Abstract
Several lines of evidence have supported a host genetic contribution to vaccine response, but genome-wide assessments for specific determinants have been sparse. Here we describe a genome-wide association study (GWAS) of protective antigen-specific antibody (AbPA) responses among 726 European-Americans who received Anthrax Vaccine Adsorbed (AVA) as part of a clinical trial. After quality control, 736,996 SNPs were tested for association with the AbPA response to 3 or 4 AVA vaccinations given over a 6-month period. No SNP achieved the threshold of genome-wide significance (p=5x10−8), but suggestive associations (p<1x10−5) were observed for SNPs in or near the class II region of the major histocompatibility complex (MHC), in the promoter region of SPSB1, and adjacent to MEX3C. Multivariable regression modeling suggested that much of the association signal within the MHC corresponded to previously identified HLA DR-DQ haplotypes involving component HLA-DRB1 alleles of *15:01, *01:01, or *01:02. We estimated the proportion of additive genetic variance explained by common SNP variation for the AbPA response after the 6 month vaccination. This analysis indicated a significant, albeit imprecisely estimated, contribution of variation tagged by common polymorphisms (p=0.032). Future studies will be required to replicate these findings in European Americans and to further elucidate the host genetic factors underlying variable immune response to AVA.
Keywords: Anthrax vaccines, Bacillus anthracis, bacterial vaccines, vaccination, Genome-wide association study
1. Introduction
The Anthrax Vaccine Research Program (AVRP) human clinical trial (clinicaltrials.gov identifier NCT0019067, hereafter referred to as AVA000) was a phase 4 study of the safety and immunogenicity of Anthrax Vaccine Adsorbed (AVA). Before 2008, the licensed AVA regimen consisted of subcutaneous (SQ) vaccine administration in 0.5 ml doses at weeks 0, 2, and 4 and months 6, 12, and 18 with yearly booster doses thereafter. AVA000 was designed to assess the impact on serological responses to AVA of switching from SQ to intramuscular (IM) administration and of omitting one or more doses from the licensed schedule. AVA000 was a multicenter, randomized, double-blind, placebo-controlled, noninferiority trial that enrolled 1,563 healthy adults (18 to 61 years of age at baseline) between 2002 and 2008. A planned interim analysis of the first 1,005 enrolled subjects demonstrated that a 3-dose IM regimen provided noninferior immunological priming by 7 months while producing fewer injection site adverse events [1]. The Food and Drug Administration has since modified their recommendations for the administration of the vaccine accordingly [2].
Anthrax produces cytotoxic effects through the interaction of three proteins: lethal factor, edema factor, and a binding component protective antigen (PA). No firm serologic correlate of protection in humans has been established. However, as PA is known to be an important component of an effective anthrax vaccine, antibodies to PA (AbPA) are commonly used as the primary measure of AVA immunogenicity [3]. While AVA has been judged safe and effective according to the Institute of Medicine, data from AVA000 and smaller previous studies have indicated significant inter-individual variability in the AbPA immune response[4,5]. This variability suggests potential host genetic influences, which are supported by observed differences in the AbPA response between European and African-Americans [1,6].
Twin studies have estimated that genetic effects on vaccine responses are strong, with estimated heritabilities for antibody levels ranging from 38.8% for mumps to as high as 88.5% in the case of measles [7]. The human leukocyte antigen (HLA) system has been the primary focus of research into specific genetic variants, including previous work by our group on response to AVA within the AVA000 trial population [8]. In that study, three HLA class II haplotypes with component HLA-DRB1 alleles of *01:01,*01:02, or *15:01 were associated with a decreased AbPA response among European-American participants. Candidate gene studies of other vaccines have also implicated variation outside of the major histocompatibility complex (MHC), but genome-wide assessments of host genetic variation and vaccine response are scarce. We are only aware of published studies on antibody response to hepatitis B vaccine and T cell response to the MRKAd5 HIV-1 gag/pol/nef vaccine [9,10].
Much current research is directed at the development of next-generation anthrax vaccines, including recombinant vaccines and monoclonal antibodies designed to block the anthrax toxin [11]. Understanding the host contribution to the observed variability in the AbPA response will continue to be important as PA will likely remain a major component of future candidate vaccines. In addition, the National Biodefense Science Board recently endorsed conducting a clinical trial to study the anthrax vaccine response in children [12]. Immunogenetic characterizations may also serve to inform computational models aimed at “predictive vaccinology”[13]. One such mathematical model is that proposed by Kumar et al. [14] for the inflammatory response to natural anthrax infection. Another involves ongoing AVRP efforts using aerosolized Bacillus anthracis to infer a correlate of human immune protection by extrapolating results from rhesus macaques [15]. In that model one particular component aims at predicting future post-vaccination AbPA levels in order to estimate probabilities of subsequent protection. These predictions might be improved by the identification of suitable genetic correlates to explain portions of the population variability in AbPA response. With these potential applications in mind, here we report the results of a genome-wide association study (GWAS) conducted in the AVA000 trial population.
