Abstract
The chloroplast is the site of photosynthesis in higher plants but also functions as the center of synthesis for primary and specialized metabolites including amino acids, fatty acids, starch, and diverse isoprenoids. Mutants that disrupt aspects of chloroplast function represent valuable tools for defining structural and biochemical regulation of the chloroplast and its interplay with whole-plant structure and function. The lutescent1 (l1) and l2 mutants of tomato (Solanum lycopersicum) possess a range of chlorophyll-deficient phenotypes including reduced rates of chlorophyll synthesis during deetiolation and enhanced rates of chlorophyll loss in leaves and fruits as they age, particularly in response to high-light stress and darkness. In addition, the onset of fruit ripening is delayed in lutescent mutants by approximately 1 week although once ripening is initiated they ripen at a normal rate and accumulation of carotenoids is not impaired. The l2 locus was mapped to the long arm of chromosome 10 and positional cloning revealed the existence of a premature stop codon in a chloroplast-targeted zinc metalloprotease of the M50 family that is homologous to the Arabidopsis (Arabidopsis thaliana) gene ETHYLENE-DEPENDENT GRAVITROPISM DEFICIENT AND YELLOW-GREEN1. Screening of tomato germplasm identified two additional l2 mutant alleles. This study suggests a role for the chloroplast in mediating the onset of fruit ripening in tomato and indicates that chromoplast development in fruit does not depend on functional chloroplasts.
The chloroplast is a defining organelle of plant cells, serving as the site of the biochemical reactions associated with photosynthesis as well as the synthesis of amino acids, fatty acids, carotenoids, vitamins, and a range of specialized metabolites (Armbruster et al., 2011). The chloroplast is derived from an ancient cyanobacterial endosymbiont with many of the ancestral genes of this organism incorporated into the host genome (Timmis et al., 2004). Current estimates suggest that there are between 2,000 and 3,000 chloroplast-localized proteins, the vast majority of which are encoded by the host genome (Armbruster et al., 2011). Therefore, chloroplast homeostasis requires coordination of the nuclear and plastid genomes together with the transport of these nuclear-encoded proteins into the chloroplast, their processing, folding, and assembly into functional protein complexes and their subsequent degradation. These processes must be intricately regulated to reflect changes in plastid function during development, the metabolic status of the cell, and be responsive to environmental conditions that perturb chloroplast function. Developing a systematic understanding of the function of chloroplast-localized proteins has broad implications for defining and manipulating plant metabolism and considerable progress has been made in this area, particularly in model organisms (Waters and Langdale, 2009; Ajjawi et al., 2010; Armbruster et al., 2011). Investigating the function of chloroplast-localized proteins in diverse species with novel physiology or specialized metabolism will provide additional and important insight into the functions of this unique organelle.
The onset of fruit ripening is marked by a reprogramming of cellular metabolism that often includes cell wall modifications together with associated fruit softening, the conversion of chloroplasts into chromoplasts and the synthesis of brightly colored pigments such as carotenoids, the conversion of starch into simple sugars, and the synthesis of volatile compounds that influence taste and aroma. These biochemical processes are highly regulated and contribute to the transformation of the fruit from an unpalatable and often toxic organ into one that is attractive and nutritious for seed-dispersing fauna (Barry, 2010; Klee and Giovannoni, 2011). The factors that regulate the ripening transition are not fully understood, although specific transcription factors belonging to the MADS-box and SQUAMOSA PROMOTER BINDING PROTEIN families are necessary for ripening and affect multiple aspects of the ripening process, including ethylene synthesis, softening, color change, and aroma volatile production (Herner and Sink, 1973; Sink et al., 1974; Vrebalov et al., 2002, 2009; Manning et al., 2006; Itkin et al., 2009; Kovács et al., 2009; Chung et al., 2010; Jaakola et al., 2010; Karlova et al., 2011; Martel et al., 2011; Seymour et al., 2011; Lee et al., 2012). In climacteric fruit such as tomato (Solanum lycopersicum), these transcription factors act upstream of ethylene biosynthesis and ethylene is required for full development of the ripe phenotype (Barry and Giovannoni, 2007; Klee and Giovannoni, 2011; Martel et al., 2011). However, recent research has also implicated additional hormones in mediating the onset of fruit ripening, including brassinosteroids and abscisic acid (Nakano et al., 2003; Lisso et al., 2006; Symons et al., 2006; Zhang et al., 2009; Chai et al., 2011; Jia et al., 2011). Together, these data suggest a complex interplay of transcriptional regulation and hormonal signals influence the ripening of fleshy fruits.
The chloroplast and subsequent development of the carotenoid-rich chromoplast in ripening fruits greatly impacts overall fruit quality with plastid-derived primary and secondary metabolites influencing multiple aspects of the ripe phenotype, including nutrient content, color, and aroma (Klee, 2010). Consequently, many of the enzymes involved in these ripening-induced metabolic changes are localized or associated with either the chloroplast or the chromoplast (Chen et al., 2004; Klee, 2010). Additionally, several mutants that alter different aspects of fruit quality do so through changes in either chloroplast or chromoplast biochemistry. For example, the high-pigment1 (hp-1), hp-2, and hp-3 loci possess altered chloroplast number and ultrastructure, contributing to fruits with elevated levels of chlorophyll, carotenoids, and flavonoids, together with altered patterns of aroma volatile production (Yen et al., 1997; Bino et al., 2005; Kolotilin et al., 2007; Galpaz et al., 2008; Kovács et al., 2009). Similarly, inhibition of chlorophyll degradation due to mutations in tomato and pepper (Capsicum annuum) homologs of the chloroplast-targeted STAY-GREEN protein of rice (Oryza sativa), are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper that influence fruit color (Barry et al., 2008; Borovsky and Paran, 2008). Recently, multiple aspects of the ripening process, including the ethylene climacteric and carotenogenesis, were found to be disrupted in the Orange ripening (OrrDS) mutant that encodes the M subunit of the plastidial NADH dehydrogenase complex (Nashilevitz et al., 2010). These studies illustrate the important role of the chloroplast in influencing fruit ripening and the quality of fleshy fruit.
The lutescent mutants of tomato, lutescent1 (l1) and l2, are nonallelic monogenic mutants that have identical phenotypes resulting in an early and progressive loss of chlorophyll from leaves and fruits. In fruit tissue, the loss of chlorophyll during development renders the fruit a whitish-yellow color prior to the onset of ripening (Kerr, 1956). Here we report that l1 and l2 mutants exhibit pleiotropic chlorophyll-deficient phenotypes including hypersensitivity to high-light stress and delayed deetiolation. In addition, mutation at the l1 and l2 loci leads to a delay in the onset of fruit ripening, suggesting the possibility of a chloroplast-derived signal that stimulates fruit ripening. Concomitant with the diverse chlorophyll-deficient phenotypes, positional cloning of the l2 locus revealed the presence of a point mutation that introduces a premature stop codon in a tomato homolog of ETHYLENE-DEPENDENT GRAVITROPISM DEFICIENT AND YELLOW-GREEN1 (EGY1), which encodes a chloroplast-targeted zinc metalloprotease required for chloroplast development in Arabidopsis (Arabidopsis thaliana; Chen et al., 2005).
