Abstract
CHIP, the carboxyl-terminus of Hsp70 interacting protein, is both an E3 ubiquitin ligase and an Hsp70 co-chaperone and is implicated in the degradation of cytosolic quality control and numerous disease substrates. CHIP has been shown to monitor the folding status of the CFTR protein, and we have successfully reconstituted this activity using a recombinant CFTR fragment consisting of the cytosolic NBD1 and R domains. We have found that efficient ubiquitination of substrates requires chaperone activity to either deliver the substrate to CHIP or to maintain the substrate in a ubiquitination-competent conformation. This chaperone activity can be provided by the Hsp70/Hsp40 molecular chaperone system as seen in the NBD1–R ubiquitination assay. Alternatively, heat treatment of CHIP can activate its own innate substrate-binding activity and allow for efficient ubiquitination of model substrates, such as denatured luciferase. Here, we describe methods for purifying the recombinant proteins necessary for in vitro reconstitution of CHIP ubiquitin ligase activity, as well as two methods used to monitor CHIP ligase activity. One method allows for the measurement of the Hsp70- and Hsp40-dependent CHIP activity while the other measures the Hsp40- and Hsp70-independent activity of heat-activated CHIP.
Keywords: Carboxy-terminus of Hsp70 interacting protein, Ubiquitination, Assay, In vitro reconstitution, Hsp40, E3 ubiquitin ligase
1. Introduction
CHIP, the carboxyl-terminus of Hsp70 interacting protein, is both an E3 ubiquitin ligase and an Hsp70 co-chaperone which has been implicated in the degradation of cytosolic quality control and numerous disease substrates (1–4). CHIP contains a TPR domain, which allows for interaction with the C-terminus of Hsp70, and a U-box domain, which is responsible for the E3 ubiquitination activity (5, 6). Chip has both Hsp70-dependent and -independent activities, both of which are important for maintaining cellular HSF activation in a U-box-independent manner (8) as well as the turnover of Hsp70 after a heat-shock response (4), thereby playing a crucial role in tuning the stress response. The assays described in this chapter allow for the biochemical measurement of both Hsp70-dependent and -independent ubiquitination activity of CHIP. These assays can also be adapted to test the ability of CHIP to ubiquitinate specific substrates of interest or to look for modulators of CHIP activity.
2. Materials
2.1. Protein Purifications
LB (Luria Broth): 10% (w/v) tryptone, 5% (w/v) yeast extract, 10% (w/v) NACL (Fisher, Cat. # BP1426-2).
IPTG [Isopropyl β-d-thiogalactopyranoside (Fisher, Cat. # BP1755-10)].
Lysozyme (Sigma, Cat. # L-6876).
Glutathione Sepharose (GE, Cat. # 17-5132-01)).
Sarkosyl (Fisher, Cat. # BP234-500).
Glutathione (GSH) (Sigma, Cat. # G6529-25G).
HA column (Bio-Rad, Cat. # 732-0081).
HQ column (Bio-Rad, Cat. # 732-0026).
Talon metal affinity resin (Clone Tech, Cat. # 635502).
STE buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 150 mM NaCl).
Glutathione elution buffer (50 mM Tris–HCl, pH 8.0, 20 mM GSH).
Buffer A (20 mM Hepes, pH 7.4, 20 mM NaCl).
Buffer B (20 mM Hepes, pH 7.4, 500 mM NaCl).
Buffer C (10 mM potassium phosphate buffer, pH 7.4).
Buffer D (500 mM potassium phosphate buffer, pH 7.4).
Buffer E (20 mM Hepes, 150 mM NaCl, pH 7.4).
Buffer F (20 mM Tris–HCl, pH 8.0, 150 mM NaCl).
Buffer G (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 150 mM imidazole).
Dialysis tubing (12,000–14,000 molecular weight cutoff).
Complete Protease Inhibitor (PI) Tablets (Roche).
PMSF (stock solution made at 100 mM in ethanol).
