Abstract
Metallothioneins (MTs) are low-molecular-weight, cysteine-rich proteins believed to play a role in cytosolic zinc (Zn) and copper (Cu) homeostasis. However, evidence for the functional properties of MTs has been hampered by methodological problems in the isolation and characterization of the proteins. Here, we document that barley (Hordeum vulgare) MT3 and MT4 proteins exist in planta and that they differ in tissue localization as well as in metal coordination chemistry. Combined transcriptional and histological analyses showed temporal and spatial correlations between transcript levels and protein abundance during grain development. MT3 was present in tissues of both maternal and filial origin throughout grain filling. In contrast, MT4 was confined to the embryo and aleurone layer, where it appeared during tissue specialization and remained until maturity. Using state-of-the-art speciation analysis by size-exclusion chromatography inductively coupled plasma mass spectrometry and electrospray ionization time-of-flight mass spectrometry on recombinant MT3 and MT4, their specificity and capacity for metal ion binding were quantified, showing a strong preferential Zn binding relative to Cu and cadmium (Cd) in MT4, which was not the case for MT3. When complementary DNAs from barley MTs were expressed in Cu- or Cd-sensitive yeast mutants, MT3 provided a much stronger complementation than did MT4. We conclude that MT3 may play a housekeeping role in metal homeostasis, while MT4 may function in Zn storage in developing and mature grains. The localization of MT4 and its discrimination against Cd make it an ideal candidate for future biofortification strategies directed toward increasing food and feed Zn concentrations.
Metallothioneins (MTs) are low-Mr metal-binding proteins and have since they were first discovered in the late 1950s been subject to intense studies in yeast, mammals, and plants (Margoshes and Vallee, 1957). MTs are thought to function in cytosolic zinc (Zn) and copper (Cu) homeostasis by forming coordination complexes between metal ions and thiol groups of multiple Cys residues in the protein (Sutherland and Stillman, 2011). In addition, several MTs have been reported to scavenge reactive oxygen species in planta (Wong et al., 2004; Kim et al., 2007; Xue et al., 2009; Yang et al., 2009). Plant MTs are structurally different from vertebrate MTs, as they contain Cys-rich domains in their N and C termini separated by variable spacer regions with none or a few metal-binding residues. The number of Cys groups and their distribution constitute the basis for the separation of plant MTs into four subfamilies (Kojima et al., 1999). The expression of MT genes varies between plant tissues and is ontogenetically regulated (Heise et al., 2007; Yuan et al., 2008). In addition, MT transcript levels respond to stimuli such as metals, hormones, heat, cold, drought, and salt (Cobbett and Goldsbrough, 2002; Akashi et al., 2004; Zhou et al., 2005; Xue et al., 2009).
Recent studies of plants with manipulated MT expression support a physiological role for MTs in metal ion homeostasis (Grennan, 2011). In Arabidopsis (Arabidopsis thaliana), constitutive expression of the seed-specific MT4 throughout the plant resulted in the accumulation of Cu but not Zn. However, the presence of MT4 in leaves and roots resulted in tolerance to both Cu and Zn when the metal ions were applied in excess (Rodríguez-Llorente et al., 2010). Simultaneous silencing of three Arabidopsis MT1 genes to transcript levels equivalent to 5% to 10% of the wild-type level induced cadmium (Cd) sensitivity (Zimeri et al., 2005). In rice (Oryza sativa), OsMT1a transcripts were specifically induced by Zn. Following the overexpression of OsMT1a, elemental analysis showed that Zn accumulation in the vegetative parts and the grain tissue was increased by 44% to 54% relative to the wild type (Yang et al., 2009).
Processes involved in Zn homeostasis in the cereal grain have received much attention in order to reveal bottlenecks in Zn loading and provide solutions to the global challenges associated with Zn deficiency-related diseases in more than 600 million people (Hotz and Brown, 2004). A possible target for the improvement of grain Zn may be MTs, but information about their functional properties and roles is lacking (Grennan, 2011). In plants, Zn is either incorporated into proteins or enters the labile pool of Zn2+, shuffling among transport proteins and Zn-chelating ligands. The network of transporters controlling Zn trafficking in and out of the cytosol has been partially uncovered and comprises multiple protein families, of which zinc-regulated transporter, iron-regulated transporter-like proteins mediate Zn uptake by roots (Vert et al., 2002), heavy metal ATPase and plant cadmium resistance2 proteins facilitate Zn transport into vascular tissues (Verret et al., 2004; Song et al., 2010), and heavy metal ATPase, metal tolerance protein, and zinc-induced facilitator proteins mediate Zn storage in vacuoles (Haydon and Cobbett, 2007; Kawachi et al., 2009; Morel et al., 2009). Deoxymugineic acid and nicotianamine are ligands that recently have been shown to function in the intercellular trafficking of Zn from roots to leaves and from leaves to developing grains (Suzuki et al., 2008; Lee et al., 2011). The roles of other Zn ligands such as amino acids, organic acids, phytochelatins, or MTs in the maintenance of cytosolic Zn homeostasis are still unclear (Tennstedt et al., 2009; Blindauer and Schmid, 2010).
Concerning the functional properties of MTs, it is a puzzling aspect that the metal-binding preferences of the proteins apparently cannot be attributed to unique amino acids in the secondary structure (Palacios et al., 2011). Also, the lack of protein data derived from native plant MTs with respect to localization and metal-protein stoichiometry consistently hampers the characterization of plant MTs and the clarification of their metabolic roles. To address the physiological function of MTs in the cereal grain, we have here functionally characterized a diverse family of 10 barley (Hordeum vulgare) MTs. Among these MTs, MT3 and MT4 were further investigated by heterologous expression in yeast and Escherichia coli and by state-of-the-art speciation analysis using electrospray ionization time-of-flight mass spectrometry (ESI-TOF-MS) and size-exclusion chromatography (SEC) inductively coupled plasma mass spectrometry (ICP-MS) to quantify their specificity and capacity for metal binding. Finally, we studied the histological localization of MT3 and MT4 in grain tissue, thereby providing, to our knowledge, the first in situ identification of cereal MTs in developing and mature grains.
RESULTS
Structural Relations among Barley MTs
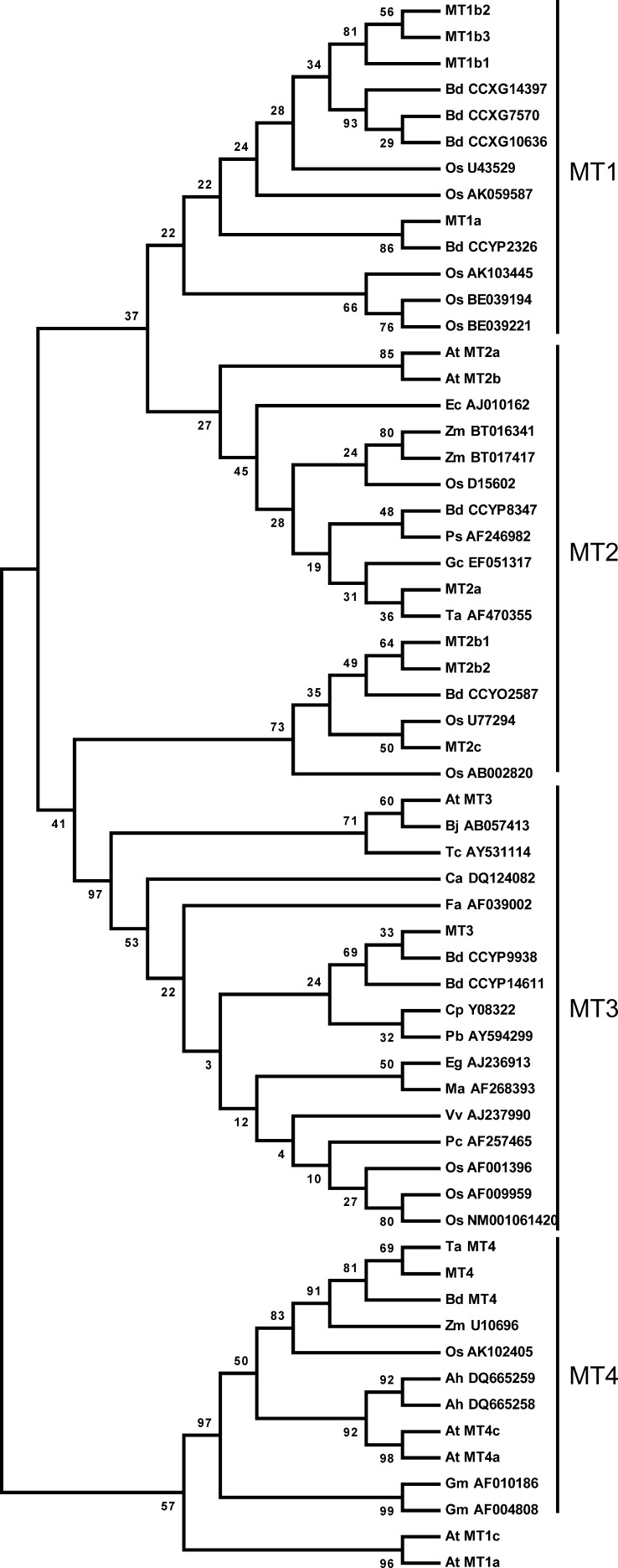
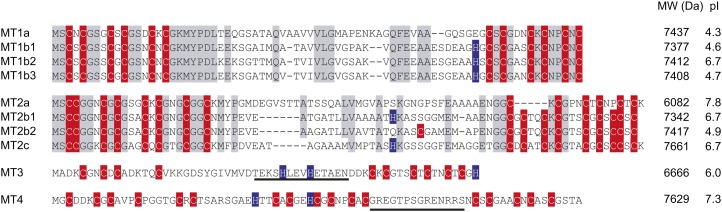
To understand the genomic basis for barley MT expression, we used The Institute for Genomic Research’s Barley Gene Indices (Lee et al., 2005) to identify 10 MT genes in barley. A phylogenetic analysis of putative protein sequences from other MTs showed that barley MTs were distributed among all four subgroups of plant MTs (Fig. 1). Analyses of the DNA arrangements of MT exons-introns (Supplemental Fig. S1) were combined with the phylogenetic analysis of MT proteins (Fig. 1) in order to identify putative MT paralogs within the barley genome. On that basis, individual MTs were designated by numbers indicating subdivision, a letter for the individual form, and a second number indicating the specific protein (Fig. 2; Kojima et al., 1999). The comparison showed 76% identity of MT1a and MT1b, 91% of MT1b1, MT1b2, and MT1b3, and 96% of MT1b2 and MT1b3 (Supplemental Table S1). The pI of the proteins within the MT1b subgroup ranged from 4.60 for MT1b1 to 6.73 for MT1b2 (Fig. 2). Among the MT2 proteins, MT2b1 and MT2b2 showed 94% identical residues but nevertheless varied in their putative metal-binding residues, MT2b1 holding one His and 17 Cys residues and MT2b2 containing 18 Cys residues (Fig. 2). There is only a single gene of each MT3 and MT4 in barley, but the derived proteins resemble the corresponding ones from other species, as evidenced by the similar internal distribution of Cys and His residues (Supplemental Fig. S2).