RESULTS
lutescent Mutants Display Pleiotropic Chlorophyll-Deficient Phenotypes
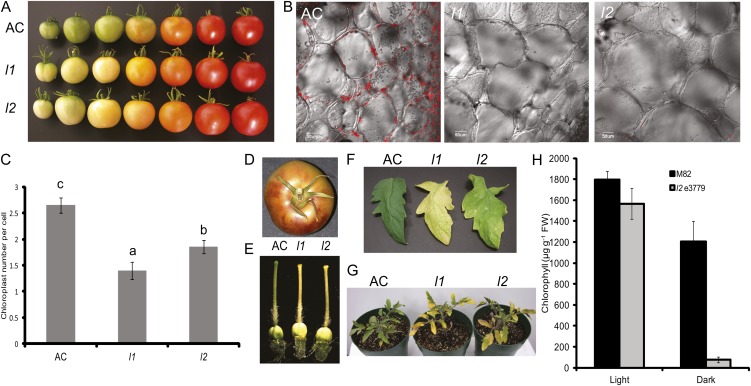
Developing fruits of the l1 and l2 mutants of tomato are characterized by reduced chlorophyll content, leading to mature fruits that are whitish yellow rather than the typical green color associated with wild-type Ailsa Craig (AC) fruit (Fig. 1A). At the cellular level mature green AC fruits viewed using confocal laser-scanning microscopy show significant amounts of chlorophyll autofluorescence and this is almost completely absent in mature fruits of l1 and l2 mutants (Fig. 1B). The chlorophyll loss observed in l1 and l2 fruits is progressive with increasing age but the average chloroplast number per cell in young developing fruits at 20 d post anthesis was considerably lower in l1 and l2 compared to AC (Fig. 1C). Growing l1 and l2 mutants over several years it was noted that the chlorophyll loss in fruits is typically more exaggerated in plants grown during the summer when light intensities are higher and photoperiods longer and was often accompanied by accumulation of purplish epidermal pigments that are likely to be anthocyanins (Fig. 1D). A lack of chlorophyll accumulation is also observed in developing pistils of l1 and l2 flowers (Fig. 1E). Progressive and early chlorophyll loss is also a feature of the leaves of l1 and l2 (Fig. 1F). This phenomenon is exaggerated in plants grown under high-light conditions where substantial chlorophyll loss was observed even in young expanding leaves but is also apparent in l2 plants held in the dark for 2 weeks (Fig. 1, G and H). Leaves of a second l2 mutant allele, designated l2 3779, which was identified in an ethane methylsulfonate (EMS) mutant screen in the M82 cultivar (Menda et al., 2004; see below for details of this allele) had 13% less chlorophyll than leaves of wild-type cv M82 when grown under light conditions. However, following 2 weeks in darkness, l2 e3779 lost 95% of the chlorophyll content whereas M82 leaves lost only 37% (Fig. 1H).
Figure 1.
Phenotypes of the lutescent mutants. A, Phenotypes of developing and ripening fruit of l1 and l2 mutants compared to wild-type (AC) fruit. B, Confocal laser-scanning microscopy images of AC, l1, and l2 fruits at the mature stage of development showing chlorophyll autofluorescence. C, Chloroplast number per cell in immature green fruit pericarp at 20 d post anthesis. Data are presented as the mean ± se of at least 103 cells. Means followed by different letters indicate statistical significance at α = 0.05. D, Image of a mature l1 fruit displaying elevated anthocyanin accumulation in the fruit peel. E, Chlorophyll-deficient phenotypes in the pistils of l1 and l2. F, Fully expanded leaflets of AC, l1, and l2 highlighting increased susceptibility to chlorophyll loss in each mutant. G, Phenotype of 3-week-old AC, l1, and l2 plants grown under high-light conditions. H, Chlorophyll breakdown in the dark in leaves of l2 e3779. Chlorophyll concentration was determined in leaves of plants of mutant l2 e3779 and its isogenic wild-type cv M82 grown for either 7 weeks in the light or 5 weeks in the light followed by 14 d under complete darkness. Data are presented as the mean ± se (n = 6).
lutescent Mutants Exhibit Reduced Rates of Deetiolation and Thylakoid Membrane Formation
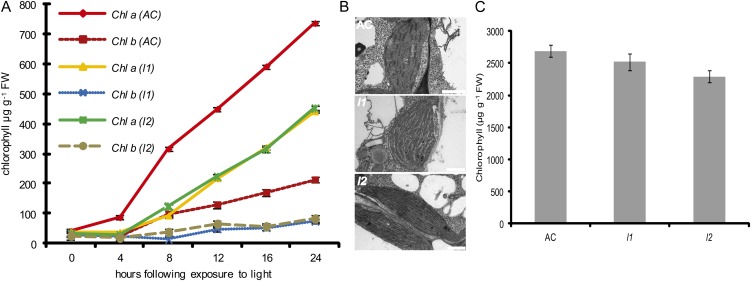
The conversion of etioplasts into chloroplasts and the subsequent development of photosynthetic competency is regulated by light (Fankhauser and Chory, 1997). To establish whether the l1 and l2 loci impact photomorphogenesis, the rate of deetiolation was determined in the cotyledons of dark-grown seedlings exposed to light. Dark-grown seedlings of l1 and l2 displayed the typical etiolated phenotype observed in AC seedlings (data not shown). However, upon exposure to light, a delay in chlorophyll accumulation was observed in both mutants (Fig. 2A). Cotyledons of wild-type AC seedlings initiated chlorophyll synthesis within 2 h of transfer to the light and this steadily increased over the 24 h time frame of the experiment. In contrast, both l1 and l2 accumulated chlorophyll at a slower rate and achieved less than 60% of the levels attained by AC seedlings after 24 h of light exposure. Concomitant with the reduced rates of chlorophyll accumulation during deetiolation, examination of plastid ultrastructure revealed impaired development of the thylakoid membranes in the chloroplasts of l1 and l2 mutants compared to those of AC plastids (Fig. 2B). Although chlorophyll accumulates more slowly during deetiolation, the chlorophyll content of fully expanded leaves is similar in AC, l1, and l2 before the onset of early chlorophyll loss (Fig. 2C).
Figure 2.
Inhibition of photomorphogenesis in lutescent mutants. A, Chlorophyll a (Chl a) and chlorophyll b (Chl b) levels in cotyledons of dark-grown seedlings following exposure to light. Data are presented as the mean of 10 replicates per data point ± se. B, Transmission electron micrographs of developing chloroplasts of cotyledons following 24 h of light exposure. Scale bars are equivalent to 1 μm. C, Chlorophyll content in expanded leaves of lutescent mutants. Data are presented as the mean of eight replicates per data point ± se.
Photosynthesis and Yield in lutescent Fruit
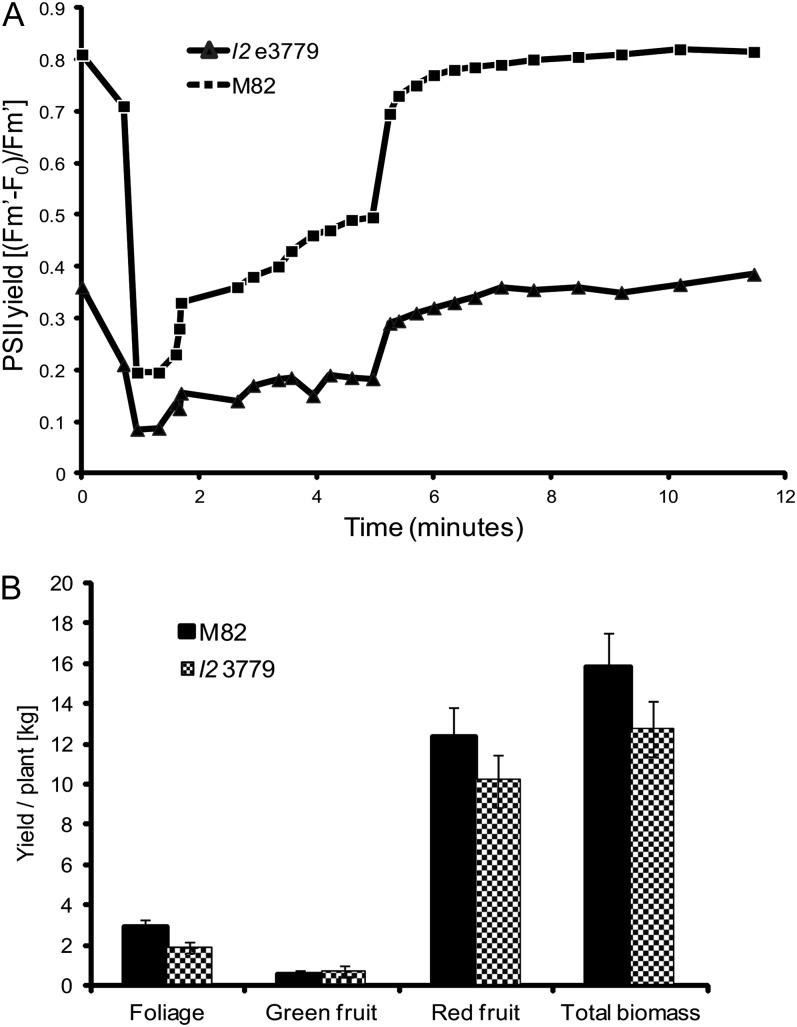
Developing tomato fruit are green due to the presence of chloroplasts that are mainly localized in the outer pericarp wall. Chlorophyll concentration in the outer pericarp tissue of fruit of cv M82 varies between fruit and with the stage of development and on average is 50- to 100-fold lower than in leaves (data not shown). The photosynthetic activity of M82 and l2 e3779 fruit at the mature green stage of development was determined by chlorophyll fluorescence (Fig. 3A). The concentration of chlorophyll was 22.2 ± 1.2 μg g−1 fresh weight in M82 and 2.8 ± 0.3 μg g−1 fresh weight in l2 e3779. PSII efficiency in fruit of M82 was similar to that of leaves (data not shown). In contrast, the fluorescence level of l2 e3779 fruit was lower than in M82 and the efficiency of PSII was significantly impaired (Fig. 3A). The determinate growth habit of the M82 variety enables quantitative analysis of fruit yield and overall biomass during harvesting. Plants of M82 and l2 3779 were grown in the field during April to July, 2011. Individual plants were analyzed at time of harvesting with the l2 3779 mutant exhibiting a 17% lower yield and biomass production than M82 (Fig. 3B).
