Abstract
Non-invasive approaches to assess tissue function could improve significantly current methods to diagnose diseases and optimize engineered tissues. In this study, we describe a two-photon excited fluorescence microscopy approach that relies entirely on endogenous fluorophores to dynamically quantify functional metabolic readouts from individual cells within three-dimensional engineered tissues undergoing adipogenic differentiation over six months. Specifically, we employ an automated approach to analyze 3D image volumes and extract a redox ratio of metabolic cofactors. We identify a decrease in redox ratio over the first two months of culture that is associated with stem cell differentiation and lipogenesis. In addition, we demonstrate that the presence of endothelial cells facilitate greater cell numbers deeper within the engineered tissues. Since traditional assessments of engineered tissue structure and function are destructive and logistically intensive, this non-destructive, label-free approach offers a potentially powerful high-content characterization tool for optimizing tissue engineering protocols and assessing engineered tissue implants.
1. INTRODUCTION
Tissue regeneration is often required after disease or trauma. To facilitate regeneration and restore tissue function, tissue grafts have traditionally been implanted, but often with suboptimal outcomes [1]. Tissue engineering offers the potential to improve repair and regeneration [2–3]. Engineered tissues are typically developed by seeding stem cells on a biomaterial scaffold and promoting proliferation and/or differentiation over the course of weeks or months. To assess tissue development and optimize culture protocols, mass spectrometry (MS), western blots, quantitative polymerase chain reaction (qPCR) techniques, immunohistochemistry (IHC), and electron microscopy have typically been employed to assess the biochemical and structural characteristics of the tissue at specific time points [4–7]. However, the destructive nature of these methods does not allow for dynamic assessments of tissue development over time. Furthermore, such techniques cannot be used to evaluate the integration of engineered tissue with native tissue following implantation. As a result, there is a need for quantitative, repeatable, non-invasive or minimally invasive methods to assess tissue biochemical status and structural integrity within three dimensional (3D) tissues.
Two-photon excited fluorescence (TPEF) microscopy offers a number of advantages over traditional approaches for imaging 3D tissues. In TPEF microscopy, molecules are brought to an excited state by the simultaneous absorption of two photons. Since each of these two photons require half the energy to excite a molecule compared to a single photon, near infrared (NIR) light can be used to excite molecules and produce emission in the visible range [8]. NIR light can penetrate deeper into tissue and the potential for out-of-focus photodamage decreases substantially compared to confocal microscopy [8–9]. Furthermore, for practical purposes, two-photon absorption is limited to the focal plane, which provides intrinsic depth sectioning without the need for confocal detection [8]. Using NIR excitation, endogenous fluorescence can be produced most efficiently from cell mitochondria by nicotinamide and flavin adenine dinucleotides (NADH and FAD, respectively) [10–11]. In addition to measures of structural organization, NADH and FAD fluorescence can be used as a sensitive measure of the metabolic status of cells [12–13]. NADH and FAD are cofactors that are directly involved in cellular respiration. They exist in either oxidized (NAD+/FAD) or reduced (NADH/FADH2) forms, but only NADH and FAD yield significant fluorescence [6, 14]. By defining a fluorescence-based redox ratio of FAD/NADH or FAD/(NADH+FAD), a number of studies have used TPEF imaging to diagnose precancerous tissue by exploiting the abnormal metabolic profiles of cancer cells [13, 15–17]. Recent work has also demonstrated dynamic changes in the redox ratio of mesenchymal stem cells undergoing osteoblastic or adipogenic differentiation over 21 days on glass cover slips [6]. However, owing to the technical challenges in maintaining and assessing tissue cultures beyond three to four weeks, few studies have assessed the long-term viability and function of engineered tissue cultures. The non-invasive and depth-resolved characteristics of TPEF make it particularly amenable to long-term evaluations of 3D engineered tissues [9, 18], but this technology has not yet been utilized to quantify the differentiation status of stem cells within engineered 3D tissues.
The goal of this study was to evaluate the utility of TPEF-derived redox ratios in detecting stem cell differentiation and to develop automated image analysis methods to assess proliferation and differentiation within 3D tissues. To this end, we evaluated the development of human adipose-derived mesenchymal stem cells (hASCs) or a co-culture of hASCs and human microvascular endothelial cells (HMVECs) seeded in 3D porous silk scaffolds over a 6 month culture period. Since non-uniform nutrient supply imposed by diffusion limitations often impedes optimal tissue development, we also aimed to characterize tissue features as a function of depth within each scaffold. Non-invasively quantifying the cell differentiation status and long-term viability of engineered adipose tissue is critical for a number of applications. Through standard TPEF imaging techniques, these non-invasive assessments can be utilized to optimize in vitro protocol development far more efficiently, assess engineered tissue quality prior to in vivo implantation, and evaluate the potential long-term status of implants. Additionally, engineered adipose tissue is increasingly being used as a drug testing platform for obesity and diabetes research [19–20], which requires long-term functional measurements. By developing a method capable of automatically identifying cells within 3D image volumes and calculating cell-specific redox ratios, we present a non-invasive quantitative imaging technique that is sensitive to the metabolic changes associated with stem cell differentiation and lipogenesis.
