RD, Beck RM, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds OR, Cardillo M, Jones KE, MacPhee RD, Beck RM, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A. The delayed rise of present-day mammals. Corrigendum. Nature. 2008;446:274. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. Telomerases. Annu. Rev. Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- Chai W, Shay JW, Wright WE. Human telomeres maintain their overhang length at senescence. Mol. Cell. Biol. 2005;25:2158–2168. doi: 10.1128/MCB.25.6.2158-2168.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Aslam S, Fletcher J, Wylie D, Clinton M, Clark AJ. Stabilization of telomere length and karyotypic stability are directly correlated with the level of hTERT gene expression in primary fibroblasts. J. Biol. Chem. 2002;277:38531–38539. doi: 10.1074/jbc.M205981200. [DOI] [PubMed] [Google Scholar]
- Drayton S, Peters G. Immortalisation and transformation revisited. Curr. Opin. Genet. Dev. 2002;12:98–104. doi: 10.1016/s0959-437x(01)00271-4. [DOI] [PubMed] [Google Scholar]
- Elmore LW, Norris MW, Sircar S, Bright AT, McChesney PA, Winn RN, Holt SE. Upregulation of telomerase function during tissue regeneration. Exp. Biol. Med. (Maywood) 2008;233:958–967. doi: 10.3181/0712-RM-345. [DOI] [PubMed] [Google Scholar]
- Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, Le S, West MD, Harley CB, Andrews WH, Greider CW, Villeponteau B. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- Flamigni F, Marmiroli S, Caldarera CM, Guarnieri C. Involvement of thiol transferase- and thioredoxin-dependent systems in the protection of ‘essential’ thiol groups of ornithine decarboxylase. Biochem. J. 1989;259:111–115. doi: 10.1042/bj2590111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth NR, Elder FF, Shay JW, Wright WE. Lagomorphs (rabbits, pikas and hares) do not use telomere-directed replicative aging in vitro. Mech. Ageing Dev. 2005;126:685–691. doi: 10.1016/j.mad.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Fradiani PA, Ascenzioni F, Lavitrano M, Donini P. Telomeres and telomerase activity in pig tissues. Biochimie. 2004;86:7–12. doi: 10.1016/j.biochi.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Freckleton RP, Harvey PH, Pagel M. Phylogenetic analysis and comparative data: a test and review of evidence. Am. Nat. 2002;160:712–726. doi: 10.1086/343873. [DOI] [PubMed] [Google Scholar]
- Garcia CK, Wright WE, Shay JW. Human diseases of telomerase dysfunction: insights into tissue aging. Nucleic Acids Res. 2007;35:7406–7416. doi: 10.1093/nar/gkm644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NM, Shay JW, Wright WE. Telomere biology in Metazoa. FEBS Lett. 2010;584:3741–3751. doi: 10.1016/j.febslet.2010.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A. Coevolution of telomerase activity and body mass in mammals: from mice to beavers. Mech. Ageing Dev. 2009;130:3–9. doi: 10.1016/j.mad.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- Hall DB, Holmlin RE, Barton JK. Oxidative DNA damage through long-range electron transfer. Nature. 1996;382:731–735. doi: 10.1038/382731a0. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Harper JM, Salmon AB, Leiser SF, Galecki AT, Miller RA. Skin-derived fibroblasts from long-lived species are resistant to some, but not all, lethal stresses and to the mitochondrial inhibitor rotenone. Aging Cell. 2007;6:1–13. doi: 10.1111/j.1474-9726.2006.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert B, Shay JW, Wright WE. Analysis of telomeres and telomerase. In: Bonifacino JS, Dasso M, Lippincott-Schwartz J, Harford JB, Yamada KM, editors. Current Protocols in Cell Biology. Hoboken, NJ: Wiley and Sons, Inc; 2003. p. 18.16.16. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Boulton ME, Kirkwood TB. Positive correlation between mammalian life span and cellular resistance to stress. Free Radic. Biol. Med. 1999;26:495–500. doi: 10.1016/s0891-5849(98)00323-2. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Takahashi T, Natoli G, Adams SR, Chen Y, Tsien RY, Karin M. Inhibition of NF-kappa B activation by arsenite through reaction with a critical cysteine in the activation loop of Ikappa B kinase. J. Biol. Chem. 2000;275:36062–36066. doi: 10.1074/jbc.M007204200. [DOI] [PubMed] [Google Scholar]