Figure 3.
Photosynthesis and yield in l2 e3779. A, Efficiency of PSII activity was determined by fluorescence measurements in fruit of l2 e3779 and M82. B, Total yield and biomass of field-grown plants of l2 e3779 and M82 (kg/plant). Data are presented as the mean of 11 replicates per data point ± se.
The Onset of Fruit Ripening Is Delayed in lutescent Mutants
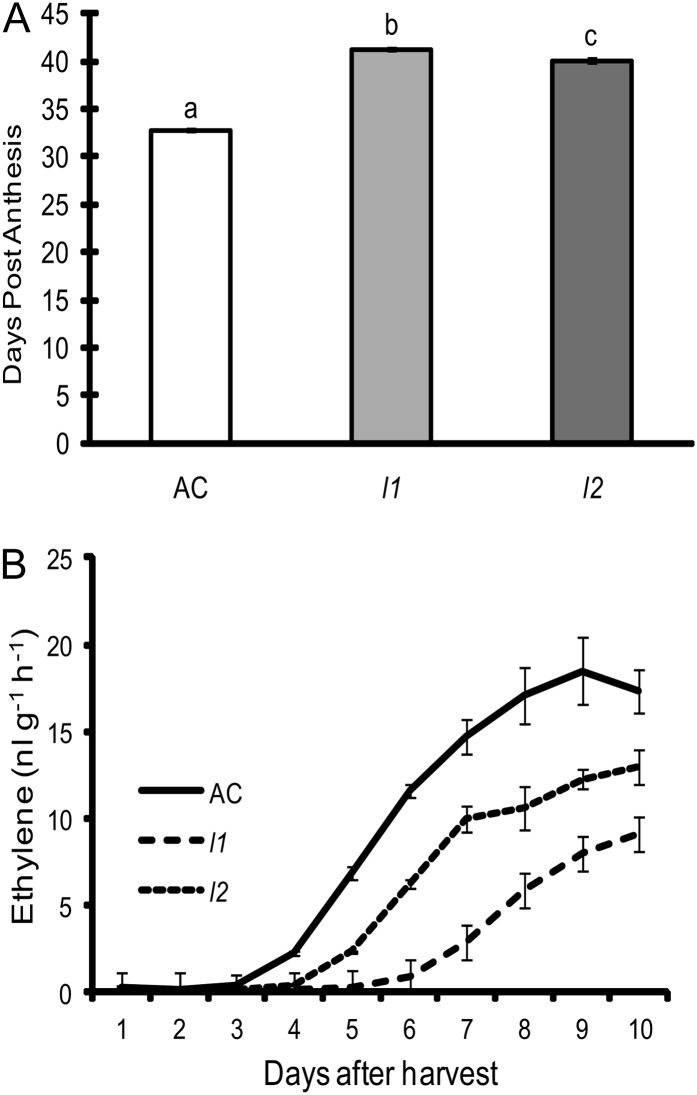
Together with the altered chlorophyll content in the fruits of the lutescent mutants (Fig. 1, A and B), it was observed that fruits of each mutant ripened slightly, but consistently, later than AC fruit. This was confirmed through determining the duration from anthesis to the onset of ripening (breaker stage) that revealed an approximate delay of 6 d in the mutants compared with wild type (Fig. 4A). This delay in the onset of ripening was also observed in detached fruits harvested at the mature green stage of development and allowed to ripen off the vine. The onset of ethylene synthesis in detached fruits was delayed in both l1 and l2 and failed to reach the same levels as that observed in AC fruits (Fig. 4B). This slow-ripening phenotype appeared to be specific to a delay in the onset of the ripening process as once ripening was initiated the rate of progression appeared identical in wild-type and mutant plants with lycopene accumulation progressing at the same rate with both mutant and wild-type fruits becoming fully ripe at approximately 7 d after the onset of ripening (Supplemental Fig. S1). Furthermore, ethylene treatment of wild-type AC, l1, and l2 fruits at the mature green stage of development led to the onset of ripening within 2 d of treatment in each genotype, indicating that ethylene responsiveness is not significantly altered in mutant fruits (Supplemental Fig. S2). Similarly, the ethylene responsiveness of l1 and l2 dark-grown seedlings was normal (Supplemental Fig. S2). Together, these data suggest that mature green l1 and l2 fruit are fully competent to ripen yet are perturbed in a signal that initiates ethylene synthesis and therefore the onset of fruit ripening.
Figure 4.
The onset of fruit ripening is delayed in lutescent mutants. A, The number of days from anthesis to the onset of ripening was determined for each genotype. Data are presented as the mean of 71 replicates per data point ± se. Means followed by different letters indicate statistical significance at α = 0.001. B, Ethylene synthesis in detached fruits of AC, l1, and l2. Data are presented as the mean of three replicates per data point ± se.
Positional Cloning of the l2 Locus
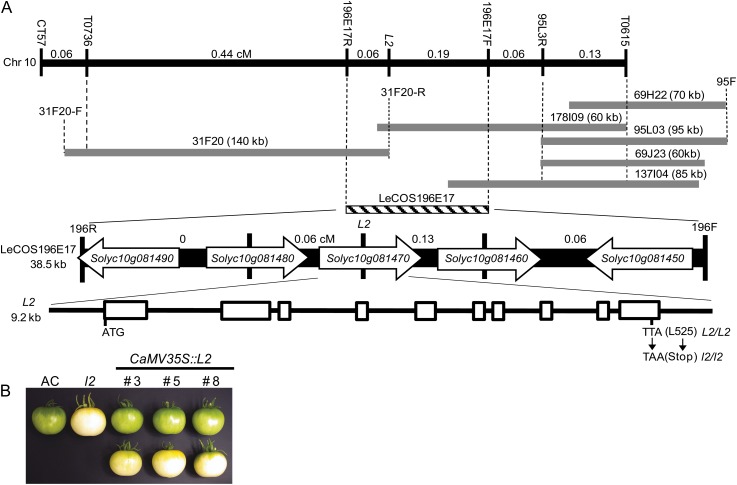
Classical linkage mapping indicated that the l2 mutant mapped in close proximity to the tangerine and hairs absent loci on linkage group VII that was subsequently identified as the long arm of chromosome 10 (Kerr, 1956). Using an F2 population of 60 individuals derived from an interspecific cross l2/l2 (tomato) × L2/L2 (Solanum galapagense), the l2 locus was mapped to a 7.5 centimorgan (cM) region of the long arm of chromosome 10 between the flanking RFLP markers T1682 and T802 (data not shown). Upon increasing the size of the F2 mapping population to 799 individuals by screening for recombinant individuals between T1682 and T0802 the position of the L2 locus was refined to a 0.88 cM interval between the RFLP markers T0615 and T0736 (Fig. 5A). T0615 and T0736 were used to screen three high Mr tomato genomic DNA libraries made from DNA extracted from Solanum pennellii and S. galapagense (Li et al., 2005; Chen et al., 2007). Five individual clones were found to hybridize to T0615 and a single 140-kb clone, 31F20, was isolated following screening with T0736. The ends of these clones were isolated by direct DNA sequencing and specific primers were designed to amplify each end by PCR. These ends were converted to RFLP probes to enhance the resolution of the genetic map and to position the genomic clones relative to one another. A probe derived from the reverse end of 31F20 (31R) hybridized to clone 178I9, indicating that the L2 locus was contained on two overlapping clones at a maximum physical distance of approximately 200 kb. 31R was used as a probe to screen a tomato cosmid library and a single clone, LeCOS196E17, was identified. The ends of this clone were isolated as previously described and converted to RFLP markers. Genetic mapping indicated that the l2 locus lies within clone LeCOS196E17, 0.06 cM from 196R and 0.19 cM from 196F.LeCOS196E17 was sequenced revealing five predicted genes, corresponding to the tomato genome locus identifiers Solyc10g081450 through Solyc10g081490 (SL2.40Ch10:61831485–61868274). Sequence similarity searches with each predicted gene revealed that Solyc10g081450 encodes an α/β-fold hydrolase-related protein, Solyc10g081460 encodes a predicted amino acid transporter, Solyc10g081470 encodes a chloroplast-targeted zinc metalloprotease, Solyc10g081480 encodes a protein of unknown function, and Solyc10g081490 encodes a MYB transcription factor. Fragments of each gene were converted into RFLP markers and were mapped in the F2 population. Solyc10g081470 cosegregated with the mutant phenotype whereas Solyc10g081480 and Solyc10g081460 were 0.06 and 0.13 cM from the L2 locus, respectively, suggesting that Solyc10g081470 is the candidate gene for the l2 locus.