Figure 1.
Phylogenetic tree of the MT family in plants. The phylogenetic tree was produced from MT cDNA sequences of subfamily 15 MTs that constitute the plant-specific MTs (Kojima et al., 1999). Plant species abbreviations are as follows: Arabidopsis (At), Arachis hypogaea (Ah), B. distachyon (Bd), Brassica juncea (Bj), Carica papaya (Cp), Coffea arabica (Ca), Eichhornia crassipes (Ec), Elaeis guineensis (Eg), Fritillaria agrestis (Fa), Glycine max (Gm), Gymnadenia conopsea (Gc), banana (Ma), rice (Os), Poa secunda (Ps), Populus beaupre (Pb), Porteresia coarctata (Pc), Thlaspi caerulescens (Tc), wheat (Ta), Vitis vinifera (Vv), and Zea mays (Zm). The phylogenetic tree was drawn using MEGA version 4.0 on MT protein sequences. Barley MTs are represented by MT1a, MT1b1, MT1b2, MT1b3, MT2a, MT2b1, MT2b2, MT2c, MT3, and MT4. Arabidopsis MTs are shown as At MT1 to At MT4. The wheat Ec protein is presented as Ta MT4 and Brachypodium MT4 as Bd MT4. Otherwise, MTs are presented by their species abbreviation followed by cDNA accession numbers. Bootstrap values are below 100 due to the size of MT proteins of approximately 60 to 85 amino acids.
Figure 2.
ClustalW alignments of barley MTs performed in T-COFFE. MT1 and MT2 clusters are aligned separately, whereas MT3 and MT4 are presented alone. Cys and His residues are shown in red and blue, respectively. Conserved sequences are presented in gray. Underlined sequences represent the peptides recognized by anti-MT3 and anti-MT4 antibodies.
Heterologous Expression of Barley MTs in Yeast
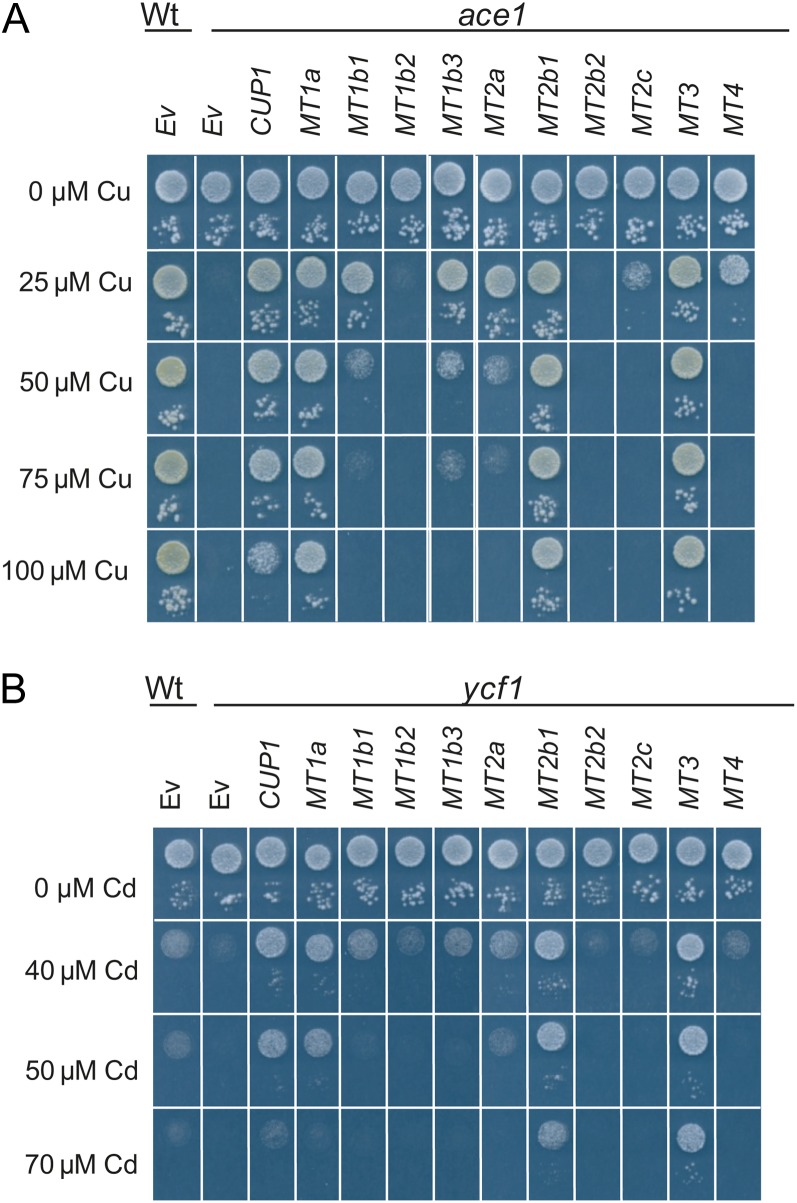
To further address the impact of MT secondary structures on the capacity to bind metal ions and to detoxify oxidative stress, we expressed barley MTs in Saccharomyces cerevisiae mutant strains. MTs were expressed in the ace1, ycf1, and skn7 yeast mutants with growth deficiencies in their tolerance toward high external concentrations of Cu, Cd, and hydrogen peroxide (H2O2), respectively. Also, cellular oxidative stress was elicited in wild-type yeast exposed to high NaCl concentrations in order to study the effects of MT expression under these conditions. Depending on the growth conditions, eight of the 10 barley MTs could complement the Cu sensitivity of the ace1 strain (Fig. 3A). Expression of MT1a, MT2b1, and MT3 in ace1 resulted in a full complementation of Cu-sensitive cells, enabling them to tolerate concentrations exceeding 100 µm Cu. MT1b1, MT1b3, MT2a, MT2c, and MT4 complemented ace1 cells to a weaker degree, allowing cells to tolerate concentrations of 25 µm Cu. Empty vector transformations of the wild type and ace1 were used as positive and negative controls, respectively. In the ycf1 strain, strong complementation of the Cd-sensitive phenotype was observed for MT1a, MT2b1, and MT3 (Fig. 3B). Here, the cells were able to outgrow even the wild type on Cd concentrations up to 50 µm. In ycf1 expressing either MT3 or MT2b1, Cd tolerance was even higher, supporting growth also on 70 µm Cd. Weaker but stable complementation was induced with the expression of MT1b1, MT1b3, and MT2a in ycf1. Expression of MT2c and MT4 in ycf1 cells did not improve cellular Cd tolerance when compared with the empty vector control. MT1b2 and MT2b2 expression did not increase metal tolerance in either ace1 or ycf1 yeast cells.
Figure 3.
Barley MT proteins mediate Cu and Cd tolerance in yeast. Yeast tolerance toward Cu or Cd following overexpression of MT1a, MT1b1, MT1b2, MT1b3, MT2a, MT2b1, MT2b2, MT2c, MT3, and MT4 in ace1 (A) or ycf1 (B). In both A and B, cells were spotted in two concentrations (OD600 = 0.05 and OD600 = 0.0005). As a negative control, the wild type (Wt), ace1, and ycf1 were transformed with the empty vector p426 (Ev). As a positive control, ace1 and ycf1 were transformed with the endogenous MT CUP1.
