Metabolic enzymes fumarate hydratase (FH) and succinate dehydrogenase (SDH) are known tumor suppressors mutated in certain human cancers. In this study, Guan and colleagues demonstrate that fumarate and succinate, two metabolites that accumulate in tumors with mutations targeting the FH or SDH genes, respectively, act as competitive inhibitors of multiple α-KG-dependent dioxygenases, including both histone demethylases and the TET family of 5-methlycytosine (5mC) hydroxylases. This study delineates a novel molecular mechanism for the tumor suppressor function of FH and SDH, whereby accumulation of their metabolites induces epigenetic modifications by inhibiting histone- and DNA-modifying enzymes.
Keywords: FH, SDH, metabolites, α-KG-dependent dioxygenases, DNA methylation, histone methylation
Abstract
Two Krebs cycle genes, fumarate hydratase (FH) and succinate dehydrogenase (SDH), are mutated in a subset of human cancers, leading to accumulation of their substrates, fumarate and succinate, respectively. Here we demonstrate that fumarate and succinate are competitive inhibitors of multiple α-ketoglutarate (α-KG)-dependent dioxygenases, including histone demethylases, prolyl hydroxylases, collagen prolyl-4-hydroxylases, and the TET (ten-eleven translocation) family of 5-methlycytosine (5mC) hydroxylases. Knockdown of FH and SDH results in elevated intracellular levels of fumarate and succinate, respectively, which act as competitors of α-KG to broadly inhibit the activity of α-KG-dependent dioxygenases. In addition, ectopic expression of tumor-derived FH and SDH mutants inhibits histone demethylation and hydroxylation of 5mC. Our study suggests that tumor-derived FH and SDH mutations accumulate fumarate and succinate, leading to enzymatic inhibition of multiple α-KG-dependent dioxygenases and consequent alterations of genome-wide histone and DNA methylation. These epigenetic alterations associated with mutations of FH and SDH likely contribute to tumorigenesis.
Several lines of evidence, including the recent identification of mutations affecting isocitrate dehydrogenase (IDH), fumarate hydratase (FH), and succinate dehydrogenase (SDH), have demonstrated that mutations in certain metabolic enzymes may play a causal role in tumorigenesis. The NADP+-dependent IDH genes IDH1 and IDH2 are frequently mutated in >75% of glioma (Parsons et al. 2008), ∼20% of acute myeloid leukaemia (AML) (Mardis et al. 2009), and several additional tumors at different frequencies (Hemerly et al. 2010; Murugan et al. 2010; Amary et al. 2011; Damato et al. 2012; Oermann et al. 2012). These mutations in IDH1/2 result in simultaneous loss and gain of activities in the production of α-ketoglutarate (α-KG) and 2-hydroxyglutarate (2-HG), respectively (Dang et al. 2009; Yan et al. 2009; Zhao et al. 2009). α-KG plays critical roles in four different metabolic and cellular pathways: as an intermediate in the Krebs cycle for energy metabolism, as a precursor of glutamine formation for the amino acid synthesis, as a nitrogen transporter for the urea cycle and ammonia detoxification, and as a cosubstrate for Fe(II)/α-KG-dependent dioxygenases. Accumulating genetic and biochemical evidence supports the notion that the alterations of Fe (II)/α-KG-dependent dioxygenases contribute to tumorigenesis (Oermann et al. 2012).
Fe(II)/α-KG-dependent dioxygenases are present in all living organisms and catalyze hydroxylation reactions on a diverse set of substrates, including proteins, alkylated DNA/RNA, lipids, antibiotics, and, most recently, 5-methylcytosine (5mC) of genomic DNA (Hausinger 2004; Loenarz and Schofield 2008; Tahiliani et al. 2009). These enzymes require Fe(II) as a cofactor metal and α-KG as a cosubstrate to catalyze the reactions in which one oxygen atom from molecular oxygen (O2) is attached to a hydroxyl group in the substrate (hydroxylation) while the other is taken up by α-KG, leading to the decarboxylation of α-KG and subsequent release of carbon dioxide (CO2) and succinate. Of the >60 estimated α-KG-dependent dioxygenases in mammalian cells (Rose et al. 2011), the JmjC domain-containing histone demethylases (KDMs) and the TET (ten-eleven translocation) family of DNA hydroxylases play central roles in epigenetic control of genomic information. While the KDMs catalyze the typical hydroxylation on the methyl group on the lysine residue (Tsukada et al. 2006), the recently discovered TET family of DNA hydroxylases catalyzes a three-step iterative oxidation reaction: converting 5mC to 5-hydroxymethylcytosine (5hmC), then converting 5hmC to 5-formylcytosine (5fC), and finally converting 5fC to 5-carboxylcytosine (5caC) (Tahiliani et al. 2009; Ito et al. 2010, 2011; He et al. 2011). A subsequent decarboxylation of 5caC by either a thymine–DNA glycosylase or other DNA repair enzymes could then lead to DNA demethylation. We and others recently demonstrated that ectopic expression of tumor-derived mutated IDH1 and IDH2 inhibits the activity of α-KG-dependent dioxygenases and, more importantly, produces 2-HG, which acts as an antagonist of α-KG to competitively inhibit the activity of α-KG-dependent dioxygenases, including both KDMs and TETs (Chowdhury et al. 2011; Xu et al. 2011). These findings provide a biochemical basis for the hypermethylation observed in human glioma with IDH1 mutation (Noushmehr et al. 2010) and the mutually exclusive manner of IDH1/2 and TET2 gene mutations in AML (Lorsbach et al. 2003; Figueroa et al. 2010).
Besides IDH1 and IDH2, six genes (FH, SDHA, SDHB, SDHC, SDHD, and SDHAF2), encoding for the subunits of two different Krebs cycle enzymes (FH and SDH), are mutated both germinally and somatically in a number of human cancers (Baysal et al. 2000; Astuti et al. 2001; Hao et al. 2009; Kaelin 2009; Bayley et al. 2010; Oermann et al. 2012). Thus far, all functionally characterized FH or SDH mutations result in either a complete loss or reduction of enzymatic activity (Tomlinson et al. 2002; Pollard et al. 2005), indicating that both FH and SDH function as tumor suppressors. The tumor suppressor roles of FH and SDH mutations have been proposed to (1) abrogate the mitochondrial function to trigger apoptosis (Scatena et al. 2007), (2) generate harmful reactive oxygen species (ROS) that induce DNA damage and genomic instability (Kaelin 2009), and (3) provoke accumulation of HIF1α, a transcription factor whose stabilization and elevation could promote cell metabolism and angiogenesis (Ivan et al. 2001; Jaakola et al. 2001; Ling et al. 2005; Selak et al. 2005). Studies into the mechanism behind how mutations in FH and SDH trigger elevated levels of HIF1α provided evidence that fumarate and succinate accumulated in cells with mutations in either FH or SDH inhibit prolyl hydroxylases (PHDs), which hydroxylates and promotes the degradation of HIF1α (Epstein et al. 2001; Yu et al. 2001; Isaacs et al. 2005; Selak et al. 2005). Structurally, both fumarate and succinate are similar to α-KG and 2-HG. They have the same acetate end and include two oxygen atoms linked to C-5 that are used by α-KG and 2-HG to interact with conserved residues in the dioxygenases, supporting the notion that fumarate and succinate may function as competitive inhibitors of α-KG-dependent dioxygenases in addition to PHDs. This study is directed toward understanding how fumarate and succinate alter epigenetic modifications and explores the underlying mechanisms of FH and SDH mutations in tumorigenesis.
