The atypical RNA-binding protein fus/TLS interacts with auxiliary splice factors of the SR family and hnRNPs and can influence alternative splice site selection. Although FUS has been well studied in cultured cells, its physiological function in whole organisms is less clear. In this study by Harland and colleagues, the function of fus/TLS during early frog development is investigated. The authors show that knockdown of fus causes mesodermal differentiation defects and epithelial dissociation and demonstrate that these defects are caused by intron retention in specific transcripts in the FGF signaling pathway and cadherin complex. The authors then use RNA-seq to confirm the splicing defects and determine that fus knockdown affects 3%–5% of all transcripts and that the affected genes are highly enriched in transcription factors and signaling components involved in developmental processes. Thus, these results provide novel insight into splicing regulators that regulate biologically coherent sets of transcripts.
Keywords: Xenopus, fus, splicing, signaling, embryo development, RNA-seq
Abstract
Here we investigated the function of the atypical RNA-binding protein fus/TLS (fused in sarcoma/translocated in sarcoma) during early frog development. We found that fus is necessary for proper mRNA splicing of a set of developmental regulatory genes during early frog development and gastrulation. Upon fus knockdown, embryos fail to gastrulate and show mesodermal differentiation defects that we connect to intron retention in fgf8 (fibroblast growth factor 8) and fgfr2 (fgf receptor 2) transcripts. During gastrulation, the animal and marginal regions dissociate, and we show that this is caused, at least in part, by intron retention in cdh1 transcripts. We confirm the specificity of splicing defects at a genomic level using analysis of RNA sequencing (RNA-seq) and show that 3%–5% of all transcripts display intron retention throughout the pre-mRNA. By analyzing gene ontology slim annotations, we show that the affected genes are enriched for developmental regulators and therefore represent a biologically coherent set of targets for fus regulation in embryogenesis. This shows that fus is central to embryogenesis and may provide information on its function in neurodegenerative disease.
Pre-mRNA splicing, alternative and constitutive, is catalyzed by the spliceosome that contains five small nuclear ribonucleoproteins (snRNPs): U1, U2, U4, U5, and U6. Splicing occurs in a stepwise fashion, and a series of distinct complexes form in the process at the 5′ and 3′ splice sites and branch point site (Wahl et al. 2009). However, these sequences are short and poorly conserved. During splicing, the spliceosome must therefore succeed in recognizing and joining splice sites that are surrounded by numerous similar sequences and often separated by considerable distances. Further complicating the task, the interactions between the snRNPs and the pre-mRNA are generally weak and rely for specificity on numerous auxiliary proteins that interact with the core snRNPs. This general principle of multiple interactions is important not only for precisely finding correct splice pairs, but also for the flexibility necessary for alternative splice site selection.
Comprehensive analysis of purified early spliceosomal complexes shows that they consist of at least 85 and perhaps as many as 150–300 different proteins with a combined mass of 2.7 MDa. Many of these function in the enzymatic and conformational changes that occur during splicing, while others are involved in exon and intron definition by binding to splicing enhancers or silencers present in the pre-mRNA. For example, members of the serine–arginine-rich splice factors (SR proteins) frequently bind sequences in exons and stimulate exon inclusion by stabilizing U1 and U2 binding to the 5′ splice site and branch point site, respectively. Other well-known splicing regulators are the heterologous nuclear RNPs (hnRNPs) that generally repress exon inclusion and hence counteract SR protein function. Adding to the complexity, splicing occurs concurrently with transcription (Perales and Bentley 2009). As pre-mRNA is synthesized by RNA polymerase II, it is expelled through a groove near the C-terminal domain (CTD) of the large subunit. The CTD is an organizing center for splice factors that ride with the polymerase and provide immediate access to the nascent RNA. However, the relationship between transcription and splicing is still poorly understood.
Regulated splicing of pre-mRNA has emerged as an important control point in gene regulation. Evolutionary conservation of alternative splicing is found in a variety of developmental regulators; for example, in the fibroblast growth factor (FGF) signaling pathway. In this pathway, both ligand and receptor pre-mRNAs are alternatively spliced, resulting in proteins with dramatically different biological properties. For instance, FGF receptor 2 (Fgfr2) gives rise to two major splice forms, the b and c forms, by use of mutually exclusive cassette exons. The b and c forms differ in their ligand-binding domains, resulting in receptors with distinct agonist preferences: FGFR2b is a high-affinity receptor for the FGF7/10 group ligands, whereas FGFR2c binds members of the FGF4/FGF8b group (Ornitz et al. 1996; Blunt et al. 1997). FGF ligands themselves are subject to alternative splicing: An alternative 3′ splice site in the second intron of fgf8 creates two splice forms with distinct activities in Xenopus; fgf8a posteriorizes the neural plate, whereas fgf8b is a potent mesoderm inducer and is required for mesoderm development (Fletcher et al. 2006).
Among potential regulators of splicing, the FET protein family consists of fus (fused in sarcoma; also known as translocated in sarcoma [TLS], Pigpen, and hnRNP-P2), TATA-box-binding-associated factor 15 (TAF15; also known as TAFII68), and Ewing's sarcoma breakpoint region 1 protein (EWSR1). FET proteins are atypical RNA-binding proteins (RBPs) that share similar domains, including an RNA recognition motif (RRM) flanked by Arg–Gly–Gly-rich regions, a Cys2–Cys2 zinc finger, and an N-terminal transcriptional activation domain (TAD) (Tan and Manley 2009). FET proteins have primarily been studied in the context of deleterious chromosomal translocations that lead to distinct soft tissue tumors. The exact functions of normal FET proteins remain unclear, but the presence of both RRM and TAD domains has led to the proposal that they act as a link between transcription and splicing (Law et al. 2006).
In SELEX experiments, FUS binds the RNA motif GGUG; it also binds single-stranded G and U homopolymers as well as ssDNA and dsDNA (Lerga et al. 2001). FUS interacts with auxiliary splice factors of the SR family and hnRNPs and can influence alternative splice site selection in cell-free systems, possibly by acting in the spliceosome assembly (Lerga et al. 2001). Furthermore, FUS has been shown to inhibit RNA polymerase III transcription (Tan and Manley 2010) and bind noncoding RNAs transcribed from the promoter region of Ccnd1, thus repressing its transcription (Wang et al. 2008). While FUS has been studied in some detail in vitro and in cultured cells, less is known about its physiological function in whole organisms. In a mouse gene trap screen, a Fus mutant with an insertion in exon 12 suffered perinatal death, possibly due to lymphocyte defects and chromosomal instability (Hicks et al. 2000).
Recently, FUS research was re-energized after mutations in the gene were found to associate with the familial form of amyolotrophic lateral sclerosis (fALS) (Kwiatkowski et al. 2009; Vance et al. 2009). Most fALS mutations identified are missense and cluster near the C terminus. These change the distribution of FUS between the nucleus and cytoplasm, but the precise function of Fus mutations in the pathophysiology of ALS remains elusive.
We identified fus in an expression cloning screen for mRNAs that affect early frog development (Dichmann et al. 2008). We show that morpholino oligonucleotide (MO)-mediated knockdown causes mesodermal differentiation defects and epithelial dissociation. These defects are caused by intron retention in specific transcripts in the FGF signaling pathway and cadherin complex. We used RNA sequencing (RNA-seq) to confirm the splicing defects and determined that fus knockdown affects 3%–5% of all transcripts and that the affected genes are highly enriched in transcription factors and signaling components involved in developmental processes. These results support the idea that splicing regulators may control biologically coherent sets of transcripts, as has also been described for the alternative splicing regulator NOVA (Ule et al. 2005).
Results
fus is expressed in the animal region and marginal zone
We isolated fus in an expression cloning screen (Dichmann et al. 2008). fus is expressed zygotically in the blastula (Fig. 1A) and strongly in the animal and marginal zone of gastrulating embryos (Fig. 1B,C). Expression was also detected in vegetal cells during late gastrulation (Fig. 1C). There are two fus homeologs/alloalleles in the pseudotetraploid Xenopus laevis that are expressed by RT–PCR in the same profile during development (data not shown).
Figure 1.