Possible roles of barley MTs in oxidative stress handling were tested by expression in the skn7 yeast mutant, which is defective in the induction of several defense genes against oxidative stress and therefore sensitive to externally applied H2O2. Unexpectedly, the presence of MTs in skn7 resulted in increased sensitivity toward H2O2 (Supplemental Fig. S3A). A similar effect was observed when the endogenous yeast MT gene CUP1 was expressed in skn7. The decline in H2O2 tolerance induced by MT expression may be related to competition for metals between the MT and the endogenous Cu/Zn superoxide dismutase SOD1. NaCl-elicited oxidative stress handling was not affected by the expression of barley MTs in wild-type yeast (Supplemental Fig. S3B).
Transcriptional Profile of MT3 and MT4 during Grain Filling
MT3 and MT4 were selected for further expression studies during grain development. The pollination time of single ears of soil-grown barley was recorded, and grains were harvested at 7, 14, 21, and 28 d after pollination (dap). For each time point, ears from four independent plants were collected and the middle part of each row of grains was separated to accommodate reverse transcription-quantitative (RT-q)PCR and immunolocalization studies. As RNA was purified from whole grains, the integrity of the produced complementary DNA (cDNA) was verified by monitoring expression data from GAPDH and the following seed-specific controls: the endosperm-specific transcripts ITR1 and HOR2 and the aleurone-specific LTP2 (Table I; Kalla et al., 1994; Opsahl-Sorteberg et al., 2004; Diaz et al., 2005).
Table I. MT3 and MT4 expression during grain filling in barley.
RT-qPCR was performed on transcripts derived from developing grains harvested at 7, 14, 21, and 28 dap. GAPDH was used as a reference. Expression levels are presented as fold change relative to grains at 7 dap. Transcripts of aleurone-specific (LTP2) and endosperm-specific (ITR1 and HOR2) genes were included as positive controls. The fold change values are shown as means ± se (n = 4). Between time points, values for each gene followed by varying letters are significantly different (P < 0.05).
MT3 transcripts were found throughout grain development at very stable levels when compared with the expression of GAPDH (Table I). In contrast to the ubiquitous expression of MT3, only a weak signal from MT4 was detected in the grain in the early phases of development. However, in grains harvested at 14 dap, the abundance of MT4 transcripts started to increase, and at 28 dap, MT4 expression was found to be 300-fold higher than at 7 dap (Table I).
Cellular Localization of MT3 and MT4 Proteins in Developing and Mature Grains
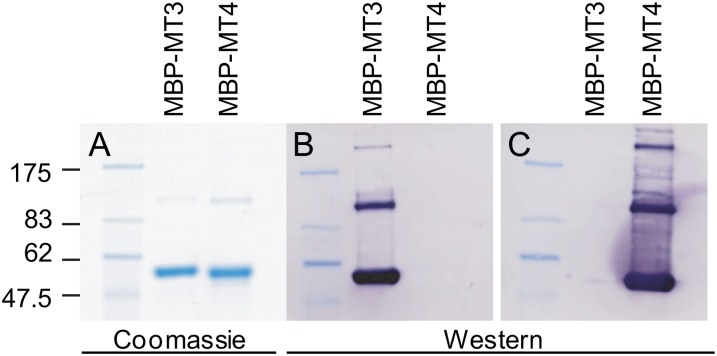
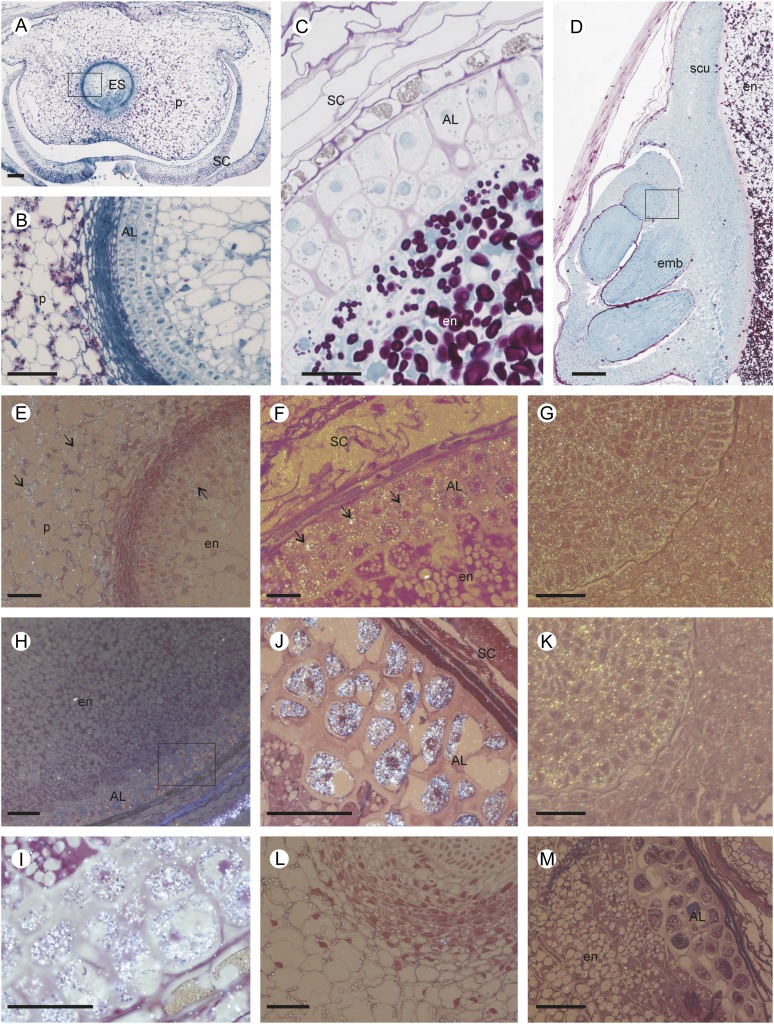
Western blots using either recombinant MT3 or MT4 (Fig. 4) or barley soluble protein (data not shown) confirmed that antibodies raised against MT3 and MT4 did not cross react with other MTs or other barley grain proteins. Histological localization of MT3 and MT4 was first investigated in developing grain using immunogold labeling followed by silver enhancement. A bright-field microscope emitting epipolarized light was used for these experiments, and the silver-enhanced gold particles appeared as a bright yellow color (Fig. 5). Samples were taken at premilk stage (7 dap), start of grain filling (14 dap), and rapid grain growth (through 21–28 dap). The seeds were allowed to desiccate prior to analysis of the mature grains. Sections of 7-dap-old grain treated with gold-labeled anti-MT3 antibody allowed the detection of a diffuse fluorescent signal (arrows) mainly located in the parenchyma cells of the pericarp and the outer cell layers of the embryo sac (Fig. 5E). In the same tissue, no signal was detected with anti-MT4 antibody (data not shown). Later during grain development (21 dap), MT3 label was clearly observed in the aleurone layer (Fig. 5F) and in the embryo (Fig. 5G). Also, labeling of anti-MT4 antibodies was observed in the aleurone cell layer of grains harvested at 21 dap (Fig. 5, H and I) and at maturity (Fig. 5J) as well as in the growing cells of the embryo (Fig. 5K). Anti-MT4 antibody signal was never observed in the inner starchy endosperm or in the outer grain testa. Control sections treated with MT3 or MT4 preimmune serum were free of label (for representative images, see Figure 5, L and M, and Supplemental Fig. S4, A–I).
Figure 4.
Verification of N-terminal fusions of recombinant MBP-MT3 and MBP-MT4. A, PageBlue protein stains of recombinant MBP-MT3 and MBP-MT4 proteins separated by SDS-PAGE. B and C, Immunoblotting using antibodies specific for barley MT3 (B) and MT4 (C). Purified MBP-MT (5 µg) was used on the immunoblots. The calculated sizes of MBP-MT3 and MBP-MT4 are 48.9 and 49.7 kD, respectively. MBP fusion proteins were consistently found to migrate approximately 8 kD higher than the calculated mass.
Figure 5.
Morphological and structural observations of developing barley grain, and histological localization of MT proteins. A, General view of a transverse section of developing grain at 7 dap showing the maternal pericarp surrounding the central embryo sac. Polysaccharides and soluble proteins were stained by periodic acid-Schiff (dark pink) and naphthol blue-black (blue). B, Magnification of the 7-dap-old embryo sac wall including the developing aleurone layer between the young endosperm (inside) and the starch-loaded parenchyma cells of the pericarp (outside). C, Median transverse section of a grain at 21 dap showing the aleurone layer and starch grains in the endosperm. D, Longitudinal section of a mature grain including the embryo. The square in D indicates the area corresponding to G and K. E to K, Histological immunolocalization of MT3 and MT4 in developing grain transverse sections stained by fuchsin and examined under bright-field epipolarized light. The silver-enhanced gold particles used for the immunolocalization appear as a bright yellow color. Immunolocalization of MT3 (E–G) and MT4 (H–K) in 7-dap (E), 21-dap (F, H, and I), and mature (G, J, and K) barley grains is shown. MT3 was located in the maternal pericarp, the aleurone cell layer, and the embryo, while MT4 was detected in grains at 21 dap in the aleurone layer and the growing cells of the embryo. L and M, Examples of control sections of grain treated with MT3 (7-dap-old grain; L) or MT4 (21-dap-old grain; M) preimmune serum. AL, Aleurone cell layer; emb, embryo; en, endosperm; ES, embryo sac; P, pericarp; SC, seed coat; scu, scutellum. Bars = 100 µm.