Results
Both fumarate and succinate inhibit the activity of α-KG-dependent KDMs in vitro
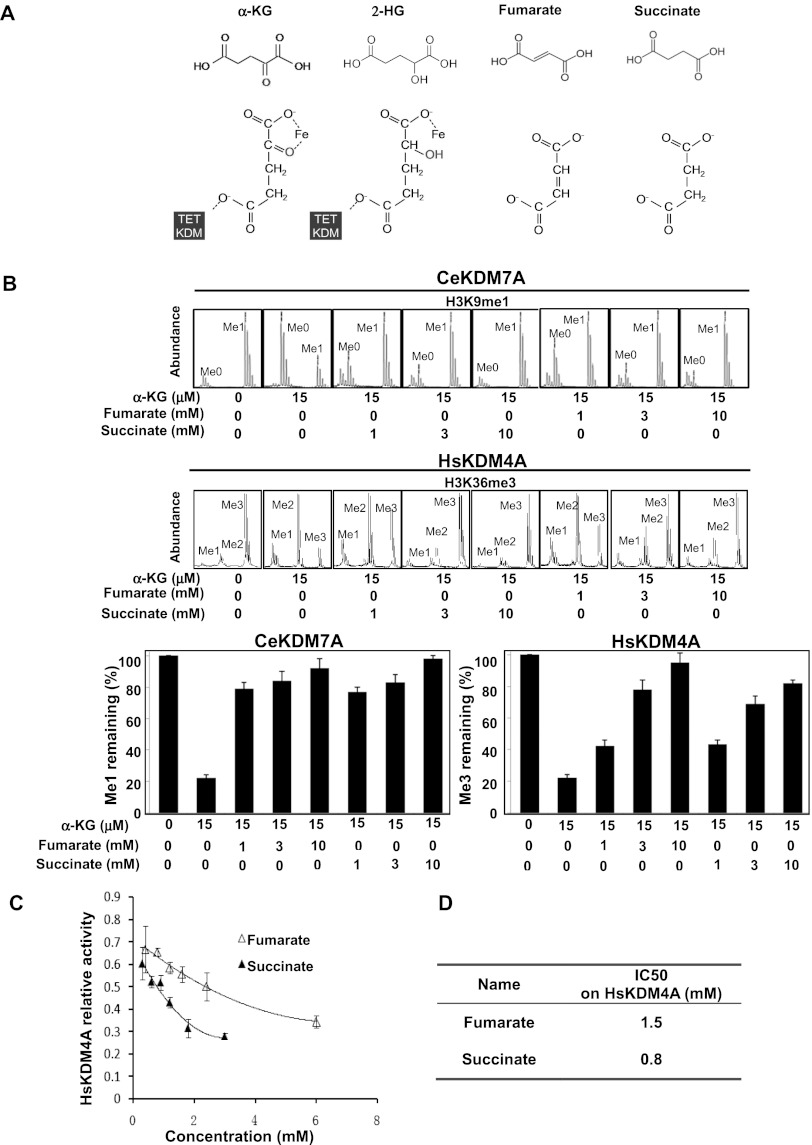
We showed previously that 2-HG inhibits α-KG-dependent dioxygenases by acting as a competitive inhibitor of α-KG (Xu et al. 2011). Likewise, fumarate and succinate also share structural similarity with α-KG, except that C2 with its linked oxygen atom (i.e., the ketone group) in α-KG is absent in fumarate and succinate. In addition, fumarate and succinate differ by only one ethylenic linkage (Fig. 1A). Such structural similarities make fumarate and succinate potential antagonists of α-KG.
Figure 1.
Fumarate and succinate inhibit the activity of α-KG-dependent KDMs in vitro. (A) Structural comparison among fumarate, succinate, α-KG, and 2-HG. (B) Fumarate and succinate inhibit the demethylase activity of C. elegans KDM7A (CeKDM7A). CeKDM7A activity toward monmethylated H3K9 was assayed in the presence or absence of 15 μM α-KG with increasing concentrations of either fumarate or succinate as indicated. The demethylated products were analyzed by mass spectrometry (MS), and mean activity values of duplicated assays, represented by percentage of remaining methylated peptides, are shown. (C) Fumarate and succinate inhibit the demethylase activity of human KDM4A (HsKDM4A) toward trimethylated H3K36, determined by mass spectrometry (MS) assay as in B. Error bars represent standard deviation (SD) for triplicate experiments. (D) Succinate is more potent to inhibit HsKDM4A than fumarate. The half maximal inhibitory concentration (IC50) of fumarate and succinate on HsKDM4A was determined by MS assay as in C. Error bars represent standard deviation (SD) for triplicate experiments. Additional MS results are shown in Supplemental Figure S1.
To test this hypothesis, we first examined the effect of fumarate and succinate on CeKDM7A, a Caenorhabditis elegans dual-specificity KDM that recognizes methylated H3K9, using a synthetic monomethylated H3K9 (H3K9me1) peptide as a substrate. Mass spectrometric analysis demonstrated that a near-physiological α-KG concentration of 15 μM could support substrate demethylation (from H3K9me1 to H3K9me0) by CeKDM7A (Fig. 1B). The physiological concentrations of fumarate and succinate are ∼100 μM and 0.5–1 mM, respectively (Bennett et al. 2009), and can be accumulated to high millimolars in tumor samples with mutation of FH or SDH (Pollard et al. 2005). Addition of 1 mM, 3 mM, and 10 mM fumarate resulted in >80% inhibition of CeKDM7A (Fig. 1B). Similar results were obtained using succinate (Fig. 1B). In addition, inhibition of CeKDM7A by fumarate or succinate became less effective, along with increased concentrations of α-KG (Supplemental Fig. S1A).
We next examined the effect of fumarate and succinate on HsKDM4A, a human histone H3K36 demethylase KDM4A/JHDM2A, using synthetic trimethylated H3K36 (H3K36me3) as a substrate. Addition of 1 mM, 3 mM, and 10 mM fumarate led to 22%, 41%, and 73% inhibition of HsKDM4A, respectively (Fig. 1B). Addition of the same concentrations of succinate resulted in 43%, 68%, and 85% inhibition of HsKDM4A, respectively (Fig. 1B). These results show that fumarate and succinate can directly inhibit KDM activity. To compare the potency of fumarate and succinate in KDM inhibition, we determined their half maximal inhibitory concentration (IC50) on HsKDM4A (Fig. 1C; Supplemental Fig. S1B). Our results demonstrated that succinate (IC50 = 0.8 mM) is more potent than fumarate (IC50 = 1.5 mM) in inhibiting HsKDM4A (Fig. 1D).
Together, these results suggest that both fumarate and succinate act as antagonists of α-KG to inhibit α-KG-dependent KDMs, with succinate being more potent than fumarate.