2. METHODS
2.1. 3D adipose tissue engineering
Human adipose derived mesenchymal stem cells (hASCs) were isolated from subcutaneous adipose tissue, which was approved under Tufts University IRB (Protocol #0906007). The isolated hASCs were expanded and adipogenic induction factors were added to the medium seven days prior to seeding in silk scaffolds. Porous silk scaffolds were prepared as previously published [7, 21–22], and scaffolds were either seeded with hASCs (n=4) or a co-culture of hASCs and HMVECs (n=10). For co-culture scaffolds, a total of 800,000 HMVECs were seeded per scaffold. Seven days after HMVEC seeding, the induced hASCs were added to scaffolds in both culture types at a density of 400,000 hASCs per scaffold. During TPEF imaging, scaffolds were maintained in phenol red-free DMEM/F12 containing 10% FBS but no induction factors. Additional details regarding the silk scaffold preparation, stem cell isolation, and cell culture methods can be found in the supplementary methods.
2.2. Two-photon excited fluorescence image acquisition protocol
The hASC and hASC/HMVEC co-culture scaffolds (n=4/group) were each imaged using two-photon excited fluorescence (TPEF) microscopy at 14 time points: Days 1, 4, 8, 11, 15, 21, 30, 43, 57, 78, 99, 127, 155, 183 after hASC seeding. After imaging on Day 183, the scaffolds were fixed for histological examination. An additional set of co-culture tissues were imaged and fixed for histology at Day 15 (n=2), Day 30 (n=2), and Day 57 (n=2) in order to capture changes in development at specific time points from the histology.
TPEF images were acquired using a Leica TCS SP2 confocal microscope equipped with a tunable (710–920 nm) titanium-sapphire laser (Mai Tai; Spectra Physics; Mountain View, CA). Images (512 × 512 pixels; 12-bit depth) were acquired using water-immersion 63× objective (NA 1.2; 220µm working distance). To isolate NADH fluorescence, images were acquired using 755nm excitation and a non-descanned photomultiplier tube (PMT) detector with a 460(±20)nm emission filter, which corresponds to the emission peak of NADH [6]. Because the two-photon action cross section of NADH decreases by several orders of magnitude at 860nm excitation [23], FAD fluorescence was isolated using 860nm excitation and a 525(±25)nm emission filter. As in previous work, lipofuscin fluorescence was also measured with the 460(±20)nm emission PMT at 860nm excitation, due to the negligible FAD fluorescence at that emission range [6].
Custom-built stainless steel positioners were designed to fit within multi-well plates and secure the scaffolds against the glass bottom of the wells during imaging. For each scaffold, image stacks (2.5µm steps) were taken at three depths within the tissue (0–40µm, 80–120µm, and 160–200µm). The temperature of the cultures was maintained during imaging using a microscope cage incubator (OkoLab; Naples, Italy). Imaging sessions did not exceed 90 minutes and samples were returned to their original incubator following acquisition. The laser power was measured at the objective for 755nm and 860nm excitation after image acquisition each day.
2.3. Spatial isolation of cell fluorescence within engineered tissues
In order to isolate and quantify cellular fluorophore intensities, an automated cell segmentation technique was developed to identify and classify fluorescent cell and silk objects within each acquired image volume using Matlab (Mathworks; Natick, MA) (Supplementary Fig. S1). Following the application of an initial intensity threshold to binarized the 3D image volume, objects within the volume were identified based on pixel connectivity. An object size-based adaptive thresholding algorithm was employed to separate individual cell and silk objects that may have been connected to each other using the initial binarization threshold. Following segmentation, objects were classified as either a cell or silk based on a linear discriminant analysis (LDA) of the object fluorescence (Supplementary Fig. S1). The cell segmentation and classification algorithms summarized above are described in greater detail in the supplementary methods.
2.4. Computation of cell-specific redox ratio measurements
All fluorescence images were normalized by detector gain and laser power as described in the supplementary methods. The average normalized fluorescence corresponding to NADH and FAD for each cell was computed from the pixels contained within each cell volume. Because lipofuscin fluorescence is isolated to lysosomes within the cell rather than the mitochondria, any pixels exceeding a normalized lipofuscin fluorescence value of 2 were removed from the calculation of mean cell NADH and FAD fluorescence in order to ensure excessive localized lipofuscin fluorescence would not affect the redox ratio metric [6]. For each cell, the redox ratio was calculated from the normalized fluorescence as FAD/(NADH+FAD).