2. Materials and Methods
2.1. Population Genetics Analysis Program (PopGen) study of the AVA000 trial population
The design and participant characteristics of the AVA000 trial population have been described previously [1,8]. Briefly, 1,563 subjects were randomized to seven study arms: group 1 received the licensed regimen (8 doses, SQ), while group 2 also received 8 doses but with IM administration. Groups 3 through 5 received between 4 and 7 doses via IM administration, while groups 6a (SQ) and 6b (IM) received saline placebo.
2.2. Measurement of IgG antibody to protective antigen
Methods for the measurement of AbPA using a quantitative enzyme-linked immunosorbent assay (ELISA) are described in Semenova et al.[16] As in our previous analysis of HLA polymorphisms [8], we focus here on responses during the initial 7 months of the AVA000 trial, with AbPA measured at 4 time points (4, 8, 26, and 30 weeks post-baseline) in response to either 3 or 4 doses of AVA. A full discussion of the impact of route of administration and number of vaccinations on the AbPA response has been described elsewhere [1]. We excluded AbPA measurements taken after missed vaccinations or at times that deviated significantly from those specified in the study protocol (Supplementary Table 1).
2.3. Genotyping and quality control
Of the 1,303 AVA000 participants randomized to AVA-containing regimens, 1,070 individuals were genotyped (sample call rate > 95%, 1 sample excluded) using the Affymetrix Genome-Wide 6.0 array (Affymetrix Inc., Santa Clara, CA, USA). Due to the multi-ethnic composition of the trial population, we inferred familial relationships (kinship coefficients) using the algorithm implemented in KING, which is designed to be robust against population structure [17]. We examined sample pairs with kinship coefficients greater than or equal to 2−4.5 = 0.04419 (threshold for third-degree relatives suggested by the KING developers), removing both samples for pairs estimated to be monozygotic twins (8 pairs) and randomly retaining one individual from any other type of pair-wise familial relationship (73 samples excluded in total). On the basis of a multidimensional scaling (MDS) analysis of the remaining 997 samples (See Supplementary Figure 1), we retained individuals in the present analysis that clustered into the majority racial/ethnic group representing European-Americans (n=726). The numbers of participants in other racial/ethnic population groups available from AVA000 (including African-Americans and Hispanic-Americans) precluded a meaningful analysis of these subgroups. Using PLINK [18], we excluded SNPs with individual genotyping call rates < 99%, minor allele frequencies (MAF) < 5%, or evidence of deviation from Hardy-Weinberg Equilibrium (HWE; p-value < 0.0001).
2.4. Supplementary Genotyping with the ImmunoChip
Of the 726 European-Americans included in the GWAS analysis, 671 (due to sample availability) were also genotyped on the Immunochip, a custom Illumina Infinium array (196,524 SNPs) designed for replication and fine mapping of established GWAS loci and strong candidate genes for autoimmune and inflammatory diseases [19]. We applied the same quality control criteria as with the Affymetrix 6.0 array data. In total, we evaluated a total of 736,996 autosomal SNPs (651,201 from the Affymetrix array and 85,795 non-overlapping SNPs from the Immunochip) for association with the AbPA response to AVA.
2.5. Statistical Modeling of AbPA Response
As in our previous analysis of HLA polymorphisms, we modeled the association of each SNP (assuming an additive mode of inheritance) with the longitudinal profile of log10 transformed AbPA measured at 4, 8, 26, and 30 weeks from baseline [8]. Briefly, parametric regression models were implemented using the survival package in the R Statistical Computing Environment [20] to account for left-censoring induced by the detection limit of the ELISA assay (3.7 μg/ml). Generalized Estimating Equations (GEE) were used to account for multiple AbPA measurements on the same individual. All models included age at enrollment (<30, 30–39, 40–49, and 50–61 years of age), sex, study site, cumulative number of AVA doses (1, 2, 3, or 4), route of administration (SQ vs IM), time since last AVA vaccination, and the top four principal components (computed on the entire multi-ethnic sample) as covariates. SNP annotation was performed using SNPDOC (https://wakegen.phs.wfubmc.edu/public/snpdoc/index.cfm).
2.6. Genome-Wide Estimation of Explained Variance for Week 30 AbPA Response
We also estimated the cumulative variance explained by common SNP variation through linkage disequilibrium (LD) with causative variation using a variance components approach [21,22]. We used the AbPA response measured at week 30 as the primary quantitative phenotype so as to avoid left-censoring issues with the detection limit of the ELISA assay; virtually all individuals displayed AbPA levels above the detection limit at that measurement time point. The variance components model was estimated using restricted maximum likelihood as implemented in the GCTA software package [23]. We applied the definition of unrelated individuals used in Yang et al. [21], removing individuals with estimated relationships of greater than 0.025.