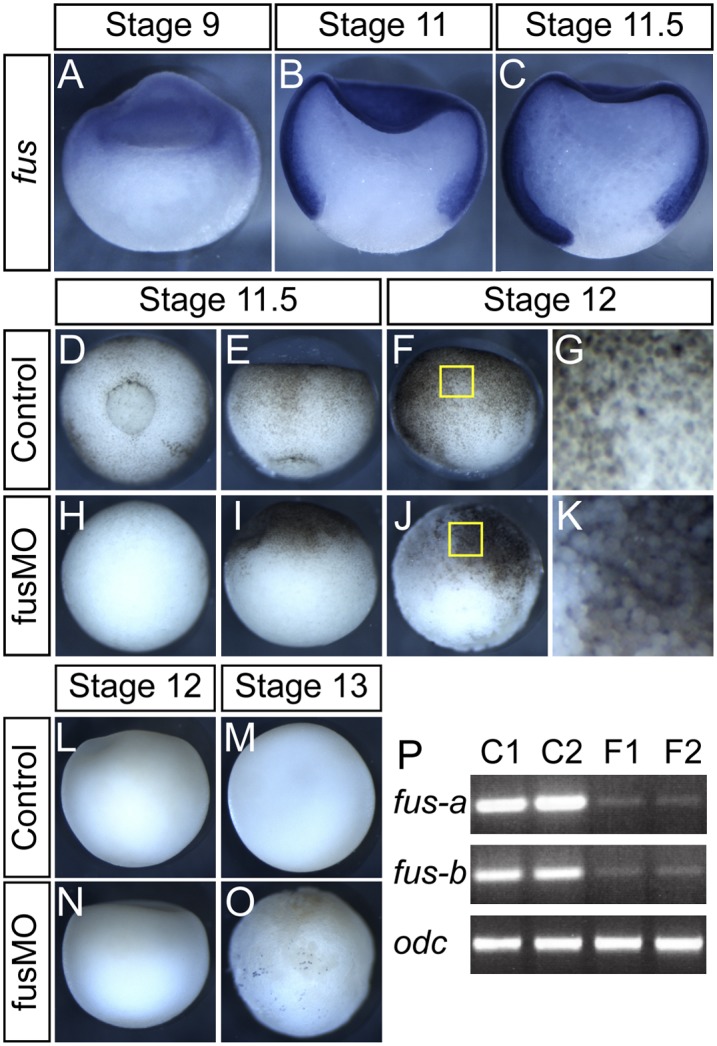
fus is expressed in the animal region, and knockdown results in gastrulation defects and animal dissociation. All pictures show a lateral view with the animal region up, except for D and H, which show vegetal view. (A–C) Normal embryos bisected and subjected to ISH with fus antisense probe. (A) Initial fus staining at stage 9 is confined to the animal region. (B) At mid-gastrulation, fus is expressed strongly throughout the prospective mesodermal and ectodermal regions but is excluded from the endoderm. (C) This pattern persists through gastrulation, although some weak staining can be observed in vegetal cells. (D–K) fus knockdown results in lack of blastopore formation and cell dissociation. (D,E) Normal embryos at mid-gastrulation, with prospective mesendoderm involuting at the blastopore. (H,I) fus morphants show complete lack of blastopore formation and gastrulation movements. (F) Normal embryo showing intact epithelia. (J) Individual cells detach from the animal and marginal regions in fus morphants. (G,K) Magnified view of the yellow box in F and J, respectively. (L–O) Absence of apoptosis during gastrulation in fus morphants. Control (L,M) and fusMO (N,O) embryos TUNEL-stained for apoptotic cells at cell dissociation (L,N) or after cell dissociation occurs (M,O). No apoptotic cells were detected at stage 12, when cell dissociation happens. At stage 13, some apoptotic cells that were detected in fus have dissociated and therefore mostly likely show a secondary effect. (P) Single-embryo RT–PCR spanning the first four exons of fus on embryos injected with fusMO4 showing aberrant splicing of the first intron in fus transcripts. Normal transcripts of both homeologs (fus-a and fus-b) in X. laevis are significantly reduced relative to control odc transcripts.
fus knockdown prevents gastrulation and causes dissociation
To characterize the function of fus, we used antisense MOs against fus RNA. One MO (fusMO1) targeted the start codon of both homeologs so as to prevent translation. Another MO (fusMO4) targeted the first splice donor site and is predicted to cause inclusion of intron 1 and premature termination. Injection of either fusMO alone or in combination resulted in identical phenotypes. We also tested two homeolog-specific MOs in X. laevis that produced no phenotype individually, but when injected in combination, they caused a phenotype similar to MO1 and MO4. We observed the strongest and most consistent effects using MO1 and MO4 in combination and used this unless otherwise stated. MO-injected embryos were normal during cleavage stages but failed to form a blastopore or show gastrulation movements, although they appeared otherwise healthy (Fig. 1D,E,H,I). From stage 12 onward, cells in the animal and marginal zone progressively dissociated from the embryo (Fig. 1F,G,J,K). TUNEL labeling was used to test whether apoptosis was the cause of the cell dissociation. There is almost no apoptosis in normal frog embryos prior to tadpole stages. Accordingly, we found no apoptotic cells in fus morphants at stage 12, when the cells dissociate (Fig. 1L,N). At stage 13, when dissociation is extensive, we detected some apoptosis in some embryos (seven of 18), whereas the rest showed no signs of apoptosis (Fig. 1M,O). We conclude that apoptosis is an occasional result of dissociation rather than a cause of it. To evaluate MO efficiency, embryos injected with the splice-blocking fusMO4 were assayed by RT–PCR with primers covering the first four exons; we detected significant reduction of the normal transcript in fus morphants (Fig. 1P).
Mesodermal differentiation defects in fus morphants
To address developmental defects in fusMO-injected embryos, we assayed tissue-specific gene expression. The nonneural ectoderm marker tfap2a was expressed normally in fus morphants (Fig. 2A,E), as was endodermal sox17b (Fig. 2B,F). Similarly, chd was robustly expressed and restricted to dorsal mesoderm, although the tissue did not involute (Fig. 2C,G). Also, fgf8 was expressed normally in the marginal zone (Fig. 2D,H). In contrast, expression of the T-box transcription factors bra and eomes was dramatically changed in fus morphants. bra is normally expressed strongly in the mesoderm from early blastula stages but was strikingly reduced in fus morphants (Fig. 2I,J). In contrast, eomes, which is normally restricted to the preinvoluting mesoderm, was expressed continuously throughout the marginal zone (Fig. 2L,M). Partial rescue of bra and quenching of eomes expression in fus morphants were obtained when they were injected with Δ5′utr-fus mRNA that did not contain the MO target sequence (Fig. 2K,N). Together, these results show that fus morphants correctly initiate zygotic transcription in all germ layers and maintain dorsal–ventral polarity but show defects in mesoderm development. Importantly, all MOs tested produced this phenotype and were rescuable with a synthetic fus mRNA, supporting the specificity of fus knockdown.
Figure 2.
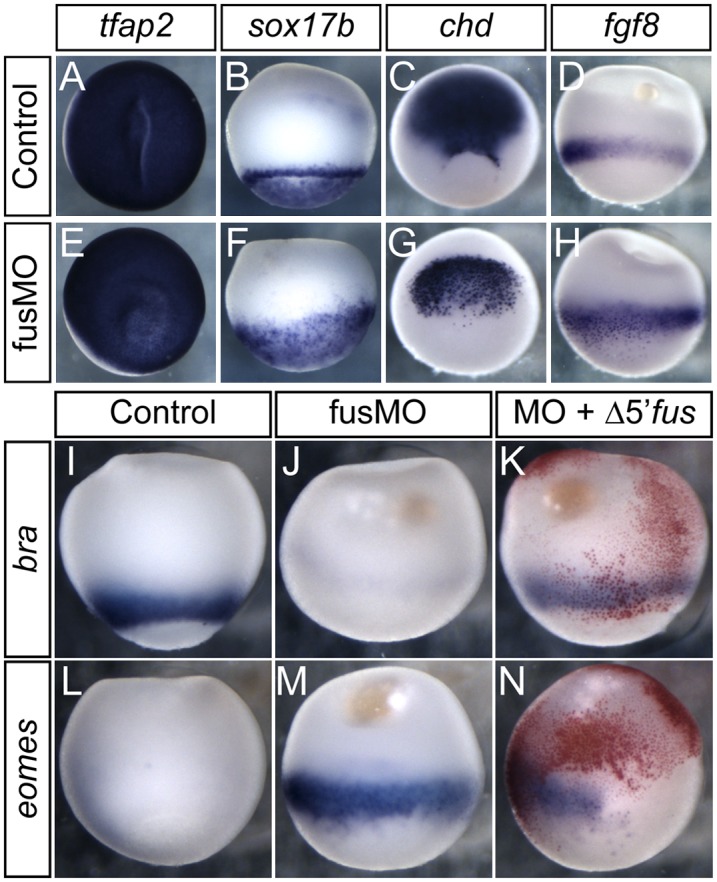
fus morphants form all germ layers, but have mesodermal differentiation defects. All pictures show lateral view with animal region up, except for A and E, which show an animal view, and C and G, which show a dorso-vegetal view. (A–H) Normal zygotic and localized gene expression in fus morphants. (A,E) Zygotic ectodermal tfap2 expression is normal in fus morphants. (B,F) Zygotic endodermal sox17b expression is normal in fus morphants. (C,G) Normal dorsal mesoderm expression of chd in fus morphants. Notice the superficial staining in fus morphants caused by lack of invagination of the mesoderm. (D,H) Normal marginal zone fgf8 expression in fus morphants. (I–N) Mesodermal differentiation defects in fus morphants. (I,J) bra is expressed circumferentially in the marginal zone in normal embryos but is essentially absent in fus morphants. (L,M) eomes expression is restricted to a narrow region of the dorsal mesoderm in normal embryos but is significantly elevated in fus morphants. (K) Rescue of bra expression in fus morphants by injection of MO-resistant fus mRNA. (N) Partial rescue of blastopore formation and eomes repression upon fus mRNA injection.
