The HMG-box is an approximately 75-amino acid residue protein domain that occurs in all eukaryotic organisms and was first identified as a characteristic feature of the chromosomal high mobility group (HMG) proteins of the HMGB type. Structural studies have demonstrated that the L-shaped fold of the domain formed by three α-helices is conserved to a greater extent than expected from amino acid sequence similarity between different HMG-boxes. The long arm consists of helix III and the N-terminal extended strand, whereas the short arm of the L-shape is composed of helices I and II forming an angle of approximately 80° between the arms (Thomas and Travers, 2001; Stros et al., 2007). The HMG-box domain mediates DNA binding primarily through the minor groove of DNA. Hydrophobic residues of the concave face of the L-shaped molecule partially intercalate between the DNA bases, thereby widening the minor groove, which results in unwinding and remarkable bending of the DNA helix. Thus, the HMG-box domain binds the outside of the DNA bend, compressing the major groove (Thomas and Travers, 2001; Stros, 2010). Some HMG-box proteins can interact with DNA sequence specifically (e.g. mammalian transcription factors such as SEX DETERMINING REGION OF Y [SRY] and LYMPHOID ENHANCHER-BINDING FACTOR1 [LEF-1]), whereas other HMG-box proteins bind DNA sequence independently (e.g. chromosomal HMGB proteins and Structure-Specific Recognition Protein1 [SSRP1]). A typical feature of both types of HMG- box domains is their selective binding to certain DNA structures, including four-way junctions and DNA minicircles (Bustin, 1999; Thomas and Travers, 2001; Stros et al., 2007; Wegner, 2010). Because HMG-box proteins induce DNA bending upon binding to linear DNA, they often act as architectural facilitators in the assembly of nucleoprotein complexes involved in transcription, recombination, or other DNA-dependent processes (Bustin, 1999; Thomas and Travers, 2001; Stros et al., 2007).
HMG-box domains are found in a variety of proteins that interact with DNA. In these proteins, the HMG-box domain(s) occurs in combination with various other protein domains of different function. Accordingly, because of this structural variability and their interaction with various other proteins, HMG-box proteins are involved in different nuclear functions. There are HMG-box proteins, for instance, that act as architectural chromosomal proteins (HMGB proteins), whereas others are transcription factors or subunits of chromatin-remodeling complexes, or they modulate DNA recombination/repair (Bustin, 1999; Stros et al., 2007). In addition to the cell nucleus, HMG-box proteins are found in mitochondria of animals and yeast, where they serve as transcriptional regulators and contribute to the organization of the mitochondrial DNA (Bonawitz et al., 2006; Kucej and Butow, 2007). Currently, there is no evidence for the occurrence of HMG-box proteins in plant mitochondria. However, an unusual HMG-box protein from Physcomitrella localizes to plastids in tobacco (Nicotiana tabacum) BY-2 cell protoplasts (Kiilerich et al., 2008), but no higher plant HMG-box protein has been reported to occur in plastids.
Various plant genomes were found to encode HMG-box proteins, suggesting that they commonly occur in plants (Riechmann et al., 2000; Stros et al., 2007). Higher plant genomes encode 10 to 15 different HMG-box proteins that range from approximately 13 to 72 kD. When compared with the human genome, which encodes 47 HMG-box proteins ranging from approximately 15 to 193 kD, HMG-box proteins are less diversified in plants (Stros et al., 2007). Whereas in humans, HMG-box-containing transcription factors represent the largest subgroup (Stros et al., 2007; Wegner, 2010), to date, it is unclear whether any of the plant HMG-box proteins act as transcription factors. No sequence-specific DNA interactions have been reported for any of the plant HMG-box proteins. In plants, the family of small chromosomal HMGB proteins represents the most diversified subgroup of HMG-box proteins (Stros et al., 2007).
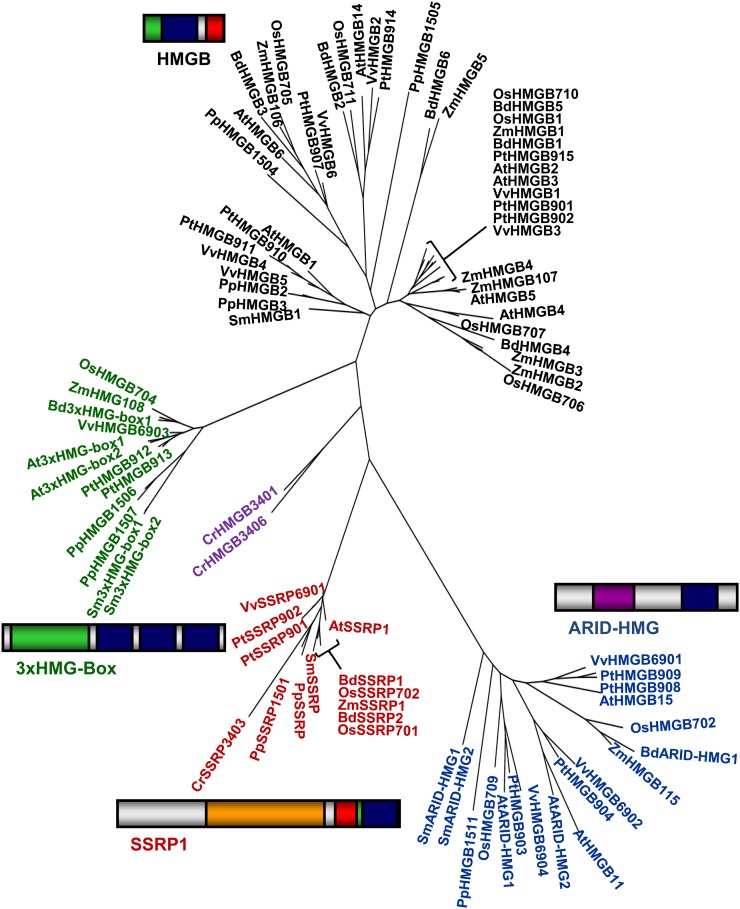
We have searched various databases, including the Plant Chromatin Database (www.chromdb.org/) and that of MIPS (database from the Munich Information Center for Protein Sequences; http://mips.helmholtz-muenchen.de/plant/), for plant proteins containing the HMG-box motif. In addition to flowering plants, we investigated the extent to which different types of HMG-box proteins are encoded in the genome sequences of the lycophyte Selaginella moellendorfii, the moss Physcomitrella patens, and the algae Chlamydomonas reinhardtii and Volvox carteri that became available in recent years. The filtered results of these searches were used to compile a relatively comprehensive list of plant HMG-box proteins (Supplemental Table S1). Based on their overall structure and amino acid sequence similarity (Fig. 1), plant HMG-box proteins can be subdivided into four distinct families: chromosomal HMGB proteins, AT-rich interaction domain (ARID)-HMG proteins, 3xHMG-box proteins, and SSRP1. We used the amino acid sequences of HMG-box proteins from nine species (three monocots, three dicots, Selaginella, Physcomitrella, and Chlamydomonas) for a multiple sequence alignment that served for the construction of a neighbor-joining tree (Fig. 1). It clearly illustrates the four distinct families of plant HMG-box proteins. Various studies performed in the past few years suggest that members of the four HMG-box protein families have different cellular functions, and we discuss here the present knowledge about the structural and functional characteristics of these proteins.