3. Results
Characteristics of the European-American AVA000 participants included in the GWAS are displayed in Table 1. Compared with individuals randomized to an AVA-containing regimen excluded from genetic analyses, included participants were slightly older (mean age of 41.0 ± 11.1 (SD) years compared to 36.6 ± 11.0). The percentage of male participants and the distribution across each of the vaccination regimens are generally similar. The top SNP associations (p≤1x10−5) from the GWAS analysis are displayed in Table 2. Supplementary Table 2 presents a complete listing of SNPs displaying evidence of association with p<1x10−4. No single SNP achieved the commonly utilized threshold for genome-wide significance (p ≈ 5 × 10−8)[24], although the genome-wide distribution of p-values did exhibit some evidence of deviation from the null expectation of no association (Supplementary Figure 2).
Table 1.
Characteristics of European-American (EA) Anthrax Vaccine Research Program human clinical trial participants included in GWAS of the immune response to AVA
| EA’s included in GWAS N = 726 |
Remaining Participants N = 577 |
† | |
|---|---|---|---|
|
| |||
| Variable | Frequency (%)‡ | Frequency (%)‡ | p-value |
| Sex | 0.850 | ||
| Male | 356 (0.49) | 279 (0.48) | |
| Age at Enrollment | 5.29 x 10−9 | ||
| < 30 years | 160 (0.22) | 204 (0.35) | |
| 30 to 39 years | 160 (0.22) | 141 (0.24) | |
| 40 to 49 years | 234 (0.32) | 153 (0.27) | |
| 50 to 61 years | 172 (0.24) | 79 (0.14) | |
| Clinical Site | < 2.2 x 10−16 | ||
| Rochester, MN | 204 (0.28) | 47 (0.08) | |
| Atlanta, GA | 122 (0.17) | 60 (0.10) | |
| Washington, DC | 106 (0.15) | 106 (0.18) | |
| Houston, TX | 135 (0.19) | 135 (0.23) | |
| Birmingham, AL | 159 (0.22) | 154 (0.27) | |
| AVA Vaccination Regimen | 0.829 | ||
| Group 1 (4-SQ) | 145 (0.20) | 114 (0.20) | |
| Group 2 (4-IM) | 144 (0.20) | 118 (0.20) | |
| Group 3 (3-IM) | 142 (0.20) | 114 (0.20) | |
| Group 4 (3-IM) | 138 (0.19) | 120 (0.21) | |
| Group 5 (3-IM) | 157 (0.20) | 111 (0.19) | |
(GWAS) Genome-wide association study (AVA) Anthrax Vaccine Adsorbed (4-SQ) Subcutaneous administration at 0, 2, 4, and 26 weeks. (4-IM) Intramuscular administration at 0, 2, 4, and 26 weeks. (3-IM) Intramuscular administration at 0, 4, and 26 weeks.
Of the 1,303 participants randomized to AVA (non-placebo) regimens (See methods).
Percentages may not sum to one due to rounding.
Table 2.
Top associations (p ≤ 1 × 10−5) with the immunoglobulin G antibody to protective antigen (AbPA) response to AVA
| SNP | Chr | Position | Array | Gene | Allele | MAF | Beta (SE) | p |
|---|---|---|---|---|---|---|---|---|
| rs11121382 | 1 | 9,334,056 | Affy 6.0 | SPSB1 | C | 0.309 | -0.099 (0.022) | 4.08 x 10−6 |
| rs2647264 | 4 | 106,488,043 | Affy 6.0 | TET2 | PPA2 | G | 0.081 | 0.165 (0.037) | 8.67 x 10−6 |
| rs634308 | 5 | 134,591,056 | Affy 6.0 | PITX1 | H2AFY | G | 0.349 | -0.100 (0.022) | 3.51 x 10−6 |
| rs9269821 | 6 | 32,657,786 | ImmunoChip | HLA-DRB1 | G | 0.280 | -0.109 (0.024) | 7.06 x 10−6 |