2× SDS-PAGE sample buffer [(100 mM Tris base, pH 6.8, 4% SDS, 20% glycerol, and Coomassie blue (2 mg/50 ml or to desired darkness)].
2.2. Ubiquitination Reaction
Reaction buffer (20 mM Hepes, pH 7.4, 50 mM NaCl, 5 mM MgCl2, 2.5 mM ATP, 2 mM DTT).
Bovine ubiquitin.
Rabbit E1.
Purified recombinant human UbcH5a protein.
Purified recombinant human CHIP protein.
Purified recombinant human Hsp70 protein.
Purified recombinant human Hsp40 (Hdj-2) protein.
Luciferase (Promega, 14.4 mg/ml).
Gdm–HCl (6 M).
2× SDS-PAGE sample buffer.
SDS-PAGE gel.
Western blot transfer buffer (20 mM Tris–HCl, 150 mM glycine, 20% methanol, 0.038% SDS).
Nitrocellulose.
PBS-T (1% Triton) (for 1 l PBS: 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, 0.24 g of KH2PO4).
Blocking solution (10% nonfat milk in PBS-T).
Primary antibody for immunoblotting (α-CFTR R-domain (mouse, R&D Systems) or α-luciferase (goat, Chemicon International) (diluted 1:1,000 in 1% nonfat milk made in PBS-T).
Secondary horseradish peroxidise-conjugated goat anti-mouse (Bio-Rad) or rabbit anti-goat antibody (Chemicon International) (diluted 1:3,000 in 1% nonfat milk made in PBS-T).
ECL (GE Healthcare).
Ubiquitination buffer (20 mM Hepes, pH 7.4, 50 mM NaCl, M bovine ubiq- 5 mM MgCl2, 2.5 mM ATP, 2 mM DTT, 10 μM bovine ubiquitin) (see Note 13).
Recombinant firefly luciferase (Promega).
3. Methods
The methods described herein include the purifications of recombinant proteins necessary for in vitro reconstitution of CHIP ubiquitin ligase activity, as well as two methods used to monitor CHIP ligase activity. One method allows for the measurement of the Hsp70- and Hsp40-dependent CHIP activity (5, 6) while the other measures the Hsp40- and Hsp70-independent activity of heat-activated CHIP (5). CHIP has been shown to monitor the folding status of the CFTR protein, and we have successfully reconstituted this activity using a recombinant CFTR fragment consisting of the cytosolic NBD1 and R domain (5). We have found that efficient ubiquitination of substrates requires chaperone activity to either deliver the substrate to CHIP or to maintain the substrate in a ubiquitination-competent conformation. This chaperone activity can be provided by the Hsp70/Hsp40 molecular chaperone system as seen in the NBD1-R ubiquitination assay. Alternatively, heat treatment of CHIP can activate its own innate substrate-binding activity and allow for efficient ubiquitination of model substrates, such as denatured luciferase (9).
3.1. Protein Purification
3.1.1. Purification of GST–NBD1–R Fusion Protein from E. coli Strain BL21(DE3)
Day 1: Prepare a 50-ml overnight culture by adding 100 μg/ml ampicillin and inoculating with BL21(DE3) E. coli harbouring the GST–NBD1–R expression vector. Incubate overnight at 37°C, shaking.
Day 2: Seed 20 ml of overnight culture into 600 ml LB with 100 μg/ml ampicillin. Grow at 37°C until the culture reaches an OD600 of 0.6 (see Note 1).
Induce with 0.2 mM of IPTG. Grow for 16 h at 30°C (see Note 1).
Harvest cells by centrifugation at 3,000 × g for 10 min at 4°C. Decant media (see Note 2).
Resuspend pellet in 10 ml of ice-cold STE buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 150 mM NaCl) with fresh PI (1×) and PMSF (1 mM).
Add 100 μl of freshly prepared lysozyme solution (10 mg/ml in water) and incubate on ice for 30 min.
Just before sonication, add 100 μl of 1 M DTT and 1.4 ml of 10% sarkosyl. Mix and sonicate for four times, each for a period of 15 s (see Note 3).