Figure 5.
Positional cloning of the L2 locus. A, Genetic and physical map of the l2 locus derived using an interspecific F2 population of 799 plants. Distances between adjacent RFLP markers are given in cM. BAC clones isolated using the RFLP markers T0615 and T0736 are shown as gray bars. BAC ends were isolated as described in the text and given F and R designations. A cosmid clone, LeCOS196E17, isolated using 31F20-R is represented as a striped bar. Vertical dashed lines are shown to indicate relative positions of genetic markers and genomic clones. LeCOS196E17 contains five predicted genes (Solyc10g081450–Solyc10g081490). The genetic distance between these genes is shown in cM. Solyc10g081470 cosegregates with the l2 phenotype and has a structure comprised of 10 exons separated by nine introns. The T → A substitution in exon 10 that converts Leu 525 to a stop codon in l2 is shown. B, Complementation of the l2 mutant phenotype. Mature green fruit from three independently transformed segregating T1 lines containing the transgene (top section) and without the transgene (bottom section). AC and l2 fruits are shown for phenotypic comparison.
Sequencing of the predicted full-length cDNA of Solyc10g081470 amplified by reverse transcription (RT)-PCR from L2/L2 and l2/l2 genotypes revealed a single T → A substitution at base 1574 of the cDNA clone (exon 10 in the genomic clone) in the l2 mutant. The substitution results in the conversion of Leu 525 into a stop codon, truncating the predicted protein by 23 amino acids. Confirmation that the single base pair substitution in Solyc10g081470 is the underlying cause of the l2 mutant phenotype was achieved through complementation analysis. The full-length Solyc10g081470 cDNA was expressed under the control of the cauliflower mosaic virus 35S promoter in the l2 mutant background. Nineteen out of 22 independently transformed primary transformants recovered showed complementation of the l2 phenotype, determined by chlorophyll content of mature green fruit. Three lines were selected for subsequent analysis in the T1 generation (Fig. 5B). Typical phenotypes of AC and l2 mature green fruits are shown along with three independent T1 lines segregating for the NPTII selectable marker. Fruits from plants in the top section contained NPTII and showed complementation of the l2 mutant phenotype. Conversely the fruit on the bottom section came from plants that had segregated out the NPTII gene and had reverted to the l2 mutant phenotype.
L2 Encodes a Tomato Homolog of Arabidopsis EGY1
L2 (Solyc10g081470) encodes a predicted protein of 547 amino acids with a molecular mass of 58.4 kD. A BlastP search using the predicted L2 protein as the query sequence revealed homology to proteins from several higher plant species and other photosynthetic organisms, including bryophytes, algae, and cyanobacteria. Significantly, the Arabidopsis protein displaying the highest similarity to L2 (73% identical/82% similarity) was encoded by the EGY1 locus, a chloroplast-targeted member of the zinc-dependent M50 family of metalloproteases (http://merops.sanger.ac.uk) that is required for normal plastid development and gravitropic response in Arabidopsis (Chen et al., 2005). The M50 family is diverse, with members from bacteria, archaea, protozoa, plants animals, and some fungi. The family is characterized by two conserved domains, HEXXH and NXXPXXXLDG that are present in L2 and related homologs (Fig. 6). These two domains are predicted to form the active site with Glu acting as the catalytic residue and the histidines and Asp residues coordinating the zinc ions. Mutant studies have demonstrated the importance of these domains for catalytic activity (Rawson et al., 1997; Rudner et al., 1999). A second feature of this family is that they are integral membrane proteins with the active site domains lying within, or in close proximity to, the membrane-spanning domains (Fig. 6). The Aramemnon Plant Membrane Database (http://aramemnon.botanik.uni-koeln.de/index.ep), which collates the membrane topology predictions of several algorithms, predicts that Arabidopsis EGY1 contains eight transmembrane-spanning domains (Fig. 6). A search utilizing the TMHMM server version 2.0 (http://www.cbs.dtu.dk) revealed that the predicted L2 protein has six transmembrane-spanning domains with an additional two domains lying slightly below the threshold of probability. Similarly, the selection of EGY1/L2 homologs depicted in Figure 6 are all predicted to contain between six and eight transmembrane-spanning domains lying in relatively conserved positions (data not shown). Localization experiments utilizing an Arabidopsis EGY1-GFP fusion demonstrated that EGY1 is targeted to the chloroplast (Chen et al., 2005). Similarly, in silico prediction of L2 subcellular localization using the Predotar server (http://urgi.versailles.inra.fr/predotar/predotar.html) resulted in a probability estimate of 0.79 for plastid localization.
Figure 6.
Amino acid alignment of L2-related proteins. Alignments based on the conserved region of L2 related proteins were generated using MUSCLE. Conserved amino acids are indicated by shaded squares. The highly conserved motifs characteristic of these proteins, GNLR, HEXXH, and NXXPXXLDG are underlined with solid black lines. The solid gray lines depict the predicted transmembrane helices for EGY1 as determined by the TmConsens algorithm (http://aramemnon.botanik.uni-koeln.de/index.ep). Accession numbers of the sequences are as follows: L2 (JQ683149), Vitis vinifera GSVIVT01029948001 (XP_002269447), Populus trichocarpa POPTRDRAFT_773851 (XP_002319720), rice Os03g0792400 (EEE60079), Synechocystis sp. PCC 6803 sll0862 (NP_440944). The sequence of Arabidopsis EGY1 is based on annotation available at The Arabidopsis Information Resource (http://www.arabidopsis.org) and the sequences of Cre19.g754700 and Ppls517_1V6.2 are based on annotations of the C. reinhardtii and Physcomitrella patens genomes available through the Phytozome database (http://www.phytozome.net/).