2.5. Histology
After the final imaging time point, scaffolds were hemisected, placed in formalin, and set aside for either cryo-sectioning or paraffin embedding. Constructs were processed according to standard histology protocols. H&E staining was used to evaluate general tissue organization. CD31 staining with hematoxylin counterstaining was used to identify HMVEC morphology and organization within the tissue. Oil Red O stained sections were used to assess lipid droplet accumulation. Details regarding the reagents used, histological protocols, and the imaging methods for stained sections can be found in the supplementary methods.
2.6. Statistical analyses
To assess changes in redox ratio, a three-factor ANOVA (culture group, time point, depth zone) of cell redox ratios was used to assess significant differences using JMP 8. The ANOVA design considered individual samples as a random effect nested within each group, and all possible effect interactions were included. Post-hoc Tukey HSD tests were used to evaluate differences within each effect. A similar ANOVA structure was used to evaluate the normalized NADH and FAD intensity, and the number of cells in each image volume. Significance was defined as α=0.05.
3. RESULTS
3.1. Evaluation of cell differentiation status through redox ratio
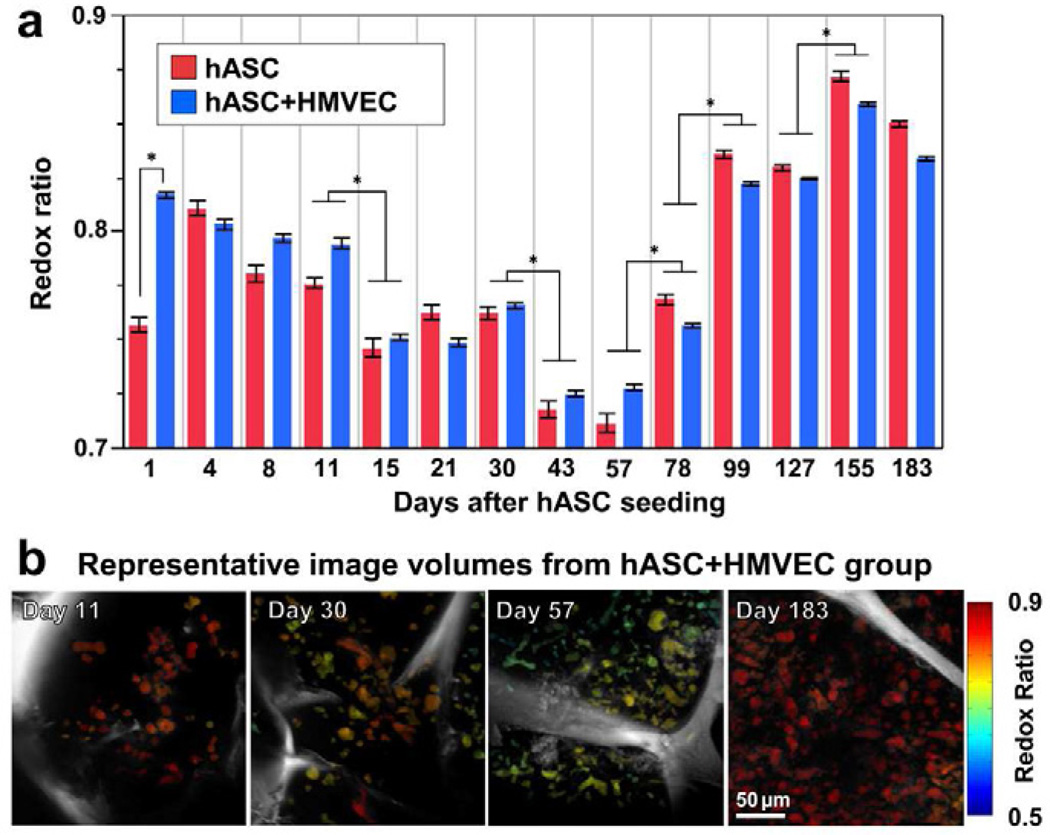
From a total of 354 image volumes, 25,299 cells were automatically identified and segmented. By computing a redox ratio of FAD/(NADH+FAD) for each cell, dynamic changes in the metabolic activity of the stem cells populating the porous silk scaffolds were detected over 183 days in culture (Fig. 1). Scaffolds containing a co-culture of hASCs and HMVECs exhibited a higher redox ratio (p=0.0386) at Day 1 after hASC seeding compared to the hASC mono-culture. After Day 1, no differences in the average redox ratio were detected between the groups. However, between Days 4 and 57, both groups exhibited a decrease in redox ratio, with significant decreases detected at Day 15 (p=0.0500) and 43 (p=0.0145) relative to the previous time point (Fig. 1). After Day 57, significant increases in redox ratio were measured at Days 78 (p=0.0002), 99 (p<0.0001), and 155 (p=0.0018).