Both fumarate and succinate increase genome-wide histone methylations, accumulate HIF1α, and reduce endostatin in cultured cells
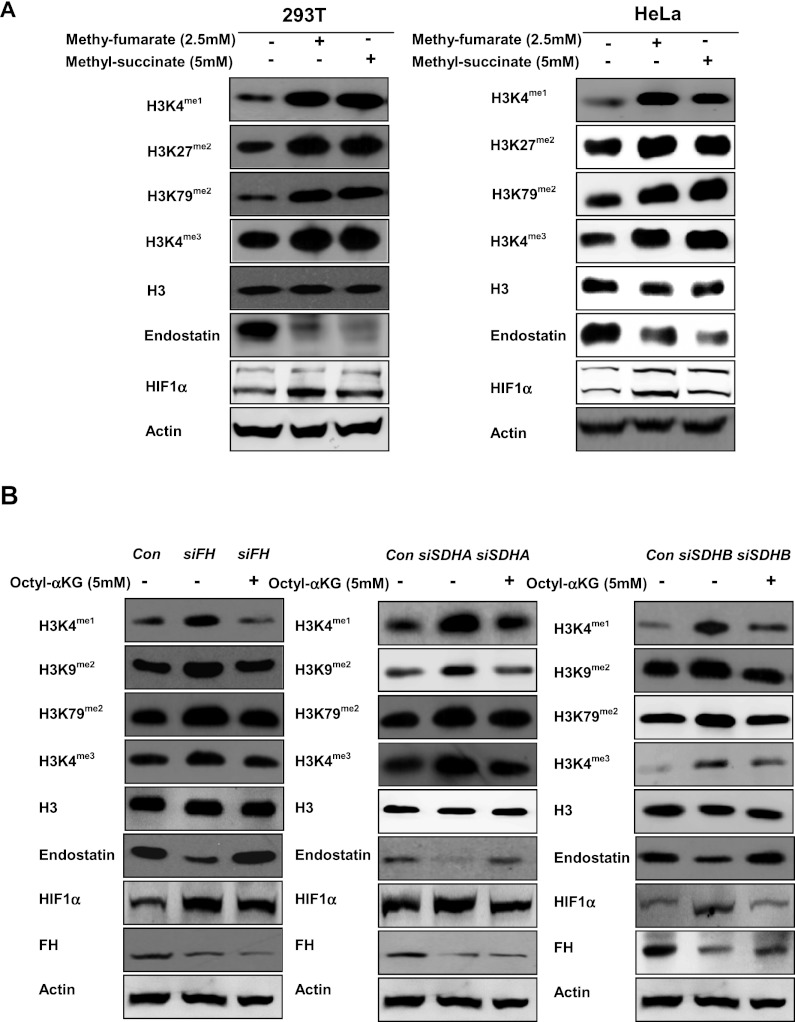
Inhibition of α-KG-dependent KDMs by fumarate and succinate in vitro led us to determine the effect of both metabolites on genome-wide histone methylations. To this end, we synthesized cell-permeable methyl-fumarate and methyl-succinate, which can rapidly enter cells and be hydrolyzed by endogenous esterases (MacKenzie et al. 2007). As expected, addition of 2.5 mM methyl-fumarate and 5 mM methyl-succinate to the cultured HeLa cells resulted in a significant increase of intracellular fumarate and succinate, respectively, as determined by gas chromatography–mass spectrometry (GC-MS) (Supplemental Fig. S2A). A similar increase of fumurate and succinate was observed in HEK293T cells treated with cell-permeable fumarate and succinate, respectively. The fumarate/α-KG and succinate/α-KG ratios were elevated in these cells (Supplemental Fig. S2B). Cell-permeable fumarate and succinate increased H3K4 monomethylation (by fourfold), H3K27 and H3K79 dimethylation (by fourfold and threefold, respectively), and H3K4 trimethylation (by twofold) (Fig. 2A). Similar results were obtained in the cultured HeLa cells after treatment with the same concentrations of cell-permeable fumarate and succinate analogs (Fig. 2A).
Figure 2.
Fumarate and succinate increase genome-wide histone methylations, accumulate HIF1α, and reduce endostatin in cultured cells. (A) Cell-permeable fumarate (2.5 mM) or succinate (5 mM) increases histone methylation and HIF1α and reduces endostatin in HEK293T cells (left) and HeLa cells (right), as determined by Western-blot. (B) Cell-permeable octyl-α-KG (5mM) diminishes the effect of FH or SDH suppression on increasing histone methylations, accumulating HIF1α, and decreasing endostatin in HEK293T cells with knockdown of FH (left panel), SDHA (middle panel), and SDHB (right panel), as determined by Western blot. See also Supplemental Figure S3.
In addition, the effect of fumarate and succinate on the activity of two additional α-KG-dependent dioxygenases, PHDs and collagen prolyl-4-hydroxylase (C-P4H) (Mussini et al. 1967), was also indirectly determined by detecting their corresponding substrate/product, HIF1α and endostatin. We found that cell-permeable fumarate and succinate increased HIF1α and decreased endostatin in both cell types (Fig. 2A), indicating that fumarate and succinate can impair the hydroxylation of prolyl residues in HIF1α and collagen via inhibiting the enzymatic activity of PHD2 and C-P4H, respectively. The effect on endostatin was particularly dramatic, suggesting that C-P4H is rather sensitive to inhibition by fumarate and succinate.
We next determined how endogenous FH and SDH activity would affect the level of fumarate and succinate and the activities of α-KG-dependent dioxygenases. We found that depletion of FH and SDHA/B by siRNA resulted in elevated levels of fumarate and succinate in HeLa cells, as determined by GC-MS assay (Supplemental Fig. S3A). Knocking down FH and SDHA/B resulted in a significant increase of histone methylations on H3K4, H3K9, and H3K79 (by threefold to fivefold); accumulation of HIF1α (by fivefold); and a decrease of endostatin (by sevenfold) (Fig. 2B). To further test the antagonistic relationship between α-KG and fumarate or succinate, cells with FH or SDHA/B knockdown were incubated with cell-permeable α-KG. Addition of 5 mM octyl-α-KG diminished the effect of FH or SDHA/B suppression on increasing histone methylations, accumulating HIF1α, and decreasing endostatin (Fig. 2B), and these effects of octyl-α-KG were in a dose-dependent manner (Supplemental Fig. S3B).
Together, our results demonstrate that fumarate and succinate act as competitors of α-KG to broadly inhibit the activity of α-KG-dependent dioxygenases, including KDMs, PHDs and C-P4Hs.
Suppression of FH or SDH expression reduces TET-catalyzed 5hmC production in cultured cells
In addition to KDMs, PHDs and C-P4Hs, another class of Fe(II)- and α-KG-dependent dioxygenases is the TET family of DNA hydroxylases. Given the dependence of TET catalytic activity on α-KG and its inhibition by 2-HG (Xu et al. 2011), we sought to determine whether fumarate and succinate could affect TET activity and DNA cytosine hydroxymethylation.
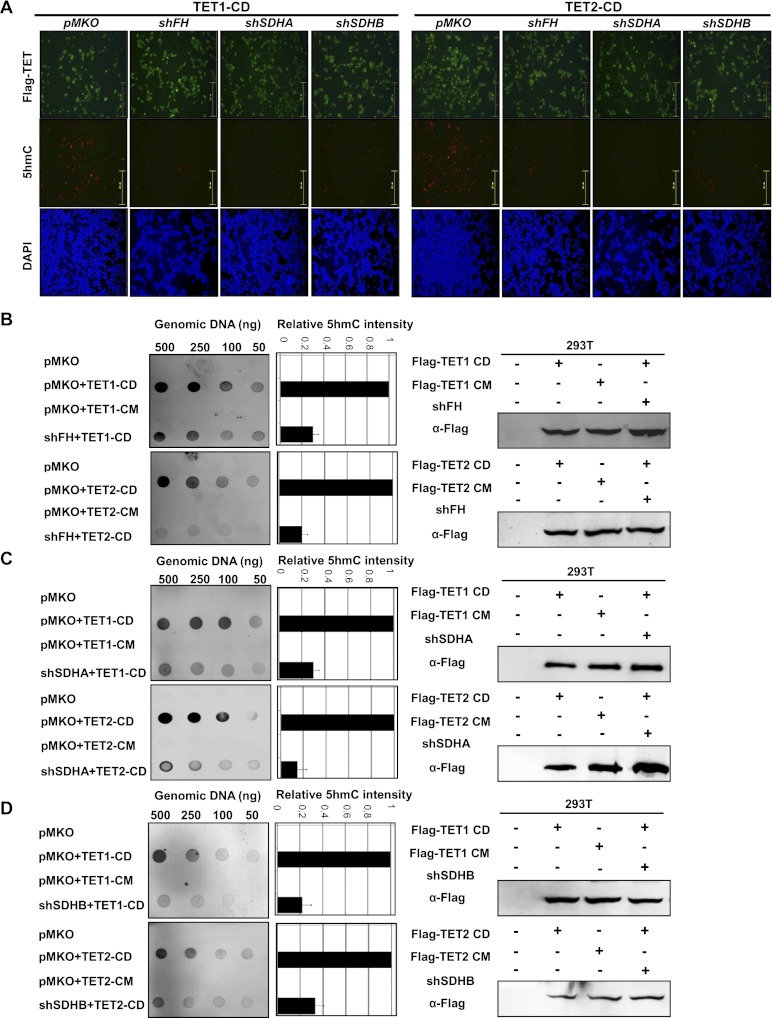
The 5hmC level in most cultured cells is undetectable, but is substantially increased in cells transiently expressing the wild-type catalytic domain of TET1 and TET2 proteins (TET1-CD and TET2-CD) and can be easily detected by immunofluorescence using an antibody specifically recognizing 5hmC (Fig. 3A; Tahiliani et al. 2009; Ito et al. 2010, 2011; He et al. 2011). HEK293T cells with stable knockdown of FH or SDHA/B were generated by retrovirus infection, and the knockdown efficiency was confirmed by Western blot (Supplemental Fig. S4A). Notably, overexpression of TET1-CD or TET2-CD was ineffective to increase 5hmC in HEK293 cells with stable knockdown of FH or SDHA/B, as determined by immunofluorescence staining with the 5hmC-specific antibody (Fig. 3A; Supplemental Fig. S4B,C).