Identification of Additional Alleles at the l2 Locus
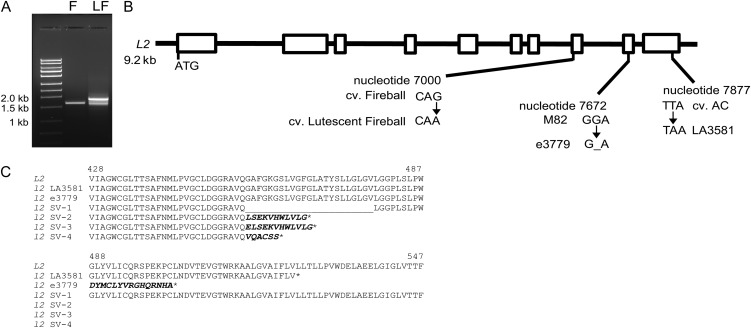
A second putative l2 allele was reported as a spontaneous mutation in a trial plot of the cultivar Fireball in 1960 and was designated Lutescent Fireball (Kerr et al., 1975; Supplemental Fig. S3). An allelism test confirmed that F1 progeny of a cross between Lutescent Fireball and the original l2 allele in the Long Red variety displayed a typical lutescent phenotype (Kerr et al., 1975). RT-PCR amplification of L2 from cv Fireball yielded the expected transcript of 1,641 bp that was identical to that obtained from cv AC. In contrast, amplification of the L2 cDNA from Lutescent Fireball revealed multiple products of a slightly altered size (Fig. 7A). Sequence analysis of these RT-PCR products revealed the existence of four distinct L2 transcripts in the Lutescent Fireball cultivar ranging in size between 1,572 and 1,846 bp, each with aberrant splicing occurring between intron 7 and exon 8. Three of the splice variants (SV2, SV3, and SV4) cause frame-shift mutations, leading to incorporation of several spurious amino acids followed by premature stop codons that truncate the predicted L2 protein between 79 and 85 amino acids (Fig. 7B). In contrast the SV1 splice variant causes an in-frame deletion of the entire eighth exon, leading to the removal of 23 amino acids between Q455 and L479 (Fig. 7C). Furthermore, the wild-type transcript was not detected in the Lutescent Fireball cultivar, suggesting that the L2 protein may be nonfunctional in this variety. To further explore the possibility that a splicing defect in L2 is the underlying cause of the Lutescent Fireball phenotype, a 750-bp genomic fragment spanning exons 7 and 8 from in both Fireball and Lutescent Fireball revealed a G → A nucleotide change at position 7000 of the L2 genomic clone in the Lutescent Fireball cultivar (Fig. 7B). This single nucleotide substitution disrupts the consensus splice acceptor site at the boundary between intron 7 and exon 8 (CAG → CAA). Together, these data suggest that the phenotype of Lutescent Fireball is caused by aberrant splicing, resulting in the failure to produce a mature L2 transcript. A third mutant resembling l2, e3779 that figures prominently in the characterization described here, was recovered from an EMS mutagenized population of the M82 cultivar (Menda et al., 2004; Supplemental Fig. S2). Sequence analysis of the L2 cDNA amplified from e3779 revealed the deletion of a G nucleotide at position 1575 that results in a frame-shift mutation that removes Gly 488, resulting in the incorporation of 15 spurious amino acids before terminating in a premature stop codon (Fig. 7, B and C).
Figure 7.
Identification of additional l2 mutant alleles. A, Aberrant splicing of L2 in the Lutescent Fireball cultivar. RT-PCR amplification of cDNA fragments corresponding to L2 from Fireball (F) and Lutescent Fireball (LF) cultivars. The approximate size of the DNA fragments is indicated. B, Relative positions and nucleotide changes observed in the l2 mutant alleles identified in the Lutescent Fireball cultivar and in the EMS derived mutant e3779. White boxes correspond to exons. C, Consequences of l2 mutant alleles on the predicted L2 protein sequence.
DISCUSSION
The Role of L2 in Chloroplast Function
The nonallelic l1 and l2 mutants display pleiotropic phenotypes that are consistent with defects in chloroplast development (Figs. 1 and 2). These data are supported by the finding that L2 encodes an ortholog of Arabidopsis EGY1, mutations that disrupt plastid number and development, leading to impaired formation of the thylakoid membranes and reduced accumulation of chlorophyll a/b-binding proteins of the light-harvesting complexes I and II (Chen et al., 2005; Guo et al., 2008). However, a notable difference between l2 and egy1 mutant alleles is that while egy1 mutant alleles have reduced chlorophyll content and are pale throughout development (Chen et al., 2005), leaves of l2 mutants are slow to deetiolate (Fig. 2) but eventually accumulate close to wild-type levels of chlorophyll prior to undergoing premature chlorophyll loss later in development (Fig. 2C). Furthermore, altered patterns of chlorophyll accumulation are evident between leaf and reproductive tissues of lutescent mutants. For example, while l1 and l2 leaves accumulate chlorophyll, the pistils and fruits of the mutants display greatly impaired chlorophyll accumulation from early in development and therefore resemble the pale phenotype of egy1 mutant alleles (Fig. 1). The basis for these differences, particularly the seemingly less significant role that L2 plays in chlorophyll accumulation in tomato leaves, is currently not understood.
In addition to a role in mediating chlorophyll accumulation during development, evidence suggests that EGY1/L2 and related homologs may play a role in mediating chloroplast responses to environmental conditions. For example, both l1 and l2 mutants are hypersensitive to darkness and high-light stress, leading to enhanced chlorosis (Fig. 1, G and H) and EGY1 protein levels in Arabidopsis are influenced by dark/light cycles with increased abundance evident in light-grown tissues (Chen et al., 2005). Similarly, iron deficiency, a stress known to reduce photosynthetic electron transfer, resulted in increased accumulation of the EGY1/L2 homolog, C_770040, from Chlamydomonas reinhardtii (Naumann et al., 2007). Furthermore, publically available Arabidopsis gene expression data revealed that EGY1 is induced under both high-light treatments and iron deficiency together with other biotic and abiotic stresses (Zimmermann et al., 2004). Together these data suggest a role for EGY1/L2 in both the early steps of chloroplast development and the maintenance of chloroplast homeostasis in response to environmental perturbation.
EGY1 and L2 belong to the M50 family of metalloproteases and each contains motifs known to be required for the activity of functionally characterized members of this family (Fig. 6). While considerable progress has been made on elucidating the role of chloroplast-localized proteases (Kato and Sakamoto, 2010), the exact biochemical function of EGY1/L2 and other plant M50 proteases, including the identity of their in vivo substrates remains unknown. Members of the M50 family of proteases are integral membrane proteins with their active sites residing within or adjacent to the membrane (Fig. 6). The M50 family belong to a category of proteases referred to as intramembrane cleaving proteases that are involved in the regulation of cellular responses to metabolic status and stress that participate in a general phenomenon known as regulated intramembrane proteolysis (RIP). For example, the founder member of the M50 family, the sterol-regulatory element binding protein (SREBP) site-2 protease regulates sterol biosynthesis and uptake in mammalian cells through release of the membrane-bound transcription factor SREBP in a pathway that involves trafficking of the sequestered SREBP from the endoplasmic reticulum to the Golgi under low-sterol conditions (Osborne and Espenshade, 2009). Similar two-step proteolysis occurs in response to stress in bacteria, allowing the release of membrane-bound transcription factors (Rudner et al., 1999; Alba et al., 2002; Kanehara et al., 2002). Far less is known about RIP in plants. Membrane-tethered transcription factors have been identified in plants and in a few instances proteolytic cleavage of these transcription factors has been demonstrated but none of these have been reported to involve transcription factors that are sequestered in the chloroplast (Seo et al., 2008; Kim et al., 2010; Liu and Howell, 2010). Therefore, it is possible that EGY1/L2 proteases participate in chloroplast-localized phenomena such as processing or degradation of proteins at the thylakoid membrane during photosystem assembly or in response to environmental perturbation and disruption of photosynthetic function. Examples of proteolysis and processing in the chloroplast are already well characterized and are involved in the turnover and repair of photosystem proteins in response to photoinhibition and abiotic stress (Kato and Sakamoto, 2010; Sun et al., 2010).
lutescent Mutants and Their Impact on Fruit Development and Ripening
Both l1 and l2 mutants exhibited a delay in the onset of fruit ripening (Fig. 4). However, once ripening is initiated in l1 and l2 fruits the rate of ripening appears normal and mutant fruits accumulate wild-type levels and composition of carotenoids (Supplemental Fig. S1), suggesting that a fully functional chloroplast is not required for chromoplast development in tomato. The concept of separation of chloroplast and chromoplast development is also observed in stay-green mutants of tomato and pepper that undergo typical chromoplast development in the absence of chlorophyll breakdown and thylakoid membrane dissolution (Cheung et al., 1993; Roca et al., 2006).