Figure 1. Transient decrease in redox ratio was detected during adipose tissue development over 6 months.
a) In both hASC and hASC/HMVEC co-culture groups, significant decreases in redox ratio were detected at Days 15 and 43 (* p≤0.05), while significant increases in redox ratio were detected at Days 78, 99, and 155 (* p≤0.0018). Significant differences between groups were only detected at Day 1 (p=0.0386).
Error bars represent standard error. b) Cell-specific redox ratios were color-coded in image volume projections in the axial direction and demonstrate the dynamic changes in redox state over time. The silk scaffold is visible within each image volume in gray-scale.
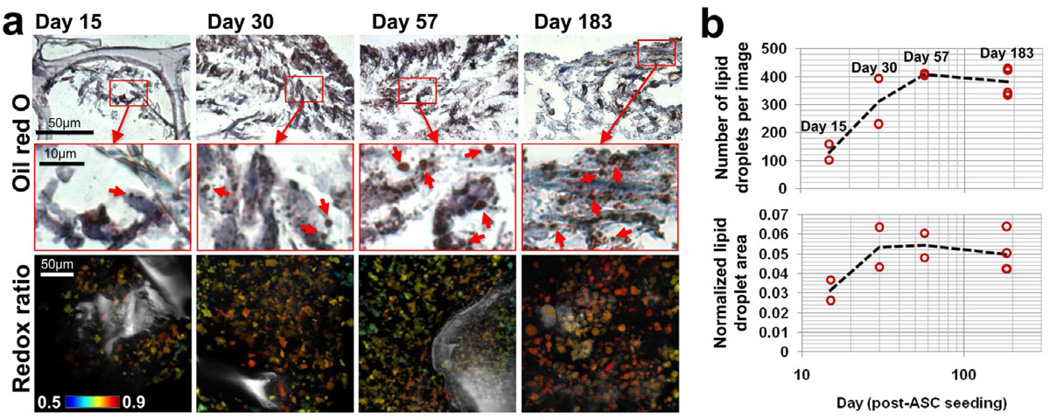
To assess the differentiation status and provide context for the changes in redox ratio, each scaffold was fixed and sectioned for histological evaluation at Day 183. An additional set of scaffolds containing a co-culture of ASCs and HMVECs were imaged and sacrificed for histology at Days 15, 30, and 57 (n=2 per time point). Lipid droplet numbers and total lipid droplet area, measured through Oil Red O staining, increased between Days 15, 30, and 57 (Fig. 2). However, droplet accumulation did not increase between Days 57 and 183, and in fact, a slightly smaller lipid droplet area and lower droplet count was detected at Day 183 (Fig. 2b). These findings demonstrated that the lipid droplet accumulation in hASCs coincided with a decrease in redox ratio between Days 11–57 (Fig. 1 and 2).
Figure 2. Lipid droplet accumulation measured by oil red O stain coincides with a decrease in redox ratio.
a) Small lipid droplets (1–3µm) were evident from oil red O staining within cells at all histological time points. An increase in droplet numbers between Day 15 and 57 coincided with a decrease in redox ratio between those time points. b) The average number of lipid droplets was quantified in each image and suggests lipid droplets began forming around Day 10 but did not increase in number after the redox ratio reached a minimum at Day 57. The total lipid droplet area was normalized by cell area in each image and also demonstrates an increase in lipid accumulation over the first 57 days, with no significant difference between Day 57 and Day 183.