Figure 3.
Knocking down FH or SDH reduces the TET-catalyzed 5hmC production in cultured cells. (A) HEK293T cells with stable knockdown of FH or SDHA/B were transiently transfected with plasmids expressing the indicated proteins. Thirty-six hours to 40 h after the transfection, cells were fixed and stained with antibodies specific to Flag to determine the expression of TET protein, with antibodies specific to 5hmC to determine the levels of 5hmC, and with antibodies specific to DAPI to view the cell nuclei. Bars, 251 μm. See also Supplemental Figure S4. (B–D) HEK293T cells were transiently transfected as described in A. Genomic DNAs were isolated from cells and immunoblotted with an antibody specific to 5hmC. Quantification of 5hmC was calculated from three independent assays. The expression of individual proteins was determined by immunoblotting as shown at the right. One representative quantification of the 5hmC level determined from the assays using 100 ng of genomic DNA is included. Error bars represent standard deviation (SD) for triplicate experiments.
To confirm the immunofluorescence data, we determined the 5hmC levels by dot blot analysis that allows for more quantitative measurement of 5hmC (Fig. 3B–D). Consistent with immunofluorescence results, ectopic expression of the wild-type TET (TET1-CD and TET2-CD), but not the catalytic mutant TET (TET1-CM and TET2-CM), dramatically increased 5hmC levels. In cells with stable knockdown of FH, SDHA, or SDHB, however, overexpression of TET1-CD or TET2-CD showed significantly less 5hmC as compared with control cells with normal FH or SDH expression (Fig. 3B–D). Knockdown of FH, SDHA, and SDHB reduced TET1-induced 5hmC levels by 71%, 73%, and 82%, respectively, as compared with control cells coinfected with retrovirus containing the control shRNA vector pMKO. A virtually identical result was obtained for TET2-catalyzed 5hmC production, which was also decreased by 80%, 83% and 69% in cells with stable knockdown of FH, SDHA, and SDHB, respectively. The DNA amount used for dot blot analysis was determined by methylene blue staining (Supplemental Fig. S8A).
Taken together, these data indicate that knockdown of FH and SDHA/B leads to elevated levels of intracellular fumarate and succinate, respectively, which act as competitors of α-KG to inhibit TET-catalyzed hydroxylation of 5mC.
Knocking down Fh or Sdha inhibits multiple α-KG-dependent dioxygenases and regulates target gene expression in vivo
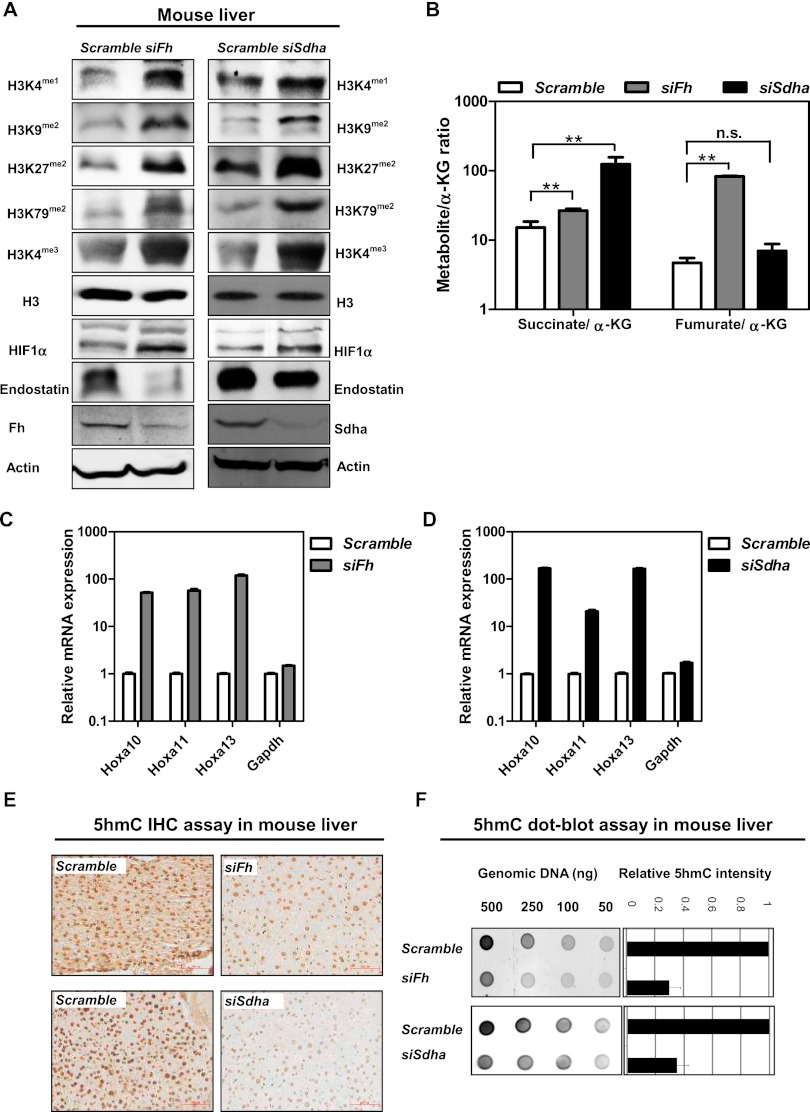
Given our findings that knockdown of FH or SDH gene expression broadly inhibits α-KG-dependent dioxygenases in cultured cells, we sought to investigate whether reduced expression of Fh and Sdh could affect α-KG-dependent dioxygenases in vivo. To this end, the RNAi approach was used to transiently knock down Fh and Sdha in mouse livers. The siRNAs directed against Fh and Sdha and the corresponding nontargeting scramble siRNAs were delivered via the hydrodynamic tail vein injection procedure to mice. At 12-h after siRNA injection, we were able to achieve 62% and 78% reduction of Fh and Sdha mRNA expression, respectively, in mouse livers (Supplemental Fig. S7A). As a result, hepatic expression of Fh and Sdha proteins was decreased by threefold and twofold, respectively (Fig. 4A).
