Abstract
Protein-induced DNA bending is an important element in the structure of many protein-DNA complexes, including those involved in replication, transcription, and recombination. To understand these structures, the path followed by the DNA in each complex must be established. We have generated an empirical relation between the degree of bending and the altered electrophoretic mobility in polyacrylamide gels that allows estimation of protein-induced bends. This technique has been used to analyze 17 different protein-DNA complexes formed by six proteins including the four proteins involved in lambda site-specific recombination. The simplicity of this technique should make it useful in estimating angles for the construction of models of protein-DNA complexes and readily applicable to many systems where questions of higher-order structure are important for understanding function.
Full text
PDF


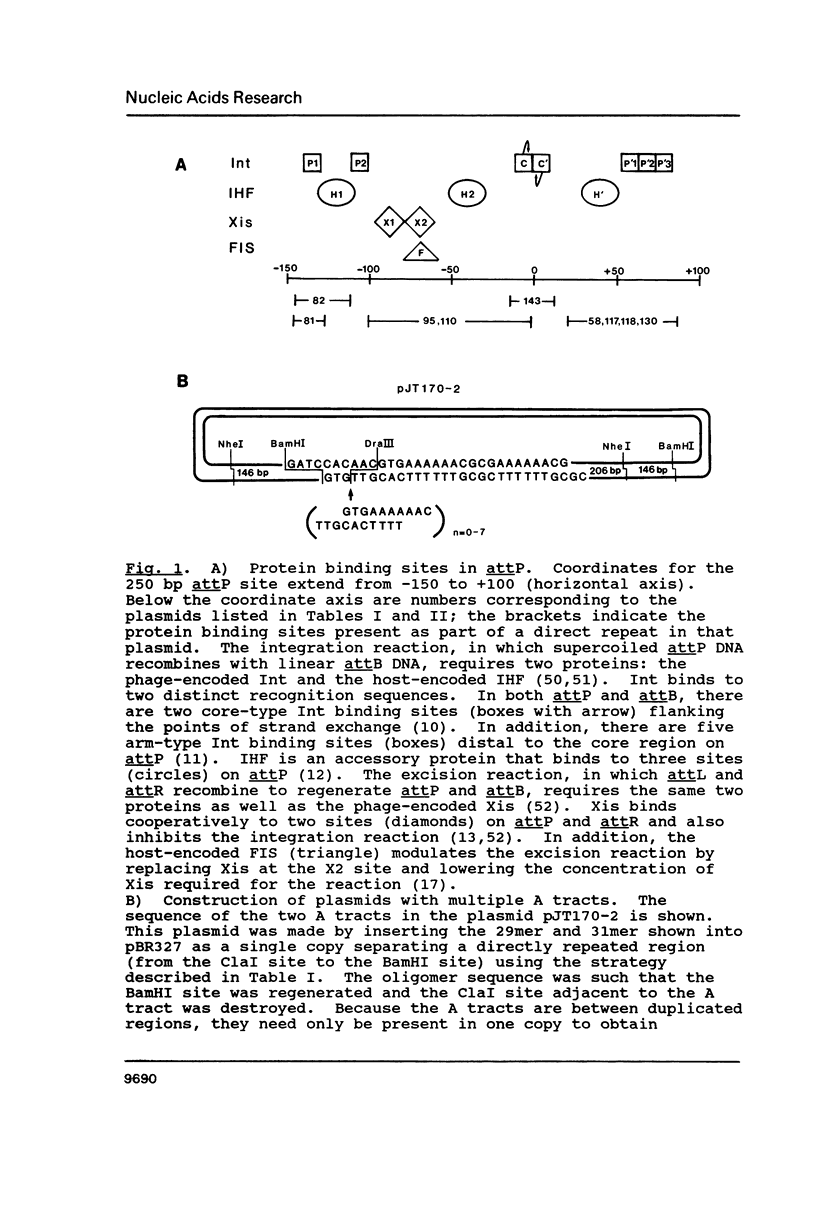







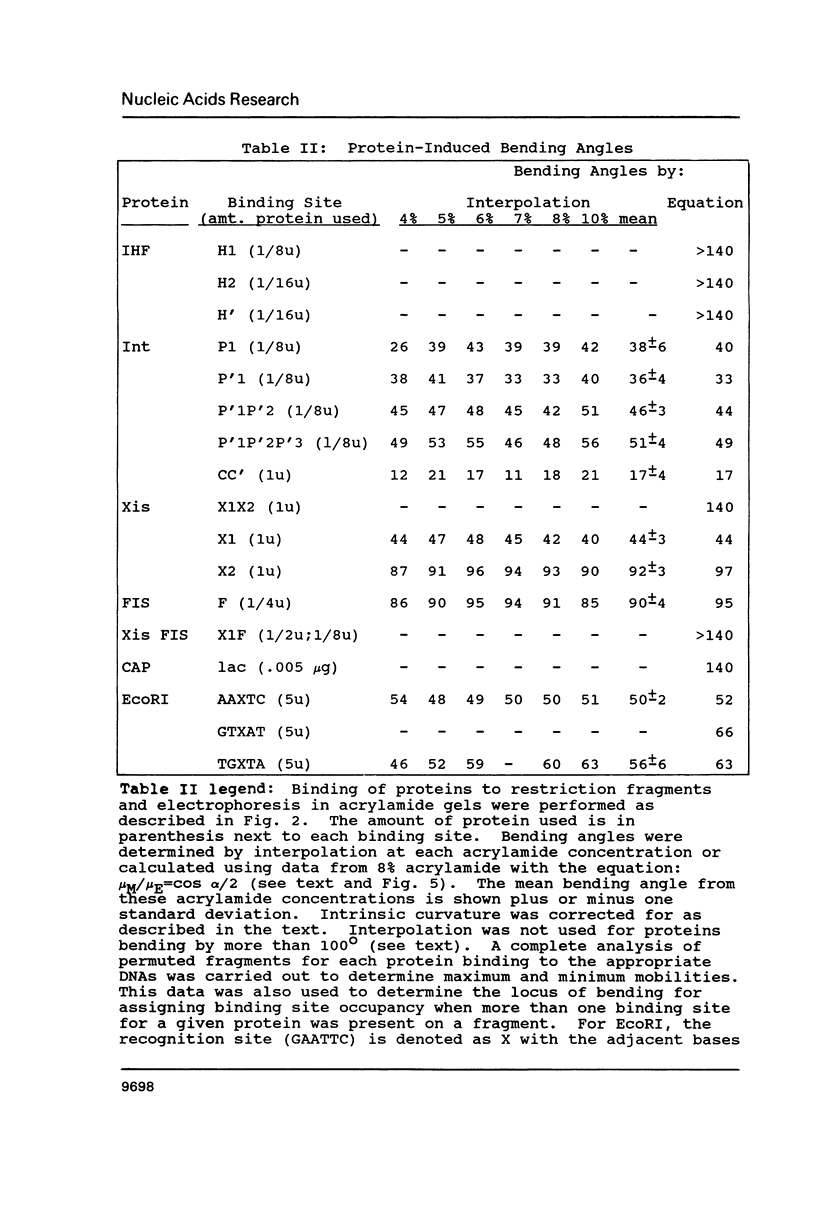







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abremski K., Gottesman S. Purification of the bacteriophage lambda xis gene product required for lambda excisive recombination. J Biol Chem. 1982 Aug 25;257(16):9658–9662. [PubMed] [Google Scholar]
- Bauer C. E., Hesse S. D., Gumport R. I., Gardner J. F. Mutational analysis of integrase arm-type binding sites of bacteriophage lambda. Integration and excision involve distinct interactions of integrase with arm-type sites. J Mol Biol. 1986 Dec 5;192(3):513–527. doi: 10.1016/0022-2836(86)90273-1. [DOI] [PubMed] [Google Scholar]
- Better M., Lu C., Williams R. C., Echols H. Site-specific DNA condensation and pairing mediated by the int protein of bacteriophage lambda. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5837–5841. doi: 10.1073/pnas.79.19.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Burlingame R. W., Love W. E., Wang B. C., Hamlin R., Nguyen H. X., Moudrianakis E. N. Crystallographic structure of the octameric histone core of the nucleosome at a resolution of 3.3 A. Science. 1985 May 3;228(4699):546–553. doi: 10.1126/science.3983639. [DOI] [PubMed] [Google Scholar]
- Bushman W., Thompson J. F., Vargas L., Landy A. Control of directionality in lambda site specific recombination. Science. 1985 Nov 22;230(4728):906–911. doi: 10.1126/science.2932798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman W., Yin S., Thio L. L., Landy A. Determinants of directionality in lambda site-specific recombination. Cell. 1984 Dec;39(3 Pt 2):699–706. doi: 10.1016/0092-8674(84)90477-x. [DOI] [PubMed] [Google Scholar]