Centrifuge at 16,000 rpm for 20 min to pellet debris (see Note 4).
Transfer supernatant to a 50-ml conical tube and discard the pellet. Add 4 ml of 10% Triton X-100 and bring up to a final volume of 20 ml with STE buffer. The final concentrations of sarkosyl and Triton X-100 are 0.7 and 2%, respectively. Incubate at room temperature for 30 min.
Pour the lysate into a tube with 1 ml of prepared glutathione sepharose (50% slurry) in PBS. Incubate at 4°C for 1 h with rotation.
Wash the beads three times with 30 ml of cold PBST.
Remove last portion of PBS-T and add 1 ml of elution buffer (50 mM Tris–HCl, pH 8.0, 20 mM GSH).
Incubate at 4°C for 10 min with rotation.
Centrifuge at 6,000 rpm for 1 min, remove supernatant to 1.6-ml tube (see Note 4).
Determine the concentration of protein with protein assay, such as Bio-Rad DC Assay (see Note 5).
Add DTT to final concentration of 10 mM. Aliquot protein and store at −80°C (see Note 6).
3.1.2. Purification of Recombinant Human Hsp70, Hdj-2, or CHIP Protein from E. coli Strain BL21(DE3)
Day 1: Prepare a 50-ml overnight culture by adding 100 μg/ml ampicillin and inoculating with BL21(DE3) E. coli harbouring the appropriate expression vector. Incubate overnight at 37°C, shaking.
Day 2: Seed 20 ml of overnight culture into 600 ml LB with 100 μg/ml ampicillin. Grow at 37°C until the culture reaches an OD600 of 0.6 (see Note 1).
Induce with 0.2 mM of IPTG. Grow for 3 h at 30°C.
Centrifuge at 3,000 × g at 4°C for 10 min. Decant media (see Note 2).
Resuspend pellet in 30 ml of cold buffer A (20 mM Hepes, pH 7.4, 20 mM NaCl), put on ice, and sonicate ten times for a period of 20 s each time (see Note 3).
Centrifuge at 12,000 rpm for 10 min to pellet debris (see Note 4).
Load supernatant on HQ column (Bio-Rad), and then wash column with buffer A for 4-column volumes.
Perform gradient elution with buffer A and buffer B (0 → 100% buffer B) (20 mM Hepes, pH 7.4, 500 mM NaCl). Collect 1.3-ml fractions and run SDS-PAGE on each fraction (see Note 7).
Combine fractions that contain your protein of interest and dialyze in buffer C (10 mM KH2PO4, pH 7.4) (see Note 7). Alternatively, you can dilute the combined fractions tenfold in buffer C.
Load dialyzed protein on HA column (Bio-Rad), wash column with 4-column volumes of buffer C.
Perform gradient elution with buffer C and buffer D (0 → 100% buffer D) (500 mM KH2PO4), and collect 1.3-ml fractions.
Run SDS-PAGE to check protein purity, and combine appropriate fractions (see Note 7). Dialyze combined fractions in buffer E (20 mM Hepes, 150 mM NaCl, pH 7.4) (see Note 8).
Determine the concentration of protein with protein assay, such as Bio-Rad DC Assay (see Note 4).
Aliquot protein and store at −80°C (see Note 6).
3.1.3. Purification of Recombinant His-UbcH5a Protein from E. coli Strain BL21(DE3)
Collect bacteria pellets as above in Subheading 3.1.2 (steps 1–4).
Resuspend pellet in 30 ml of ice-cold buffer F (20 mM Tris–HCl, pH 8.0, 150 mM NaCl). Sonicate on ice ten times for a period of 20 s each time (see Note 3).
Centrifuge at 12,000 rpm for 10 min to pellet debris and then collect supernatant (see Note 4).
Load supernatant on column containing Talon affinity residue (Covance). Wash column with buffer F (20 mM Tris–HCl, pH 8.0, 150 mM NaCl) for 4-column volumes.