| rs482044 | 6 | 32,684,042 | ImmunoChip | HLA-DRB1 | HLA-DQA1 | G | 0.388 | 0.091 (0.021) | 9.80 x 10−6 |
| rs3104402 | 6 | 32,789,654 | ImmunoChip | HLA-DRB1 | HLA-DQA1 | A | 0.073 | -0.165 (0.036) | 5.83 x 10−6 |
| rs10758161 | 9 | 32,745,028 | Affy 6.0 | TAF1L | TMEM215 | G | 0.106 | -0.159 (0.036) | 8.23 x 10−6 |
| rs6478282 | 9 | 118,964,916 | Affy 6.0 | ASTN2 | A | 0.262 | 0.101 (0.022) | 5.59 x 10−6 |
| rs12683718 | 9 | 118,982,293 | Affy 6.0 | ASTN2 | G | 0.262 | 0.099 (0.022) | 7.25 x 10−6 |
| rs732949 | 13 | 52,415,885 | ImmunoChip | PCDH8 | OLFM4 | C | 0.066 | 0.166 (0.037) | 7.73 x 10−6 |
| rs7230711 | 18 | 46,944,617 | Affy 6.0 | - | - | C | 0.075 | -0.162 (0.033) | 1.22 x 10−6 |
| rs1539857 | 18 | 46,951,934 | Affy 6.0 | SMAD4 | MEX3C | C | 0.076 | -0.158 (0.033) | 2.49 x 10−6 |
| rs16952919 | 18 | 46,989,825 | Affy 6.0 | MEX3C | - | T | 0.071 | -0.140 (0.031) | 7.28 x 10−6 |
| rs16952953 | 18 | 46,999,850 | Affy 6.0 | MEX3C | - | G | 0.074 | -0.162 (0.034) | 1.93 x 10−6 |
| rs7238856 | 18 | 47,000,228 | Affy 6.0 | MEX3C | - | A | 0.077 | -0.156 (0.033) | 2.38 x 10−6 |
| rs7241194 | 18 | 47,004,131 | Affy 6.0 | MEX3C | - | C | 0.079 | -0.151 (0.033) | 4.51 x 10−6 |
(AVA) Anthrax Vaccine Adsorbed. Beta denotes the estimated regression coefficient corresponding to the additive mean change in log10[AbPA] for the displayed allele based on a longitudinal model fit to measurements at 4, 8, 26, and 30 weeks. All models were adjusted for age, sex, study site, route of administration (SQ vs IM), cumulative number of AVA doses, time since last vaccination, and the top 4 principal components (See Methods). For SNPs in intergenic regions, Gene lists the nearest gene upstream and downstream within ± 500 kb.
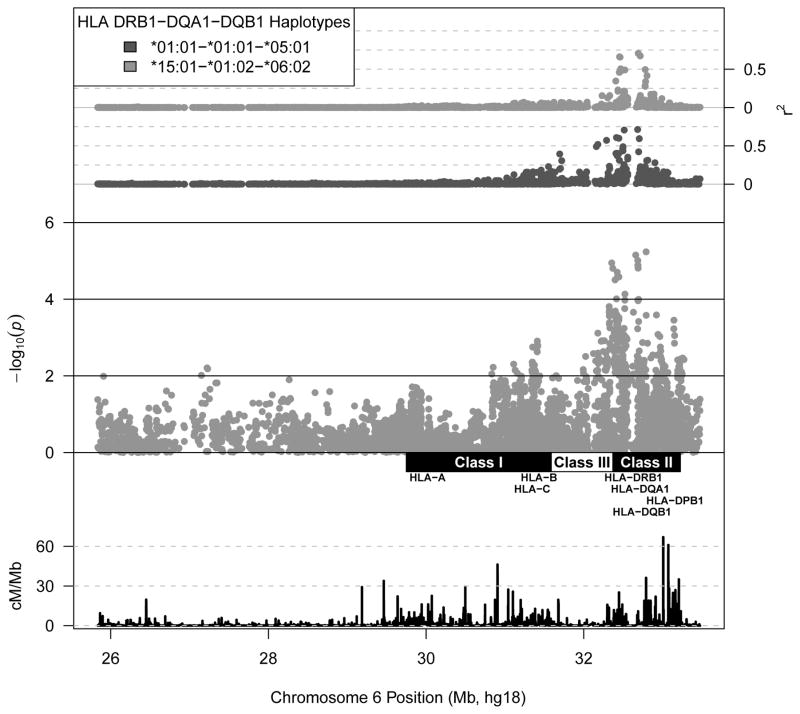
Among the strongest associations observed were those of three SNPs located in or near HLA-DRB1— rs9269821 (p=7.06 x 10−6), rs482044 (p=9.80 x 10−6), and rs3104402 (p=5.83 x 10−6) (Figure 2 and Table 2). As shown in Figure 1, these SNPs are in moderate LD with the common HLA DR-DQ haplotypes identified in our previous analysis, HLA-DRB1-DQA1-DQB1 *01:01-*01:01-*05:01 (r2=0.359 with rs9269821) and *15:01-*01:02-*06:02 (r2=0.456 with rs3104402) [8]. We subsequently performed additional regression analyses including these haplotypes (along with *01:02-*01:01-*05:01) as additional covariates (Figure 2). While most of the observed association localizing to the class II region was attenuated by adjusting for the effect of these haplotypes, we did observe a modest associations independent of variation linked to HLA-DRB1. For example, the A allele at rs2301226, which is in LD with the HLA-DPB1 *02:01 allele (r2=0.622), was associated with a higher AbPA response (conditional β=0.099, SE=0.031, p=0.001). We observed similar associations localizing to the class I region, with the T allele at rs2735035 (conditional β =-0.063, SE=0.021, p=0.003, r2=0.814 with HLA-A *02) and the G allele at rs4947306 (conditional β=0.084, SE=0.028, p=0.003, r2=0.216 and 0.133 with HLA-C *06 HLA-B *57 and HLA-Cw *06 – HLA-B*13 haplotypes respectively).