Figure 1.
Schematic representation of the overall structure of the four families of plant HMG-box proteins and their amino acid sequence similarity. Whereas HMGB proteins, ARID-HMG proteins, and SSRP1 contain a single HMG-box domain, the 3xHMG-box proteins have three copies of the HMG-box domain. The overall domain structure of the four groups of HMG-box proteins that were identified in plants are presented schematically: HMG-box domain (blue), basic region (green), acidic region (red), SSR domain of SSRP1 (orange), and ARID (violet). The amino acid sequences of HMG-box proteins from Brachypodium distachyon (Bd), rice (Os), maize (Zm), Arabidopsis (At), P. trichocarpa (Pt), grape (Vv), S. moellendorfii (Sm), P. patens (Pp), and C. reinhardtii (Cr; compare with Supplemental Table S1) were aligned by multiple sequence alignment that served for the construction of a neighbor-joining tree using the software package SeaView (http://pbil.univ-lyon1.fr/software/seaview.html). The four families of plant HMG-box proteins, HMGB (in black), ARID-HMG (in blue), 3xHMG-box (in green), and SSRP1 (in red), occur as distinct groups. Whereas the proteins of Selaginella and Physcomitrella group with their counterparts from flowering plants, the two Chlamydomonas HMGB-type sequences (in violet) group separately.
CHROMOSOMAL HMGB PROTEINS
Originally, HMG proteins were identified as proteins with unusual physicochemical properties when calf thymus chromatin was extracted with 0.35 m NaCl (Goodwin et al., 1973). Subsequently, based on their characteristic amino acid sequences, they were subdivided into three structurally distinct families termed HMGA, HMGB, and HMGN (Bustin and Reeves, 1996; Grasser et al., 2007a). In this article, we concentrate exclusively on the HMG-box containing HMGB family. The HMGB proteins (13–27 kD) of different organisms exhibit a diverse overall structure. The vertebrate proteins, for instance, consist of two HMG-box domains, a basic linker region and an acidic C-terminal domain, whereas plant HMGB proteins (Fig. 1) contain a single HMG-box domain that is flanked by basic N-terminal and acidic C-terminal domains (Thomas and Travers, 2001; Stros et al., 2007). Database analyses revealed that HMGB-type proteins apparently occur in all plants and also in algae (Supplemental Table S1).
Plant HMGB proteins are structurally more diversified than their animal counterparts (Stros et al., 2007). Thus, the Arabidopsis (Arabidopsis thaliana) genome encodes eight proteins that, according to their amino acid sequences, can be classified as HMGB proteins. However, experimental analyses demonstrate that the protein encoded by the Arabidopsis Genome Initiative locus At5g23405, despite marked sequence similarity to well-characterized HMGB proteins, does not share the features of bona fide HMGB proteins, including predominant nuclear localization and DNA-binding activity (Grasser et al., 2006). Therefore, it is required to test experimentally the functionality of predicted HMG-box domains, which may be of particular importance for domains that display a lower degree of sequence conservation such as the putative algal HMG-box proteins. Plant HMGB proteins and their counterparts from other sources essentially have in common the characteristic DNA interactions such as low affinity, sequence-independent binding to linear DNA, but high-affinity interaction with DNA structures (four-way junctions, DNA minicircles, supercoiled DNA) and pronounced DNA bending upon binding linear DNA. We have previously reviewed these aspects of DNA binding (Grasser et al., 2007a) and pointed out that various HMGB proteins (e.g. those occurring in maize [Zea mays] and Arabidopsis) display differences in their DNA interactions and posttranslational modifications. These differences indicate that plants have a repertoire of architectural chromatin-associated proteins. DNA-binding experiments with full-length and truncated proteins also indicated that interactions between the N-terminal basic domain (which increases DNA binding) and the acidic C-terminal domain (which reduces DNA binding) regulate the DNA interactions of maize and rice (Oryza sativa) HMGB proteins (Ritt et al., 1998; Wu et al., 2003). Spectrometric measurements and cross-linking experiments confirmed intramolecular interactions between the terminal domains of plant HMGB proteins and that the interactions are enhanced by the phosphorylation of Ser residues within the acidic tail (Thomsen et al., 2004). Consistent with these findings, removal of the acidic tail of Arabidopsis HMGB1 and HMGB5 reduced the remarkable mobility of the proteins within cell nuclei in living cells as measured by fluorescence recovery after photobleaching (FRAP) experiments (Launholt et al., 2006). The intramolecular domain interactions appear to influence various properties of plant HMGB proteins, including DNA interactions, mobility within the nucleus, and subcellular localization (Ritt et al., 1998; Wu et al., 2003; Launholt et al., 2006; Pedersen et al., 2010).
Although it is well documented that mammalian HMGB1 can be detected also outside the nucleus, acting as a kind of cytokine (Müller et al., 2004; Yang and Tracey, 2010), chromosomal HMGB proteins are generally considered nuclear proteins (Grasser et al., 2007a; Reeves, 2010). In line with that, plant HMG proteins traditionally were purified from chromatin or isolated nuclei (Spiker, 1984; Grasser et al., 1991). Systematic examination of the subcellular localization of Arabidopsis HMGB1, HMGB5, and HMGB6 proteins as well as of the HMGB-type protein AtHMGB14 by analyzing the distribution of GFP fusions and by immunofluorescence microscopy confirmed the nuclear localization (Grasser et al., 2004, 2006; Launholt et al., 2006; Lildballe et al., 2008). In contrast to these findings, recent experiments revealed that Arabidopsis HMGB2/3 and HMGB4, in addition to being found in the nucleus, are detected to different extents in the cytosol. Monitoring the distribution of photoactivatable GFP fused to HMGB2 and HMGB4 demonstrated that both proteins can shuttle between nucleus and cytoplasm, whereas HMGB1 remained nucleus localized (Pedersen et al., 2010). Currently, it is unclear why some plant HMGB proteins are strictly nuclear whereas others can shuttle between nucleus and cytosol. An extranuclear/extracellular role like that of HMGB1, which acts as a specific mediator in injury and inflammation of mammals (Müller et al., 2004; Yang and Tracey, 2010), appears unlikely for plants. In view of the interplay between HMGB proteins and linker histones (see below), the nucleocytosolic partitioning of HMGB proteins may serve as a means to regulate the nuclear content of these architectural proteins and thereby modulate the balance with linker histones that may influence chromatin structure.