Perform gradient elution with buffer F and buffer G (0 → 100% buffer G) (20 mM Tris–HCl, pH 8, 150 mM NaCl, 150 mM imidazole), and collect 1.3-ml fractions.
Run SDS-PAGE to check protein purity, and combine appropriate fractions. Dialyze in buffer E (20 mM Hepes, 150 mM NaCl, pH 7.4) (see Note 8).
Determine the concentration of protein with protein assay, such as Bio-Rad DC Assay (see Note 5).
Aliquot protein and store at −80°C (see Note 6).
3.1.4. In Vitro Reconstitution of the Hsp70/Hsp40-Dependent Ubiquitination Activity of the E3 Ubiquitin Ligase, CHIP
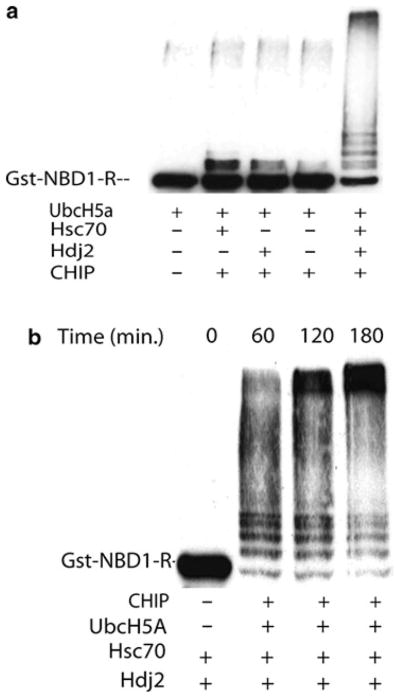
This assay reconstitutes the ubiquitination of a CFTR fragment (NBD1-R) and requires the action of the Hsp70 and Hsp40 molecular chaperones. The Hsp40/Hsp70 chaperone pair is thought to deliver substrate to CHIP. CHIP binds the Hsp70 chaperone through its TPR domains, and then promotes polyubiquitination of the substrate through action of the U-box domain (Fig. 1).
Fig. 1.
Hsp40/Hsp70-dependent ubiquitination of GST–NBD1–R. (a) Efficient ubiquitination of GST–NBD1–R is only achieved in the presence of the Hsp40 and Hsp70 chaperone pair. All reactions contain E1 enzyme and E2 (UbcH5a) while the reaction in lane 1 is lacking the E3 CHIP as a negative control. (b) Time course of ubiquitination. A time-dependent increase in ubiquitination can be observed over the course of 3 h. Lane 1 represents the negative control in which CHIP and UbcH5a are not added to the reaction mixture.
Combine appropriate amounts of a 10× reaction buffer (200 mM Hepes, pH 7.4, 500 mM NaCl, 50 mM MgCl2, 25 mM ATP, and 20 mM DTT) (see Notes 9 and 13) with 10 μM bovine ubiquitin, 0.1 μM rabbit E1, and 1 μM GST–NBD1–R.
Add purified E2 (4 μM UbcH5a), E3 (3 μM CHIP), chaperones (4 μM of Hdj2 and 2 μM of Hsp70), or buffer controls to appropriate samples (see Note 11). Make sure that proteins are concentrated enough so that total reaction volume can be kept at 25 μl (see Note 5).
Incubate reaction at 37°C for 2 h.
Terminate reaction by adding 25 μl of 2× SDS sample buffer with BME (see Note 12) to each 25 μl reaction.
Load 25 μl of reaction on 7% SDS-PAGE gels and after electrophoresis, transfer to nitrocellulose membranes.
Block membranes with blocking solution for 1 h at room temperature with gentle agitation.
Rinse membranes with PBS-T to remove the blocking solution.
Incubate membranes with αR domain antibody (1:1,000 dilution in 1% nonfat milk) for 1 h at room temperature with gentle agitation.
Wash membranes three times for 5 min each with PBS-T.
Add HRP-conjugated secondary antibody (1:3,000 dilution in 1% nonfat milk) and incubate for 1 h at room temperature with gentle agitation.