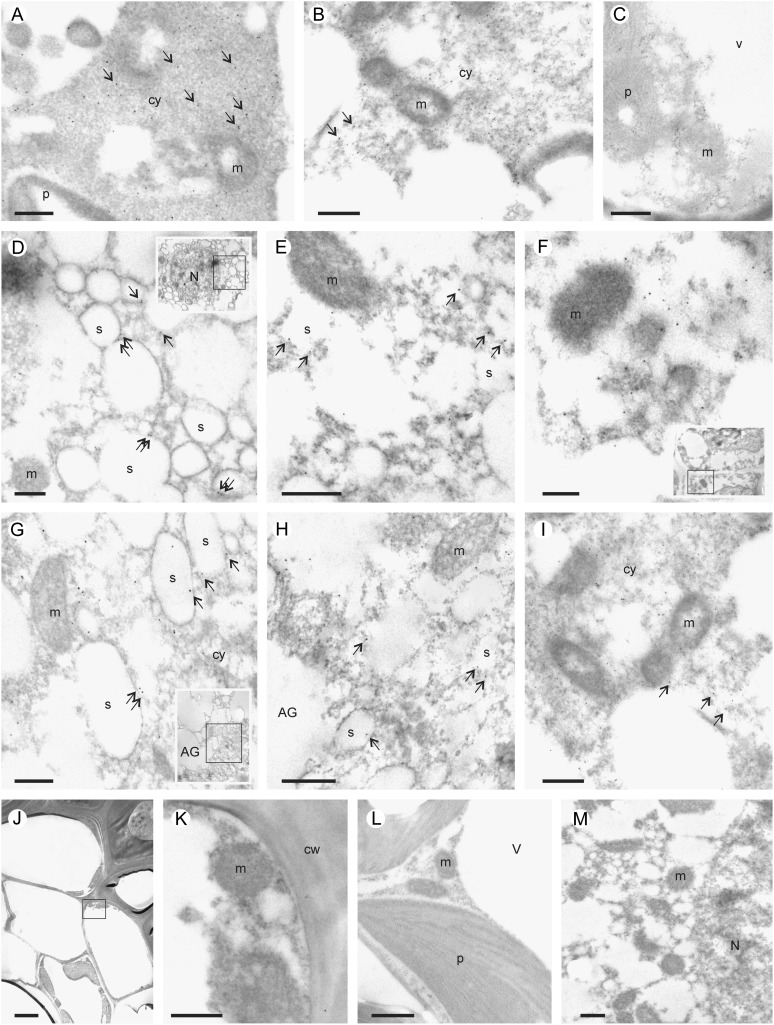
Transmission electron microscopic immunolocalization of MT3 and MT4 supported that MT3 was localized in tissues of both maternal and filial origin, whereas MT4 was present in the embryo and aleurone layer (Fig. 6). MT3 and MT4 were located in the cytosol in embryonic cells and MT3 also in the cytosol of parenchyma cells (Fig. 6, A and B). In the aleurone layer of grains harvested at 21 dap, both MT proteins localized to the membranes of enclosed compartments closely associated with aleurone grains known as spherosomes, oleosomes, or lipid bodies (Fig. 6, D and G). Finally, both MT3 and MT4 were readily detected in tissues of the aleurone layer and embryo in mature grains (Fig. 6, E, F, H, and I). No signal were detected in sections treated with MT3 or MT4 preimmune serum (Fig. 6, L and M, Supplemental Fig. S4, J–R).
Figure 6.
Transmission electron microscopic immunolocalization of MT3 and MT4 in developing grains. A and B, Transverse sections showing the immunolocalization of MT3 in grains at 7 dap including parenchyma cells of the pericarp (A) and the young aleurone layer (B). Arrows indicate the presence of MT3 in the cytosol. C, The same tissues were not labeled when treated with anti-MT4 antibodies. D to I, Localization of MT3 (D–F) and MT4 (G–I) in grain at 21 dap (D and G) and at maturity (E, F, H, and I). In the aleurone layer (D, E, G, and H), MT proteins were localized in the membrane of the spherosomes. In the embryo (F and I), MTs were detected in the cytosol. J and K, General view (J) and detail (K) of the seed coat of a grain at 21 dap showing the absence of MT4. L and M, Control sections of grain treated with MT3 (7-dap-old grain; L) or MT4 (21-dap-old grain; M) preimmune serum. AG, Aleurone grain; cw, cell wall; cy, cytosol; m, mitochondria; N, nucleus; P, plastid; S, spherosome; V, vacuole. Bars = 0.5 µm (A–I, K–M) and 2 µm (J).
SEC-ICP-MS Analysis of Recombinant MT3 and MT4
The stoichiometry of the coordination complexes between metal ions and MTs was analyzed using recombinant MT3 and MT4 produced in bacterial cells cultivated without the addition of metals to the growth medium (TBglu), followed by protein purification and speciation analysis by SEC-ICP-MS (Table II). When MT3 was produced under these conditions, the molar Zn-MT3 ratio was Zn0.32MT3, whereas that of Cu-MT3 was Cu0.006MT3 (i.e. more than 50-fold lower). Zinc spiking of the bacterial culture increased the metal-ligand stoichiometry almost 3-fold to Zn0.81MT. Likewise, when the bacterial cultures were Cu and Cd spiked, metal-MT3 ratios increased to Cu0.34MT3 and Cd0.19MT3, corresponding to 50- and 450-fold increases, respectively (Table II).
Table II. Metal binding of recombinant MT3 and MT4 protein expressed as the number of metal ions per MT protein.
E. coli cultures were supplemented to concentrations of 120 µm Zn, 52.5 µm Cu, or 10 µm Cd. As a control, cells grown without metal supplements were used (70 µm Zn, 2.5 µm Cu, or 0 µm Cd). Zn, Cu, Cd, and sulfur were measured by SEC-ICP-MS based on the detection of 66Zn, 63Cu, 112Cd, and 48SO, respectively. Values are shown as means ± se (n = 4). Between metal treatments, the metal-MT ratios for each protein followed by varying letters are significantly different (P < 0.05).
| Protein | Culture Medium | Zn/MT | Cu/MT | Cd/MT | Metal/MT |
|---|---|---|---|---|---|
| MT3 | Control | 0.32 ± 0.01 b | 0.006 ± 0.0006 b | – | 0.33 ± 0.0 b |
| MT3 | +Zn | 0.81 ± 0.14 a | 0.042 ± 0.019 b | – | 0.85 ± 0.14 a |
| MT3 | +Cu | 0.44 ± 0.04 b | 0.34 ± 0.02 a | – | 0.78 ± 0.05 a |
| MT3 | +Cd | 0.40 ± 0.09 b | 0.0078 ± 0.0006 b | 0.19 ± 0.06 | 0.42 ± 0.09 b |
| MT4 | Control | 2.63 ± 0.03 b | 0.011 ± 0.001 b | – | 2.64 ± 0.03 b |
| MT4 | +Zn | 2.66 ± 0.14 b | 0.010 ± 0.001 b | – | 2.67 ± 0.14 b |
| MT4 | +Cu | 3.26 ± 0.08 a | 0.19 ± 0.01 a | – | 3.45 ± 0.09 a |
| MT4 | +Cd | 3.15 ± 0.11 a | 0.007 ± 0.0004 b | 0.12 ± 0.02 | 3.17 ± 0.11 a |
In contrast to MT3, recombinant MT4 was highly selective for Zn when exposed to a multiple-ion solution spiked with Zn, Cu, or Cd. MT4 purified from cells grown in nonspiked TBglu yielded Zn2.63MT4, whereas the corresponding ratio for Cu was Cu0.011MT4 and no coordination to Cd could be detected (Table II). Spiking TBglu with Zn, Cu, and Cd did not produce a higher Zn-MT4 ratio, indicating that MT4 was already saturated when produced from the TBglu cells. The molar ratios with Cu and Cd were Cu0.19MT4 and Cd0.12MT4 (i.e. much lower than that for the Zn-MT4 complex). This shows that MT4 has a strong preference for Zn and that the complex saturates in vivo when three Zn are coordinated with each MT4.
DISCUSSION
Classification of MTs
MTs in plants are ubiquitous and elusive at the same time. The complete inventory of barley MTs presented here illustrates their complexity at both the genomic and functional levels. Family subgroups 1 to 4 (Fig. 1) separate MTs according to their number and sequence of internal Cys residues and the separating linker regions. In barley, the conserved arrangement of exon-intron positions and high protein identities (Supplemental Figs. S1 and S2) suggests that subgroups MT1b and MT2b originate from gene duplications. Duplication within MT genes is a well-known phenomenon that has been characterized in Drosophila melanogaster (Maroni et al., 1987) and different oyster species (Tanguy et al., 2002). In humans, MT1 has undergone extensive duplications rendering 13 MT1 paralogs of which five have developed into pseudogenes (Moleirinho et al., 2011), and also in yeast, the copy number of the endogenous MT gene CUP1 varies from one to multiple tandem repeats and correlates positively with yeast Cu and Cd tolerance, showing that the duplicated genes encode functional MT proteins (Karin et al., 1984).