Within the cell nucleus, HMGB proteins are highly dynamic proteins that bind DNA/chromatin only transiently before moving on to the next binding site (Bianchi and Agresti, 2005; Catez and Hock, 2010). Whereas linker histone H1 increases chromatin compaction, HMGB (and other HMG proteins) that share chromatin-binding sites with H1 (but bind chromatin more loosely than H1) reduce chromatin packaging, facilitating the access of regulatory factors to chromatin targets (Bustin et al., 2005; Catez and Hock, 2010; Thomas and Stott, 2012). To study the interplay between H1 and HMGB proteins, Catez et al. (2004) microinjected potential competitor proteins (e.g. HMGB1) into cells expressing the other tested protein as a GFP fusion (e.g. H1-GFP). The mobility (related to that chromatin interaction) of the GFP fusion was measured by FRAP in injected cells and uninjected control cells. Interestingly, these experiments revealed that in living mammalian cells, HMGB proteins (and other HMG proteins) compete with linker histone H1 for chromatin binding. In line with that, mammalian HMGB1 interacts in vitro with linker histones via its acidic C-terminal domain, which may result in an enhanced DNA binding of HMGB1 (Cato et al., 2008). In the chromatin context, this could promote the displacement of H1 by HMGB proteins, resulting in a more accessible chromatin structure that is competent for transcription (Bustin et al., 2005; Catez and Hock, 2010; Thomas and Stott, 2012). As determined by FRAP experiments, Arabidopsis HMGB1 and HMGB5 are highly mobile nuclear proteins with a high turnover on chromatin and a very short residence time. Their chromatin-binding dynamics is clearly higher than that of linker histone H1.2. HMGB proteins lacking the acidic C-terminal domain displayed in these experiments a reduced mobility, suggesting that the increased affinity for DNA of the truncated proteins stabilized their chromatin interactions (Launholt et al., 2006). Therefore, in plant cell nuclei, there is a range of mobile HMGB proteins that, in collaboration with other chromosomal proteins, can modulate chromatin structure.
In addition to the interplay with linker histones, HMGB proteins as chaperones are involved in the assembly of specific nucleoprotein complexes such as enhanceosomes regulating transcription (Agresti and Bianchi, 2003; Grasser et al., 2007a). The potential of plant HMGB proteins to promote the formation of specific nucleoprotein structures is evident from their stimulatory architectural role in site-specific recombination reactions, which was observed in vitro and in vivo (Stemmer et al., 2002). Moreover, HMGB proteins can facilitate the binding of certain plant transcription factors (basic Leu zipper, DNA binding with one finger [DOF]) to their DNA recognition sites by direct interaction (in several instances via their HMG-boxes) with these factors (Schultz et al., 1996; Yanagisawa, 1997). In the case of maize DOF2, the specificity of the interaction is regulated by protein kinase CK2-mediated phosphorylation and is mainly determined by the terminal domains of the HMGB proteins (Krohn et al., 2002; Grasser et al., 2007b). HMGB5 (which represents the most potent facilitator of DOF2 DNA binding) can assist target site recognition of the transcription factor also in the nucleosomal context (Cavalar et al., 2003). Studies with HMGB proteins of different sources have indicated that they are involved as architectural factors in various DNA-dependent nuclear processes, including transcription, recombination, and DNA repair. Due to their abundance, dynamics, and versatile interaction with DNA and other proteins, HMGB proteins have the ability to facilitate these processes by modulating chromatin structure and/or by assisting the efficient formation of critical nucleoprotein complexes (Reeves, 2010; Stros, 2010).
In plants, it has become apparent that chromatin-mediated regulation of gene expression plays an important role in the response to abiotic stress conditions, and it appears that linker histones and HMGB proteins are involved in the stress-induced reactions (Kim et al., 2010). The expression of some Arabidopsis HMGB genes is differentially regulated by abiotic stress treatment (Kwak et al., 2007). Importantly, transgenic Arabidopsis plants that overexpress Arabidopsis HMGB2 (or a cucumber [Cucumis sativus] HMGB protein) upon different abiotic stress treatments showed reduced seed germination (and subsequent growth), whereas overexpression of HMGB4 had no marked influence. The observed effects (in part) may be due to the altered expression of a number of germination-responsive genes (Kwak et al., 2007; Jang et al., 2008). Both the absence and overexpression of HMGB1 in Arabidopsis resulted in an increased sensitivity toward methyl methanesulfonate. When exposed to elevated NaCl concentrations, the germination of HMGB1 overexpressors was reduced, whereas the hmgb1 mutant germinated normally. Transcript profiling of hmgb1 plants compared with control plants revealed that a large number of genes were differentially expressed. Among the down-regulated genes, the Gene Ontology category of stress-responsive genes was overrepresented (Lildballe et al., 2008). Exposure of plants to abiotic stress conditions requires a switch in the gene expression program. Chromatin rearrangements, presumably involving HMGB proteins, linker histones, and other chromatin modifiers may coordinate plant gene expression in response to the different types of environmental stress conditions (Kim et al., 2010). In cotton (Gossypium hirsutum), a role of HMGB proteins in differentiation and proliferation was reported. Reduced expression of cotton HMGB3 (which is preferentially expressed in embryonic tissue) altered the potential of cells to differentiate and dedifferentiate during somatic embryogenesis. Some of the genes differentially expressed in the HMGB3-deficient cells (versus control cells) were found to encode putative plant components of the pathway that in mammals is involved in β-catenin signaling (Hu et al., 2011). The significance of this interesting finding remains to be clarified. Another novel role of plant HMGB proteins was identified in the maintenance of Arabidopsis chromosome ends. Plants lacking HMGB1 displayed shortened telomeres, whereas in plants overexpressing HMGB1, elongated telomeres were observed. In contrast to mouse cells lacking HMGB1, in Arabidopsis, shortening of the telomeres apparently is not caused by reduced telomerase activity (Schrumpfová et al., 2011). Currently, the mechanism of action of HMGB1 in Arabidopsis telomere maintenance is obscure, but it is possible that HMGB proteins other than HMGB1 influence the telomeric chromatin structure. This adds another example to the versatile functions of HMGB proteins in plant cell nuclei.