Figure 4.
Knocking down Fh or Sdha inhibits multiple α-KG-dependent dioxygenases and regulates gene expression in vivo. (A) Mice with transient knockdown of Fh or Sdha in the liver were generated by using the hydrodynamic tail vein-based delivery technique (n = 3 per group). The Fh or Sdha knockdown efficiency and levels of histone methylation, HIF1α, and endostatin were determined by Western blot. (B) The ratios of succinate/α-KG and fumarate/α-KG in mouse livers after suppression of Fh or Sdha were determined by GC-MS. Error bars represent standard deviation (SD) for triplicate experiments. (**) P < 0.01 versus Scramble; (n.s.) not significant. See also Supplemental Figure S5. (C,D) The mRNA expression of Hoxa genes in mouse livers after suppression of Fh or Sdha was determined by quantitative RT–PCR. Error bars represent standard deviation (SD) for triplicate experiments. (E,F) The 5hmC levels in mouse livers after suppression of Fh or Sdha were determined by immunohistochemistry (E) and dot blot assay (F). Bars, 334 μm. Error bars represent standard deviation (SD) for triplicate experiments. See also Supplemental Figure S6.
Transient knockdown of Fh or Sdha in mouse livers led to accumulation of intracellular fumarate or succinate, respectively, as determined by GC-MS (Supplemental Fig. S5A,B). Notably, the succinate/α-KG ratio was also elevated in Fh knockdown liver cells, although less dramatically than Sdha knockdown. In contrast, the fumarate/α-KG ratio was increased only in Fh knockdown, but not in Sdha knockdown, liver cells (Fig. 4B).
We next determined how the accumulation of fumarate and succinate would affect the activities of α-KG-dependent dioxygenases. Transient knockdown of Fh in mouse livers significantly increased monomethylation on H3K4 (by 2.5-fold); dimethylation on H3K9 (by 4.2-fold), H3K27 (by 4.3-fold), and H3K79 (by 5.2-fold); and trimethylation on H3K4 (by 2.8-fold). Likewise, knockdown of Sdha showed significant increases of monomethylation on H3K4 (by 1.9-fold); dimethylation on H3K9 (by fourfold), H3K27 (by 3.8-fold), and H3K79 (by 4.8-fold); and trimethylation on H3K4 (by 4.3-fold) (Fig. 4A; Supplemental Fig. S6A).
To further substantiate the above results, we determined mRNA expression of Hoxa genes whose up-regulation is associated with increased H3K79 dimethylation in MLL rearranged mouse leukemia and human AML patients (Krivtsov et al. 2008). In line with the observed increase of H3K79 dimethylation, mRNA expression of several Hoxa genes (e.g., Hoxa10, Hoxa11, and Hoxa13) was up-regulated by >10-fold in mouse livers after suppression of the Fh or Sdha gene (Fig. 4C,D). These data suggest that accumulation of fumarate and succinate can inhibit KDMs in mouse liver cells in vivo.
Immunohistochemical staining showed that 5hmC was readily detectable in mouse livers (Fig. 4E). Transient knockdown of Fh or Sdha by siRNA significantly decreased 5hmC in liver cells, especially near hepatic portal veins (Fig. 4E; Supplemental Fig. S6B). In agreement, a dot blot assay demonstrated that 5hmC production was decreased by 72% and 63% in the livers of Fh and Sdha siRNA knockdown mice, respectively (Fig. 4F; Supplemental Fig. S6C). Suppression of Fh or Sdha did not significantly affect the hepatic mRNA levels of Tet genes (Supplement Fig. S7B), indicating that transient knockdown of Fh or Sdha would inhibit the activity of TETs by not down-regulating the transcription of TET genes.
Taken together, the above findings provide in vivo evidence supporting the notion that decreased activity of FH and SDHA leads to accumulation of fumarate and succinate, which inhibit α-KG-dependent dioxygenases and regulate target gene expression.
Tumor-derived FH and SDH mutants inhibit α-KG-dependent dioxygenases
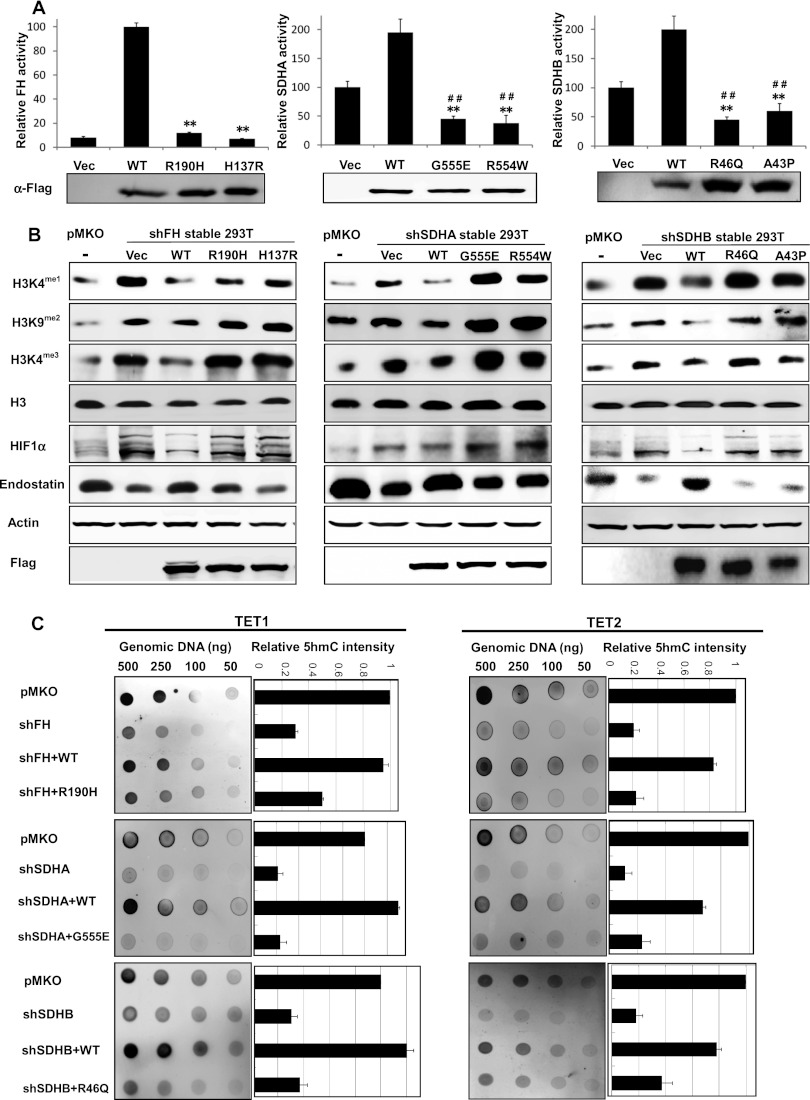
Most tumor-derived mutations targeting FH or SDH genes cause obvious loss of function such as truncation or frameshift (Alam et al. 2005; Bayley et al. 2008; Bardella et al. 2011), which would eliminate the activity of FH or SDH enzymes. There are also many missense mutations whose exact functional consequences have not been characterized. We next determined whether ectopic expression of tumor-derived FH or SDH mutants would affect the activity of endogenous α-KG-dependent dioxygenases. To avoid the potential effect from endogenous proteins, cells with stable depletion of endogenous FH or SDHA/B were generated. Then, ectopic expression of wild-type and tumor-associated mutants were introduced by retroviral vectors. Some of the tumor-derived FH or SDHA/B mutants displayed a dramatic decrease of enzyme activity. For instance, the activity of FHR190H (the most frequent mutation in hereditary leiomyomatosis and renal cell cancer [HLRCC] patients) and FHH137R is only 17% and 10%, respectively, of wild-type FH (Fig. 5A). The activity of tumor-derived mutants SDHAG555E and SDHAR554W is 51% and 46%, respectively, of wild-type SDHA, and the activity of SDHBR46Q (the most frequent SDHB mutation in paraganglioma) and SDHBA43P mutants is 48% and 61%, respectively, of wild-type SDHB.