As l1 and l2 mutants have disrupted chloroplast function, it is likely that the delayed fruit ripening phenotypes observed in mutant fruit arise as an indirect consequence of an altered chloroplast-derived signal that promotes the onset of ripening. The recent finding that disruption of Glu-1-semialdehyde aminotransferase activity in tomato fruits alters early seed development (Lytovchenko et al., 2011) provides a possible mechanism that could ultimately lead to a delay in the onset of ripening. It will be of interest to examine the rates of ripening in Glu-1-semialdehyde aminotransferase-silenced lines and early seed development in l1 and l2 mutants, particularly as fruit of lutescent mutants are also compromised in their photosynthetic capacity (Fig. 3A).
Alternatively, lower chloroplast numbers in developing fruits of l1 and l2 could be supportive of reduced levels of a factor that promotes ripening. The nature of the signals that stimulate ripening are not fully understood, although multiple factors including ethylene are known to play a role in ripening induction (Srivastava and Handa, 2005; Barry and Giovannoni, 2007; Zhang et al., 2009; Barry, 2010). The ability of ethylene to trigger rapid and normal ripening of l1 and l2 fruit at the mature green stage of development, coupled with the delay in the onset of ethylene synthesis suggests that l1 and l2 fruit possess a typical response to ethylene but may be perturbed in the ability to initiate the ripening-related increase in ethylene production (Fig. 4; Supplemental Fig. S1). Recent characterization of the OrrDS mutant of tomato that encodes the M subunit of the chloroplast-localized NADH dehydrogenase complex, also supports a role for a chloroplast-derived signal that promotes ripening (Nashilevitz et al., 2010). The OrrDSmutant exhibits a delay in the onset of fruit ripening and ripening-related ethylene biosynthesis. Although the reason for this delay is not fully understood, the OrrDS mutant has a 30% to 40% reduction in the levels of both Ser and Asp, two chloroplast-derived amino acids that are precursors of Met, which serves as the precursor of ethylene (Nashilevitz et al., 2010). It will be of interest to determine whether chloroplast metabolism in l1 and l2 fruit influences seed development and precursor pools for the synthesis of hormones and to define whether a link exists between these factors and the observed delay in the onset of fruit ripening.
CONCLUSION
Cloning of the L2 gene has identified a link between chloroplast development and as-yet-unidentified factors that influence the onset of fruit ripening. The mechanism of this interaction, together with the specific biochemical activity of EGY1/L2 and the potential role of these proteins in RIP remain to be identified. In this regard, we have shown that the l1 mutant, which resides at a distinct locus on chromosome 8 (Tanksley et al., 1992), possesses an identical phenotype to that of l2, suggesting the strong possibility that the underlying gene may be functionally related to L2. Therefore, identification of the gene underlying the l1 mutant phenotype may provide insight into the molecular function of EGY1/L2 in plants.
MATERIALS AND METHODS
Plant Material and Treatments
Tomato (Solanum lycopersicum) seeds homozygous for the l1 (LA3717) and l2 (LA3581) mutations and Solanum galapagense (LA0483) were obtained from the Tomato Genetics Resource Center, University of California, Davis. The parental cultivar AC was originally obtained from the Glasshouse Crops Research Institute (Littlehampton, Sussex, UK). Seeds of the cultivars Fireball (CN 18026) and Lutescent Fireball (CN 1586) were obtained from Plant Gene Resources of Canada. Seeds of the EMS mutant, e3779 and the M82 parental cultivar were provided by Dani Zamir (Hebrew University of Jerusalem). Tomato plants were grown in peat-based compost supplemented with fertilizer in greenhouses equipped with heating and cooling systems and supplemental lighting either at Cornell University, Ithaca, New York or at Michigan State University, East Lansing, Michigan and under greenhouse conditions at Hebrew University of Jerusalem, Israel or in the field at an experimental station in Akko, Israel.
Evaluation of the deetiolation response of tomato seedlings was performed as follows. Surface-sterilized seeds were sown on 1% water agar supplemented with 0.5× Murashige and Skoog salts pH 5.6 and incubated in the dark for 7 d at 25°C. The germinated seedlings were then exposed to light for up to 24 h and the cotyledons excised and either immediately frozen in liquid N2 or immersed in fixative for transmission electron microscopy (described below). Experiments to evaluate the triple-response phenotype in dark-grown tomato seedlings were performed as previously described (Barry et al., 2005) with the exception that seedlings were measured at 8 d after sowing. Ethylene treatments were performed by sealing fruits at the mature green stage of development in glass canning jars of a known volume and exposing to 10 μL L−1 of ethylene for 24 h. Following ethylene treatments fruits were held at room temperature for 72 h and ripening evaluated. For evaluation of response to high-light conditions plants were grown under a 16 h photoperiod at 27°C/20°C day/night temperature and a light intensity of 500 μmol m−2 s−1. Plants were photographed 3 weeks after germination.
Ethylene Measurements
Mature green fruit were harvested and held at room temperature for 24 h. For ethylene determination, fruits were sealed each day for 2 h in glass jars and a 1-mL headspace sample was analyzed by gas chromatography using a Carle AGC series 100 gas chromatograph equipped with an activated alumina column and flame ionization detector, as previously described (Watkins et al., 2004).
Measurements of Photosynthetic Efficiency
The efficiency of PSII was estimated by measuring chlorophyll fluorescence with a computer-controlled pulse amplitude modulated fluorometer IMAG-MAX/L (Walz). Detached fruit were dark adapted for 20 min. A 2-cm-thick slice was cut and placed in the measuring chamber with the fruit surface facing up toward the measuring beam. A low (0.1 μmol photons m−2 s−1) modulated (1.6 kHz) measuring light was used to determine the minimal fluorescence level, followed by a 1 s of saturating white light pulse of 1,500 μmol photons m−2 s−1 to obtain the maximal fluorescence level. The fruit was then illuminated with continuous actinic red light (650 nm, 40-μmol photons m−2 s−1), until a steady-state fluorescence was reached (up to 10 min). A second saturating pulse was then applied to determine the maximal fluorescence level in the light-adapted state.
Pigment Analysis
Chlorophyll was extracted from tissues in acetone and quantified as previously described (Lichtenthaler, 1987). Fruit carotenoids were analyzed by HPLC as previously described (Ronen et al., 1999).
Confocal Laser-Scanning Microscopy
The chloroplasts of tomato fruit were imaged using an Olympus FluoView FV1000 laser-scanning confocal microscope. Chlorophyll autofluorescence was detected with a 650 to 750 nm band pass filter following excitation at 488 nm with an argon laser line. Cells were visualized using differential interference contrast transmitted light microscopy using a 20× UPlan SApo objective with a numerical aperture of 0.75. Images were merged using Olympus FluoView software.
Transmission Electron Microscopy
Cotyledons were fixed in a mixture of 2.5% glutaraldehyde and 2.5% paraformaldehyde in 0.1 m cacodylate buffer at 4°C for 24 h, postfixed in 1% osmium tetroxide, and dehydrated in a graded acetone series. Samples were infiltrated and embedded in Poly/Bed 812 resin (Polysciences Inc.). Thin sections (70-nm thickness) were obtained with a PTXL ultramicrotome (RMC, Boeckeler Instruments) on 200 mesh copper grids stained with uranyl acetate and lead citrate. Sections were imaged using a JEOL 100CX transmission electron microscope (JEOL) at a 100-kV accelerating voltage.