Compromised FGF-dependent gene expression in fus morphants
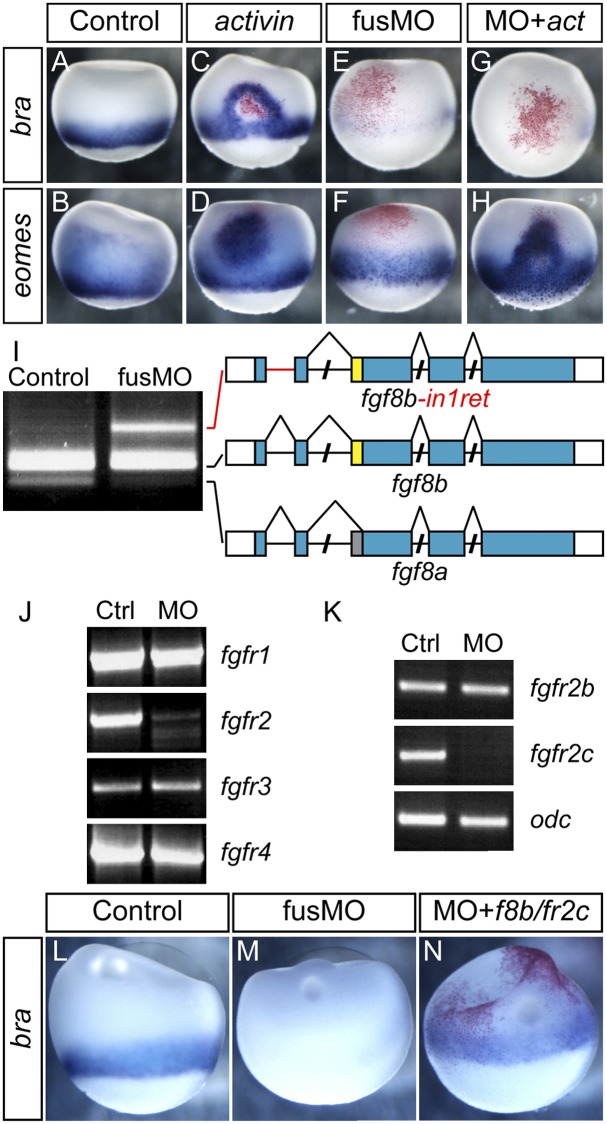
Several signaling pathways converge in the frog embryo to specify and pattern mesoderm. Signaling by nodal is the predominant mesoderm inducer, but FGF signaling is required to maintain and pattern mesodermal fates (LaBonne and Whitman 1994; Jones et al. 1995; Fletcher and Harland 2008). To address which pathways might be perturbed in fus morphants, we injected activin mRNA. Normally, activin mRNA induces both bra and eomes, but if FGF signaling is inhibited, only eomes is induced. At stage 10.5, both genes are expressed in the marginal zone (Fig. 3A,B). Injection with 0.33 pg of activin mRNA ectopically induced both bra and eomes (Fig. 3C,D). High levels of activin induce gsc, while lower levels induce bra, resulting in a halo of bra induction around the injection site. Strikingly, when we injected activin mRNA into fus morphants, bra was not induced, whereas eomes induction occurred as normal (Fig. 3E–H). This experiment suggests that activin signaling functions in fus morphants but that FGF signaling might be compromised.
Figure 3.
Defective FGF-dependent mesodermal gene expression and aberrant splicing of fgf8 and fgfr2. (A–H) Activin can induce eomes but not bra in fus morphants. (A,B) Normal expression of bra and eomes at stage 10.5. (C,D) activin mRNA injection induces ectopic bra and eomes at the site of injection (red staining) in normal embryos. (E,F) fusMO alone represses bra and, at this stage, modestly enhances eomes expression. (G,H) activin mRNA in combination with fusMO fails to induce ectopic bra expression, whereas activin potently induces ectopic eomes in combination with fusMO. (I) Defective splicing of fgf8 transcripts in fus morphants. RT–PCR showing fgf8a and fgf8b transcripts. In the fusMO lane, an aberrant transcript, fgf8b-in1ret, retains intron 1. (J) Selective decrease of fgfr2 transcript in fusMO morphants. RT–PCR using primers to amplify full-length fgfr1–4 transcripts. (K) RT–PCR using primers for individual splice forms of fgfr2 shows selective absence of the fgfr2c splice form in fus morphants, whereas fgfr2b is unaffected. odc was used as RT–PCR control. (L–N) Rescue of bra expression pattern by fgf8b and fgfr2c mRNA injection in fus morphants. (L) Normal bra expression at stage 10.5. (M) fus knockdown represses bra expression. (N) Coinjection of fgf8b and fgfr2c mRNA into two blastomeres at the two-cell stage of fus morphants partly restores endogenous bra expression.
FGF-mediated mesoderm induction requires fgf8, which is expressed in the marginal zone even in fus morphants (Fig. 2D,H). We tested the integrity of fgf8 transcripts by RT–PCR using primers covering the first three exons, including the alternatively spliced exon 3 acceptor site. In control embryos, the two isoforms are readily detected, with fgf8b being the major form (Fig. 3I). In contrast, the fgf8a isoform was almost completely absent in fus morphants, and instead a higher-molecular-weight fragment was detected (Fig. 3I). Sequencing of the fragment showed that it corresponded to an aberrant splice form (fgf8b-in1ret), in which the 87-base-pair (bp) intron1 was retained (Fig. 3I, red line). Normal fgf8b translation would be prevented by a stop codon in intron1, but the transcript could potentially produce an N-terminal-truncated protein originating from an internal start codon. We cloned and tested fgf8b-in1ret activity by mRNA injection but did not detect any activity, suggesting that it behaves as a null. Thus, since fgf8b-in1ret did not act as a dominant negative and we detected normal fgf8b, the defect in FGF signaling is not primarily at the ligand level.
We then analyzed the expression of FGFRs by RT–PCR using primers that amplify the full-length transcript. While three of the four receptor mRNAs were detected at equivalent levels after fus knockdown, we observed a dramatic decrease in full-length fgfr2 (Fig. 3J). Since fgfr2 is expressed as two major isoforms, we examined their expression by RT–PCR using isoform-specific primers. Using this semiquantitative approach, we found a complete loss of fgfr2c, whereas the levels of fgfr2b appeared normal (Fig. 3K). However, fgfr2b is maternally deposited, and it is likely that we detected perduring maternal transcripts in these experiments. fgfr2c binds fgf8 ligands, and we hypothesized that the failure of fus morphants to express bra might result from defects in fgfr2c expression. To test this, we injected fus morphants with a combination of fgf8b and fgfr2c and examined the expression of bra. Indeed, when injecting fgf8b together with fgfr2c, we partly restored normal bra expression (Fig. 3L–N). In rescue attempts with fgf8b or fgfr2c individually, fgf8b failed, while fgfr2c provided only modest rescue (data not shown), suggesting that both mRNAs are required for bra restoration, and perhaps other signaling components might be compromised. Together, these results strongly suggest that the failure of fus morphants to express bra during gastrulation is caused in part by defects in FGF signaling components, principally fgfr2c.
cdh1 splicing defects in fus morphants cause epithelial dissociation
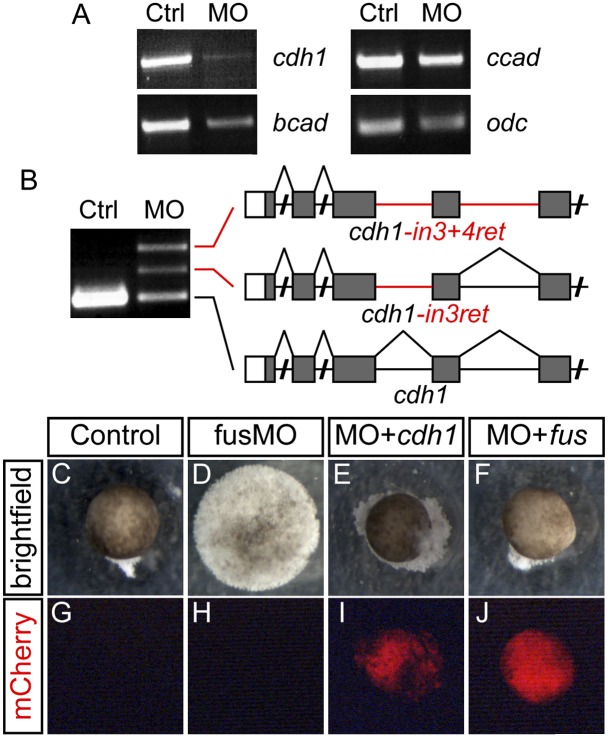
While we could restore bra expression with the FGF ligand and receptor, cell dissociation persisted, suggesting that this defect is an independent result of fus knockdown. Cadherins are critical to early cell adhesion, so we used RT–PCR to analyze the three early-expressed cadherin transcripts (Choi and Gumbiner 1989; Herzberg et al. 1991; Brieher and Gumbiner 1994). Using primers amplifying the entire coding DNA sequence (CDS), we saw a dramatic reduction in full-length cdh1 (Fig. 4A). We detected a smaller reduction in bcad, whereas ccad was relatively normal. To determine whether the reduction in full-length cdh1 was due to improperly spliced transcripts similar to that in fgf8, we targeted RT–PCR to smaller regions of the transcript. Indeed, using primers covering the first five exons of cdh1, we detected three transcripts in fus morphants (Fig. 4B). Sequencing of the two larger fragments showed that they were partially spliced transcripts that retained either intron 3 or introns 3 and 4 (Fig. 4B). Extending this analysis across cdh1, we found that several other introns were retained. These observations suggest that intron retention is widespread along the cdh1 transcript in fus morphants. Similar analysis of bcad and ccad transcript splicing showed no change in fus morphants (data not shown), suggesting that the minor reduction in bcad is not due to missplicing.
Figure 4.