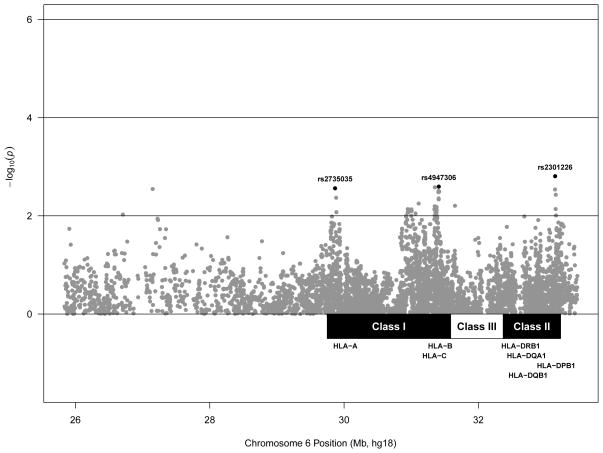
Figure 2. SNP associations with the immunoglobulin G antibody to protective antigen (AbPA) response to AVA within the extended major histocompatibility complex conditional on previously identified HLA DR-DQ haplotypes.
(AVA) Anthrax Vaccine Adsorbed. Values denote the association with the longitudinal AbPA response measured at 4, 8, 26, and 30 weeks adjusted for age, sex, study site, route of administration (SQ vs IM), cumulative number of AVA doses, time since last vaccination, top 4 principal components, and for the effect of HLA DR-DQ haplotypes *01:01-*01:01-*05:01, *01:02-*01:01-*05:01, and *15:01-*01:02-*06:02 (See Methods). (rs2301226) r2 =0.622 with HLA-DPB1 *02:01. (rs2735035) r2 =0.814 with HLA-A *02 (rs4947306) r2=0.216 and 0.133 with HLA-C *06 HLA-B*57 and HLA-C *06- HLA-B *13 haplotypes respectively.
Figure 1. SNP associations with the immunoglobulin G antibody to protective antigen (AbPA) response to AVA within the extended major histocompatibility complex.
(AVA) Anthrax Vaccine Adsorbed. Middle axis displays SNP associations with longitudinal AbPA response measured at 4, 8, 26, and 30 weeks adjusted for age, sex, study site, route of administration (SQ vs IM), cumulative number of AVA doses, time since last vaccination, and the top 4 principal components (See Methods). Bottom axis displays local recombination rate estimates from the International HapMap Project [44]. Top axis displays linkage disequilibrium (r2) between each SNP and selected HLA DRB1-DQA1-DQB1 haplotypes.
Beyond the human MHC, we also observed comparable associations for a cluster of intergenic SNPs on chromosome 18q21.2 near the mex-3 homolog C (MEX3C) gene. The strongest association was observed for rs16952953 (p=1.93x10−6), with the G allele associated with a lower AbPA response (β=-0.162, SE=0.034). We also note the association of rs11121382 (p=4.08x10−6), which lies in the promoter/regulatory region of the splA/ryanodine receptor domain and suppressor of cytokine signaling (SOCS) box containing 1 (SPSB1) gene on chromosome 1p36.22, where the C allele was similarly associated with a lower AbPA response (β=-0.099, SE=0.022). Finally, using a variance components modeling approach, we estimated that common SNP variation explained90.12% (SE=0.4864, p=0.032) of the AbPA response at 30 weeks (following 3 or 4 AVA doses). Although this analysis suggested a statistically significant genetic contribution to variability in 30-week AbPA response, the large standard error clearly indicates an imprecise estimate. Thus, we would caution against the interpretation that common SNP variation can account for the majority of the residual variability in the AbPA response (after accounting for age, number of vaccinations, etc.), as predictive accuracy based on a variance components model would be expected to be substantially lower [25].