Genetic and Physical Mapping of the l2 Locus
An interspecific F2 mapping population was generated from the following cross tomato (l2/l2) (LA3581) × S. galapagense (L2/L2; LA0483). Genomic DNA isolation and molecular mapping using RFLP markers was performed as previously described (Barry et al., 2005). Details of tomato genetic maps and the chromosome 10 molecular markers can be accessed through the Solanaceous Genomics Network (http://solgenomics.net/). Restriction enzymes yielding RFLPs between tomato and S. galapagense for given DNA probes are as follows; T736: HaeIII, CT57: NdeI, T615: NlaIV, 95R: AvaII, 196E17F: XbaI, 196E17R: EcoRV, cLET10H23: RsaI, L2: ApoI, cTOF24I6: NsiI. A physical contig spanning the L2 locus was obtained via screening and characterization of ordered bacterial artificial chromosome (BAC) and cosmid libraries derived from tomato, Solanum pennellii, and S. galapagense (Li et al., 2005; Chen et al., 2007). Clones LA0483BAC69H22, LA0483BAC69J23, and LA0483BAC178I9 were isolated from an S. galapagense library. Clones LpenCOS31F20, LpenCOS95L3, and LpenBAC137I4 were isolated from two S. pennellii libraries. The approximate size of each BAC clone was determined by pulsed-field gel electrophoresis. BAC and cosmid ends were isolated by direct DNA sequencing and converted to RFLP markers for further genetic and physical mapping. Subcloning of the tomato cosmid clone LeCOS196E17 was achieved by three separate restriction enzyme digests (BamHI, EcoRI, and HinDIII) followed by ligation into pBluescript SK− (Stratagene), previously linearized with the same restriction enzymes. The resultant clones were sequenced with vector primers to yield a number of anchor points that were subsequently utilized to extend and complete the sequence of LeCOS196E17 using multiple rounds of primer extension.
RNA Isolation and Cloning of L2 cDNA and Genomic Fragments
RNA was extracted from leaf and fruit tissues using either previously described methods or the RNeasy plant mini kit (Qiagen; Griffiths et al., 1999; Barry et al., 2005). cDNA synthesis was performed using the transcriptor first-strand cDNA synthesis kit (Roche Applied Science). The coding region of L2 was amplified by RT-PCR using cDNA synthesized from RNA extracted from leaf of either wild-type (L2/L2) or mutant (l2/l2) genotypes using the primers L2PROT-F: 5′-GGACTTTTATCAAACAGCTAACTTGCA-3′ and L2PROT-R: 5′-GGCACAACCCAACTTACAATAATTGTA-3′. Detection of the point mutation in the l2Fireball allele was achieved through amplification of a genomic fragment spanning exons 7 and 8 from the Fireball and Lutescent Fireball cultivars using the primers L2GEN-1F: 5′-GGTAATTTTCAGCATGCTTGGTCTA-3′ and L2GEN-1R: 5′-GAGAATCACAGGCATTTGATCTAC-3′. Fragments were amplified using the Pfu Ultra DNA polymerase (Agilent Technologies) and cloned into either EcoRV linearized pBluescript SK− (Agilent Technologies) using standard cloning techniques or into pCR4Blunt-TOPO vector (Invitrogen Corporation). Sequencing of inserts was accomplished using vector primers and the gene-specific primers L2PROT-1: 5′-GAGCTAGGAATTGCGTCTCA-3′, L2PROT-2: 5′-TGAGACGCAATTCCTAGCTC-3′, L2PROT-3: 5′-TGCTACCAGTTGGATGTCTT-3′, and L2PROT-4: 5′-AAGACATCCAACTGGTAGCA-3′.
DNA Constructs and Plant Transformation
The L2 cDNA insert was released from pBluescript SK− by digestion with XbaI and SacI. The fragment was ligated downstream of the cauliflower mosaic virus 35S promoter in the binary vector pBI121, previously modified by removal of the UidA coding region by digestion with the same restriction enzymes. Transgenic tomato plants were generated through cotyledon-derived explants via Agrobacterium tumefaciens mediated transformation (strain LBA4404), using previously described methods (Fillatti et al., 1987). Transgene integration was determined by a combination of gel-blot and PCR analysis.
DNA Sequence Analysis and Bioinformatics Resources
DNA sequences were assembled using Sequencher version 4.7 (Genecodes Corporation). The prediction of genes within genomic DNA sequence was performed using the FGENESH hidden Markov model-based gene structure prediction program (http://linux1.softberry.com/berry.phtml). Amino acid alignments were generated using MUSCLE (Edgar, 2004) and were decorated using the Boxshade server version 3.2.1 (http://www.ch.embnet.org). Analysis of membrane-spanning domains was accomplished using the Aramemnon database (http://aramemnon.uni-koeln.de/; Schwacke et al., 2003) and prediction of chloroplast transit peptides was accomplished using the Predotar prediction server (http://urgi.versailles.inra.fr/predotar/predotar.html).
Statistical Analysis
Statistical analyses were performed using SAS (SAS Institute, www.sas.com). The genotypic constituents were evaluated using least squares means.
Sequences reported in this manuscript have been deposited in GenBank under the following accession number: JQ683149.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Carotenoid composition in ripe fruits of the lutescent mutants.
Supplemental Figure S2. The lutescent mutants have a normal ethylene response.
Supplemental Figure S3. Phenotypes of additional l2 alleles.
Supplementary Material
Acknowledgments
We thank Drs. Alicia Pastor and Melinda Frame (Center for Advanced Microscopy, Michigan State University) for assistance with transmission electron microscopy and confocal laser-scanning microscopy, respectively.
Glossary
- AC
Ailsa Craig
- cM
centimorgan
- RT
reverse transcription
- RIP
regulated intramembrane proteolysis
- SREBP
sterol-regulatory element binding protein
- BAC
bacterial artificial chromosome
References
- Ajjawi I, Lu Y, Savage LJ, Bell SM, Last RL. (2010) Large-scale reverse genetics in Arabidopsis: case studies from the Chloroplast 2010 Project. Plant Physiol 152: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. (2002) DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev 16: 2156–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U, Pesaresi P, Pribil M, Hertle A, Leister D. (2011) Update on chloroplast research: new tools, new topics, and new trends. Mol Plant 4: 1–16 [DOI] [PubMed] [Google Scholar]
- Barry CS. (2010) Factors influencing the ripening and quality of fleshy fruits. In L Ostergaard, ed, Annual Plant Reviews, Vol 38. Wiley-Blackwell, Oxford, UK, pp 296–326 [Google Scholar]
- Barry CS, Giovannoni JJ. (2007) Ethylene and fruit ripening. J Plant Growth Regul 26: 143–159 [Google Scholar]
- Barry CS, McQuinn RP, Chung M-Y, Besuden A, Giovannoni JJ. (2008) Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol 147: 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry CS, McQuinn RP, Thompson AJ, Seymour GB, Grierson D, Giovannoni JJ. (2005) Ethylene insensitivity conferred by the Green-ripe and Never-ripe 2 ripening mutants of tomato. Plant Physiol 138: 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bino RJ, Ric de Vos CH, Lieberman M, Hall RD, Bovy A, Jonker HH, Tikunov Y, Lommen A, Moco S, Levin I. (2005) The light-hyperresponsive high pigment-2dg mutation of tomato: alterations in the fruit metabolome. New Phytol 166: 427–438 [DOI] [PubMed] [Google Scholar]
- Borovsky Y, Paran I. (2008) Chlorophyll breakdown during pepper fruit ripening in the chlorophyll retainer mutation is impaired at the homolog of the senescence-inducible stay-green gene. Theor Appl Genet 117: 235–240 [DOI] [PubMed] [Google Scholar]
- Chai YM, Jia HF, Li CL, Dong QH, Shen YY. (2011) FaPYR1 is involved in strawberry fruit ripening. J Exp Bot 62: 5079–5089 [DOI] [PubMed] [Google Scholar]
- Chen G, Bi YR, Li N. (2005) EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. Plant J 41: 364–375 [DOI] [PubMed] [Google Scholar]
- Chen GP, Hackett R, Walker D, Taylor A, Lin ZF, Grierson D. (2004) Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol 136: 2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KY, Cong B, Wing R, Vrebalov J, Tanksley SD. (2007) Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science 318: 643–645 [DOI] [PubMed] [Google Scholar]
- Cheung A-Y, McNellis T, Piekos B. (1993) Maintenance of chloroplast components during chromoplast differentiation in the tomato mutant green flesh. Plant Physiol 101: 1223–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J. (2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64: 936–947 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. (1997) Light control of plant development. Annu Rev Cell Dev Biol 13: 203–229 [DOI] [PubMed] [Google Scholar]