- Calladine C. R., Drew H. R., McCall M. J. The intrinsic curvature of DNA in solution. J Mol Biol. 1988 May 5;201(1):127–137. doi: 10.1016/0022-2836(88)90444-5. [DOI] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984 Dec;39(3 Pt 2):707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- Diekmann S., Wang J. C. On the sequence determinants and flexibility of the kinetoplast DNA fragment with abnormal gel electrophoretic mobilities. J Mol Biol. 1985 Nov 5;186(1):1–11. doi: 10.1016/0022-2836(85)90251-7. [DOI] [PubMed] [Google Scholar]
- Echols H. Multiple DNA-protein interactions governing high-precision DNA transactions. Science. 1986 Sep 5;233(4768):1050–1056. doi: 10.1126/science.2943018. [DOI] [PubMed] [Google Scholar]
- Gardner J. F., Nash H. A. Role of Escherichia coli IHF protein in lambda site-specific recombination. A mutational analysis of binding sites. J Mol Biol. 1986 Sep 20;191(2):181–189. doi: 10.1016/0022-2836(86)90255-x. [DOI] [PubMed] [Google Scholar]
- Griffith J. D., Nash H. A. Genetic rearrangement of DNA induces knots with a unique topology: implications for the mechanism of synapsis and crossing-over. Proc Natl Acad Sci U S A. 1985 May;82(10):3124–3128. doi: 10.1073/pnas.82.10.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J., Bleyman M., Rauch C. A., Kitchin P. A., Englund P. T. Visualization of the bent helix in kinetoplast DNA by electron microscopy. Cell. 1986 Aug 29;46(5):717–724. doi: 10.1016/0092-8674(86)90347-8. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Evidence for the existence of stable curvature of DNA in solution. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4632–4636. doi: 10.1073/pnas.81.15.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull G. F., Noble S. M., Grindley N. D. The gamma delta resolvase induces an unusual DNA structure at the recombinational crossover point. Cell. 1987 Apr 10;49(1):103–110. doi: 10.1016/0092-8674(87)90760-4. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Crothers D. M. Calibration of DNA curvature and a unified description of sequence-directed bending. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1763–1767. doi: 10.1073/pnas.85.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Levene S. D., Crothers D. M. A computer graphics study of sequence-directed bending in DNA. J Biomol Struct Dyn. 1983 Oct;1(2):429–435. doi: 10.1080/07391102.1983.10507452. [DOI] [PubMed] [Google Scholar]
- Levene S. D., Wu H. M., Crothers D. M. Bending and flexibility of kinetoplast DNA. Biochemistry. 1986 Jul 15;25(14):3988–3995. doi: 10.1021/bi00362a003. [DOI] [PubMed] [Google Scholar]