Heterologous Expression of MTs in Yeast Facilitates Cu and Cd Tolerance
To obtain an overview of the functionality among barley MTs in metal chelation, yeast was used as a heterologous expression system. MT1a, MT2b1, and MT3 were able to efficiently complement ace1 and ycf1 growth deficiencies related to Cu and Cd hypersensitivity, showing that these proteins participate in both Cu and Cd chelation (Fig. 3). For the other MTs, diverse growth responses toward Cu or Cd were observed even among closely related proteins (Fig. 3). This may reflect that gene duplications in combination with a stringent selection pressure toward one copy of the duplicated genes have allowed for spontaneous mutations to alter specific physiological functions or even cause loss of function (Zhou and Goldsbrough, 1995; Lynch and Conery, 2000). The variable performance in yeast drop tests among MT1b and MT2b paralogs (Fig. 3) may also be related to varying MT expression levels in yeast or differences in the plant-specific linker region between metal-binding residues among MTs. In the MT1b group, three amino acid substitutions, E25K, A30T, and G60S, are present in MT1b2 as compared with MT1b3. E25K results in a change from the acidic Glu to the basic Lys, a change that also shifts the pI from 4.7 in MT1b3 to 6.7 in MT1b2 (Fig. 2). All substitutions within MT1b2 are located in the linker region of the MT. In MT2b1 and MT2b2, three amino acid changes, H44Q, S48C, and A55P (Fig. 2), could result in changes in the metal coordination or steric rearrangements. These results clearly illustrate that metal chelation and coordination abilities of barley MTs in vivo are complex and cannot exclusively be predicted based on Cys distribution or the occurrence of His residues, which form the basis for classification into MT subgroups.
MT3 and MT4 Transcripts and Proteins Correlate in Developing Grains
To investigate the physiological role of MTs during cereal grain development and maturity, MT3 and MT4 were selected for further studies. The selection was primarily based on previous reports of MT3 transcripts in aboveground tissues of plants, such as leaves, fruits, and seeds (Liu et al., 2002; Brkljacić et al., 2004), and the seed-specific nature of MT4 transcripts from Arabidopsis and wheat (Triticum aestivum; Kawashima et al., 1992; Guo et al., 2003). Both MT3 and MT4 are unique proteins in barley, thus facilitating MT-specific immunolocalizations in planta.
MT3 transcript abundance was rather constant at four different stages of grain filling (Table I). This limited ontogenetic fluctuation agrees with the fact that MT3 transcript levels were present in both maternal and filial tissues of the grain, embracing embryo, aleurone, and endosperm, as documented by immunohistochemistry using gold labeling followed by silver enhancement (Figs. 5 and 6). The widespread abundance of MT3 in all seed tissues points to a function for MT3 as a housekeeping or a general metal homeostasis protein rather than as a seed-specific storage protein. The abundance of both MT4 transcript (Table I) and MT4 protein (Figs. 5 and 6) differed from the pattern of MT3 by strictly following the development of seed-specific tissues such as embryo and aleurone layers, thus showing similarities with other grain-specific proteins, such as LTP2, ITR1, and HOR2 (Kalla et al., 1994; Opsahl-Sorteberg et al., 2004; Diaz et al., 2005). The localization of MT3 and MT4 in embryo and aleurone layers of developing grains places the MTs in tissues with high concentrations of Cu and Zn (Lombi et al., 2011). The exact speciation of grain Cu and Zn is not well characterized; however, previous work using SEC-ICP-MS indicated that the embryonic Zn fraction is coupled to protein ligands and not to phytate (Persson et al., 2009).
Identification of Native Plant MTs
To date, only a few native plant MTs have been isolated. The early Cys-labeled protein (Ec), which is the MT4 homolog of wheat, was purified from mature wheat embryos (Hanley-Bowdoin and Lane, 1983) and is a Zn-binding MT (Lane et al., 1987). The Ec protein is the only native plant MT that has been studied biochemically with respect to MT structure and metal-binding dynamics (Peroza et al., 2009; Leszczyszyn et al., 2010). In Arabidopsis, MT2 and MT3 have been purified using sequential size-exclusion, Cu-affinity, and thiol-affinity chromatography; however, these proteins have not been biochemically characterized (Murphy et al., 1997). Finally, a recent proteomic approach in rice embryos directed toward translational responses to Cu stress found rice MT2c in a pool of Cu-inducible proteins (Zhang et al., 2009). Our data show that MT protein and transcript levels correlate well both over time and space in developing grains (Table I; Figs. 5 and 6), thereby directly coupling plant MT3 and MT4 transcript levels with their corresponding proteins and showing that low protein abundance may not constitute the main obstacle for purifications. The MT3- and MT4-specific antibodies developed here (Fig. 4) enable tracking of MTs during the purification process, thereby clarifying the conditions needed to stabilize MTs during extraction and storage. In addition, as the MT3 and MT4 antibodies are specific for the Cys-free linker regions of the proteins, this suggests that the linker region is oriented toward the surface of the protein and may provide an efficient position for antibodies toward other plant MTs.
Differential Metal Speciation by MT3 and MT4
To clarify if recombinant MT3 and MT4 are metal loaded in vivo, state-of-the-art SEC-ICP-MS was used to quantify metal-MT stoichiometry. The recovered metal-MT3 complexes were found to consist of a combination of apo-MT3 and MT3 loaded with a single metal ion without discrimination among Zn, Cu, or Cd (Table II). Seen in combination with the ability to complement ace1 and ycf1 yeast mutants (Fig. 3), MT3 could physiologically function as a Zn/Cu/Cd homeostasis protein responding to cytosolic changes in metal concentrations.
The metal-binding stoichiometry of thiol-rich peptides, including MTs, is generally very sensitive to the composition and concentration of ions in the medium to which they are exposed. In our work, barley MT3 expressed in E. coli was loaded in vivo with Zn at low to moderate concentrations (70–120 µm Zn), which resulted in a mixture of apo-MT3 and non-Zn-saturated MT3. Freisinger (2007) studied purified banana (Musa acuminata) MT3 expressed in E. coli exposed to 100 µm Zn during cultivation (i.e. similar to our conditions) but used a very Zn-rich buffer containing 1,000 µm Zn during cell lysis. This approach will inevitably increase the Zn load to MTs during lysis and purification and consequently lead to much higher Zn-MT ratios. In agreement, Freisinger (2007) found a Zn:MT ratio of 3:1 relative to 1:1 observed using our procedure. Freisinger (2007) also determined the saturated binding capacity of banana MT3 to be four metal ions per protein under in vitro conditions. Combining this ratio with our results underlines that the Zn-MT ratio changes markedly with the Zn concentrations in the environment of the living cell. Under conditions where the cell is being exposed to low to moderate Zn concentrations, the Zn-MT ratio is significantly lower than the maximum Zn-MT ratio of a fully Zn-loaded complex, indicating a considerable capacity of MTs to participate in Zn homeostasis. A physiological role as a housekeeping protein, ready to respond to cellular fluctuations in Zn/Cu/Cd levels, also fits with the localization in developing grains, where MT3 operates in both maternal and filial tissues (Figs. 5 and 6).
Recombinant MT4 operates differently from MT3. MT4 metal loaded in bacterial cells was consistently found as a mixture of Zn2-MT4 and Zn3-MT4 species (Table II). MT4 did not respond to additional metal supplementation of Zn, Cu, or Cd. Previous results revealed that recombinant MT4 from sesame (Sesamum indicum; SiMT4) purified and subjected to elemental analysis by inductively coupled plasma atomic emission spectroscopy behaved similarly to barley MT4, as SiMT4 was found to coordinate two Zn ions (Chyan et al., 2005). However, extensive studies on both native and recombinant wheat Ec protein have shown that this protein has the capacity to coordinate up to six Zn ions (Leszczyszyn et al., 2007; Peroza and Freisinger, 2007).
Due to the seed-specific localization and the stable binding of Zn, we propose that the physiological function of MT4 is as a storage protein. Heterologous expression in yeast did not indicate a role for MT4 in metal chelation or homeostasis of Cu or Cd (Fig. 3); however, SEC-ICP-MS established that Zn2-MT4, Zn3-MT4, and possibly also Zn4-MT4 exist in vivo. These contrasting results indicate that metal-loaded MT4 does not undergo ligand exchange with the surrounding multiple-ion environment. If MT4 exists as a stable complex with Zn in yeast, the potential for MT4 as a metal chelator during Cu or Cd toxicity would be limited. During grain filling in barley, anti-MT4 antibodies stained specifically the embryo and aleurone layer, and MT4 could not be detected when these tissues were not fully developed. Additionally, MT4 colocalizes with the high Zn concentrations in embryo and aleurone layers found during grain development and at maturity (Lombi et al., 2011).