- Fillatti JJ, Kiser J, Rose R, Comai L. (1987) Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium-tumefaciens vector. Bio-Technology 5: 726–730 [Google Scholar]
- Galpaz N, Wang Q, Menda N, Zamir D, Hirschberg J. (2008) Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J 53: 717–730 [DOI] [PubMed] [Google Scholar]
- Griffiths A, Barry C, Alpuche-Solis A-G, Grierson D. (1999) Ethylene and developmental signals regulate expression of lipoxygenase genes during tomato fruit ripening. J Exp Bot 50: 793–798 [Google Scholar]
- Guo D, Gao XR, Li H, Zhang T, Chen G, Huang PB, An LJ, Li N. (2008) EGY1 plays a role in regulation of endodermal plastid size and number that are involved in ethylene-dependent gravitropism of light-grown Arabidopsis hypocotyls. Plant Mol Biol 66: 345–360 [DOI] [PubMed] [Google Scholar]
- Herner RC, Sink KC. (1973) Ethylene production and respiratory behavior of the rin tomato mutant. Plant Physiol 52: 38–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. (2009) TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Jaakola L, Poole M, Jones MO, Kämäräinen-Karppinen T, Koskimäki JJ, Hohtola A, Häggman H, Fraser PD, Manning K, King GJ, et al. (2010) A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol 153: 1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY. (2011) Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol 157: 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehara K, Ito K, Akiyama Y. (2002) YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev 16: 2147–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA. (2011) Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23: 923–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W. (2010) New insights into the types and function of proteases in plastids. Int Rev Cell Mol Biol 280: 185–218 [DOI] [PubMed] [Google Scholar]
- Kerr EA. (1956) lutescent 2, l2. Report of the Tomato Genetics Cooperative 6: 17 [Google Scholar]
- Kerr EA, Lyall LH, Graham TO. (1975) A mutation to lutescent-2 in “Fireball”. Report of the Tomato Genetics Cooperative 25: 10 [Google Scholar]
- Kim SG, Lee S, Seo PJ, Kim SK, Kim JK, Park CM. (2010) Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics 95: 56–65 [DOI] [PubMed] [Google Scholar]
- Klee HJ. (2010) Improving the flavor of fresh fruits: genomics, biochemistry, and biotechnology. New Phytol 187: 44–56 [DOI] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ. (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Kolotilin I, Koltai H, Tadmor Y, Bar-Or C, Reuveni M, Meir A, Nahon S, Shlomo H, Chen L, Levin I. (2007) Transcriptional profiling of high pigment-2dg tomato mutant links early fruit plastid biogenesis with its overproduction of phytonutrients. Plant Physiol 145: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács K, Fray RG, Tikunov Y, Graham N, Bradley G, Seymour GB, Bovy AG, Grierson D. (2009) Effect of tomato pleiotropic ripening mutations on flavour volatile biosynthesis. Phytochemistry 70: 1003–1008 [DOI] [PubMed] [Google Scholar]
- Lee JM, Joung J-G, McQuinn R, Chung M-Y, Fei Z, Tieman D, Klee H, Giovannoni J. (2012) Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J 70: 191–204 [DOI] [PubMed] [Google Scholar]
- Li CY, Schilmiller AL, Liu GH, Lee GI, Jayanty S, Sageman C, Vrebalov J, Giovannoni JJ, Yagi K, Kobayashi Y, et al. (2005) Role of beta-oxidation in jasmonate biosynthesis and systemic wound signaling in tomato. Plant Cell 17: 971–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids—pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Lisso J, Altmann T, Müssig C. (2006) Metabolic changes in fruits of the tomato dx mutant. Phytochemistry 67: 2232–2238 [DOI] [PubMed] [Google Scholar]
- Liu JX, Howell SH. (2010) Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22: 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytovchenko A, Eickmeier I, Pons C, Osorio S, Szecowka M, Lehmberg K, Arrivault S, Tohge T, Pineda B, Anton MT, et al. (2011) Tomato fruit photosynthesis is seemingly unimportant in primary metabolism and ripening but plays a considerable role in seed development. Plant Physiol 157: 1650–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ. (2011) The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol 157: 1568–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menda N, Semel Y, Peled D, Eshed Y, Zamir D. (2004) In silico screening of a saturated mutation library of tomato. Plant J 38: 861–872 [DOI] [PubMed] [Google Scholar]
- Nakano R, Ogura E, Kubo Y, Inaba A. (2003) Ethylene biosynthesis in detached young persimmon fruit is initiated in calyx and modulated by water loss from the fruit. Plant Physiol 131: 276–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashilevitz S, Melamed-Bessudo C, Izkovich Y, Rogachev I, Osorio S, Itkin M, Adato A, Pankratov I, Hirschberg J, Fernie AR, et al. (2010) An orange ripening mutant links plastid NAD(P)H dehydrogenase complex activity to central and specialized metabolism during tomato fruit maturation. Plant Cell 22: 1977–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann B, Busch A, Allmer J, Ostendorf E, Zeller M, Kirchhoff H, Hippler M. (2007) Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7: 3964–3979 [DOI] [PubMed] [Google Scholar]
- Osborne TF, Espenshade PJ. (2009) Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it’s been. Genes Dev 23: 2578–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL. (1997) Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell 1: 47–57 [DOI] [PubMed] [Google Scholar]
- Roca M, Hornero-Méndez D, Gandul-Rojas B, Mínguez-Mosquera MI. (2006) Stay-green phenotype slows the carotenogenic process in Capsicum annuum (L.) fruits. J Agric Food Chem 54: 8782–8787 [DOI] [PubMed] [Google Scholar]
- Ronen G, Cohen M, Zamir D, Hirschberg J. (1999) Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J 17: 341–351 [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Fawcett P, Losick R. (1999) A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci USA 96: 14765–14770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge UI, Kunze R. (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Kim SG, Park CM. (2008) Membrane-bound transcription factors in plants. Trends Plant Sci 13: 550–556 [DOI] [PubMed] [Google Scholar]
- Seymour GB, Ryder CD, Cevik V, Hammond JP, Popovich A, King GJ, Vrebalov J, Giovannoni JJ, Manning K. (2011) A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria x ananassa Duch.) fruit, a non-climacteric tissue. J Exp Bot 62: 1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sink KC, Herner RC, Knowlton LL. (1974) Chlorophyll and carotenoids of the rin tomato mutant. Can J Bot 52: 1657–1660 [Google Scholar]
- Srivastava A, Handa AK. (2005) Hormonal regulation of tomato fruit development: a molecular perspective. J Plant Growth Regul 24: 67–82 [Google Scholar]
- Sun XW, Fu TJ, Chen N, Guo JK, Ma JF, Zou MJ, Lu CM, Zhang LX. (2010) The stromal chloroplast Deg7 protease participates in the repair of photosystem II after photoinhibition in Arabidopsis. Plant Physiol 152: 1263–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons GM, Davies C, Shavrukov Y, Dry IB, Reid JB, Thomas MR. (2006) Grapes on steroids: brassinosteroids are involved in grape berry ripening. Plant Physiol 140: 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB, et al. (1992) High density molecular linkage maps of the tomato and potato genomes. Genetics 132: 1141–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5: 123–135 [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJM, McQuinn R, Chung M, Poole M, Rose JKC, Seymour G, Grandillo S, Giovannoni J, et al. (2009) Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Waters MT, Langdale JA. (2009) The making of a chloroplast. EMBO J 28: 2861–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins CB, Nock JF, Weis SA, Jayanty S, Beaudry RM. (2004) Storage temperature, diphenylamine, and pre-storage delay effects on soft scald, soggy breakdown and bitter pit of ‘Honeycrisp’ apples. Postharvest Biol Technol 32: 213–221 [Google Scholar]
- Yen HC, Shelton BA, Howard LR, Lee S, Vrebalov J, Giovannoni JJ. (1997) The tomato high-pigment (hp) locus maps to chromosome 2 and influences plastome copy number and fruit quality. Theor Appl Genet 95: 1069–1079 [Google Scholar]
- Zhang M, Yuan B, Leng P. (2009) The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J Exp Bot 60: 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.