4. Discussion
GWAS have been a popular tool for investigating human genetic correlates of various disease outcomes. In our evaluation of variable AbPA responses induced by AVA vaccination, no SNP displayed an association surpassing the generally accepted, but conservative, threshold of genome-wide significance. Some of the strongest associations mapped to the MHC class II region near the adjacent HLA-DRB1 and HLA-DQA1 loci—those previously associated with lower AbPA [8]. While many of the SNP signals in this region appeared to tag previously identified HLA DR-DQ haplotypes, some evidence remains for modest residual association within the MHC (Figure 4). The small size of the AVA000 trial and the lack of an immediate replication cohort prevented us from teasing out independent MHC association signals. A recent GWAS of response to hepatitis B vaccine indicated likely independent association signals corresponding to HLA-DR, HLA-DP, and the class III region [10]. The conditional analyses performed here similarly suggested a possible association with SNPs linked to HLA-DPB1 independent of the effect of HLA DR-DQ. As with the other associations reported here, this result naturally requires replication.
None of the other polymorphisms with the strongest associations detected (Table 2) occur at loci for which a clear role in vaccine immune response is currently known. For example, there is little previous work implicating variation in or near MEX3C (also designated as the RING finger and KH domain-containing protein 2, RKHD2) with any trait; no associations have been reported in the NHGRI GWAS catalog [26]. Guzman et al. [27] reported the lone genetic association of two SNPs in RKHD2 with essential hypertension in a case-control study from Spain. Two recent systems biology studies have both reported significant changes in MEX3C expression in peripheral blood mononuclear cells following influenza vaccination [28,29], however the paucity of experimental data on MEX3C/RKHD2 function makes any suggestion of a mechanistic role in vaccine response highly speculative.
Perhaps more plausible was the association of rs11121382, which lies in the promoter/regulatory region of SPSB1. The SPSB1 protein interacts with MET, a receptor protein-kinase for hepatocyte growth factor (HGF) [30]. The SPSB1 protein also regulates inducible nitric-oxide synthase levels (iNOS), which in turn regulates the production of nitric oxide (NO) in activated macrophages [31]. Lewis et al. recently demonstrated that the induction of iNOS by SPSB1 is controlled by a negative feedback loop involving SPSB2, downstream of regulation by TLR3 and TLR4 [32]. The role of SPSB1 in the production of NO suggests a potential pathway through which genetic variation might influence the host immune response. No other SNP in this region that passed quality control appeared associated with AbPA variation, though none was even in moderate LD with rs11121382 (maximum r2 =0.18 within a 50 kb window). Visual inspection of the intensity data for this SNP did not suggest any technical problem with genotyping. As with the SNPs near MEX3C, interpretation of the association with rs11121382 must await further research on vaccine response
The current investigation only had adequate power (>80%) to detect very large effects (~5.3% of the variance for a continuous trait) at the genome-wide significance level of 5 × 10−8. While a suitable replication cohort is not immediately available, an effort to replicate our findings here is underway, including a component of functional/systems biology research. Furthermore, the design of the AVA000 trial and the sensitivity of the ELISA assay used for AbPA could have obscured associations of host genetic variants contributing to the innate immune response to anthrax vaccine. The first available AbPA measurement occurred at 4 weeks, at which time all participants randomized to 4 dose regimens had already received 2 AVA doses; and at that point a large majority (~75%) of individuals randomized to the 3 dose regimen had AbPA levels below the assay detection limit. More informative research on host genetic factors that influence variability in the innate immune response to AVA may therefore require carefully timed observations soon after the first dose.
In view of the high estimated heritabilities for vaccine-induced antibody levels mentioned earlier, our failure to discern effects attributable to individual SNPs here raises the “missing heritability” question facing the genetics community at large [33,34]. The absence of large effects and our results using variance-components modeling are both consistent with a highly polygenic genetic architecture underlying the response to AVA. This architecture resembles the infinitesimal model of quantitative genetics proposed by Fisher (summarized in References [35] and [36]). This model envisioned a large number of subtle additive effects on trait variation, whereas large deleterious effects on the immune response (or other traits that bear on reproductive fitness) would be eliminated from the population through natural selection.
Although response to as “simple” an agent as a vaccine might be more homogeneous than would be expected for a naturally occurring infection, as a quantitative trait the response is nevertheless relatively complex. Both theoretical and empirical evidence [37] have supported a highly polygenic, largely additive genetic basis for complex traits in humans, including evidence from recent large-scale studies for schizophrenia/bipolar disorder [38], height [39], body mass index [40], and blood lipid levels [41]. Of course, a polygenic host contribution would complicate the search for individual causative polymorphisms, but analyses using variance component models have suggested that the aggregate contribution of multiple SNPs can be accurately estimated with sufficient sample sizes [21,22]. The implication for predictive vaccinology is that it may be possible to construct predictive models of vaccine response without directly mapping all associated SNPs. This modeling approach would require both substantial sample sizes to achieve a useful degree of predictive accuracy [25,42] and longitudinal measurements of outcomes less sensitive to assay detection limits. New approaches including “systems vaccinology” could perhaps provide mechanistic and predictive insight into the immune response to AVA, while alleviating some (though not all) of the requirement for large sample size [43]. Whether these approaches can identify specific genetic characteristics that can guide the design of future candidate anthrax vaccines remains to be seen.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the other site principal investigators from the AVA000 trial for their efforts in data collection: Janiine Babcock, M.D. (Walter Reed Army Institute of Research), Wendy Keitel, M.D. (Baylor College of Medicine), and Harry Keyserling, M.D. (Emory University School of Medicine).