ARID-HMG PROTEINS
Another group of HMG-box proteins that was identified by sequence database searches are the proteins containing in addition to an HMG-box a so-called ARID (Riechmann et al., 2000; Stros et al., 2007). The ARID is a DNA-binding module that was first identified in the mouse B-cell transcription factor Bright and the Drosophila Dead ringer protein. Later on, ARIDs were found in a variety of animal transcription factors that regulate cell proliferation and differentiation. The ARID consensus sequence spans approximately 100 amino acid residues that are organized in a modified helix-turn-helix fold (Kortschak et al., 2000; Wilsker et al., 2002). ARIDs bind DNA through a novel mechanism that involves major groove immobilization of a large loop, which connects the helices of the helix-turn-helix motif, and through concomitant structural rearrangements adding stabilizing contacts from a β-hairpin. The initially characterized ARID proteins were found to bind preferentially to AT-rich sequences, prompting the name of the domain. Analysis of protein-DNA interactions of other ARIDs by various methods revealed that ARIDs of different proteins bind DNA nonsequence specifically (Iwahara et al., 2002; Patsialou et al., 2005). Plant genomes encode various ARID proteins, and in most cases, the ARID occurs in combination with other protein domains, including Myb, PHD, and HMG-box domains (Riechmann et al., 2000). Experimental analysis of the ARID protein SIP1 from Lotus japonicus that contains an ARID (but no other known domain) demonstrated that it binds AT-rich elements of the NIN gene promoter. It has been suggested that SIP1 plays a role in nodule formation related to the Rhizobium-legume plant symbiosis (Zhu et al., 2008).
The combination of an N-terminal ARID and a C-terminal HMG-box in the ARID-HMG proteins (34–56 kD, except a Physcomitrella protein with 82 kD; Fig. 1) is specific for plants, occurring in flowering plants as well as Selaginella and Physcomitrella, but apparently not in algae (compare with Supplemental Table S1). ARID-HMG proteins seem to be more diversified in dicot species than in monocots. Comparing the amino acid sequences of ARID-HMG proteins from various species indicated the existence of distinct structural subgroups (Supplemental Fig. S1). Two of the four Arabidopsis ARID-HMG genes, termed ARID-HMG1/2, were found to be widely expressed in the plant, albeit at different levels. In BY-2 protoplasts, ARID-HMG1 and ARID-HMG2 localize primarily to the nucleus. ARID-HMG1 displays a slightly preferential binding to AT-rich DNA (when compared with GC-rich DNA), and mediated by its HMG-box domain it can bind DNA structure specifically. Both the ARID and the HMG-box domain contribute to the DNA interactions of ARID-HMG1 (Hansen et al., 2008). Because both the ARID and the HMG-box occur in a variety of proteins with very different functions, the role of the ARID-HMG proteins in plants is unknown and requires further experimentation.
3xHMG-BOX PROTEINS
The 3xHMG-box proteins (43–60 kD) are composed of an N-terminal basic domain (which shows no similarity to other proteins) and three copies of the HMG-box domain in the C-terminal part (Fig. 1). According to database analyses, the 3xHMG-box proteins occur exclusively in plants, but they are relatively conserved among plants, because they are encoded in genomes of lower plants and higher plants (compare with Supplemental Table S1). Whereas some species encode two proteins (e.g. Arabidopsis and Populus trichocarpa), other species encode only a single 3xHMG-box protein (e.g. rice and grape [Vitis vinifera]). We were unable to find 3xHMG-box sequences from algae, but interestingly, Chlamydomonas apparently encodes proteins with two putative HMG-box domains, which do not occur in plants. In the absence of experimental data, it is unknown whether these proteins belong to the HMGB family (vertebrate HMGB proteins have two HMG-boxes) or whether they represent the “algal version” of the 3xHMG-box proteins. We have used the amino acid sequences of the individual HMG-box domains of the 3xHMG-box proteins (BoxA–BoxC) along with the sequences of the HMG-box domains of HMGB, SSRP1, and ARID-HMG proteins to construct a neighbor-joining tree. This analysis revealed that the various HMG-box sequences, despite their overall conservation, group strictly according to the protein family they originate from (Supplemental Fig. S2). Moreover, the different HMG-box sequences (termed BoxA, BoxB, and BoxC) of 3xHMG-box proteins are clearly distinguished, forming separate branches.
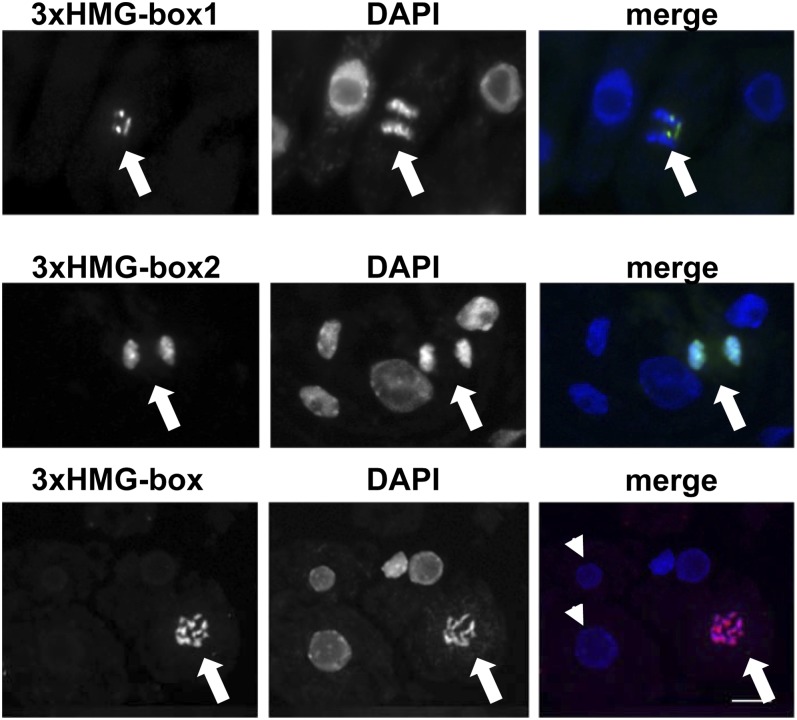