Wash membranes three times for 5 min each with PBS-T.
Add ECL and expose to film.
3.1.5. In Vitro Reconstitution of the Hsp70- and Hsp40-Independent CHIP Ubiquitin Ligase Activity
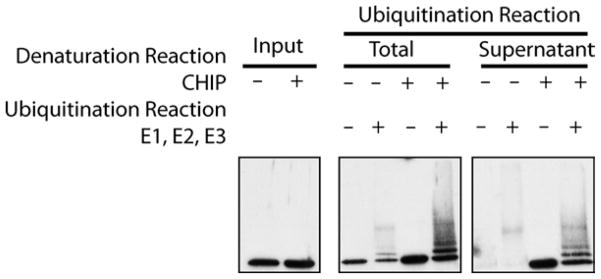
Recently, we have demonstrated that CHIP can act as a chaperone by binding to denatured substrates and preventing their aggregation (9). This activity of CHIP is enhanced after heat shock, as is seen for many other chaperone proteins. The following assay allows one to test for the ability of CHIP to ubiquitinate substrates independent of the Hsp40/Hsp70 chaperone pair, can be used to test the ability of proteins to modulate CHIP’s activity, or can be adapted to directly test the ability of CHIP to ubiquitinate a specific substrate. In this assay, when luciferase is heat denatured in the presence of CHIP, the CHIP protein is able to prevent the aggregation of the luciferase which maintains the luciferase in a ubiquitination-competent state. When the luciferase is denatured in the absence of the CHIP protein, the majority aggregates, and is not ubiquitinated (Fig. 2).
Fig. 2.
Hsp40/Hsp70-independent ubiquitination of denatured luciferase. When CHIP is present during the reaction in which luciferase is heat denaturated by incubation at 42°C for 15 min, the luciferase is maintained in a soluble, ubiquitination-competent state. The total ubiquitination reaction (lanes 3–6) represents the amount of ubiquitination that occurs when CHIP is included or excluded from the denaturation reaction. In lanes 7–10, the denaturation reaction is spun at 20,000 rpm for 10 min at 4°C and the supernatant is removed to a different tube. Ubiquitination enzymes are then added to this supernatant fraction. As apparent from lanes 7 and 8, when luciferase is denatured in the absence of CHIP, the majority aggregates, pellets after centrifugation, and is therefore not detectable in the ubiquitination reactions performed on the supernatant fraction alone. Alternatively, when CHIP is present during the initial denaturation reaction (lanes 9 and 10 ), the CHIP is maintained soluble and efficiently ubiquitinated in the presence of the ubiquitination enzymes.
Make 50 μl of a luciferase cocktail containing the following: 10 mM ATP, 20 mM Hepes, pH 7.4, 50 mM NaCl, 5 mM g/ml luciferase. MgCl2, 10 mM DTT, and 28 μg/ml luciferase.
Make two different denaturation buffers: A: 40 μl of dialysis buffer from CHIP purification (see Note 8) and B: 40 μl of CHIP diluted in dialysis buffer to a concentration of 0.25 mg/ml.
Add 10 μl of the luciferase cocktail to denaturation reaction A or denaturation reaction B to make your final volume 50 μl. Mix gently by pipetting.
-
Take three 10-μl aliquots of each denaturation reaction. You have six reactions (A1, A2, A3, B1, B2, B3).
Heat at 42°C for 15 min – this sample is your denatured input.
Heat at 42°C for 15 min – this sample receives ubiquitination enzymes.
Store on ice – this represents your nondenatured input.
After the 42°C heat denaturation, put tubes on ice.
-
Add 10 μl of 2× ubiquitination buffer 1 (40 mM Hepes, pH 7.4, 100 mM NaCl, 10 mM MgCl2, 5 mM ATP, 4 mM DTT, and 20 μM bovine ubiquitin) to samples A1 and B1, and 10 μl of 2× ubiquitination buffer 2 (40 mM Hepes, pH 7.4, 100 mM M bovine NaCl, 10 mM MgCl2, 5 mM ATP, 4 mM DTT, 20 μM bovine ubiquitin, 0.2 μM rabbit E1, 8 μM UbcH5a, and 6 μM CHIP) to samples A2 and B2 (see Note 13).