Missplicing of cdh1 underlies the cell adherence defect in fus morphants. (A) RT–PCR of full-length cadherin transcripts showing significant and specific reduction of cdh1 in fus morphants, whereas bcad and ccad levels are normal. odc was used as RT–PCR control. (B) Intron retention of cdh1 in fus morphants causes a decrease in properly spliced transcripts. RT–PCR using primers covering the first five exons of cdh1 detects two aberrant transcripts retaining intron 3 (cdh1-in3ret) or introns 3 and 4 (cdh1-in3+4ret), respectively. (C–J) Restoration of cell adherence by cdh1 and fus mRNA in fusMO-injected animal caps. (C) Control animal cap forms an epidermal ball (24 of 24 caps had epithelial integrity). (D) The fusMO-injected animal cap completely dissociates into individual cells (zero of 22 showed integrity). (E) Epithelial integrity in animal cap coinjected with fusMO and cdh1 mRNA (14 of 19). (F) Epithelial integrity in animal cap coinjected with fusMO and MO-resistant fus mRNA (13 of 16). (G–J) Red fluorescence emission from coinjected mCherry mRNA used as tracer in E and F.
Expression of a dominant-negative cdh1 isoform results in blistering of the epithelium, resembling the cell dissociation observed in fus morphants (Levine et al. 1994). To determine whether normal cdh1 could prevent the fus morphant dissociation, we cut animal caps from fus morphants injected with cdh1 mRNA along with mCherry as tracer. Uninjected animal caps developed into ectodermal epithelium, whereas caps cut from fus morphants completely dissociated to single cells at stage 13 (Fig. 4C,D). In contrast, caps cut from fus morphants injected with full-length cdh1 mRNA retained their epithelial structure for several more stages (Fig. 4E,I). In addition, we were able to postpone dissociation of fus morphants by injecting Δ5′utr-fus (Fig. 4F,J). These experiments suggest that dissociation of fus morphants is caused mainly by intron retention in cdh1 mRNA, resulting in loss of epithelial integrity.
The C-terminal tail of fus is not necessary for splicing
Multiple human mutations clustering in the last 12 amino acids of fus have been associated with fALS (Kwiatkowski et al. 2009; Vance et al. 2009). We assessed the role of this domain using a splice-blocking MO (fus-fALS-MO5) that prevents inclusion of this part of the transcript (Supplemental Fig. 1). Embryos where the C-terminal part of fus is missing develop normally and do not show missplicing of cdh1, fgf8, or fgfr2, indicating that the C-terminal tail is dispensable for proper splicing. We did, however, notice strong defects in the morphants' ability to swim, although the basis for this is unclear. Axial muscle pattern was normal, as was gross neural anatomy, as judged by staining with the antibodies 12/101 (muscle), 3A10, and 6F11 (neurofilament and NCAM) (data not shown).
Replication of the fus morphant phenotype in Xenopus tropicalis
Since the dramatic phenotype we observed upon fus knockdown has not been demonstrated in other organisms, we tested whether fus function is conserved between frog species. Injection of fusMO1 and fusMO4 alone or in combination into X. tropicalis embryos robustly reproduced all of the phenotypic changes observed in X. laevis, including failure to gastrulate, cell dissociation, and down-regulation of bra at gastrulation as well as concurrent up-regulation of eomes by in situ hybridization (ISH) (data not shown). Using RT–PCR, we also detected severely diminished levels of full-length fgfr2 and cdh1 as well the abnormal fgf8 and cdh1 transcripts with retained introns. Together, these observations support the argument that the effects are specific, and the function of fus is conserved.
RNA-seq to determine global abundance of missplicing in fus morphants
To assess the global impact of fus knockdown, we compared transcripts between X. tropicalis wild-type embryos and fus morphants using RNA-seq. While X. laevis is a tractable organism for developmental analysis, its pseudo-tetraploid genome complicates genomic analysis in contrast to the simple diploid X. tropicalis (Hellsten et al. 2010). We therefore prepared paired-end Illumina libraries from fus morphants and control embryos at stage 10 and stage 12 (Supplemental Table1). Using RNA-seq, we addressed three questions: (1) Is intron retention confined to specific introns, or are all introns in an affected transcript retained? (2) How pervasive is intron retention in fus morphants? (3) Do transcripts with retained introns encode a biologically coherent set of proteins? Our strategy for mapping and analysis is outlined in Supplemental Figure 2, and general mapping statistics are in Supplemental Figure 3 and Supplemental Table 1.
fus knockdown affects all introns in a subset of transcripts
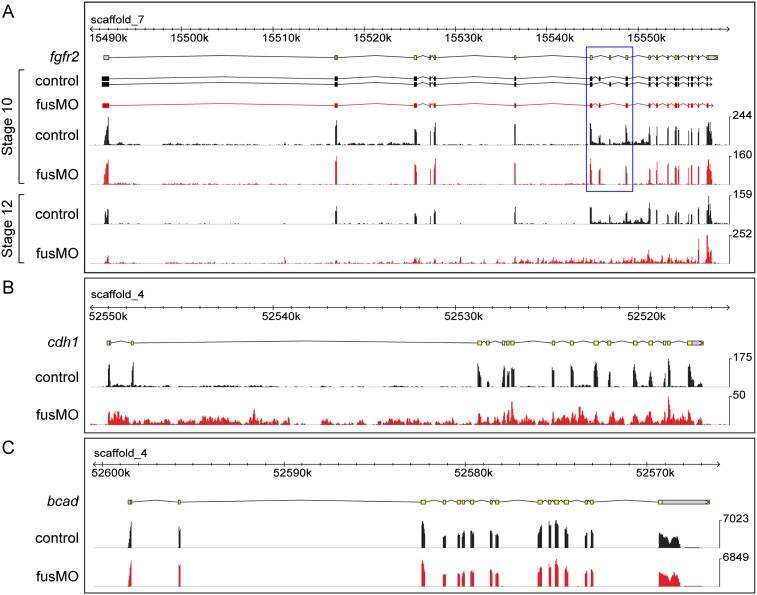
To determine whether all introns in affected transcripts are retained, we used TopHat (Trapnell et al. 2009) to align mate pairs to the X. tropicalis genome assembly v7.1 and Cufflinks (Trapnell et al. 2010) to assemble de novo transcripts for visualization. At stage 10, Cufflinks assembled transcripts corresponding to the b and c forms of fgfr2 in the control samples but only predicted the presence of the maternal b form in fus morphants, confirming our RT–PCR analysis (Fig. 5A). Surprisingly, at stage 12, we observed flattening of the transcript profile, indicating general failure to splice fgfr2 in fus morphants. Next, we examined the cdh1 locus. In agreement with our RT–PCR, we again found pervasive flattening of the transcript profile (Fig. 5B), showing that splice defects are not confined to specific introns but affect all introns in the transcript.
Figure 5.
Pervasive intron retention in fus morphants detected by RNA-seq. In all panels, the top yellow and gray track shows the JGI v7.1 gene models for reference. The black tracks show the control read profile, and the red tracks show read profile for fus morphants. (A) Two splice forms, fgfr2b and fgfr2c, are detected in control lanes at stage 10, whereas only fgfr2b is detected in fus morphants. The alternatively spliced region is boxed in blue. At stage 12, the control read distribution is similar to that at stage 10, whereas the fusMO lane shows a complete flattening of the transcript profile, indicating that all introns are retained. (B) Normal exon profile of cdh1 in control lane at stage 10; dramatic intron retention is apparent in fus morphants. (C) The bcad transcript is normal in fus morphants at stage 10.
To assess specificity, we tested bcad, which is adjacent to cdh1 in the genome but was found to be unperturbed in our RT–PCR analysis. RNA-seq confirmed that this transcript was normal in fus morphants (Fig. 5C), as were most other transcripts that we inspected. In addition, we confirmed other RT–PCR data, including the dramatic decrease of bra and the increase of eomes expression, as well as the retention of intron1 of fgf8 (Supplemental Fig. 4). These three affected developmental genes all showed dramatic inclusion of all introns, showing again that missplicing affects the entire transcript. Inspection of these and other selected genes also suggested that developmental regulatory genes might be the principal targets of fus regulation. To address whether these transcripts might be translated into potentially dominant-negative isoforms or were retained in the nucleus, we did ISH and observed strong punctate staining consistent with nuclear retention of misspliced transcripts (Supplemental Fig. 5).
Intron retention affects a limited number of transcripts
To determine the extent of intron retention, we trimmed all reads to 76 bp and aligned them as individual reads to 182,834 unique introns extracted from JGI v7.1 gene models as well as to genomic scaffolds. We used the number of reads aligning to introns and scaffolds to calculate the percentage of intron reads. At stage 10, we detected a slight increase in intronic reads, from 1.2% in control to 1.5% in fus morphants (Supplemental Table2). At stage 12, the difference in intronic reads increased to 9.7% in fus morphants versus 5.6% in controls. These observations support that the increase in intron retention in fus morphants is limited and may be highly specific.