Recently, the two Arabidopsis 3xHMG-box proteins, termed 3xHMG-box1 and 3xHMG-box2, which share 77% amino acid sequence identity, were analyzed experimentally. The 3xHMG-box proteins are widely expressed in the plant, but in contrast to other HMG-box proteins, they are expressed in a proliferation-dependent manner preferentially in mitotic cells. Examination of GFP fusion proteins and immunofluorescence studies demonstrated that 3xHMG-box1 and 3xHMG-box2 associate with condensed chromosomes during different stages of M phase (Pedersen et al., 2011). This finding clearly discriminates the 3xHMG-box proteins from other (plant) HMG-box proteins (e.g. Arabidopsis HMGB proteins and SSRP1) that interact with interphase chromatin but do not associate with mitotic chromosomes (Duroux et al., 2004; Launholt et al., 2006; Lildballe et al., 2008; Pedersen et al., 2010). 3xHMG-box2 generally binds the condensed chromosomes, whereas 3xHMG-box1 associates with specific chromosomal regions, representing the ribosomal DNA loci (Fig. 2). In addition to mitotic cells, the 3xHMG-box proteins are expressed in meiotic cells and interact with condensed chromosomes in pollen mother cells (Pedersen et al., 2011). The association of the 3xHMG-box proteins with both mitotic and meiotic chromosomes argues for a general role in chromosome function related to cell division, such as chromosome condensation and/or segregation. The reorganization of chromatin into compact mitotic chromosomes is of fundamental importance for faithful chromosome segregation (Ohta et al., 2011). In line with a role in DNA/chromatin condensation, the three HMG-box domains as well as the basic N-terminal domain contribute synergistically to the DNA interactions of 3xHMG-box2 (Pedersen et al., 2011). Therefore, basically the entire 3xHMG-box protein can interact with DNA and, perhaps in combination with other factors, may contribute to the condensation of DNA and chromatin during mitosis/meiosis. Alternatively, it was suggested (Jerzmanowski and Kotlinski, 2011) that during mitosis/meiosis, 3xHMG-box proteins may compete with linker histone H1 (similar as described above for HMGB proteins in interphase nuclei). This could relax the chromosome binding of H1, enhancing its function in microtubule nucleation, a feature that was described for plant H1. However, there are other possible functions and open questions that need to be addressed, such as the issue of why some plants have two 3xHMG-box proteins and other plants have only a single one.
Figure 2.
Arabidopsis 3xHMG-box proteins associate with mitotic chromosomes. Root cells of Arabidopsis plants expressing 3xHMG-box1-GFP (top row) or 3xHMG-box2 (middle row) under the control of the cauliflower mosaic virus 35S promoter were examined by immunofluorescence using an anti-GFP antibody (left) and for comparison a 4′,6-diamino-phenylindole (DAPI) stain (middle), and the merge of the two images is shown at right. Whereas 3xHMG-box2 generally associates with the condensed chromosomes of a metaphase cell (arrows), 3xHMG-box1 is only detected at restricted chromosomal regions (that by fluorescent in situ hybridization were identified as ribosomal DNA loci; Pedersen et al., 2011). In wild-type cells (bottom row), an antibody raised against 3xHMG-box2 (that also recognizes 3xHMG-box1) detects 3xHMG-box proteins bound to the condensed chromosomes of a mitotic metaphase cell (arrows), but no signal is observed in interphase cells (arrowheads).
SSRP1
SSRP1 together with the SPT16 protein forms the dimeric facilitates chromatin transcription (FACT) complex that was originally identified in yeast and mammalian cells (Brewster et al., 1998; Orphanides et al., 1999; Wittmeyer et al., 1999). The FACT complex modulates the repressive barrier that chromatin/nucleosomes represent to transcription and other DNA-dependent processes. It acts as a histone chaperone that assists RNA polymerase II-catalyzed transcript elongation of chromatin templates by disassembling nucleosomes in the path of the polymerase. Importantly, FACT is also involved in transcript elongation-dependent reassembly of nucleosomes, thereby maintaining the original chromatin state repressing cryptic transcript initiation from within the coding sequences. Evidence from various systems indicates that these dynamic chromatin changes are mediated by FACT in collaboration with other histone chaperones, ATP-dependent remodeling complexes, and histone-modifying enzymes (Reinberg and Sims, 2006; Winkler and Luger, 2011; Formosa, 2012).
Genes coding for SSRP1 are conserved in flowering plants as well as Selaginella and Physcomitrella and occur also in algae (compare with Supplemental Table S1). Some plant species (e.g. Arabidopsis and maize) have a single gene encoding SSRP1, whereas others (rice and P. trichocarpa) have two genes encoding closely related proteins (approximately 80% amino acid sequence identity). Plant SSRP1 (61–78 kD) has an overall structure (Fig. 1) similar to that of its animal counterparts, with sequence similarity throughout the N-terminal, middle, and acidic domains as well as the C-terminal HMG-box domain, but lacking the approximately 80 amino acid residues that form the C-terminal region of animal SSRP1 (Röttgers et al., 2000). Interestingly, in yeast, the situation is somewhat different, as POB3 (the yeast SSRP1) lacks an HMG-box domain, whose function is provided by a separate small HMGB-type protein termed NHP6 (Winkler and Luger, 2011; Formosa, 2012). A short basic region N terminal to the HMG-box domain is essential for the nuclear localization of maize SSRP1. SSRP1 binds DNA sequence independently, but mediated by the HMG-box domain it recognizes the structure of supercoiled and minicircle DNA as well as nucleosome particles, and the DNA interactions are modulated by the phosphorylation of SSRP1 by protein kinase CK2 (Röttgers et al., 2000; Lichota and Grasser, 2001; Krohn et al., 2003).