- Lanza RP, Cibelli JB, Blackwell C, Cristofalo VJ, Francis MK, Baerlocher GM, Mak J, Schertzer M, Chavez EA, Sawyer N, Lansdorp PM, West MD. Extension of cell life-span and telomere length in animals cloned from senescent somatic cells. Science. 2000;288:665–669. doi: 10.1126/science.288.5466.665. [DOI] [PubMed] [Google Scholar]
- Lee HW, Blasco MA, Gottlieb GJ, Horner JW, II, Greider CW, DePinho RA. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- Luebeck EG, Moolgavkar SH. Multistage carcinogenesis and the incidence of colorectal cancer. Proc. Natl Acad. Sci. USA. 2002;99:15095–15100. doi: 10.1073/pnas.222118199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- Martin AP, Palumbi SR. Body size, metabolic rate, generation time, and the molecular clock. Proc. Natl Acad. Sci. USA. 1993;90:4087–4091. doi: 10.1073/pnas.90.9.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathon NF, Malcolm DS, Harrisingh MC, Cheng L, Lloyd AC. Lack of replicative senescence in normal rodent glia. Science. 2001;291:872–875. doi: 10.1126/science.1056782. [DOI] [PubMed] [Google Scholar]
- Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, Bacchetti S, Haber DA, Weinberg RA. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Capellini I, Barton RA, Mundy NI. Reconstructing the ups and downs of primate brain evolution: implications for adaptive hypotheses and Homo floresiensis. BMC Biol. 2010;8:9. doi: 10.1186/1741-7007-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and humans. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Oh HY, Jin X, Kim JG, Oh MJ, Pian X, Kim JM, Yoon MS, Son CI, Lee YS, Hong KC, Kim H, Choi YJ, Whang KY. Characteristics of primary and immortalized fibroblast cells derived from the miniature and domestic pigs. BMC Cell Biol. 2007;8:20. doi: 10.1186/1471-2121-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa S, Kawanishi S. Site-specific DNA damage at GGG sequence by oxidative stress may accelerate telomere shortening. FEBS Lett. 1999;453:365–368. doi: 10.1016/s0014-5793(99)00748-6. [DOI] [PubMed] [Google Scholar]
- Organ CL, Janes DE, Meade A, Pagel M. Genotypic sex determination enabled adaptive radiations of extinct marine reptiles. Nature. 2009;461:389–392. doi: 10.1038/nature08350. [DOI] [PubMed] [Google Scholar]
- Ouellette MM, Aisner DL, Savre-Train I, Wright WE, Shay JW. Telomerase activity does not always imply telomere maintenance. Biochem. Biophys. Res. Commun. 1999;254:795–803. doi: 10.1006/bbrc.1998.0114. [DOI] [PubMed] [Google Scholar]
- Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- Petersen S, Saretzki G, von Zglinicki T. Preferential accumulation of single-stranded regions in telomeres of human fibroblasts. Exp. Cell Res. 1998;239:152–160. doi: 10.1006/excr.1997.3893. [DOI] [PubMed] [Google Scholar]
- Rand DM. Thermal habit, metabolic rate and the evolution of mitochondrial DNA. Trends Ecol. Evol. 1994;9:125–131. doi: 10.1016/0169-5347(94)90176-7. [DOI] [PubMed] [Google Scholar]
- Seluanov A, Chen Z, Hine C, Sasahara TH, Ribeiro AA, Catania KC, Presgraves DC, Gorbunova V. Telomerase activity coevolves with body mass not lifespan. Aging Cell. 2007;6:45–52. doi: 10.1111/j.1474-9726.2006.00262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur. J. Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26:867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, DePinho RA. Cellular senescence: mitotic clock or culture shock? Cell. 2000;102:407–410. doi: 10.1016/s0092-8674(00)00046-5. [DOI] [PubMed] [Google Scholar]
- Simmons NB, Seymour KL, Habersetzer J, Gunnell GF. Primitive Early Eocene bat from Wyoming and the evolution of flight and echolocation. Nature. 2008;451:818–821. doi: 10.1038/nature06549. [DOI] [PubMed] [Google Scholar]
- Steinert S, White DM, Zou Y, Shay JW, Wright WE. Telomere biology and cellular aging in nonhuman primate cells. Exp. Cell Res. 2002;272:146–152. doi: 10.1006/excr.2001.5409. [DOI] [PubMed] [Google Scholar]
- Wright WE, Shay JW. Telomere dynamics in cancer progression and prevention: fundamental differences in human and mouse telomere biology. Nat. Med. 2000;6:849–851. doi: 10.1038/78592. [DOI] [PubMed] [Google Scholar]
- Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Zou Y, Yi X, Wright WE, Shay JW. Human telomerase can immortalize Indian muntjac cells. Exp. Cell Res. 2002;281:63–76. doi: 10.1006/excr.2002.5645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.