CONCLUSION
This study demonstrates that the barley MT family is composed of at least eight, potentially 10, functional metal-binding MTs. Protein data combined with heterologous MT gene expression in yeast showed that MT metal-binding abilities cannot be predicted by secondary protein structures alone, as putative MT paralogs perform in a variable manner in yeast. Furthermore, MT3 and MT4 seem to play different physiological roles during grain filling and in mature seeds, with MT3 primarily acting as a Zn and Cu housekeeping protein maintaining metal homeostasis, whereas MT4 coordinates Zn with a putative storage function. Immunological data for MT3 and MT4 locate both MTs to the embryo and aleurone layer of developing grains. MT protein and transcript levels correlate well both over time and space in developing grains. The physiological position in grains and the high discriminative behavior toward Zn and against Cd illustrated here by MT4 particularly make this MT an obvious candidate for future biofortification approaches directed toward increasing Zn seed concentrations for food and feed.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Barley (Hordeum vulgare ‘Golden Promise’) was used for all experiments. For cultivation on soil to maturity, plants were grown during autumn in a greenhouse (20°C day/18°C night, relative humidity of 70%, 16-h photoperiod, with a total light intensity of approximately 250 μmol m−2 s−1). Individual spikes were labeled at pollination and harvested for RT-qPCR and immunolocalization of MTs at 7, 14, 21, and 28 dap. The elemental composition of the mature grains from the central part of the spikelet was measured using ICP-MS with data validation against certified reference material of durum wheat (Triticum aestivum) grains (Hansen et al., 2009). Iron, manganese, Cu, and Zn concentrations in the mature grains were 27.1 ± 1.9, 24.9 ± 2.2, 6.3 ± 0.3, and 28.4 ± 2.1 µg g−1 dry matter, respectively (n = 4).
Cloning of Barley MTs
BLAST searches with Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) MT sequences against barley EST data were performed against TIGR’s Barley Gene Indices (http://compbio.dfci.harvard.edu/tgi/; Lee et al., 2005). This resulted in 10 unique EST sequences representing MT homologs. These MTs were subsequently cloned from both genomic DNA and cDNA purified from the barley cv Golden Promise. Primers (Supplemental Table S2) were designed to span the untranslated region bordering the MT open reading frame to obtain maximal template specificity. Cloning was done using uracil excision-based cloning into a user-compatible pYES2 vector designed according to Nour-Eldin et al. (2006). To maximize and make uniform MT expression in yeast, each MT open reading frame was subcloned into the p426 vector containing a Gal-inducible promoter (Mumberg et al., 1994) using primers containing BglII/XhoI restriction sites directly bordering the open reading frame. As MT2b1 has an endogenous BglII site, BamHI was used in the primer. As a positive control, the yeast endogenous MT gene CUP1 was cloned from BY4741 genomic DNA with BglII and XhoI restriction sites in the primers to facilitate expression from p426. For protein expression in Escherichia coli, forward primers of MT3 and MT4 contained an XmnI restriction site. MT3 and MT4 flanked by XmnI/XhoI were then subcloned into pMAL-c2 for N-terminal protein fusion to maltose-binding protein (MBP; New England Biolabs). User PCRs were performed by PfuTurbo Cx polymerase (Stratagene), whereas other proofreading PCRs were done using LA Taq (Takara Bio). PCR products were verified by sequencing.
Phylogenetic Analysis
The phylogenetic tree was drawn with MEGA version 4.0 (http://www.megasoftware.net; Tamura et al., 2007). Barley MT accession numbers are presented below. Rice and Brachypodium distachyon MT homologs were obtained through BLAST searches using barley MT1a, MT2a, MT3, and MT4 against EST databases of the individual species, whereas other MTs were identified with general BLAST. Other MTs were presented by their species abbreviations followed by cDNA accession numbers. Brachypodium MT4 (BdMT4) represents an intronless genomic sequence (Bd3.34663243-497). The ClustalW alignments of MT proteins presented in Figure 2 were performed in T-COFFE (Notredame et al., 2000).
Yeast Complementation Experiments
The Saccharomyces cerevisiae strains used in this study were the Cu-regulated transcription factor mutant ace1 (Mata his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 YGL166w::kanMX4), the Cd-hypersensitive mutant ycf1 (Mata his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 YDR135c::kanMX4), the oxidative stress-regulated transcription factor mutant skn7 (Mata his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 YHR206w::kanMX4), and the wild-type strain BY4741 (Mata his3Δ1 leu2Δ0 ura3Δ0 met15Δ0). All strains were purchased from Euroscarf (Brachmann et al., 1998). The Zn-sensitive strain zrc1cot1 in the BY4741 yeast background did not produce conclusive results with the expression of barley MTs (data not shown). As Euroscarf BY strains are derivatives of S288c, the endogenous yeast MT CRS5 is not expressed due to a stop codon in YOR031w (accession no. NM_001183450). Vector p426 was used for Gal-inducible MT expression and as a negative control. Yeast strains were transformed as described by Gietz and Schiestl (2007). Selection medium with supplements was 2% Gal, yeast nitrogen base without amino acids, and 3 mg L−1 Leu and Met. All yeast strains contained the centromeric vector pRS313 with a functional copy of HIS3 to avoid the use of His supplements. Yeast cells from fresh plates were resuspended in water to a final optical density at 600 nm (OD600) = 1.0 and spotted in serial dilutions for drop tests.
MT Expression, Purification, and Identification
The pMAL-c2 vector containing N-terminal fusions of MT3 or MT4 to MBP was expressed in E. coli strain BL21(DE3)pLysS (Novagen) by standard procedures. BL21 cultures in TBglu (1.2% tryptone, 2.4% yeast extract, 0.5% glycerol, and 0.2% Glc) with or without metal were started at OD600 = 0.05. Metal supplements were given as 50 µm Zn, 50 µm Cu, or 10 µm Cd to yield final concentrations of 120 µm Zn, 52.5 µm Cu, or 10 µm Cd, respectively. After 2 h, MT production was induced by isopropylthio-β-galactoside and allowed to proceed for 3 h. Harvested cells were frozen at −80°C. Recombinant proteins were affinity purified using amylose resin according to the manufacturer’s recommendations (New England Biolabs). As column buffer, 20 mm Tris-HCl, pH 7.4, and 200 mm NaCl were used. During cell sonication, protease activities were inhibited by a protease inhibitor cocktail (Roche). To avoid ligand exchange and the loss of coordinated metal ions during purifications, EGTA, dithiothreitol, and β-mercaptoethanol were omitted from the extraction buffers. Recombinant proteins were documented by SDS-PAGE using Tris-Tricine 8% gels and stained with PageBlue protein stain (Fermentas). MT was cleaved from MBP using Factor Xa as recommended (New England Biolabs) and concentrated using YM30 and YM3 microcon centrifugal filter devices (Millipore) with 30- and 3-kD cutoffs, respectively. Desalting was done on YM3 filters using ultrapure degassed 25 mm Tris-HCl, pH 7.5. Purified MT was stored at −80°C.
Antibody Design and Western Blotting
Polyclonal antibodies against MT proteins were obtained by immunization of rabbits (Agrisera) with two synthetic peptides, H2NC-TEKSHLEVHETAEN-CONH2 and H2NC-GREGTPSGRENRRS-CONH2, corresponding to regions in the linker regions of barley MT3 and MT4 proteins, respectively (Supplemental Fig. S2). The linker region-specific peptides used in antibody production were selected to minimize cross-reactions to other MTs. Also, the unique composition of the synthetic peptides was verified by BLAST analysis against other barley proteins.
The specificity of the antibodies raised against the synthetic peptides was tested using SDS-PAGE followed by western blotting of barley grain proteins or purified MT3/MT4. Proteins were immobilized on a polyvinylidene difluoride membrane (Millipore). The blotting was performed in phosphate-buffered saline buffer, pH 6.8 (8 g L−1 NaCl, 0.2 g L−1 KCl, 1.44 g L−1 Na2HPO4·2H2O, and 0.24 g L−1 KH2PO4), using standard procedures. Primary MT3- and MT4-specific antibodies were diluted 1:5,000.
RNA Purification, cDNA Synthesis, and RT-qPCR
Total RNA was isolated using the Fast RNA Pro Green Kit (MP Biomedicals) and treated with TURBO DNase (Applied Biosystems). RNA was converted to cDNA with Moloney murine leukemia virus reverse transcriptase (New England Biolabs), oligo(dT), random hexamer primers, and deoxyribonucleotide triphosphates as the manufacturer recommends. cDNA was normalized by dilution and amplified by RT-qPCR using DyNAmo Flash Master Mix and ROX Passive Reference dye from the DyNAmo Flash SYBR Green qPCR Kit (Finnzymes) as the manufacturer recommends and run on an Mx3000P Real-Time PCR System (Stratagene). Primers are presented in Supplemental Table S1. The quantitative PCR program was as follows: 95°C for 7 min, then 40 cycles at 95°C for 10 s, followed by 60°C for 30 s (SYBR Green kit; Finnzymes). Samples were run in four biological replicates with two technical replicates each. Quantifications of MT gene expression were done using HvGAPDH as reference (Tauris et al., 2009). As positive controls LTP2 (TC258870), ITR1 (TC139046), and HOR2 (TC138699) were used (Kalla et al., 1994; Diaz et al., 2005). Expression levels were calculated using primer-specific standard curves and the Pfaffl equation (Pfaffl, 2001). Univariate statistical analysis was performed using Student’s t test on fold change mean values of biological replicates with SAS version 9.1 (SAS Institute).
MT Identification and Speciation Analysis
The identities of the purified recombinant MT3 and MT4 peptides were confirmed by their accurate masses using ESI-TOF-MS (Micromass LCT; Waters). The total ESI-TOF-MS spectra were acquired in the range from 100 to 2,000 D. The instrument was calibrated with polyethylene glycol standards (Sigma). Recombinant proteins were verified by ESI-TOF-MS using the calculated masses for MT3 and MT4. The multiple charged ions for MT3 (6,866.68 D) were found as (mass-to-charge ratio): 1,720.96 [M+4H]4+, 1,374.30 [M+5H]5+, 1,144.75 [M+6H]6+, 980.93 [M+7H]7+, 858.78 [M+8H]8+, 763.18 [M+9H]9+, and 686.95 [M+10H]10+; those for MT4 (7,730.60 D) were found as (mass-to-charge ratio): 1,546.95 [M+5H]5+, 1,289.30 [M+6H]6+, 1,105.25 [M+7H]7+, 967.38 [M+8H]8+, 859.73 [M+9H]9+, 773.87 [M+10H]10+, and 703.71 [M+11H]11+.