Figure 5.
Ectopic expression of tumor-derived FH or SDH mutants inhibits α-KG-dependent dioxygenases in cultured cells. (A) HEK293T cells with stable FH or SDHA/B knockdown were transiently transfected with plasmids expressing the indicated proteins, and the enzyme activity of wild-type and mutant FH or SDH was measured as described in the Material and Methods. Error bars represent standard deviation (SD) for triplicate experiments. (**) P < 0.01 versus wild type; (##) P < 0.01 versus vector. (B) Overexpression of tumor-derived FH or SDHA/B mutants inhibits histone demethylation and hydroxylation of prolyl residues. Levels of histone methylation, HIF1α, and endostatin were determined by Western blot. (C) Overexpression of tumor-derived FH or SDHA/B mutants inhibits the TET-catalyzed 5mC oxidation. 5hmC levels were determined by dot blot. Error bars represent standard deviation (SD) for triplicate experiments.
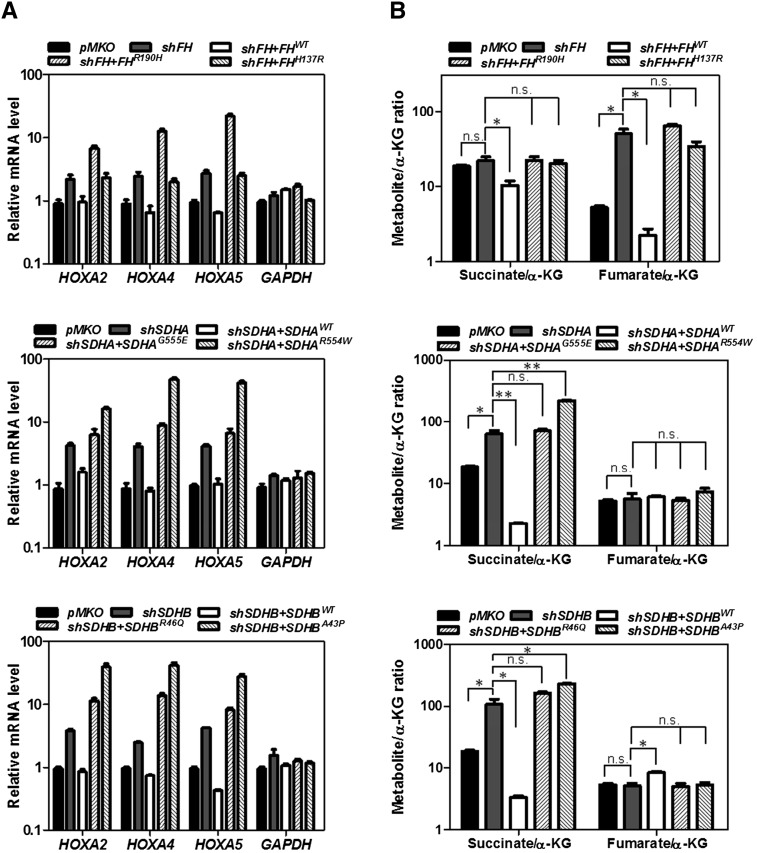
Next, we set forth to determine whether the activities of α-KG-dependent dioxygenases would be affected in cells expressing tumor-derived FH or SDH mutants. We found that ectopic expression of wild-type FH or SDHA/B resulted in decreases of dimethylation on H3K9 (by 1.5-fold) as well as monomethylation and trimethylation on H3K4 (by 2.5-fold and twofold, respectively), consistent with increased KDM activity (Fig. 5B). However, these changes in genome-wide histone methylations were not observed in cells re-expressing tumor-derived FH or SDHA/B mutants (Fig. 5B). Since alterations of histone methylation will likely have a broad impact on gene expression, we determined mRNA expression of HOXA genes in these cells and found that knockdown of FH or SDHA/B resulted in up-regulation of several HOXA genes (e.g., HOXA2, HOXA4, and HOXA5). Re-expression of wild-type FH or SDH reduced HOXA gene expression to the control levels. In contrast, re-expression of mutant FH or SDH had no effect or even increased HOXA gene expression (Fig. 6A).
Figure 6.
Tumor-derived FH or SDH mutants are not functional in fumarate or succinate metabolism. (A) HEK293T cells with stable FH or SDHA/B knockdown were transfected with the indicated plasmids. Quantitative RT–PCR was performed to determine mRNA expression of HOXA genes in these cells. Error bars represent standard deviation (SD) for triplicate experiments. (B) The ratios of succinate/α-KG and fumarate/α-KG were determined by GC-MS. Error bars represent standard deviation (SD) for triplicate experiments. (*) P < 0.05 versus Scramble;; (**) P < 0.01 versus Scramble; (n.s.) not significant. See also Supplemental Figure S9.
In addition, ectopic expression of wild-type FH or SDHA/B decreased HIF1α and increased endostatin in cells with depletion of endogenous FH or SDHA/B as compared with cells expressing empty pMKO vector. These changes in HIF1α and endostatin were, again, not found in cells re-expressing tumor-derived FH or SDHA/B mutants (Fig. 5B). Furthermore, we investigated the impact of tumor-derived FH and SDH mutations on TET-catalyzed 5mC oxidation. Stable cells with depletion of endogenous FH, SDHA, and SDHB displayed much weaker 5hmC signal (29%, 27%, and 18%, respectively) as compared with control pMKO cells after transfection with TET1-CD or TET2-CD. Cotransfection with wild-type FH, SDHA, or SDHB, but not their mutants (i.e., FHR190H, SDHAG555E, or SDHBR46Q), could rescue the reduction of 5hmC levels in FH or SDH stable knockdown cells (Fig. 5C; Supplemental Fig. S8B). These findings are consistent with the notion that accumulation of fumarate or succinate in the FH or SDH knockdown cells inhibits TET activity, thereby lowering 5hmC levels. Re-expression of wild-type FH and SDH dramatically reduced fumarate and succinate, respectively, in the FH or SDH knockdown cells and thus increased 5hmC by relieving inhibition on TETs. In contrast, re-expression of the tumor-derived mutants had no effect or even increased fumarate and/or succinate levels (Fig. 6B; Supplemental Fig. S9A–C). Together, these results indicate that the tumor-derived FH and SDH mutants are not functional in fumarate or succinate metabolism. Accumulation of fumarate or succinate in cancer cells containing FH or SDH mutations may contribute to alterations of epigenetic DNA modification via inhibiting TETs.