3.2. Evaluation of differences in cell proliferation over time in 3D adipose cultures
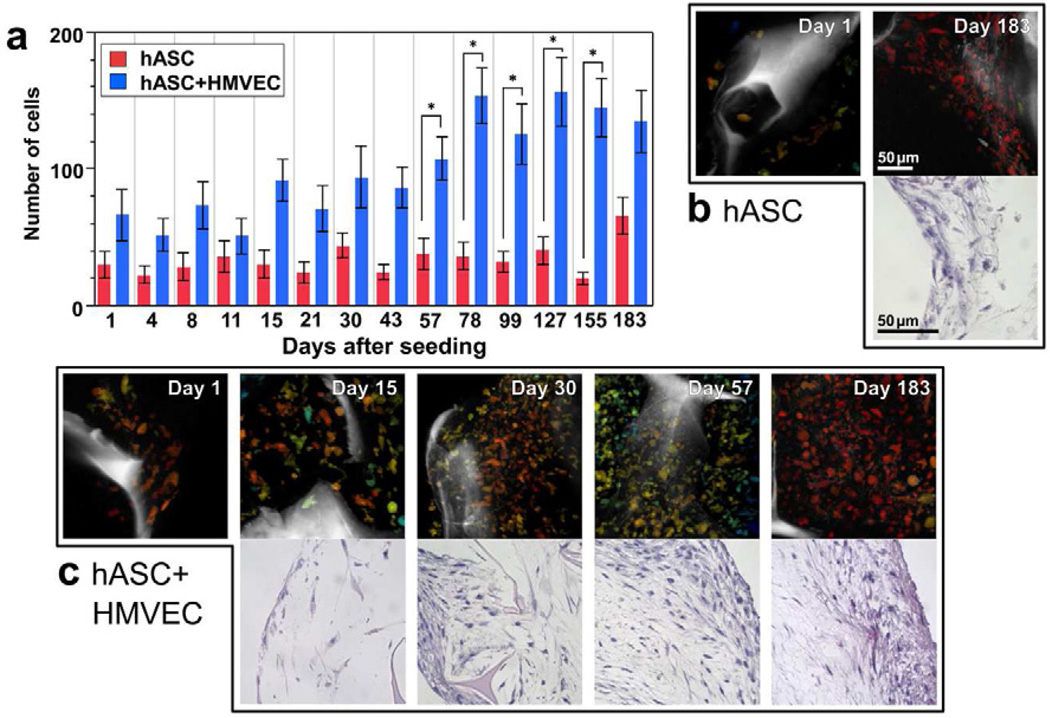
Although the redox ratio did not significantly differ between the hASC/HMVEC co-culture and the hASC mono-culture tissues, differences in cell proliferation were detected over time (Fig. 3). Overall, more cells were found in the co-culture group compared to the mono-culture group at all time points (p=0.0034), which is consistent with the additional 800,000 HMVECs seeded in the co-culture scaffolds. However a significant interaction between groups and time (p=0.0065) indicated cell numbers in the co-culture group increased over time at a greater rate than the mono-culture group. In fact, significantly more cells were detected in the co-culture group compared to the hASC mono-culture at Days 57 through 155 (p≤0.0162). The increases in cell numbers over time in the co-culture group matched the trends observed at static time points using hematoxylin and eosin (H&E) stained sections (Fig. 3b). Additionally, fewer cells were observed in H&E stained sections of the mono-culture tissue compared to the co-culture group (Fig. 3b and 3c).
Figure 3. Differences in cell proliferation were detected among the groups through automated cell segmentation of TPEF images and correspond to differences in H&E histology.
a) The average number of cells in the image volumes of the co-culture (hASC+HMVEC) group significantly increased with significant differences detected at Days 57 through 155 relative to the hASC mono-culture group (* p≤0.0162). Error bars represent standard error. b) The number of cells detected in TPEF image volume projections coincided with trends in the H&E stained sections. Fewer cells were visible in both TPEF volumes and H&E sections within the hASC mono-culture group at Day 183 compared to the co-culture group. c) An increase in cell numbers over time is evident in the co-culture group through both TPEF image volumes and H&E stained sections. The TPEF and H&E image scale bars in (b) apply to the respective images in (c) as well.
3.3. Cell proliferation and differentiation as a function of depth within 3D tissue
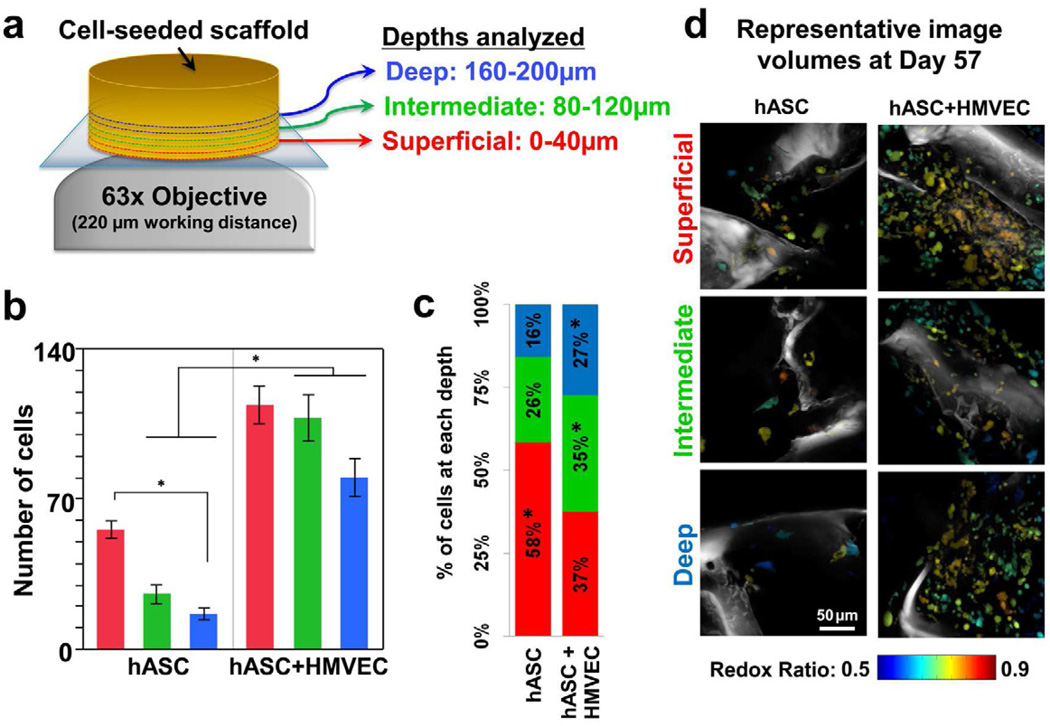
In addition to temporal changes in redox ratio and cell numbers, differences with respect to distance from the scaffold surface were assessed through the acquisition of image volumes at three specific depth zones (0–40µm, 80–120µm, and 160–200µm) (Fig. 4a). More cells were detected in the co-culture group at all depths compared to the hASC mono-culture, with significant differences (p≤0.0281) detected