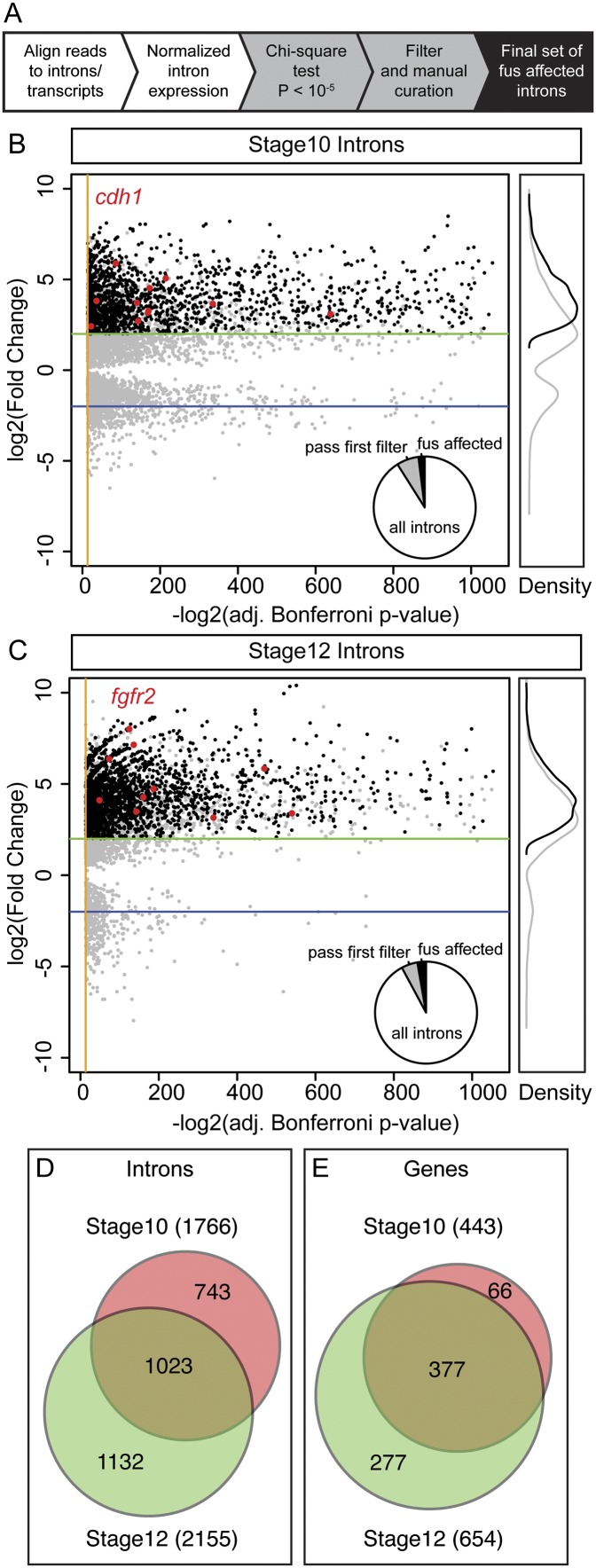
To survey the genes whose intron retention is affected in fus morphants, we mapped individual reads to the unique introns as well as to mature transcripts and calculated normalized fold changes in intron reads and their associated P-values using a Bonferroni-corrected χ2 test for independence. As an initial filter, we selected introns with a change in expression level with a P-value of <10−5, which constituted 7.7% (stage 10) and 5.5% (stage 12) of all introns in expressed genes. However, the majority of introns showed only small changes, likely to be noise, and were unlikely to be physiologically significant. In addition, inspection of transcript profiles with large changes on the genome browser showed that they contained a proportion of false negatives, primarily caused by nested or overlapping gene models. We therefore applied a second, stricter set of filters that required a more than fourfold increase in intron reads relative to the control sample and validated these by inspecting the transcript profile of the paired read alignment on the genome browser. We set a transcript abundance cutoff of more than six RPKM (reads per kilobase pair per million mapped reads), since transcripts expressed below this threshold did not display interpretable transcript profiles (outlined in Fig. 6A; Supplemental Fig. 2). After this final filter, we identified 1766 and 2155 introns that were affected by fus knockdown at stage 10 and stage 12, respectively (Fig. 6B,C). This set included all introns of the genes we previously found to be misspliced by RT–PCR, such as cdh1 and fgfr2. Based on these criteria, between 2% and 3% of all introns in expressed genes are affected by fus knockdown (Fig. 6D; Supplemental Table 2). The retained introns were distributed along 443 and 654 genes in the stage 10 and stage 12 set, respectively (Fig. 6E), confirming that retained introns cluster in specific transcripts, rather than being distributed across all transcripts. Indeed, an average of 3.3 (stage 10) and 4.0 (stage 12) introns were retained per affected gene, corresponding to an 18-fold to 30-fold increase, respectively, above what would be expected if the distribution was even across all expressed genes (Supplemental Table2). In addition, there was a substantial overlap in both introns and affected genes at the two stages: 377 of the 443 genes (85%) that were found to be affected at stage 10 were also affected at stage 12. These results show that fus knockdown affects intron splicing in a limited number of specific genes.
Figure 6.
Intron retention in fus morphants affects a small fraction of all introns and transcripts. (A) Brief outline of the strategy for identifying affected introns. Scatter plot showing stage 10 (B) and stage 12 (C) introns with an adjusted P-value of <10−5. The distribution of the first pass introns (gray) indicates that the P-value is not sufficient to identify retained introns in fus morphants, as a significant number of the passed introns display only small changes. Introns changed more than fourfold and those that passed validation are shown in black. As a reference, introns from cdh1 at stage 10 (B) and from fgfr2 at stage 12 (C) are plotted as red dots. Pie charts in the bottom right of the scatter plots show the proportion of introns that pass the first filter (gray slice) and the final validated set of fus-affected introns (black slice). The white slice represents unchanged introns from expressed genes. Unchanged introns (P > 10−5) are not plotted in the scatter plots. Orange lines indicate the cutoff P-value = 10−5, and green and blue lines indicate thresholds for fourfold increase and decrease, respectively. The right panel shows relative kernel density plots of first-pass introns (gray line) and the final curated set (black line). (D) Area-proportional Venn diagrams showing the number of retained introns in fus morphants at stages 10 (red) and 12 (green), showing significant overlap between the stages. The number in parentheses indicates the total number of introns retained at the given stage. (E) Venn diagram similar to D, but showing the number of genes with retained introns in fus morphants. A significant majority (85%) of genes that are affected at stage 10 are also affected later, whereas almost half (42%) are only affected at stage 12.
In principle, fus could be necessary for transcription of one or more splicing factors, which in turn is necessary for proper splicing. We tested our RNA-seq data for differential gene expression using DEGseq (Wang et al. 2010). We found that hnrnpa1 and hnrnpdl were the only known splice factors that had significantly decreased expression (2.5-fold and 2.3-fold, respectively). However, both of these transcripts are misspliced in fus morphants, and their repression is likely a reflection of transcript instability. Indeed, we observed a frequent decrease in expression levels of intron-retained transcripts (data not shown).
fus knockdown affects developmental regulatory genes
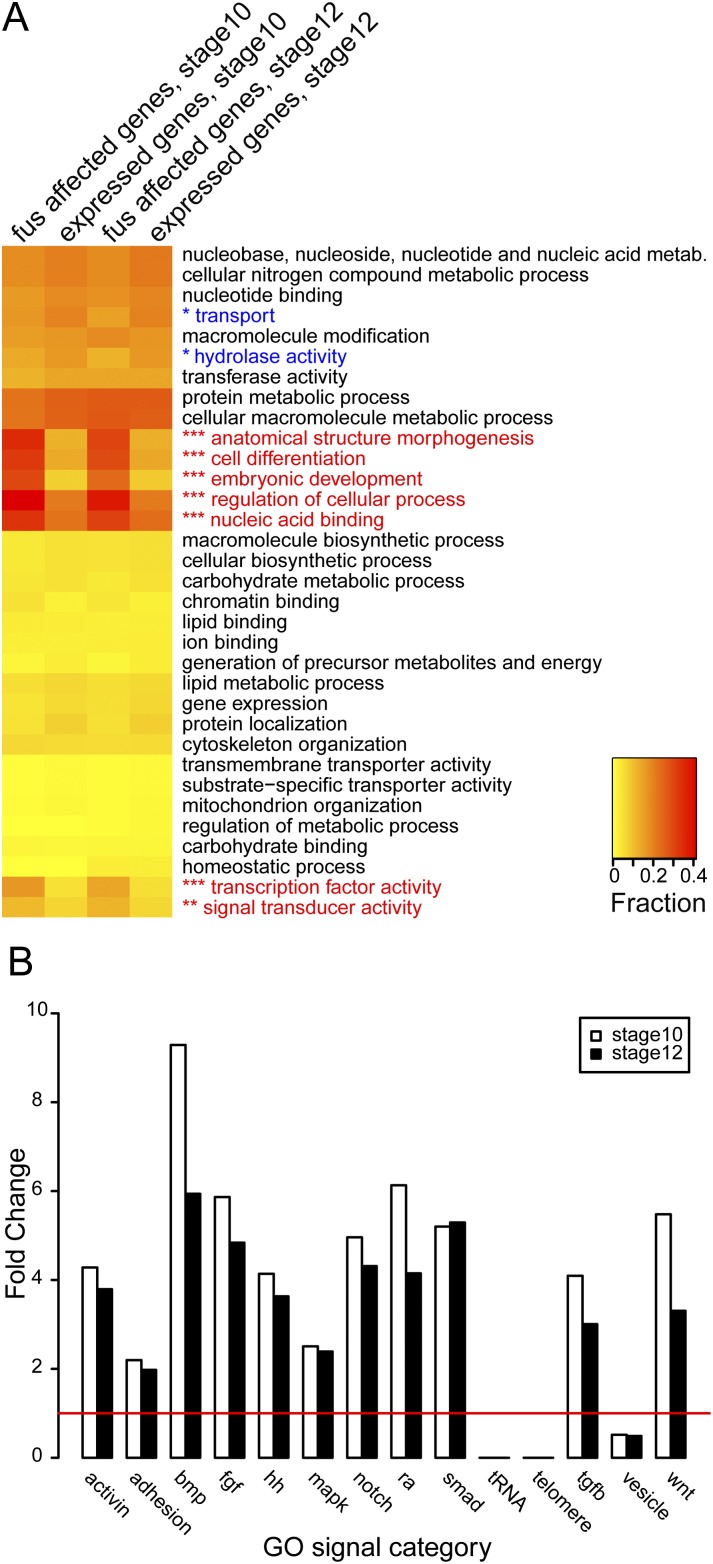
A list of genes affected by fus missplicing (Supplemental Table 4) indicated that there was a strong representation of developmental regulators, in particular signaling components and transcription factors. To perform a systematic and unbiased assessment of potential enrichment of functionally related proteins, we used Blast2GO (Gotz et al. 2008) to assign gene ontology (GO) slim terms to the fus-affected genes and compared them with GO slims assigned to all expressed genes. GO slim terms collapse multiple GO terms into broader categories that can be useful when comparing large sets of proteins. The GO slim categories most enriched in fus-affected genes were involved in embryo development, anatomical morphogenesis, and cell differentiation (Fig. 7A). In fact, approximately one-third of all fus-affected genes were assigned one or more of these categories compared with only one-tenth in the expressed gene sets. In addition, categories containing transcription factor activity, nucleic acid binding, and signal transduction activity were also enriched by as much as threefold. Together, this systematic annotation of functional terms shows that fus-affected genes frequently are regulators of developmental processes as either transcription factors or receptors or are otherwise involved in intracellular signaling.