- Liu-Johnson H. N., Gartenberg M. R., Crothers D. M. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986 Dec 26;47(6):995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- Lumpkin O. J., Déjardin P., Zimm B. H. Theory of gel electrophoresis of DNA. Biopolymers. 1985 Aug;24(8):1573–1593. doi: 10.1002/bip.360240812. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nash H. A. Integration and excision of bacteriophage lambda: the mechanism of conservation site specific recombination. Annu Rev Genet. 1981;15:143–167. doi: 10.1146/annurev.ge.15.120181.001043. [DOI] [PubMed] [Google Scholar]
- Nash H. A. Integrative recombination of bacteriophage lambda DNA in vitro. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1072–1076. doi: 10.1073/pnas.72.3.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A. Purification and properties of the Escherichia coli protein factor required for lambda integrative recombination. J Biol Chem. 1981 Sep 10;256(17):9246–9253. [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Pargellis C. A., Nunes-Düby S. E., de Vargas L. M., Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988 Jun 5;263(16):7678–7685. [PubMed] [Google Scholar]
- Pollock T. J., Nash H. A. Knotting of DNA caused by a genetic rearrangement. Evidence for a nucleosome-like structure in site-specific recombination of bacteriophage lambda. J Mol Biol. 1983 Oct 15;170(1):1–18. doi: 10.1016/s0022-2836(83)80224-1. [DOI] [PubMed] [Google Scholar]
- Prentki P., Chandler M., Galas D. J. Escherichia coli integration host factor bends the DNA at the ends of IS1 and in an insertion hotspot with multiple IHF binding sites. EMBO J. 1987 Aug;6(8):2479–2487. doi: 10.1002/j.1460-2075.1987.tb02529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Richet E., Abcarian P., Nash H. A. The interaction of recombination proteins with supercoiled DNA: defining the role of supercoiling in lambda integrative recombination. Cell. 1986 Sep 26;46(7):1011–1021. doi: 10.1016/0092-8674(86)90700-2. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Robertson C. A., Nash H. A. Bending of the bacteriophage lambda attachment site by Escherichia coli integration host factor. J Biol Chem. 1988 Mar 15;263(8):3554–3557. [PubMed] [Google Scholar]
- Ross W., Landy A. Bacteriophage lambda int protein recognizes two classes of sequence in the phage att site: characterization of arm-type sites. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7724–7728. doi: 10.1073/pnas.79.24.7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Landy A. Patterns of lambda Int recognition in the regions of strand exchange. Cell. 1983 May;33(1):261–272. doi: 10.1016/0092-8674(83)90355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuey D. J., Parker C. S. Bending of promoter DNA on binding of heat shock transcription factor. Nature. 1986 Oct 2;323(6087):459–461. doi: 10.1038/323459a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Measurement of DNA length by gel electrophoresis. Anal Biochem. 1979 Dec;100(2):319–323. doi: 10.1016/0003-2697(79)90235-5. [DOI] [PubMed] [Google Scholar]