at the intermediate (80–120µm) and deep zones (160–200µm). In addition, a significantly lower cell count (p=0.0454) was measured in the deepest zone of hASC mono-culture compared to the most superficial zone (Fig. 4b). The proportion of total cells within a scaffold residing in the superficial depths was significantly greater (p=0.0001) in the mono-culture group compared to the co-culture group (Fig. 4c).
Figure 4. Cell segmentation of TPEF images reveals depth-dependent differences between groups.
a) TPEF image volumes were acquired at three different depths within the tissue. b) Significantly more cells were identified from TPEF volumes at the intermediate and deepest depths in hASC+HMVEC co-culture scaffolds compared to hASC mono-culture scaffolds (* p≤0.0281). Fewer cells were identified in the deepest region (160–200µm) of the mono-culture tissue compared to the surface (0–40µm) (* p=0.0454). Error bars represent standard error. c) A significantly greater proportion of the total cells resided in the intermediate and deep zones within the co-culture scaffolds compared to the mono-culture scaffolds (* p≤0.0149). d) Despite maintaining larger cell populations deeper within the tissue, no depth-dependent differences in redox ratio were detected in the co-culture scaffolds as demonstrated by the representative image volume projections.
Despite significantly more cells residing deeper within scaffolds in the co-culture group, no differences in the redox ratio were observed as a function of depth and group (Fig. 4d). To provide context for the different spatial distributions of cells within the scaffolds of the two culture groups despite consistent redox ratio measurements, CD-31 staining of the tissue sections were evaluated to identify the structural organization of HMVECs within the tissue (Fig. 5). Clusters of HMVECs were present at various depths exceeding 200 µm, and appeared to promote ASC growth deeper within the tissue (Fig. 5a). In addition, the formation of rudimentary lumens by the HMVECs (Fig. 5b) may have helped to facilitate oxygen or nutrient transport to some degree.
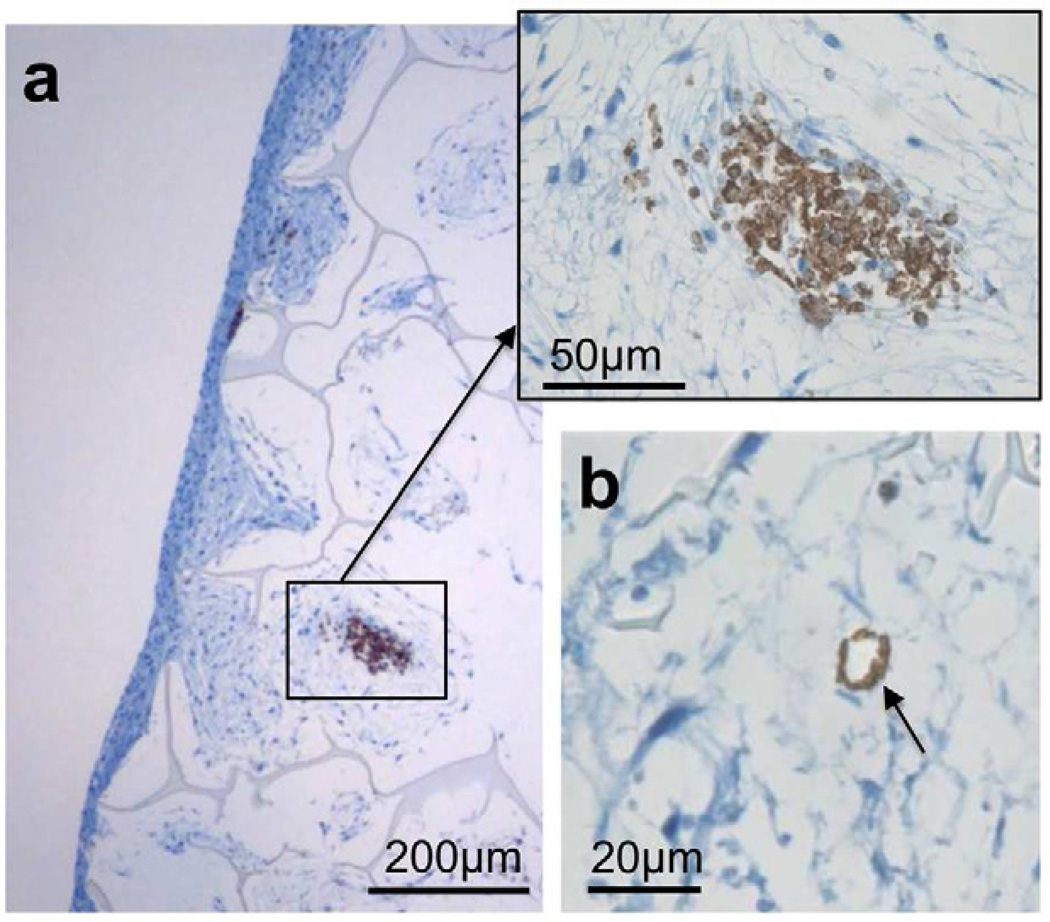
Figure 5. HMVECs may facilitate cell proliferation deeper within co-culture tissues.
a) CD-31 stained sections demonstrate the aggregation of HMVECs within the tissue, which may promote ASC growth deeper within the tissue. b) The formation of rudimentary lumens by the HMVECs (indicated by arrow) may also aid in oxygen and nutrient transport deeper within the tissue.
3.4. Changes in NADH and FAD fluorescence intensity during culture