Figure 7.
GO term analysis of fus-affected genes. (A) Intron-retained genes in fus morphants are enriched for developmental regulators. Heat map showing GO slim terms annotated to intron-retained genes in fus morphants and expressed genes at stage 10 and stage 12. GO slim categories that are significantly enriched in fus-affected genes are indicated in red, and categories that are depleted are in blue. GO slim categories that contain terms associated with embryo development, morphogenesis, and differentiation (anatomical structure morphogenesis, cell differentiation, and embryonic development) are enriched in fus-affected genes. Also enriched are terms associated with transcription (nucleic acid binding and transcription factor activity) and signal transduction (signal transducer activity). (Inset) Values are expressed as a fraction of genes that are assigned the GO slim term, with rarely represented terms in yellow and more common terms in red. (***) P < 0.001; (**) P < 0.01 at both stage 10 and stage 12. (*) P < 0.05 at stage 12 only. Only GO slim terms from levels 3 and 4 in the categories “molecular function” and “biological process,” respectively, are shown. (B) All major developmental signaling pathways are affected in fus morphants. Bar graph showing the representation of GO signal categories in fus-affected genes expressed as fold change compared with expressed genes. The major developmental pathways regulated by either activin, adhesion, bone morphogenetic protein (bmp), fgf, hedgehog (hh), MAP kinase (mapk), notch, retinoic acid (ra), smad, tgf-β (tgfb), and wnts are enriched at stages 10 and 12. In contrast, GO terms associated with transfer RNA (tRNA), telomere, and intracellular vesicles (vesicle) are depleted in fus-affected genes. The red line indicates no change.
In our embryological experiments, we used a candidate-based approach to focus on components in the FGF signaling pathways and cadherin-mediated adhesion. To test whether fus knockdown affects components of other signaling pathways, we constructed “GO signal” terms that contained biological process terms associated with all major developmental pathways. As an outgroup, we created categories that contained terms involved in transfer RNA (tRNA), telomere, and vesicle processes. We then calculated enrichment and depletion similar to that in our GO slim analysis. Surprisingly, all major signaling pathways were enriched in the fus-affected sets, including all branches of TGF-β, Hedgehog, Notch, and Wnt pathways (Fig. 7B). In addition, categories containing FGF and MAP kinase (MAPK) pathways as well as adhesion were all enriched, suggesting that the candidate genes that we tested by RT–PCR analysis were part of a larger set of genes belonging to these pathways that were affected in fus morphants. In addition, we observed some enrichment of other pathways, such as Hippo, Jak–Stat, and Toll, but excluded them from further analysis because of the scarcity of genes assigned to these classes (data not shown). Importantly, all nonsignaling outgroups were dramatically underrepresented in fus-affected genes, as documented by the complete absence of genes involved in tRNA and telomere processes as well as low representation of terms involving vesicle processes. In summary, our systematic assessment of GO terms associated with genes affected by fus knockdown show that they are developmental regulators that impact all major signaling pathways.
Discussion
In this study, we show that fus/TLS is required for splicing of a set of regulatory genes during early frog embryogenesis. We show that fus is required for gastrulation and epithelial integrity in Xenopus and that this is a result of the failure to remove introns from pre-mRNAs that encode proteins important for development. Using a candidate approach, we connect the failure to induce bra expression on schedule to missplicing of FGF signaling components and connect cell dissociation in the animal pole to missplicing of cdh1. Importantly, we could rescue these defects by injecting normal versions of the transcripts, at least partially. Previously, we and others have implicated splicing proteins in developmental regulatory events in Xenopus, but it has been difficult to identify the primary targets (Gerber et al. 2002; Liu and Harland 2005). Here we used the phenotypes of the affected embryos to identify some misregulated components and extended this using RNA-seq to show that we can analyze missplicing in an unbiased manner. We used RNA-seq to show that fus affects between 3.3% and 5.5% of all genes expressed and showed that these genes are enriched for genes functioning in all major signaling pathways. In the set of fus-affected genes, we found multiple signaling components and transcription factors with diverse functions during development. Our systematic analysis of GO slim terms assigned to fus-affected transcripts showed a clear enrichment for terms involved in regulatory functions during embryogenesis. This corroborates the examples of FGF and cadherin transcripts we found by our candidate approach and attests to fus as a central factor in early frog embryogenesis. The fact that all major developmental pathways were affected explains why we could only partially and temporarily restore normal gastrulation and adherence using our candidate approach.
Splicing of pre-mRNA is emerging as a potentially important target for large-scale regulation of genes with integrated biological functions. In the case of the NOVA family of alternative splicing regulators, alternative splicing events involved in synaptogenesis in the brain are regulated (Ule et al. 2003, 2005). Here we provide an additional example of a biologically coherent set of transcripts—in this case, regulated by fus. In addition, this study shows regulation of constitutive splicing of transcripts with a common function. This raises the possibility of novel and unexpected layers of regulation of gene expression. While embryonic development and cell differentiation were the most common categories enriched in fus-affected transcripts, not all transcripts were assigned these GO terms, including many hypothetical and uncharacterized proteins, raising the possibility that these proteins may have yet uncharacterized functions during development.
Another FET protein, EWSR1, has been shown to regulate alternative splicing of transcripts involved in DNA damage repair and MAPK signaling in a study that used a splicing-sensitive microarray for genes involved in cancer and RNA processing (Paronetto et al. 2011). It would be interesting to study EWSR1 function in a genome-wide unbiased approach to determine whether intron retention occurs upon EWSR1 knockdown as well.
FUS has primarily been studied in mammalian cell culture or cell-free systems, and the precise biological function has remained elusive, although it has been proposed as a integrator of transcription and splicing (Law et al. 2006). Both FUS and TAF15 were shown by mass spectrometry analysis to be part of the spliceosome and could therefore participate directly in the splicing reaction (Rappsilber et al. 2002; Zhou et al. 2002). We suggest that fus loads onto the polymerase at the promoter of a subset of genes and facilitates splicing as the transcript emerges from the polymerase. This would also suggest that this subset of genes has developed a dependence on fus for splicing, since other genes are spliced normally in the absence of fus. fus could ride with RNA polymerase II and either directly interact with the 5′ splice site and recruit U1-snRNP to the nascent pre-mRNA or recruit other splicing factors. In this context, it is interesting that TAF15 has been shown to interact with the RNA component of U1, which provides direct interaction with the 5′ splice site (Jobert et al. 2009). The finding that a fus-binding motif (GGUG) closely resembles a consensus 5′ splice site (GGUG/A) suggests a possible direct interaction with the 5′ splice site. However, another study shows that fus binds a different intronic AU-rich stem–loop structure at a higher affinity than the single-stranded GGUG motif identified by SELEX (Hoell et al. 2011). fus binding to intronic structures near either the 5′ or 3′ splice site is highly compatible with the splicing defects that we found in our study.
There is considerable evidence for fus interacting with both SR proteins and other types of splicing factors (Tan and Manley 2009). SR proteins are central to exon definition by binding to exonic splicing enhancers and recruiting the spliceosome to adjacent splice sites. Failure to recruit SR proteins to pre-mRNA could perceivably lead to intron inclusion. Since pre-mRNA processing is governed through a complex web of weak protein–RNA interactions, both direct interaction of fus with the 5′ splice site and recruitment of SR proteins could be necessary for proper splicing.
In our RNA-seq experiments, we detected 1.2%–5.6% reads from control samples mapping to introns. This is in general agreement with the 4% intron reads detected in the mouse transcriptome (Mortazavi et al. 2008). The lower level of intron reads in both control and fus morphants at the early stage is likely due to persistence of maternal transcripts that do not contribute pre-mRNA remnants. Stage 12 is the latest we could analyze intact embryos before they dissociate, and it is therefore unclear whether fus affects splicing of significantly more transcripts at later stages.