Discussion
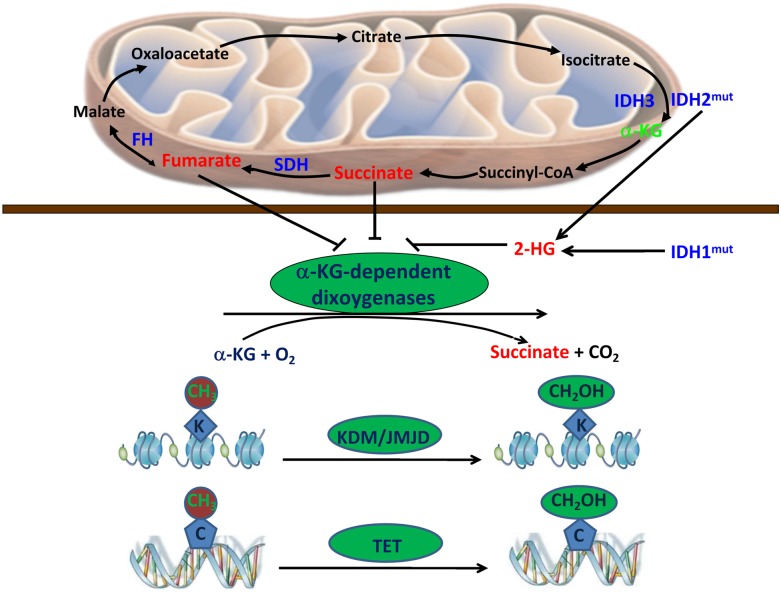
Succinate and fumarate serve important physiological functions in cell metabolism and could become oncogenic when their concentrations accumulate to abnormally high levels. It has been proposed that FH or SDH mutations lead to accumulation of their substrates, fumarate and succinate, which bind directly to and inhibit the activity of PHDs, leading to increased stability and elevated levels of HIF proteins (King et al. 2006). We show here that fumarate and succinate can also function as α-KG antagonists to broadly inhibit α-KG-dependent dioxygenases besides PHDs, including the JMJD family KDMs and the TET family of 5mC hydroxylases (Fig. 7). These observations suggest that tumor cells containing FH or SDH mutations accumulate fumarate and succinate, which then inhibit histone and DNA demethylations. Given the well-characterized tumor suppressor function of TET, we propose that inhibition of the TET family enzymes may contribute to tumorigenesis of FH or SDH mutant cancer. Moreover, alterations of histone methylation will likely have a broad impact on gene expression, which may also contribute to the tumor suppressor functions of FH and SDH.
Figure 7.
Fumarate and succinate can function as α-KG antagonists to broadly inhibit α-KG-dependent dioxygenases, including the JMJD family KDMs and the TET family of 5mC hydroxylases.
In this study, we show that the succinate/α-KG ratio is elevated in cells expressing several tumor-derived SDH mutants (e.g., SDHAR554W or SDHBA43P). Considering succinate is a product of α-KG-dependent dioxygenase reactions, an alternative model would be that the high levels of succinate accumulated in FH or SDH mutated cells may inhibit the activity of α-KG-dependent dioxygenases through product inhibition. However, the succinate/α-KG ratio is not changed in cells expressing tumor-derived FH mutants, suggesting that the product inhibition mechanism may not explain the impairment of α-KG-dependent dioxygenases in FH mutated cells. It is also possible that the loss of function of FH or SDH in tumorigenesis involves other mechanisms independent of the regulation of α-KG-dependent dioxygenases. Recently, a new function of fumarate—covalently attaching to cysteine residues—was reported. Fumarate can directly cause aberrant succination of many proteins, including KEAP1 (Kelch-like ECH-associated protein 1; an electrophile-sensitive component of an E3 ubiquitin ligase) (Adam et al. 2011). KEAP1 is the principal regulator of the nuclear factor (erythroid-derived 2)-like 2 (NRF2) via controlling its ubiquitylation and degradation, thereby activating the antioxidant pathway (Adam et al. 2011; Ooi et al. 2011). Moreover, KEAP1 and NRF2 have been implicated in tumor development (Taguchi et al. 2011), although their contributions to oncogenesis in FH mutant cancer still require further exploration.
One cannot help but notice the common mechanisms shared by mutations in the three metabolic tumor suppressor genes IDH, FH, and SDH. Mutation in IDH results in accumulation of the oncometabolite D-2-HG together with reduction of α-KG. A common paradigm emerges that alteration of metabolic intermediates caused by mutations in metabolic tumor suppressors is responsible for the tumor suppression effect of metabolic enzyme mutations. For the group of IDH, FH, and SDH, the common targets are the α-KG-dependent dioxygenases, including both KDMs and DNA demethylases. These observations suggest a possibility of manipulating metabolites and/or that metabolic enzymes may provide a potential therapeutic approach for cancer treatment. It is well established that metabolism can be regulated by transcription, which controls the levels of metabolic enzyme expression. However, our study also suggests a reciprocal mechanism: regulation of transcription by metabolism via the changing levels of metabolic intermediates—such as fumarate and succinate—that influence gene expression by epigenetic modifications.
Materials and methods
Please refer to the Supplemental Material for more detailed information about antibodies; plasmids, cell culture, and transient transfection; GC-MS analysis; retroviral infection and generation of stable cell lines; animals; immunoprecipitation and Western blot; quantitative real-time PCR; immunohistochemical analysis; immunofluorescence assay; and dot blot assay.
Preparation of siRNAs and siRNA treatment
For cell transfection, siRNAs targeting human FH (Gene ID: 2271), SDHA (Gene ID: 6389), and SDHB (Gene ID: 6390) were purchased commercially (Invitrogen). siRNAs were received as desalted, deprotected oligonucleotides. Cultured cells were transfected with siRNAs using Lipofectamin 2000 (Invitrogen). At 4–6 h post-transfection, the culture medium was changed, and the cells were harvested at 24–36 h after transfection.
For mouse injection, siRNAs targeting mouse Fh and Sdha were chemically synthesized using 2′Ome modification (Genepharma), and their sequences are listed in Supplemental Table 1. Synthetic siRNAs were delivered using a modified “hydrodynamic transfection method,” by which 1 OD of siRNA oligo dissolved in 1 mL of Ringer’s solution (0.9% NaCl, 0.03% KCl, 0.016% CaCl2) was rapidly injected into the tail vein. An equal volume of normal saline was used as control. At 12 h after siRNA injection, mouse livers were dissected, stored in 4% formalin (Shenggong) or frozen in liquid nitrogen, and stored at −80°C for further analysis.
Enzyme activity assay
To measure CeKDM7A demethylase activity toward dimethylated H3K9, synthetic monomethylated peptide corresponding to monomethylated H3K9 (ARTKQTARK (me1)STGGKA) was used as substrate. Demethylase assays were carried out in the presence of 10 μg of enzyme, 1 μg of peptide in 20 μL of buffer (20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 50 μM (NH4)2Fe(SO4)2, 100 μM α-KG, 2 mM Vc, 10 mM PMSF] for 3 h. The demethylation reaction mixture was desalted by passing through a C18 ZipTip (Millipore). To examine the inhibitory effect of fumarate or succinate on the reaction, various concentrations of fumuarate or succinate were incubated with KDM7A briefly before adding other reaction mixtures. The samples were analyzed by a MALDI-TOF/TOF mass spectrometer.
To measure HsKDM4A demethylase activity toward trimethylated H3K36, synthetic trimethylated peptide corresponding to trimethylated H3K36 (RKSAPATGGVK(me3)KPHRY) was used as substrate. A construct encoding HsKDM4A residues 1–357 was amplified by PCR and cloned into pET28a-His. After purification, the activity of the enzyme (10 μg each tube) was measured as described above.
For the FH enzyme activity assay, cells were collected at 36 h after transient transfection with wild-type or mutant FH plasmids. After immunoprecipitation with Flag beads, wild-type or mutated FH was eluted by a Flag peptide into a total volume of 100 μL. Afterward, 2–5 μL of eluted protein was added to 200 μL of enzyme assay buffer (50 mM malate, 10 mM potassium phosphate at pH 7.3), and the absorbance at OD240 was recorded.
For the SDH complex activity assay, cells were collected at 36 h after transient transfection with wild-type or mutant SDHA/B plasmids. Afterward, cells were lysed with 600 μL of enzyme assay buffer (0.1% Triton X-100, 25 mM KHPO4 at pH 7.4) and then incubated with 20 mM succinate, 50 μM decylubiquinone, 5 μM rotenone, 2 μM Antimycin A, and 10 μM NaN3 for 15 min at room temperature. The reaction was initiated by adding 50 μM DCIP, and the changes of absorbance at 600 nm were recorded.