The ionic compositions of the MT coordination complexes were analyzed using SEC-ICP-MS-based state-of-the-art speciation analysis (Husted et al., 2011). SEC was used to fractionate the metal ion-binding MTs from their apo forms and labile cations. A cross-linked dextran agarose stationary phase (Superdex Peptide 10/300 GL; Amersham Biosciences) was used to maintain the integrity of the MT coordination complexes. The mobile phase was 50 mm Tris (4 mmol of Trizma-HCl and 1 mmol of Trizma base; Sigma), pH 7.5, and operated at a flow rate of 1 mL min−1 (Persson et al., 2009). The SEC column was mass calibrated by use of a Diode Array Detector at 214 nm and with elemental detection using Cu/Zn superoxide dismutase (32 kD; containing Zn, Cu, and sulfur), myoglobin (16.95 kD; containing iron and sulfur), Zn7MT2a (6.59 kD; containing Zn and sulfur), vitamin B12 (1.36 kD; containing cobalt and phosphorus), and myoinositol-2 monophosphate (0.46 kD; containing phosphorus). A linear regression curve was constructed from the retention times and the log-transformed molecular masses of the calibrants.
The ICP-MS device was operated in oxygen mode in order to increase sulfur sensitivity, enabling simultaneous detection and quantification of all elements of interest using a method based on the procedures described by Persson et al. (2009). Initially, the ICP-MS device was tuned in standard mode, the tune settings were extrapolated to helium mode using ChemStation software (Agilent Technologies), and the flow rate of oxygen (10% in helium) to the octopole reaction system was set to 0.5 mL min−1. Through analysis, the following oxide and nonoxide ions were monitored: 48SO+, 59Co+, 63Cu+, 65Cu+, 66Zn+, 67Zn+, and 114Cd+. In order to correct for plasma instability, vitamin B12 (cobalamine) was added as an internal standard to all extracts and monitored as 59Co+. Before analysis, an eight-point external calibration was performed using a commercial multielement standard (P/N 4400-132565; CPI International) by ICP-MS-based flow-injection analysis. The calibration was checked on column (SEC-ICP-MS) using Cys and Zn7MT2a from rabbit liver (Bestenbalt), and the quantified Zn-sulfur stoichiometries always deviated less than 10% from the theoretical ratios.
Cytoimmunochemical Studies
Grains were fixed in 2% (w/v) paraformaldehyde and 0.15% (v/v) glutaraldehyde dissolved in sodium phosphate buffer, pH 7.4, for 12 h at 4°C. The material was dehydrated in an ethanol series and embedded in LR White resin (London Resin Co.). Polymerization was carried out in gelatin capsules at 52°C. For structural investigations, thin sections of 1 μm were stained by the Periodic Acid Schiff's-Naphtol Blue-Black method as described (Sangwan et al., 1992). For immunological studies using light microscopy, 1-μm sections were treated as described by Kichey et al. (2005) but were incubated for 2 h at room temperature with anti-MT3 or anti-MT4 rabbit serum diluted 1:200 in T1 buffer containing 1% bovine serum albumin. Labeling was silver enhanced as described by British Biocell, and sections were back stained with 1% (w/v) fuchsin before microscopic observations under bright-field plus epipolarized light. In negative controls, MT preimmune rabbit sera were used.
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: MT1a (JN997428), MT1b1 (JN997429), MT1b2 (JN997430), MT1b3 (JN997436), MT2a (JN564040), MT2b1 (JN997431), MT2b2 (JN997432), MT2c (JN997433), MT3 (JN997434), and MT4 (JN997435).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Exon-intron gene organization of barley MT genes.
Supplemental Figure S2. ClustalW alignments of MT3 (A) and MT4 (B) proteins.
Supplemental Figure S3. Effect of barley MT expression on H2O2 tolerance in skn7 and on salt tolerance in wild-type yeast.
Supplemental Figure S4. Controls of grain sections observed in immunolocalizations and with the transmission electron microscope.
Supplemental Table S1. Identity scores (%) among MT1 and MT2 protein subgroups in barley.
Supplemental Table S2. Primers used in this study.
Supplementary Material
Glossary
- MT
metallothionein
- Zn
zinc
- Cu
copper
- Cd
cadmium
- H2O2
hydrogen peroxide
- dap
d after pollination
- RT-q
reverse transcription-quantitative
- SEC
size-exclusion chromatography
- ICP-MS
inductively coupled plasma mass spectrometry
- ESI-TOF-MS
electrospray ionization time-of-flight mass spectrometry
- OD600
optical density at 600 nm
- cDNA
complementary DNA
References
- Akashi K, Nishimura N, Ishida Y, Yokota A. (2004) Potent hydroxyl radical-scavenging activity of drought-induced type-2 metallothionein in wild watermelon. Biochem Biophys Res Commun 323: 72–78 [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Blindauer CA, Schmid R. (2010) Cytosolic metal handling in plants: determinants for zinc specificity in metal transporters and metallothioneins. Metallomics 2: 510–529 [DOI] [PubMed] [Google Scholar]
- Brkljacić JM, Samardzić JT, Timotijević GS, Maksimović VR. (2004) Expression analysis of buckwheat (Fagopyrum esculentum Moench) metallothionein-like gene (MT3) under different stress and physiological conditions. J Plant Physiol 161: 741–746 [DOI] [PubMed] [Google Scholar]
- Chyan CL, Lee TT, Liu CP, Yang YC, Tzen JT, Chou WM. (2005) Cloning and expression of a seed-specific metallothionein-like protein from sesame. Biosci Biotechnol Biochem 69: 2319–2325 [DOI] [PubMed] [Google Scholar]
- Cobbett C, Goldsbrough P. (2002) Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol 53: 159–182 [DOI] [PubMed] [Google Scholar]
- Diaz I, Martinez M, Isabel-LaMoneda I, Rubio-Somoza I, Carbonero P. (2005) The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant J 42: 652–662 [DOI] [PubMed] [Google Scholar]
- Freisinger E. (2007) Spectroscopic characterization of a fruit-specific metallothionein: M. acuminata MT3. Inorg Chim Acta 360: 369–380 [Google Scholar]
- Gietz RD, Schiestl RH. (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2: 31–34 [DOI] [PubMed] [Google Scholar]
- Grennan AK. (2011) Metallothioneins, a diverse protein family. Plant Physiol 155: 1750–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W-J, Bundithya W, Goldsbrough PB. (2003) Characterization of the Arabidopsis metallothionein gene family: tissue-specific expression and induction during senescence and in response to copper. New Phytol 159: 369–381 [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Lane BG. (1983) A novel protein programmed by the mRNA conserved in dry wheat embryos: the principal site of cysteine incorporation during early germination. Eur J Biochem 135: 9–15 [DOI] [PubMed] [Google Scholar]
- Hansen TH, Laursen KH, Persson DP, Pedas P, Husted S, Schjoerring JK. (2009) Micro-scaled high-throughput digestion of plant tissue samples for multi-elemental analysis. Plant Methods 5: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon MJ, Cobbett CS. (2007) A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiol 143: 1705–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise J, Krejci S, Miersch J, Krauss G-J, Humbeck K. (2007) Gene expression of metallothioneins in barley during senescence and heavy metal treatment. Crop Sci 47: 1111–1118 [Google Scholar]
- Hotz C, Brown KH. (2004) International Zinc Nutrition Consultative Group (IZiNCG): assessment of the risk of zinc deficiency in populations and options for its control. Food Nutr Bull 25: S91–S202 [PubMed] [Google Scholar]
- Husted S, Persson DP, Laursen KH, Hansen TH, Pedas P, Schiller M, Hegelund JN, Schjoerring JK. (2011) Review: the role of atomic spectrometry in plant science. J Anal At Spectrom 26: 52–79 [Google Scholar]
- Kalla R, Shimamoto K, Potter R, Nielsen PS, Linnestad C, Olsen O-A. (1994) The promoter of the barley aleurone-specific gene encoding a putative 7 kDa lipid transfer protein confers aleurone cell-specific expression in transgenic rice. Plant J 6: 849–860 [DOI] [PubMed] [Google Scholar]
- Karin M, Najarian R, Haslinger A, Valenzuela P, Welch J, Fogel S. (1984) Primary structure and transcription of an amplified genetic locus: the CUP1 locus of yeast. Proc Natl Acad Sci USA 81: 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawachi M, Kobae Y, Mori H, Tomioka R, Lee Y, Maeshima M. (2009) A mutant strain Arabidopsis thaliana that lacks vacuolar membrane zinc transporter MTP1 revealed the latent tolerance to excessive zinc. Plant Cell Physiol 50: 1156–1170 [DOI] [PubMed] [Google Scholar]
- Kawashima I, Kennedy TD, Chino M, Lane BG. (1992) Wheat Ec metallothionein genes: like mammalian Zn2+ metallothionein genes, wheat Zn2+ metallothionein genes are conspicuously expressed during embryogenesis. Eur J Biochem 209: 971–976 [DOI] [PubMed] [Google Scholar]