We suggest that the selective absence of fgfr2c in fus morphants is due to the persistence of maternal fgfr2b transcripts, while all zygotic fgfr2c transcripts are affected. Interestingly, we could restore bra expression in its normal domain, but not ectopic expression, by simultaneous injection of fgf8b and fgfr2c. This suggests that not all FGF signaling is restored and that we overcame a partial downstream defect by boosting the upstream signaling. This is supported by our RNA-seq analysis that identified several misspliced downstream MAPK components.
While the RNA-seq showed a global reduction of splicing across the specifically affected transcripts, RT–PCR analysis showed that there was still some correct splicing. This could be a result of incomplete knockdown. Alternatively, there could be a varying degree of dependence on fus, depending on the precise splicing context. By RT–PCR, we could detect proper splicing of introns that were clearly retained by RNA-seq; for example, introns 2–4 in fgf8. This is likely caused by selective amplification of short sequences by RT–PCR relative to long introns.
Mutations in the C terminus of fus has been implicated in the familial variant of the neurodegenerative disease ALS (Kwiatkowski et al. 2009; Vance et al. 2009). ALS and other neurodegenerative disorders have altered RNA metabolism (for review, see Cooper et al. 2009; Strong 2010). However, aberrant intron inclusion has not been reported in fALS patients. Interestingly, a study used exon arrays to probe changes in sporadic ALS patients and found aberrant splicing in cell adhesion and cell matrix genes (Rabin et al. 2010). However, the exact nature of the missplicing was not determined, nor is it known whether Fus was mutated in the patients. We attempted to determine the function of the C-terminal tail in fus by a targeted splice MO that cause exclusion of this region. Our experiments suggest that the C-terminal tail of fus is dispensable for splicing, but may be important for nervous system differentiation or function.
A mouse gene trap is the only other vertebrate loss-of-function study published (Hicks et al. 2000). This study found no developmental defects, but instead found perinatal death and genomic instability. It is unclear why frog knockdown caused such a dramatic phenotype compared with the mouse mutant. While the gene trap disrupted exon 12 and therefore left most of the CDS intact, very little truncated protein was detected, arguing that the mutant was close to a null. It is possible that other FET family members mask the effect of fus deficiency in mice, and compound mutants have not been reported. While we did not perform compound knockdown of FET members in frogs, our preliminary results suggest that neither ewsr1 nor taf15 is necessary for early frog development (data not shown). Alternatively, the differences may reflect differences in the biological regulation by fus, with its role in Xenopus perhaps illustrating an ancestral role in splicing of developmental regulators or even a regulatory role acquired in the amphibian lineage.
In conclusion, we examine here the fus function of the RBP fus during frog embryogenesis and provide evidence that it orchestrates splicing of a group of transcripts involved in transcriptional regulation and signal transduction involved in embryogenesis and cell differentiation. Our work provides unexpected information on the function of fus and, potentially, on its contribution to neurodegenerative disease.
Materials and methods
Xenopus embryo microinjection
X. laevis eggs were collected and in vitro-fertilized and embryos were cultured and microinjected by standard procedures (Sive et al. 1999). X. tropicalis embryos were collected from natural matings and injected in both blastomeres at the two-cell stage as described (Khokha et al. 2002). Embryos were staged according to the normal table (Nieuwkoop 1994). For control injections and lineage tracing for mRNA injections, we used nuclear β-galactosidase (for staining prior to ISH) or mCherry (for live tracking in animal cap experiments). Tracing of MO injections used fluorescein-labeled control MOs (GeneTools).
Plasmids and mRNA synthesis
A full-length X. laevis fus clone (6A17), pCS108-fus-Xl, was isolated in an expression cloning screen described previously (Dichmann et al. 2008). A rescue construct (pCS108-Δ5′UTR-fus-Xl) lacking the sequences targeted by the morpholinos was synthesized by PCR. For X. tropicalis fgfr2c mRNA, IMAGE clone 7630892 was used. X. laevis cdh1 and fgf8b plasmids have been described previously (Levine et al. 1994; Fletcher et al. 2006). CS108 vectors containing the cDNA inserts were linearized with AscI, except for cdh1, which was linearized with XbaI. Tracers used were nuclear β-galactosidase (nβgal-CS2+) (Turner and Weintraub 1994) and mCherry cloned into CS108. Capped mRNA was synthesized using mMessage mMachine SP6 kit (Ambion).
Whole-mount RNA ISH
Embryos injected with mRNA were fixed for 30 min in MEMFA with 3.7% formaldehyde and washed in PBS with 0.1% Tween-20, and tracer was visualized using Red-Gal substrate (Research Organics); after staining, embryos were refixed for 2 h and dehydrated prior to ISH (Sive et al. 1999). Embryos injected only with MOs were fixed for 2 h and then dehydrated. The following antisense probes have been described previously: Xbra (Smith et al. 1991), noggin (Smith and Harland 1992), tfap2 (Luo et al. 2002), eomes (Ryan et al. 1996), sox17β (Hudson et al. 1997), and chordin (Sasai et al. 1994). fus probe was synthesized from clone 6A17 (Dichmann et al. 2008), linearized with NotI, and transcribed with T7 RNA polymerase.
MOs
The MOs (GeneTools) used were as follows: fusMO1 (translation start) (5′-ATCGTTGGTGGCCATGTTGCGGTAT-3′) and fusMO4 (first 5′ splice site junction) (5′-GTAATTCCTTACCGTTGGTGGCCAT-3′) target both X. laevis and X. tropicalis, while fusMO2 (5′-CTGTCCGGTACAGACCTCTGTCTTA-3′) and fusMO3 (5′-ATGTCCCGAAAGCACCGCCCACACA-3′) are homeolog-specific in X. laevis and target the 5′ UTR. The sequence matching the start codon is underlined. fus-fALS-MO5 (5′-CAGAATTTACCTGGAATCCATTTTC-3′) was used to prevent splicing of the last exon of fus.
RT–PCR
Total RNA was isolated using Trizol (Invitrogen), and oligo-dT-primed cDNA was synthesized using SuperScript II (Invitrogen) followed by PCR using PlatinumTaq (Invitrogen) by standard methods. Primer sets included odc (Hudson et al. 1997) and fgf8a/b (253 of 286 bp) (Fletcher et al. 2006). Primers designed for this study are listed in Supplemental Table 3.
RNA-seq library preparation
Paired-end libraries for Illumina sequencing were synthesized according to standard protocols. Briefly, 10 μg of total RNA was isolated using Trizol (Invitrogen) and poly-A-selected twice on oligo-dT-conjugated dynabeads (Invitrogen), followed by zinc ion fragmentation (Ambion). First and second strand synthesis, end repair, single <A> addition, and paired-end adaptor ligation used standard reagents from Invitrogen and New England Biolabs. Libraries were size-selected and subjected to 11 or 14 (for 2 × 101 and 2 × 76, respectively) rounds of PCR amplification using Phusion polymerase (New England Biolabs). Paired-end sequencing at the Vincent J. Coates Genomics Sequencing Laboratory at the University of California at Berkeley used Illumina Genome Analyzer II or HiSeq2000 machines (for paired-end 76- and 101-bp reads, respectively).
Bioinformatics
Transcriptome analysis
Overview of the RNA-seq analysis strategy is provided in Supplemental Figure 2A. Read pairs were aligned to the X. tropicalis genome (assembly v7.1) using TopHat version 1.2 (Trapnell et al. 2009). Predicted transcripts were assembled using Cufflinks version 1.x (Trapnell et al. 2010). Aligned reads and Cufflinks-assembled transcripts were visualized on Gbrowse2 (http://www.gmod.org).
Identification of retained introns
To determine retained introns, we calculated expression for each intron normalized to its cognate transcript expression level and total number of mapped reads in the sample. First, we excluded copies of introns that exist in more than one copy in JGI v7.1 gene models to identify “unique” introns (Supplemental Fig. 2B) and extracted their sequences from a Bio::DB::GFF database. These introns and JGI v7.1 transcripts were used as the basis for alignment and identification of retained introns. To assure consistency across samples and minimize the probability of hitting splice junctions, we trimmed all RNA-seq reads to 76 bp and aligned them as single reads using Bowtie, allowing up to seven matches from the best strata to be reported (Langmead et al. 2009). We calculated the Bonferroni-corrected χ2 test of independence based on the number of reads aligning to each intron and its cognate transcript. P-values of <10−5 acted as a first filter, followed by a normalized intron change of more than fourfold and RPKM of more than six. These algorithmically identified introns were validated by inspecting the transcript profile for intron retention and false negatives, often caused by nested or overlapping genes, or erroneous gene models. Such false negatives were eliminated from further analysis.
Annotation of v7.1 gene models
We used two strategies for assigning names to v7.1 gene models. Most genes were assigned v7.2 names based on a mutual best hit (MBH) BLASTP strategy. Briefly, following a BLASTP comparison of predicted peptides from JGI v7.1 and v7.2 gene models, MBHs were assigned using a script provided by Simon Prochnik (Joint Genome Institute). Parallel to that, we assigned names from the NCBI database to gene models not covered by the v7.2 set using an iterative MBH BLASTP strategy: Following a BLASTP comparison of predicted peptides from JGI v7.1 gene models and all X. tropicalis proteins in the NCBI database, MBHs were assigned. Successfully assigned proteins were removed, and the remaining proteins were subjected to an additional round of MBH assignment. This was repeated for a total of six rounds, after which names were assigned to 29,868 out of 34,814 gene models. Remaining gene models were assigned their top hit only if the E-value was <10−30. In total, 30,088 (86.4%) gene models were assigned names by this method. In Supplemental Table 4, IDs starting with “xetrov71” indicate that they were not covered by v7.2 and therefore are named according to their best hit in the NCBI database.