The existence of the FACT complex consisting of SSRP1 and SPT16 in Arabidopsis was demonstrated by colocalization of the two proteins and by coimmunoprecipitation with antibodies specific for SSRP1 and SPT16 (Duroux et al., 2004). In line with its role in active transcription, Arabidopsis SSRP1 (and SPT16) is detected by immunofluorescence analyses in euchromatic regions of the nucleus but not in heterochromatic chromocenters. Using chromatin immunoprecipitation, FACT was found to associate with the entire transcribed region of active genes in a transcription-dependent manner, but not with nontranscribed genes or intergenic regions (Duroux et al., 2004; Perales and Más, 2007). Arabidopsis SSRP1 is an essential gene (Lolas et al., 2010), which reflects the situation in mouse and yeast, where inactivation of the SSRP1/POB3 genes (or SPT16) proved critical for cell viability (Singer and Johnston, 2004; Formosa, 2012). Reduced levels of SSRP1 (or SPT16) cause various defects in Arabidopsis vegetative and generative development. Thus, mutant plants display an increased number of leaves and inflorescences, show early bolting, have abnormal flower and leaf architecture, and their seed production is severely affected (Lolas et al., 2010). The transcript level of the central floral repressor FLC is clearly reduced in ssrp1 and spt16 mutant plants under both long-day and short-day conditions. Consistently, the transcript level of the flowering-time integrator SOC1 (which is regulated by FLC) is increased. This indicates that the early-bolting phenotype of the ssrp1/spt16 mutants in Arabidopsis is caused by reduced FLC expression (Lolas et al., 2010). Examining purified mammalian protein complexes in a reconstituted in vitro transcription system revealed that histone H2B ubiquitination stimulated FACT function during transcript elongation (Pavri et al., 2006). Moreover, H2B ubiquitination and SPT16 collaborate in yeast to maintain the proper nucleosome occupancy of transcribed genes that is critical for the repression of cryptic transcript initiation within the coding sequences of genes (Fleming et al., 2008), but the exact mechanism of the interplay between FACT and H2B ubiquitination is still unclear (Laribee et al., 2007). In Arabidopsis, genetic interactions were observed when double mutants affected in the expression of FACT subunits and HUB1 (an H2B monoubiquitinase) were comparatively analyzed with the corresponding single mutants and control plants. Hence, FACT and HUB1 appear to act independently in the induction of flowering, but they act synergistically in modulating leaf growth, and regarding the control of the leaf venation pattern and silique development, SSRP1/SPT16 were epistatic to HUB1 (Lolas et al., 2010). These findings indicated multiple levels of interaction between Arabidopsis FACT and HUB1 during transcript elongation and that the two factors cooperate in the regulation of some target genes, whereas they act independently at others. More detailed studies of the interaction between FACT and HUB1 (and other chromatin modifiers/remodelers) may provide insight into the interplay of factors that prime chromatin for transcription.
A novel role of SSRP1 that was recently uncovered in Arabidopsis is its involvement in parent-of-origin-specific gene expression (genomic imprinting). SSRP1 is required for DNA demethylation and for the activation/repression of parentally imprinted genes in the female central cell, which represents the progenitor cell of endosperm before fertilization (Ikeda et al., 2011). The authors propose that SSRP1 may be involved in altering the chromatin state, which facilitates DNA demethylation in the central cell by the DNA demethylase DEMETER. Interestingly, this function appears to be independent of SPT16, because, in contrast to ssrp1 mutants, no comparable effect could be observed in spt16 mutants (Ikeda et al., 2011), although the overall phenotype of ssrp1 and spt16 mutants is similar (Lolas et al., 2010). It has been reported that mammalian SSRP1 also has functions in gene regulation that are independent of SPT16, for instance, as a transcriptional coactivator (Spencer et al., 1999; Zeng et al., 2002; Li et al., 2007). Still, the majority of SSRP1 functions appear to be related to the FACT complex. In accord with that, the Arabidopsis FACT subunits were found to colocalize (Duroux et al., 2004) and the mRNA levels of SSRP1 and SPT16 throughout the plant resemble each other quite substantially (Supplemental Fig. S3). Therefore, it will be interesting to examine not only the role of the HMG-box protein SSRP1 as part of the FACT complex but also its SPT16-independent functions.
STRUCTURALLY AND FUNCTIONALLY DISTINCT HMG-BOX PROTEINS
Proteins containing HMG-box domain(s) occur in very different forms and sizes. Moreover, there are the HMGB proteins and SSRP1, which apparently occur in all eukaryotes, whereas HMG-box transcription factors seem to be specific for animals and the 3xHMG-box and ARID-HMG proteins appear to be plant specific. In addition to their structural variability, the versatile interactions with DNA and other proteins contribute to the multiple functions that HMG-box proteins adopt in the cell nucleus. Their HMG-box domains often act as architectural elements mediating (transient) alterations in chromatin structure and/or facilitating the proper three-dimensional assembly of higher order nucleoprotein complexes as a prerequisite for biological functionality. Hence, many DNA-dependent processes in the nucleus are modulated or regulated by HMG-box proteins, including the initiation and elongation stages of transcription, DNA replication and repair, chromatin remodeling, and DNA recombination (Bustin, 1999; Thomas and Travers, 2001; Reeves and Adair, 2005; Stros et al., 2007; Wegner, 2010; Formosa, 2012). Still, additional functions of HMG-box proteins are discovered also from studies in plants, such as the role of SSRP1 in genomic imprinting (Ikeda et al., 2011) or the specific association of 3xHMG-box proteins with mitotic/meiotic chromosomes (Pedersen et al., 2011). Therefore, it can be expected that research in plants will continue contributing to a better understanding of the mode of action of the different HMG-box proteins as well as to the elucidation of novel functions.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence similarity of HMG-ARID proteins.
Supplemental Figure S2. Amino acid sequence similarity of the individual HMG-box domains.
Supplemental Figure S3. Similar expression of the genes encoding the FACT subunits.
Supplemental Table S1. Plant proteins containing HMG-box domain(s).
Supplementary Material
Acknowledgments
We thank Andreas Houben (Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung Gatersleben) for providing Figure 2.