The normalized NADH and FAD cell fluorescence intensity significantly increased throughout the duration of the experiment (Supplementary Fig. S2). Scaffolds containing only hASCs (n=4) exhibited an overall higher average intensity for both NADH (p=0.0080) and FAD (p=0.0033) fluorescence measurements compared to the scaffolds containing a co-culture of hASCs and HMVECs (n=4). For both groups, the average NADH fluorescence exhibited an increase in intensity over the initial time points and remained elevated, even though some fluctuations were observed (Supplementary Fig. S2). A 2.7-fold increase in fluorescence was detectable at the end of the experiment on Day 183 relative to Day 1 for both groups (p≤0.0001). Unlike NADH, FAD fluorescence did not substantially increase during the initial culture period. However, a substantial increase in FAD fluorescence was detected during Days 99 through 183, ultimately resulting in a 3.4-fold increase at Day 183 relative to Day 1 (Supplementary Fig. S2b).
4. DISCUSSION
This study provides a non-invasive, quantitative optical technique to assess spatiotemporal patterns of stem cell differentiation in developing 3D engineered tissues. Using an in vitro model of adipose tissue, a significant decrease in redox ratio was observed over the first 57 days of culture (Fig. 1). This finding is consistent with the lower optical redox ratio found in adipogenic cultures relative to propagating mesenchymal stem cells in previous short-term 2D differentiation studies [6]. In the current study, the decrease in redox ratio corresponded to an accumulation of lipid droplets (Fig. 2) that matched previous Oil Red O staining of differentiating hASC cultures [7, 22]. Interestingly, the average redox ratio of the cells began increasing between Days 57–183, and no additional lipid droplet accumulation was observed between these time points. These findings suggest that a decrease in optical redox ratio is associated with lipogenesis during adipogenic differentiation. The apparent lack of additional lipid synthesis between Day 57 and 183 suggests the increase in redox ratio observed after Day 57 corresponds to a return to a baseline metabolic state without significant anabolic demands. While only adipogenic differentiation was investigated in this study, a recent fluorescence lifetime microscopy study has reported increases in bound NADH and decreases in FAD during the differentiation of Caenorhabditis elegans germ cells [24], which suggests the decreases in redox ratio observed during hASC differentiation in this work is likely applicable to other tissue types as well.