Role of the Funding Source: This work was supported by the National Institute of Allergy and Infectious Diseases through contracts N01-AI40068 and 272201000023C-0-0-1, and in part by research computing resources acquired and managed by University of Alabama at Birmingham IT Research Computing. Samples used in this study were made available through a clinical trial funded by the Centers for Disease Control and Prevention, including a supplemental grant to GAP. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention, the National Institute of Allergy and Infectious Diseases, or the University of Alabama at Birmingham.
Footnotes
Disclosures: GAP has served as a consultant to Emergent Biosolutions Inc. (Rockville, MD, USA).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Marano N, Plikaytis BD, Martin SW, Rose C, Semenova VA, Martin SK, et al. Effects of a reduced dose schedule and intramuscular administration of anthrax vaccine adsorbed on immunogenicity and safety at 7 months: a randomized trial. The Journal of the American Medical Association. 2008;300(13):1532–43. doi: 10.1001/jama.300.13.1532. [DOI] [PubMed] [Google Scholar]
- 2. [Accessed 3/15/2010];BioThrax - Package Insert. 2010 http://www.fda.gov/downloads/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/LicensedProductsBLAs/UCM074923.pdf.
- 3.Saile E, Quinn CP. Anthrax Vaccines. In: Bergman N, editor. Bacillus Anthracis and Anthrax. Wiley-Blackwell; New Jersey: 2010. pp. 269–93. [Google Scholar]
- 4.Singer DE, Schneerson R, Bautista CT, Rubertone MV, Robbins JB, Taylor DN. Serum IgG antibody response to the protective antigen (PA) of Bacillus anthracis induced by anthrax vaccine adsorbed (AVA) among U.S. military personnel. Vaccine. 2008;26(7):869–73. doi: 10.1016/j.vaccine.2007.11.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanson JF, Taft SC, Weiss AA. Neutralizing antibodies and persistence of immunity following anthrax vaccination. Clinical and Vaccine Immunology. 2006;13(2):208–13. doi: 10.1128/CVI.13.2.208-213.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowe SR, Garman L, Engler RJM, Farris D, Ballard JB, Harley JB, et al. Anthrax vaccination induced anti-lethal factor IgG: Fine specificity and neutralizing capacity. Vaccine. 2011;29(20):3670–8. doi: 10.1016/j.vaccine.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan PL, Jacobson RM, Poland GA, Jacobsen SJ, Pankratz VS. Twin studies of immunogenicity--determining the genetic contribution to vaccine failure. Vaccine. 2001;19(17–19):2434–9. doi: 10.1016/s0264-410x(00)00468-0. [DOI] [PubMed] [Google Scholar]
- 8.Pajewski NM, Parker SD, Poland GA, Ovsyannikova IG, Song W, Zhang K, et al. The role of HLA-DR-DQ haplotypes in variable antibody responses to Anthrax Vaccine Adsorbed. Genes and Immunity. 2011;12(6):457–65. doi: 10.1038/gene.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fellay J, Frahm N, Shianna KV, Cirulli ET, Casimiro DR, Robertson MN, et al. Host genetic determinants of T cell responses to the MRKAd5 HIV-1 gag/pol/nef vaccine in the STEP trial. The Journal of Infectious Diseases. 2011;203(6):773–9. doi: 10.1093/infdis/jiq125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Png E, Thalamuthu A, Ong RT, Snippe H, Boland GJ, Seielstad M. A genome-wide association study of hepatitis B vaccine response in an Indonesian population reveals multiple independent risk variants in the HLA region. Human Molecular Genetics. 2011;20(19 ):3893–8. doi: 10.1093/hmg/ddr302. [DOI] [PubMed] [Google Scholar]
- 11.Bienek DR, Loomis LJ, Biagini RE. The anthrax vaccine: no new tricks for an old dog. Human Vaccines. 2009;5(3):184–9. doi: 10.4161/hv.5.3.7308. [DOI] [PubMed] [Google Scholar]
- 12.Couzin-Frankel J. Bioterror research. Panel endorses anthrax vaccine study in children. Science. 2011;334(6056):577. doi: 10.1126/science.334.6056.577. [DOI] [PubMed] [Google Scholar]
- 13.Poland GA, Ovsyannikova IG, Jacobson RM. Vaccine immunogenetics: bedside to bench to population. Vaccine. 2008;26(49):6183–8. doi: 10.1016/j.vaccine.2008.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar R, Chow CC, Bartels JD, Clermont G, Vodovotz Y. A mathematical simulation of the inflammatory response to anthrax infection. Shock. 2008;29(1):104–11. doi: 10.1097/SHK.0b013e318067da56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madigan D. Anthrax vaccines : bridging correlates of protection in animals to immunogenicity in humans. Center for Biologics Evaluation and Research, Food and Drug Administration, U.S. Department of Health and Human Services; Washington, DC: Nov 8, 2007. [Google Scholar]