Incubate at 37°C for 1 h.
Terminate reactions by adding 2× sample buffer with BME to samples, heat at 37°C for 5 min, and run on 10% SDS-PAGE gel.
Transfer to nitrocellulose and follow the Western blotting procedure above (steps 6–12 in Subheading 3.1.4).
Acknowledgments
DMC is supported by NIH GM056981. CP is supported by NIH AG024282 and GM61728.
Footnotes
In order to verify that the protein of interest has been induced sufficiently, you can collect a sample of bacteria before and after induction, lyse in sample buffer by sonication, and run on an SDS-PAGE gel. The induced protein should be visible by Coomassie stain.
The bacterial pellets can be frozen and stored at −20°C before proceeding with the purification.
When sonicating, ensure that the samples remain cold. In order to do this, pack the samples on ice during sonication and wait for at least 30 s in between each sonication.
Samples can be saved at each step of the purification to monitor the purity and determine if the protein of interest is being lost at any particular step.
When determining protein concentration, always make BSA standards for standard line in the same buffer as protein of interest. If the protein of interest is not as concentrated as desired, then it can be concentrated using concentrators, such as the Amicon Ultra-15 centrifugal filter device with a 10,000 molecular weight cutoff. Typically, a concentration of 1–4 mg/ml is ideal for the subsequent reactions.
The protein can be stored at −80°C for an extended period of time, but should be subjected to minimal freeze thaw cycles.
If the purification is being done with the use of a detector, then the elution can be monitored at an absorbance of 280 nm. Then, fractions which correspond to the protein peaks can be assayed by SDS-PAGE. Otherwise, every other fraction of the entire elution can be tested in order to determine which fractions are the most pure for the protein of interest.
A portion of dialysis buffer can be saved after dialysis, aliquoted, and frozen in order to have a buffer-matched control for future assays.
The pH of ATP stocks should be adjusted to 7.5 with NaOH.
Making a cocktail containing all the common reagents for a series of samples within an experiment reduces the pipetting error that would come from adding each reagent individually.
As a negative control, the E2 or E3 proteins can be left out of the assay.
Add 80 μl of BME and 10 μl PI per ml of 2× SB fresh before use.
We typically make a 10× stock of the ubiquitination buffer without ATP, DTT, and ubiquitin, then dilute it to the desired working concentration, and add ATP, DTT, and ubiquitin fresh.
References
- 1.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–5. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 2.Cyr DM, Hohfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci. 2002;27:368–75. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 3.Younger JM, Chen L, Ren HY, Rosser MNF, Fan C-Y, Patterson C, Cyr DM. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–82. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 4.Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquiti nation of substrates and Hsp70. Nature. 2006;440:551–5. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Younger JM, Ren HY, Chen L, Fields A, Patterson C, Cyr DM. A foldable CFTR{Delta}F508 biogenic intermediate accumulates upon inhibition of the Hsc70-CHIP E3 ubiquitin ligase. J Cell Biol. 2004;167:1075–85. doi: 10.1083/jcb.200410065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang J, Ballinger CA, Wu Y, Cyr DM, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938–44. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 7.Min JN, Whaley RA, Sharpless NE, Lockyer P, Portbury AL, Patterson C. CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol Cell Biol. 2008;28:4018–25. doi: 10.1128/MCB.00296-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai Q, Zhang C, Wu Y, Cyr DM, Patterson C. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22:5446–58. doi: 10.1093/emboj/cdg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosser MF, Washburn E, Muchowski PJ, Patterson C, Cyr DM. Chaperone functions of the E3 ubiquitin ligase CHIP. J Biol Chem. 2007;282:22267–77. doi: 10.1074/jbc.M700513200. [DOI] [PubMed] [Google Scholar]