Statistical analysis
Statistical analysis was performed with a two-tailed unpaired Student’s t-test. All data shown represent the results obtained from three independent experiments with standard deviations (mean ± SD). The value of P < 0.05 was considered statistically significant.
Acknowledgments
We thank the members of the Fudan MCB laboratory for discussions and support throughout this study. We appreciate Dr. Bin Wang for providing cell-permeable octyl-α-KG ester, methyl-succinate ester, and methyl-fumarate ester. This work was supported by MOST 973 (nos. 2012CB910300, 2012CB910101, 2011CB910600, and 2009CB918401) and NSFC (grant nos. 30600112, 30871255, 31071192, and 81120108016). This work was also supported by the 985 Program, Shanghai key project (grant no. 09JC1402300), the Shanghai Leading Academic Discipline Project (project no. B110), NIH grants (to Y.X. and K.L.G.), and James McDonnell Foundation and Samuel Waxman Foundation (to Y.X.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.191056.112.
References
- Adam J, Hatipoglu E, O’Flaherty L, Ternette N, Sahgal N, Lockstone H, Baban D, Nye E, Stamp GW, Wolhuter K, et al. 2011. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: Roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell 20: 524–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam NA, Olpin S, Rowan A, Kelsell D, Leigh IM, Tomlinson IP, Weaver T 2005. Missense mutations in fumarate hydratase in multiple cutaneous and uterine leiomyomatosis and renal cell cancer. J Mol Diagn 7: 437–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O’Donnell P, Grigoriadis A, Diss T, et al. 2011. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol 224: 306–308 [DOI] [PubMed] [Google Scholar]
- Astuti D, Latif F, Dallol A, Dahia PL, Douglas F, George E, Sköldberg F, Husebye ES, Eng C, Maher ER 2001. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet 69: 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardella C, Pollard PJ, Tomlinson I 2011. SDH mutations in cancer. Biochim Biophys Acta 1807: 1432–1443 [DOI] [PubMed] [Google Scholar]
- Bayley JP, Launonen V, Tomlinson IP 2008. The FH mutation database: An online database of fumarate hydratase mutations involved in the MCUL (HLRCC) tumor syndrome and congenital fumarase deficiency. BMC Med Genet 9: 20 doi: 10.1186/1471-2350-9-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley JP, Kunst HP, Cascon A, Sampietro ML, Gaal J, Korpershoek E, Hinojar-Gutierrez A, Timmers HJ, Hoefsloot LH, Hermsen MA, et al. 2010. SDHAF2 mutations in familial and sporadic paraganglioma and phaeochromocytoma. Lancet Oncol 11: 366–372 [DOI] [PubMed] [Google Scholar]
- Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PE, Rubinstein WS, Myers EN, et al. 2000. Mutations in SDHD,a mitochondrial complex II gene, in hereditary paraganglioma. Science 287: 848–851 [DOI] [PubMed] [Google Scholar]
- Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol 5: 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. 2011. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep 12: 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damato S, Alorjani M, Bonar F, McCarthy SW, Cannon SR, O’Donnell P, Tirabosco R, Amary MF, Flanagan AM 2012. IDH1 mutations are not found in cartilaginous tumours other than central and periosteal chondrosarcomas and enchondromas. Histopathology 60: 363–365 [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107: 43–54 [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, Devilee P, Cremers CW, Schiffman JD, Bentz BG, et al. 2009. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science 325: 1139–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausinger RP 2004. FeII/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol 39: 21–68 [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly JP, Bastos AU, Cerutti JM 2010. Identification of several novel non-p.R132 IDH1 variants in thyroid carcinomas. Eur J Endocrinol 163: 747–755 [DOI] [PubMed] [Google Scholar]
- Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, Merino M, Trepel J, Zbar B, Toro J, et al. 2005. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: Novel role of fumarate in regulation of HIF stability. Cancer Cell 8: 143–153 [DOI] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG Jr 2001. HIF1α targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 292: 464–468 [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. 2001. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292: 468–472 [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr 2009. SDH5 mutations and familial paraganglioma: Somewhere Warburg is smiling. Cancer Cell 16: 180–182 [DOI] [PubMed] [Google Scholar]
- King A, Selak MA, Gottlieb E 2006. Succinate dehydrogenase and fumarate hydratase: Linking mitochondrial dysfunction and cancer. Oncogene 25: 4675–4682 [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. 2008. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 14: 355–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling TS, Shi RH, Zhang GX, Zhu H, Yu LZ, Ding XF 2005. Common single nucleotide polymorphism of hypoxia-inducible factor-1α and its impact on the clinicopathological features of esophageal squamous cell carcinoma. Chin J Dig Dis 6: 155–158 [DOI] [PubMed] [Google Scholar]
- Loenarz C, Schofield CJ 2008. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol 4: 152–156 [DOI] [PubMed] [Google Scholar]
- Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR 2003. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia 17: 637–641 [DOI] [PubMed] [Google Scholar]
- MacKenzie ED, Selak MA, Tennant DA, Payne LJ, Crosby S, Frederiksen CM, Watson DG, Gottlieb E 2007. Cell-permeating α-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells. Mol Cell Biol 27: 3282–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. 2009. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361: 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan AK, Bojdani E, Xing M 2010. Identification and functional characterization of isocitrate dehydrogenase 1 (IDH1) mutations in thyroid cancer. Biochem Biophys Res Commun 393: 555–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussini E, Hutton JJ Jr, Udenfriend S 1967. Collagen proline hydroxylase in wound healing, granuloma formation, scurvy, and growth. Science 157: 927–929 [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. 2010. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17: 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oermann EK, Wu J, Guan KL, Xiong Y 2012. Alterations of metabolic genes and metabolites in cancer. Semin Cell Dev Biol. doi: 10.1016/j.semcdb.2012.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi A, Wong JC, Petillo D, Roossien D, Perrier-Trudova V, Whitten D, Min BW, Tan MH, Zhang Z, Yang XJ, et al. 2011. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell 20: 511–523 [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard PJ, Brière JJ, Alam NA, Barwell J, Barclay E, Wortham NC, Hunt T, Mitchell M, Olpin S, Moat SJ, et al. 2005. Accumulation of Krebs cycle intermediates and over-expression of HIF1α in tumours which result from germline FH and SDH mutations. Hum Mol Genet 14: 2231–2239 [DOI] [PubMed] [Google Scholar]
- Rose NR, McDonough MA, King ON, Kawamura A, Schofield CJ 2011. Inhibition of 2-oxoglutarate dependent oxygenases. Chem Soc Rev 40: 4364–4397 [DOI] [PubMed] [Google Scholar]
- Scatena R, Bottoni P, Giorgia Botta G, Martorana GE, Giardina B 2007. The role of mitochondria in pharmacotoxicology: A reevaluation of an old, newly emerging topic. Am J Physiol Cell Physiol 293: C12–C21 doi: 10.1152/ajpcell.00314.2006 [DOI] [PubMed] [Google Scholar]
- Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E 2005. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell 7: 77–85 [DOI] [PubMed] [Google Scholar]
- Taguchi K, Motohashi H, Yamamoto M 2011. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells 16: 123–140 [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, et al. 2002. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet 30: 406–410 [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. 2009. IDH1 and IDH2 mutations in gliomas. N Engl J Med 360: 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, White SB, Zhao Q, Lee FS 2001. HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc Natl Acad Sci 98: 9630–9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, et al. 2009. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science 324: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]