Glossary
- HMG
high mobility group
- FRAP
fluorescence recovery after photobleaching
- ARID
AT-rich interaction domain
- FACT
facilitates chromatin transcription
References
- Agresti A, Bianchi ME. (2003) HMGB proteins and gene expression. Curr Opin Genet Dev 13: 170–178 [DOI] [PubMed] [Google Scholar]
- Bianchi ME, Agresti A. (2005) HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev 15: 496–506 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Clayton DA, Shadel GS. (2006) Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell 24: 813–825 [DOI] [PubMed] [Google Scholar]
- Brewster NK, Johnston GC, Singer RA. (1998) Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J Biol Chem 273: 21972–21979 [DOI] [PubMed] [Google Scholar]
- Bustin M. (1999) Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol 19: 5237–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M, Catez F, Lim J-H. (2005) The dynamics of histone H1 function in chromatin. Mol Cell 17: 617–620 [DOI] [PubMed] [Google Scholar]
- Bustin M, Reeves R. (1996) High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol 54: 35–100 [DOI] [PubMed] [Google Scholar]
- Catez F, Hock R. (2010) Binding and interplay of HMG proteins on chromatin: lessons from live cell imaging. Biochim Biophys Acta 1799: 15–27 [DOI] [PubMed] [Google Scholar]
- Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. (2004) Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol 24: 4321–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cato L, Stott K, Watson M, Thomas JO. (2008) The interaction of HMGB1 and linker histones occurs through their acidic and basic tails. J Mol Biol 384: 1262–1272 [DOI] [PubMed] [Google Scholar]
- Cavalar M, Möller C, Offermann S, Krohn NM, Grasser KD, Peterhänsel C. (2003) The interaction of DOF transcription factors with nucleosomes depends on the positioning of the binding site and is facilitated by maize HMGB5. Biochemistry 42: 2149–2157 [DOI] [PubMed] [Google Scholar]
- Duroux M, Houben A, Růzicka K, Friml J, Grasser KD. (2004) The chromatin remodelling complex FACT associates with actively transcribed regions of the Arabidopsis genome. Plant J 40: 660–671 [DOI] [PubMed] [Google Scholar]
- Fleming AB, Kao C-F, Hillyer C, Pikaart M, Osley MA. (2008) H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell 31: 57–66 [DOI] [PubMed] [Google Scholar]
- Formosa T. (2012) The role of FACT in making and breaking nucleosomes. Biochim Biophys Acta 1819: 247–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin GH, Sanders C, Johns EW. (1973) A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur J Biochem 38: 14–19 [DOI] [PubMed] [Google Scholar]
- Grasser KD, Grill S, Duroux M, Launholt D, Thomsen MS, Nielsen BV, Nielsen HK, Merkle T. (2004) HMGB6 from Arabidopsis thaliana specifies a novel type of plant chromosomal HMGB protein. Biochemistry 43: 1309–1314 [DOI] [PubMed] [Google Scholar]
- Grasser KD, Launholt D, Grasser M. (2007a) High mobility group proteins of the plant HMGB family: dynamic chromatin modulators. Biochim Biophys Acta 1769: 346–357 [DOI] [PubMed] [Google Scholar]
- Grasser KD, Wurz A, Feix G. (1991) Isolation and characterisation of high-mobility-group proteins from maize. Planta 185: 350–355 [DOI] [PubMed] [Google Scholar]
- Grasser M, Christensen JM, Peterhänsel C, Grasser KD. (2007b) Basic and acidic regions flanking the HMG-box domain of maize HMGB1 and HMGB5 modulate the stimulatory effect on the DNA binding of transcription factor Dof2. Biochemistry 46: 6375–6382 [DOI] [PubMed] [Google Scholar]
- Grasser M, Lentz A, Lichota J, Merkle T, Grasser KD. (2006) The Arabidopsis genome encodes structurally and functionally diverse HMGB-type proteins. J Mol Biol 358: 654–664 [DOI] [PubMed] [Google Scholar]
- Hansen FT, Madsen CK, Nordland AM, Grasser M, Merkle T, Grasser KD. (2008) A novel family of plant DNA-binding proteins containing both HMG-box and AT-rich interaction domains. Biochemistry 47: 13207–13214 [DOI] [PubMed] [Google Scholar]
- Hu L, Yang X, Yuan D, Zeng F, Zhang X. (2011) GhHmgB3 deficiency deregulates proliferation and differentiation of cells during somatic embryogenesis in cotton. Plant Biotechnol J 9: 1038–1048 [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Kinoshita Y, Susaki D, Ikeda Y, Iwano M, Takayama S, Higashiyama T, Kakutani T, Kinoshita T. (2011) HMG domain containing SSRP1 is required for DNA demethylation and genomic imprinting in Arabidopsis. Dev Cell 21: 589–596 [DOI] [PubMed] [Google Scholar]
- Iwahara J, Iwahara M, Daughdrill GW, Ford J, Clubb RT. (2002) The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA. EMBO J 21: 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JY, Kwak KJ, Kang H. (2008) Expression of a high mobility group protein isolated from Cucumis sativus affects the germination of Arabidopsis thaliana under abiotic stress conditions. J Integr Plant Biol 50: 593–600 [DOI] [PubMed] [Google Scholar]
- Jerzmanowski A, Kotlinski M. (2011) Conserved chromatin structural proteins: a source of variation enabling plant-specific adaptations? New Phytol 192: 563–566 [DOI] [PubMed] [Google Scholar]
- Kiilerich B, Stemmer C, Merkle T, Launholt D, Gorr G, Grasser KD. (2008) Chromosomal high mobility group (HMG) proteins of the HMGB-type occurring in the moss Physcomitrella patens. Gene 407: 86–97 [DOI] [PubMed] [Google Scholar]
- Kim JM, To TK, Nishioka T, Seki M. (2010) Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ 33: 604–611 [DOI] [PubMed] [Google Scholar]
- Kortschak RD, Tucker PW, Saint R. (2000) ARID proteins come in from the desert. Trends Biochem Sci 25: 294–299 [DOI] [PubMed] [Google Scholar]
- Krohn NM, Stemmer C, Fojan P, Grimm R, Grasser KD. (2003) Protein kinase CK2 phosphorylates the high mobility group domain protein SSRP1, inducing the recognition of UV-damaged DNA. J Biol Chem 278: 12710–12715 [DOI] [PubMed] [Google Scholar]
- Krohn NM, Yanagisawa S, Grasser KD. (2002) Specificity of the stimulatory interaction between chromosomal HMGB proteins and the transcription factor Dof2 and its negative regulation by protein kinase CK2-mediated phosphorylation. J Biol Chem 277: 32438–32444 [DOI] [PubMed] [Google Scholar]
- Kucej M, Butow RA. (2007) Evolutionary tinkering with mitochondrial nucleoids. Trends Cell Biol 17: 586–592 [DOI] [PubMed] [Google Scholar]
- Kwak KJ, Kim JY, Kim YO, Kang H. (2007) Characterization of transgenic Arabidopsis plants overexpressing high mobility group B proteins under high salinity, drought or cold stress. Plant Cell Physiol 48: 221–231 [DOI] [PubMed] [Google Scholar]
- Laribee RN, Fuchs SM, Strahl BD. (2007) H2B ubiquitylation in transcriptional control: a FACT-finding mission. Genes Dev 21: 737–743 [DOI] [PubMed] [Google Scholar]