GO slim and GO signal annotation
We used Blast2GO to assign GO terms to all expressed genes at stages 10 and 12 (Gotz et al. 2008). These were then collapsed into standard GO slim terms (http://www.geneontology.org/GO.slims). We calculated the fraction of the occurrence of each term in affected genes as well as in all expressed genes as Fraction = Ngenes with term in set/Ntotal genes in set.
Statistical significance was calculated using the false discovery rate (FDR)-adjusted χ2 test of independence. For presentation in Figure 7A, we used level 3 terms from biological_process and level 4 terms from molecular_function GO slim categories, (we excluded the extremely abundant term “protein binding”). Custom GO signal terms were constructed by combining relevant GO terms identified primarily by the presence of the term in their title so that they did not overlap. For example, terms associated with SMAD signaling were not included in the categories that contained bone morphogenetic protein (BMP), activin, or TGF-β (TGF-b) signaling. Fold changes in fus morphants compared with expressed genes were calculated by pooling the number genes of each associated term.
Acknowledgments
We thank Dr. Dave Hendrix (Center for Integrative Genomics; now at Massachusetts Institute of Technology), Dr. Simon Prochnik (Joint Genome Institute), and members of Daniel Rokhsar's laboratory (Joint Genome Institute/University of California at Berkeley) for bioinformatics help, and Dr. Elizabeth Purdom (University of California at Berkeley) for guidance on statistical analysis. We also thank members of the Harland laboratory for helpful suggestions, as well as Dr. Barry Gumbiner for providing the cdh1 plasmid. This work was funded by NIH grant GM42341.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.187278.112.
References
- Blunt AG, Lawshé A, Cunningham ML, Seto ML, Ornitz DM, MacArthur CA 1997. Overlapping expression and redundant activation of mesenchymal fibroblast growth factor (FGF) receptors by alternatively spliced FGF-8 ligands. J Biol Chem 272: 3733–3738 [DOI] [PubMed] [Google Scholar]
- Brieher WM, Gumbiner BM 1994. Regulation of C-cadherin function during activin induced morphogenesis of Xenopus animal caps. J Cell Biol 126: 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Gumbiner B 1989. Expression of cell adhesion molecule E-cadherin in Xenopus embryos begins at gastrulation and predominates in the ectoderm. J Cell Biol 108: 2449–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G 2009. RNA and disease. Cell 136: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichmann DS, Fletcher RB, Harland RM 2008. Expression cloning in Xenopus identifies RNA-binding proteins as regulators of embryogenesis and Rbmx as necessary for neural and muscle development. Dev Dyn 237: 1755–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RB, Harland RM 2008. The role of FGF signaling in the establishment and maintenance of mesodermal gene expression in Xenopus. Dev Dyn 237: 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RB, Baker JC, Harland RM 2006. FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development 133: 1703–1714 [DOI] [PubMed] [Google Scholar]
- Gerber WV, Vokes SA, Zearfoss NR, Krieg PA 2002. A role for the RNA-binding protein, hermes, in the regulation of heart development. Dev Biol 247: 116–126 [DOI] [PubMed] [Google Scholar]
- Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36: 3420–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. 2010. The genome of the Western clawed frog Xenopus tropicalis. Science 328: 633–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg F, Wildermuth V, Wedlich D 1991. Expression of XBcad, a novel cadherin, during oogenesis and early development of Xenopus. Mech Dev 35: 33–42 [DOI] [PubMed] [Google Scholar]
- Hicks GG, Singh N, Nashabi A, Mai S, Bozek G, Klewes L, Arapovic D, White EK, Koury MJ, Oltz EM, et al. 2000. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat Genet 24: 175–179 [DOI] [PubMed] [Google Scholar]
- Hoell JI, Larsson E, Runge S, Nusbaum JD, Duggimpudi S, Farazi TA, Hafner M, Borkhardt A, Sander C, Tuschl T 2011. RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol 18: 1428–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson C, Clements D, Friday RV, Stott D, Woodland HR 1997. Xsox17α and -β mediate endoderm formation in Xenopus. Cell 91: 397–405 [DOI] [PubMed] [Google Scholar]
- Jobert L, Pinzón N, Van Herreweghe E, Jády BE, Guialis A, Kiss T, Tora L 2009. Human U1 snRNA forms a new chromatin-associated snRNP with TAF15. EMBO Rep 10: 494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Kuehn MR, Hogan BL, Smith JC, Wright CV 1995. Nodal-related signals induce axial mesoderm and dorsalize mesoderm during gastrulation. Development 121: 3651–3662 [DOI] [PubMed] [Google Scholar]
- Khokha MK, Chung C, Bustamante EL, Gaw LWK, Trott KA, Yeh J, Lim N, Lin JCY, Taverner N, Amaya E, et al. 2002. Techniques and probes for the study of Xenopus tropicalis development. Dev Dyn 225: 499–510 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. 2009. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323: 1205–1208 [DOI] [PubMed] [Google Scholar]
- LaBonne C, Whitman M 1994. Mesoderm induction by activin requires FGF-mediated intracellular signals. Development 120: 463–472 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186-gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law WJ, Cann KL, Hicks GG 2006. TLS, EWS and TAF15: A model for transcriptional integration of gene expression. Brief Funct Genomics Proteomics 5: 8–14 [DOI] [PubMed] [Google Scholar]
- Lerga A, Hallier M, Delva L, Orvain C, Gallais I, Marie J, Moreau-Gachelin F 2001. Identification of an RNA binding specificity for the potential splicing factor TLS. J Biol Chem 276: 6807–6816 [DOI] [PubMed] [Google Scholar]
- Levine E, Lee CH, Kintner C, Gumbiner BM 1994. Selective disruption of E-cadherin function in early Xenopus embryos by a dominant negative mutant. Development 120: 901–909 [DOI] [PubMed] [Google Scholar]
- Liu KJ, Harland RM 2005. Inhibition of neurogenesis by SRp38, a neuroD-regulated RNA-binding protein. Development 132: 1511–1523 [DOI] [PubMed] [Google Scholar]
- Luo T, Matsuo-Takasaki M, Thomas ML, Weeks DL, Sargent TD 2002. Transcription factor AP-2 is an essential and direct regulator of epidermal development in Xenopus. Dev Biol 245: 136–144 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD. 1994. Normal table of Xenopus laevis (Daudin). Garland Publishing, Inc., New York. [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M 1996. Receptor specificity of the fibroblast growth factor family. J Biol Chem 271: 15292–15297 [DOI] [PubMed] [Google Scholar]
- Paronetto MP, Miñana B, Valcárcel J 2011. The Ewing sarcoma protein regulates DNA damage-induced alternative splicing. Mol Cell 43: 353–368 [DOI] [PubMed] [Google Scholar]
- Perales R, Bentley D 2009. ‘Cotranscriptionality': The transcription elongation complex as a nexus for nuclear transactions. Mol Cell 36: 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin SJ, Kim JMH, Baughn M, Libby RT, Kim YJ, Fan Y, Libby RT, La Spada A, Stone B, Ravits J 2010. Sporadic ALS has compartment-specific aberrant exon splicing and altered cell-matrix adhesion biology. Hum Mol Genet 19: 313–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res 12: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Garrett N, Mitchell A, Gurdon JB 1996. Eomesodermin, a key early gene in Xenopus mesoderm differentiation. Cell 87: 989–1000 [DOI] [PubMed] [Google Scholar]
- Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont LK, De Robertis EM 1994. Xenopus chordin: A novel dorsalizing factor activated by organizer-specific homeobox genes. Cell 79: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM 1999. Early development of Xenopus laevis: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- Smith WC, Harland RM 1992. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell 70: 829–840 [DOI] [PubMed] [Google Scholar]
- Smith JC, Price BM, Green JB, Weigel D, and Herrmann BG 1991. Expression of a Xenopus homolog of Brachyury (T) is an immediate-early response to mesoderm induction. Cell 67: 79–87 [DOI] [PubMed] [Google Scholar]
- Strong MJ 2010. The evidence for altered RNA metabolism in amyotrophic lateral sclerosis (ALS). J Neurol Sci 288: 1–12 [DOI] [PubMed] [Google Scholar]
- Tan AY, Manley JL 2009. The TET family of proteins: Functions and roles in disease. J Mol Cell Biol 1: 82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AY, Manley JL 2010. TLS inhibits RNA polymerase III transcription. Mol Cell Biol 30: 186–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DL, Weintraub H 1994. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev 8: 1434–1447 [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302: 1212–1215 [DOI] [PubMed] [Google Scholar]
- Ule J, Ule A, Spencer J, Williams A, Hu J-S, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, et al. 2005. Nova regulates brain-specific splicing to shape the synapse. Nat Genet 37: 844–852 [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. 2009. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323: 1208–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R 2008. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454: 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Wang X, Zhang X 2010. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26: 136–138 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185 [DOI] [PubMed] [Google Scholar]