- 16.Semenova VA, Schiffer J, Steward-Clark E, Soroka S, Schmidt DS, Brawner MM, et al. Validation and long term performance characteristics of a quantitative enzyme linked immunosorbent assay (ELISA) for human anti-PA IgG. Journal of Immunological Methods. 2012;376(1–2):97–107. doi: 10.1016/j.jim.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–73. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Research & Therapy. 2011;13(1):101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2010. http://www.R-project.org. [Google Scholar]
- 21.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nature Genetics. 2010;42(7):565–9. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. American Journal of Human Genetics. 2011;88(3):294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. American Journal of Human Genetics. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genetic Epidemiology. 2008;32(3):227–34. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makowsky R, Pajewski NM, Klimentidis YC, Vazquez AI, Duarte CW, Allison DB, et al. Beyond missing heritability: prediction of complex traits. PLoS Genetics. 2011;7:e1002051. doi: 10.1371/journal.pgen.1002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindorff LA, MacArthur J, Wise A, Junkins H, Hall P, Klemm A, et al. [Accessed 1/15/2010];A Catalog of Published Genome-Wide Association Studies. Available at www.genome.gov/gwastudies.
- 27.Guzmán B, Cormand B, Ribasés M, González-Núñez D, Botey A, Poch E. Implication of chromosome 18 in hypertension by sibling pair and association analyses: putative involvement of the RKHD2 gene. Hypertension. 2006;48(5):883–91. doi: 10.1161/01.HYP.0000244085.52918.a0. [DOI] [PubMed] [Google Scholar]
- 28.Bucasas KL, Franco LM, Shaw CA, Bray MS, Wells JM, Niño D, et al. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. The Journal of Infectious Diseases. 2011;203(7):921–9. doi: 10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, et al. Systems biology of vaccination for seasonal influenza in humans. Nature Immunology. 2011;12(8):786–95. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang D, Li Z, Messing EM, Wu G. The SPRY domain-containing SOCS box protein 1 (SSB-1) interacts with MET and enhances the hepatocyte growth factor-induced Erk-Elk-1-serum response element pathway. The Journal of Biological Chemistry. 2005;280(16):16393–401. doi: 10.1074/jbc.M413897200. [DOI] [PubMed] [Google Scholar]
- 31.Nishiya T, Matsumoto K, Maekawa S, Kajita E, Horinouchi T, Fujimuro M, et al. Regulation of inducible nitric-oxide synthase by the SPRY domain- and SOCS box-containing proteins. The Journal of Biological Chemistry. 2011;286(11):9009–19. doi: 10.1074/jbc.M110.190678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis RS, Kolesnik TB, Kuang Z, D’Cruz AA, Blewitt ME, Masters SL, et al. TLR Regulation of SPSB1 Controls Inducible Nitric Oxide Synthase Induction. Journal of Immunology. 2011;187(7):3798–805. doi: 10.4049/jimmunol.1002993. [DOI] [PubMed] [Google Scholar]
- 33.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nature Reviews Genetics. 2010;11(6):446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farrall M. Quantitative genetic variation: a post-modern view. Human Molecular Genetics. 2004;13(Spec No 1):R1–7. doi: 10.1093/hmg/ddh084. [DOI] [PubMed] [Google Scholar]
- 36.Hill WG. Understanding and using quantitative genetic variation. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences. 2010;365(1537):73–85. doi: 10.1098/rstb.2009.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill WG, Goddard ME, Visscher PM. Data and Theory Point to Mainly Additive Genetic Variance for Complex Traits. PLoS Genetics. 2008;4(2):e1000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–8. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics. 2010;42(11):937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meuwissen TH. Accuracy of breeding values of “unrelated” individuals predicted by dense SNP genotyping. Genetics, Selection, Evolution. 2009;41:35. doi: 10.1186/1297-9686-41-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakaya HI, Pulendran B. Systems vaccinology : its promise and challenge for HIV vaccine development. Current Opinion in HIV and AIDS. 2012;7 :24–31. doi: 10.1097/COH.0b013e32834dc37b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3. 1 million SNPs. Nature. 2007;449(7164):851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.