- Launholt D, Merkle T, Houben A, Schulz A, Grasser KD. (2006) Arabidopsis chromatin-associated HMGA and HMGB use different nuclear targeting signals and display highly dynamic localization within the nucleus. Plant Cell 18: 2904–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zeng SX, Landais I, Lu H. (2007) Human SSRP1 has Spt16-dependent and -independent roles in gene transcription. J Biol Chem 282: 6936–6945 [DOI] [PubMed] [Google Scholar]
- Lichota J, Grasser KD. (2001) Differential chromatin association and nucleosome binding of the maize HMGA, HMGB, and SSRP1 proteins. Biochemistry 40: 7860–7867 [DOI] [PubMed] [Google Scholar]
- Lildballe DL, Pedersen DS, Kalamajka R, Emmersen J, Houben A, Grasser KD. (2008) The expression level of the chromatin-associated HMGB1 protein influences growth, stress tolerance, and transcriptome in Arabidopsis. J Mol Biol 384: 9–21 [DOI] [PubMed] [Google Scholar]
- Lolas IB, Himanen K, Grønlund JT, Lynggaard C, Houben A, Melzer M, Van Lijsebettens M, Grasser KD. (2010) The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J 61: 686–697 [DOI] [PubMed] [Google Scholar]
- Müller S, Ronfani L, Bianchi ME. (2004) Regulated expression and subcellular localization of HMGB1, a chromatin protein with a cytokine function. J Intern Med 255: 332–343 [DOI] [PubMed] [Google Scholar]
- Ohta S, Wood L, Bukowski-Wills J-C, Rappsilber J, Earnshaw WC. (2011) Building mitotic chromosomes. Curr Opin Cell Biol 23: 114–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Wu W-H, Lane WS, Hampsey M, Reinberg D. (1999) The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400: 284–288 [DOI] [PubMed] [Google Scholar]
- Patsialou A, Wilsker D, Moran E. (2005) DNA-binding properties of ARID family proteins. Nucleic Acids Res 33: 66–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. (2006) Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125: 703–717 [DOI] [PubMed] [Google Scholar]
- Pedersen DS, Coppens F, Ma L, Antosch M, Marktl B, Merkle T, Beemster GTS, Houben A, Grasser KD. (2011) The plant-specific family of DNA-binding proteins containing three HMG-box domains interacts with mitotic and meiotic chromosomes. New Phytol 192: 577–589 [DOI] [PubMed] [Google Scholar]
- Pedersen DS, Merkle T, Marktl B, Lildballe DL, Antosch M, Bergmann T, Tönsing K, Anselmetti D, Grasser KD. (2010) Nucleocytoplasmic distribution of the Arabidopsis chromatin-associated HMGB2/3 and HMGB4 proteins. Plant Physiol 154: 1831–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M, Más P. (2007) A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19: 2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. (2010) Nuclear functions of the HMG proteins. Biochim Biophys Acta 1799: 3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R, Adair JE. (2005) Role of high mobility group (HMG) chromatin proteins in DNA repair. DNA Repair (Amst) 4: 926–938 [DOI] [PubMed] [Google Scholar]
- Reinberg D, Sims RJ., III (2006) De FACTo nucleosome dynamics. J Biol Chem 281: 23297–23301 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Ritt C, Grimm R, Fernández S, Alonso JC, Grasser KD. (1998) Basic and acidic regions flanking the HMG domain of maize HMGa modulate the interactions with DNA and the self-association of the protein. Biochemistry 37: 2673–2681 [DOI] [PubMed] [Google Scholar]
- Röttgers K, Krohn NM, Lichota J, Stemmer C, Merkle T, Grasser KD. (2000) DNA-interactions and nuclear localisation of the chromosomal HMG domain protein SSRP1 from maize. Plant J 23: 395–405 [DOI] [PubMed] [Google Scholar]
- Schrumpfová PP, Fojtová M, Mokroš P, Grasser KD, Fajkus J. (2011) Role of HMGB proteins in chromatin dynamics and telomere maintenance in Arabidopsis thaliana. Curr Protein Pept Sci 12: 105–111 [DOI] [PubMed] [Google Scholar]
- Schultz TF, Spiker S, Quatrano RS. (1996) Histone H1 enhances the DNA binding activity of the transcription factor EmBP-1. J Biol Chem 271: 25742–25745 [DOI] [PubMed] [Google Scholar]
- Singer RA, Johnston GC. (2004) The FACT chromatin modulator: genetic and structure/function relationships. Biochem Cell Biol 82: 419–427 [DOI] [PubMed] [Google Scholar]
- Spencer JA, Baron MH, Olson EN. (1999) Cooperative transcriptional activation by serum response factor and the high mobility group protein SSRP1. J Biol Chem 274: 15686–15693 [DOI] [PubMed] [Google Scholar]
- Spiker S. (1984) High-mobility group chromosomal proteins of wheat. J Biol Chem 259: 12007–12013 [PubMed] [Google Scholar]
- Stemmer C, Fernández S, Lopez G, Alonso JC, Grasser KD. (2002) Plant chromosomal HMGB proteins efficiently promote the bacterial site-specific β-mediated recombination in vitro and in vivo. Biochemistry 41: 7763–7770 [DOI] [PubMed] [Google Scholar]
- Stros M. (2010) HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta 1799: 101–113 [DOI] [PubMed] [Google Scholar]
- Stros M, Launholt D, Grasser KD. (2007) The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Sci 64: 2590–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JO, Stott K. (2012) H1 and HMGB1: modulators of chromatin structure. Biochem Soc Trans 40: 341–346 [DOI] [PubMed] [Google Scholar]
- Thomas JO, Travers AA. (2001) HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem Sci 26: 167–174 [DOI] [PubMed] [Google Scholar]
- Thomsen MS, Franssen L, Launholt D, Fojan P, Grasser KD. (2004) Interactions of the basic N-terminal and the acidic C-terminal domains of the maize chromosomal HMGB1 protein. Biochemistry 43: 8029–8037 [DOI] [PubMed] [Google Scholar]
- Wegner M. (2010) All purpose Sox: the many roles of Sox proteins in gene expression. Int J Biochem Cell Biol 42: 381–390 [DOI] [PubMed] [Google Scholar]
- Wilsker D, Patsialou A, Dallas PB, Moran E. (2002) ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ 13: 95–106 [PubMed] [Google Scholar]
- Winkler DD, Luger K. (2011) The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J Biol Chem 286: 18369–18374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmeyer J, Joss L, Formosa T. (1999) Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase α. Biochemistry 38: 8961–8971 [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhang W, Pwee K-H, Kumar PP. (2003) Rice HMGB1 protein recognizes DNA structures and bends DNA efficiently. Arch Biochem Biophys 411: 105–111 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. (1997) Dof DNA-binding domains of plant transcription factors contribute to multiple protein-protein interactions. Eur J Biochem 250: 403–410 [DOI] [PubMed] [Google Scholar]
- Yang H, Tracey KJ. (2010) Targeting HMGB1 in inflammation. Biochim Biophys Acta 1799: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng SX, Dai M-S, Keller DM, Lu H. (2002) SSRP1 functions as a co-activator of the transcriptional activator p63. EMBO J 21: 5487–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Chen T, Zhu M, Fang Q, Kang H, Hong Z, Zhang Z. (2008) A novel ARID DNA-binding protein interacts with SymRK and is expressed during early nodule development in Lotus japonicus. Plant Physiol 148: 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.