- Spengler S. J., Stasiak A., Cozzarelli N. R. The stereostructure of knots and catenanes produced by phage lambda integrative recombination: implications for mechanism and DNA structure. Cell. 1985 Aug;42(1):325–334. doi: 10.1016/s0092-8674(85)80128-8. [DOI] [PubMed] [Google Scholar]
- Stellwagen N. C. Anomalous electrophoresis of deoxyribonucleic acid restriction fragments on polyacrylamide gels. Biochemistry. 1983 Dec 20;22(26):6186–6193. doi: 10.1021/bi00295a023. [DOI] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Moitoso de Vargas L., Koch C., Kahmann R., Landy A. Cellular factors couple recombination with growth phase: characterization of a new component in the lambda site-specific recombination pathway. Cell. 1987 Sep 11;50(6):901–908. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Waechter-Brulla D., Gumport R. I., Gardner J. F., Moitoso de Vargas L., Landy A. Mutations in an integration host factor-binding site: effect on lambda site-specific recombination and regulatory implications. J Bacteriol. 1986 Dec;168(3):1343–1351. doi: 10.1128/jb.168.3.1343-1351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., de Vargas L. M., Skinner S. E., Landy A. Protein-protein interactions in a higher-order structure direct lambda site-specific recombination. J Mol Biol. 1987 Jun 5;195(3):481–493. doi: 10.1016/0022-2836(87)90177-x. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L. E., Trifonov E. N. Estimation of wedge components in curved DNA. Nature. 1987 Apr 16;326(6114):720–722. doi: 10.1038/326720a0. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L., Bodner M., Trifonov E. N., Choder M. Curved DNA: design, synthesis, and circularization. Proc Natl Acad Sci U S A. 1986 Feb;83(4):862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwicker J., Engelman B. P., Steitz T. A. Electrostatic calculations and model-building suggest that DNA bound to CAP is sharply bent. Proteins. 1987;2(4):283–289. doi: 10.1002/prot.340020404. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Yin S., Bushman W., Landy A. Interaction of the lambda site-specific recombination protein Xis with attachment site DNA. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1040–1044. doi: 10.1073/pnas.82.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Direct evidence for DNA bending at the lambda replication origin. Science. 1987 Apr 24;236(4800):416–422. doi: 10.1126/science.2951850. [DOI] [PubMed] [Google Scholar]