- Kichey T, Le Gouis J, Sangwan B, Hirel B, Dubois F. (2005) Changes in the cellular and subcellular localization of glutamine synthetase and glutamate dehydrogenase during flag leaf senescence in wheat (Triticum aestivum L.). Plant Cell Physiol 46: 964–974 [DOI] [PubMed] [Google Scholar]
- Kim SH, Lee HS, Song WY, Choi KS, Hur Y. (2007) Chloroplast-targeted BrMT1 (Brassica rapa type-1 metallothionein) enhances resistance to cadmium and ROS in transgenic Arabidopsis plants. J Plant Biol 50: 1–7 [Google Scholar]
- Kojima Y, Binz P-A, Kägi JHR. (1999) Nomenclature of metallothionein: proposal for revision. In C Klaassen, ed, Metallothionein IV. Birkhäuser Verlag, Basel, pp 3–6 [Google Scholar]
- Lane B, Kajioka R, Kennedy T. (1987) The wheat-germ-Ec protein is a zinc-containing metallothionein. Biochem Cell Biol 65: 1001–1005 [Google Scholar]
- Lee S, Persson DP, Hansen TH, Husted S, Schjoerring JK, Kim Y-S, Jeon US, Kim Y-K, Kakei Y, Masuda H, et al (2011) Bio-available zinc in rice seeds is increased by activation tagging of nicotianamine synthase. Plant Biotechnol J 9: 865–873 [DOI] [PubMed] [Google Scholar]
- Lee Y, Tsai J, Sunkara S, Karamycheva S, Pertea G, Sultana R, Antonescu V, Chan A, Cheung F, Quackenbush J. (2005) The TIGR Gene Indices: clustering and assembling EST and known genes and integration with eukaryotic genomes. Nucleic Acids Res 33: D71–D74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczyszyn OI, Schmid R, Blindauer CA. (2007) Toward a property/function relationship for metallothioneins: histidine coordination and unusual cluster composition in a zinc-metallothionein from plants. Proteins 68: 922–935 [DOI] [PubMed] [Google Scholar]
- Leszczyszyn OI, White CRJ, Blindauer CA. (2010) The isolated Cys2His2 site in EC metallothionein mediates metal-specific protein folding. Mol Biosyst 6: 1592–1603 [DOI] [PubMed] [Google Scholar]
- Liu P, Goh C-J, Loh C-S, Pua E-C. (2002) Differential expression and characterization of three metallothionein-like genes in Cavendish banana (Musa acuminata). Physiol Plant 114: 241–250 [DOI] [PubMed] [Google Scholar]
- Lombi E, Smith E, Hansen TH, Paterson D, de Jonge MD, Howard DL, Persson DP, Husted S, Ryan C, Schjoerring JK. (2011) Megapixel imaging of (micro)nutrients in mature barley grains. J Exp Bot 62: 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Margoshes M, Vallee BL. (1957) A cadmium protein from equine imaging hyperintensity in Alzheimer’s disease: correlation with kidney cortex. J Am Chem Soc 79: 4813–4814 [Google Scholar]
- Maroni G, Wise J, Young JE, Otto E. (1987) Metallothionein gene duplications and metal tolerance in natural populations of Drosophila melanogaster. Genetics 117: 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moleirinho A, Carneiro J, Matthiesen R, Silva RM, Amorim A, Azevedo L. (2011) Gains, losses and changes of function after gene duplication: study of the metallothionein family. PLoS ONE 6: e18487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, Vavasseur A, Richaud P. (2009) AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol 149: 894–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22: 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A, Zhou J, Goldsbrough PB, Taiz L. (1997) Purification and immunological identification of metallothioneins 1 and 2 from Arabidopsis thaliana. Plant Physiol 113: 1293–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217 [DOI] [PubMed] [Google Scholar]
- Nour-Eldin HH, Hansen BG, Nørholm MH, Jensen JK, Halkier BA. (2006) Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res 34: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opsahl-Sorteberg H-G, Divon HH, Nielsen PS, Kalla R, Hammond-Kosack M, Shimamoto K, Kohli A. (2004) Identification of a 49-bp fragment of the HvLTP2 promoter directing aleurone cell specific expression. Gene 341: 49–58 [DOI] [PubMed] [Google Scholar]
- Palacios Ò, Atrian S, Capdevila M. (2011) Zn- and Cu-thioneins: a functional classification for metallothioneins? J Biol Inorg Chem 16: 991–1009 [DOI] [PubMed] [Google Scholar]
- Peroza EA, Freisinger E. (2007) Metal ion binding properties of Triticum [corrected] aestivum Ec-1 metallothionein: evidence supporting two separate metal thiolate clusters. J Biol Inorg Chem 12: 377–391 [DOI] [PubMed] [Google Scholar]
- Peroza EA, Kaabi AA, Meyer-Klaucke W, Wellenreuther G, Freisinger E. (2009) The two distinctive metal ion binding domains of the wheat metallothionein Ec-1. J Inorg Biochem 103: 342–353 [DOI] [PubMed] [Google Scholar]
- Persson DP, Hansen TH, Laursen KH, Schjoerring JK, Husted S. (2009) Simultaneous iron, zinc, sulfur and phosphorus speciation analysis of barley grain tissues using SEC-ICP-MS and IP-ICP-MS. Metallomics 1: 418–426 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Llorente ID, Pérez-Palacios P, Doukkali B, Caviedes MA, Pajuelo E. (2010) Expression of the seed-specific metallothionein mt4a in plant vegetative tissues increases Cu and Zn tolerance. Plant Sci 178: 327–332 [Google Scholar]
- Sangwan RS, Bourgeois Y, Brown S, Vasseur G, Sangwan-Norreel B. (1992) Characterization of competent cells and early events of Agrobacterium-mediated genetic transformation in Arabidopsis thaliana. Planta 188: 439–456 [DOI] [PubMed] [Google Scholar]
- Song W-Y, Choi KS, Kim Y, Geisler M, Park J, Vincenzetti V, Schellenberg M, Kim SH, Lim YP, Noh EW, et al (2010) Arabidopsis PCR2 is a zinc exporter involved in both zinc extrusion and long-distance zinc transport. Plant Cell 22: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland DEK, Stillman MJ. (2011) The “magic numbers” of metallothionein. Metallomics 3: 444–463 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Tsukamoto T, Inoue H, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. (2008) Deoxymugineic acid increases Zn translocation in Zn-deficient rice plants. Plant Mol Biol 66: 609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tanguy A, Boutet I, Bonhomme F, Boudry P, Moraga D. (2002) Polymorphism of metallothionein genes in the Pacific oyster Crassostrea gigas as a biomarker of response to metal exposure. Biomarkers 7: 439–450 [DOI] [PubMed] [Google Scholar]
- Tauris B, Borg S, Gregersen PL, Holm PB. (2009) A roadmap for zinc trafficking in the developing barley grain based on laser capture microdissection and gene expression profiling. J Exp Bot 60: 1333–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennstedt P, Peisker D, Böttcher C, Trampczynska A, Clemens S. (2009) Phytochelatin synthesis is essential for the detoxification of excess zinc and contributes significantly to the accumulation of zinc. Plant Physiol 149: 938–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verret F, Gravot A, Auroy P, Leonhardt N, David P, Nussaume L, Vavasseur A, Richaud P. (2004) Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett 576: 306–312 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat J-F, Curie C. (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HL, Sakamoto T, Kawasaki T, Umemura K, Shimamoto K. (2004) Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol 135: 1447–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T, Li X, Zhu W, Wu C, Yang G, Zheng C. (2009) Cotton metallothionein GhMT3a, a reactive oxygen species scavenger, increased tolerance against abiotic stress in transgenic tobacco and yeast. J Exp Bot 60: 339–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wu YR, Li Y, Ling HQ, Chu CC. (2009) OsMT1a, a type 1 metallothionein, plays the pivotal role in zinc homeostasis and drought tolerance in rice. Plant Mol Biol 70: 219–229 [DOI] [PubMed] [Google Scholar]
- Yuan J, Chen D, Ren YJ, Zhang XL, Zhao J. (2008) Characteristic and expression analysis of a metallothionein gene, OsMT2b, down-regulated by cytokinin suggests functions in root development and seed embryo germination of rice. Plant Physiol 146: 1637–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lian C, Shen Z. (2009) Proteomic identification of small, copper-responsive proteins in germinating embryos of Oryza sativa. Ann Bot (Lond) 103: 923–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou GK, Xu YF, Liu JY. (2005) Characterization of a rice class II metallothionein gene: tissue expression patterns and induction in response to abiotic factors. J Plant Physiol 162: 686–696 [DOI] [PubMed] [Google Scholar]
- Zhou J, Goldsbrough PB. (1995) Structure, organization and expression of the metallothionein gene family in Arabidopsis. Mol Gen Genet 248: 318–328 [DOI] [PubMed] [Google Scholar]
- Zimeri AM, Dhankher OP, McCaig B, Meagher RB. (2005) The plant MT1 metallothioneins are stabilized by binding cadmiums and are required for cadmium tolerance and accumulation. Plant Mol Biol 58: 839–855 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.