The initial decrease in the redox ratio of the differentiating hASCs may be explained, in part, by evidence of mitochondrial biogenesis upon differentiation. Proteomic analysis of hASCs has demonstrated that although mitochondrial proteins make up only 8% of the total protein content, they consisted of 18% of upregulated proteins 9 days after the addition of adipogenic induction factors [25]. The 2.7-fold increase in NADH fluorescence and 3.4-fold increase in FAD over the duration of the culture period in this study also suggest substantial mitochondrial biogenesis. Using a 3T3-L1 cell line, previous work has also demonstrated a 5- to 8-fold increase in mitochondrial fluorescence staining during adipogenesis, in addition to a 20-fold increase in the expression of a variety of mitochondrial proteins [26]. Interestingly, only a two-fold increase in the rate of oxygen consumption was observed following adipogenic differentiation in that study [26]. Treatment with FCCP suggested adipocytes were only consuming oxygen at a rate that was 25% of the cell’s mitochondrial capability, compared to a rate that was approximately 60% of the capability for undifferentiated cells [26]. A lower proportion of mitochondria undergoing oxidative phosphorylation following adipogenic differentiation in that study would likely produce a lower NAD+/NADH ratio, which is consistent with the initial decrease in redox ratio observed during differentiation in this study (Fig. 1).
Interestingly, previous work has identified substantial mitochondrial biogenesis during myogenic and osteogenic differentiation [39–42], which suggests a transient decrease in redox ratio may provide a suitable biomarker for differentiation across a range of cell types. In fact, previous and ongoing studies in our lab have identified transient decreases in the redox ratio of differentiating osteoblasts and keratinocytes [6, 16]. These previous findings, and the different carbon and ATP demands across differentiated cell types, suggest differences in the magnitude and duration of redox ratio changes may enable a sensitive detection of the differentiating cell type.
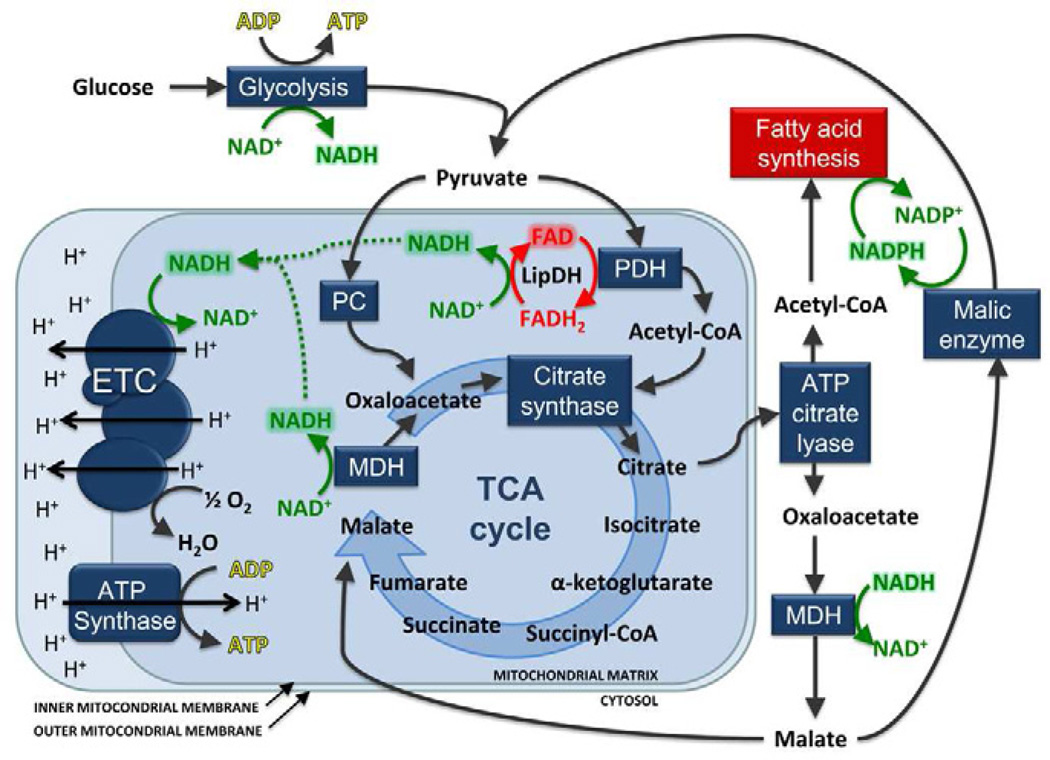
Although the physiological mechanisms that lead to mitochondrial biogenesis and a lower redox ratio during differentiation are not well understood, mitochondrial proteins play key roles in fatty acid synthesis (Fig. 6). The de novo synthesis of fatty acids requires that pyruvate be shuttled into the mitochondria, so that it can be converted to either oxaloacetate by pyruvate carboxylase (PC) or acetyl-CoA by pyruvate dehydrogenase complex (PDHC). Acetyl-CoA and oxaloacetate are condensed into citrate by citrate synthase during the first step of the tricarboxylic acid (TCA) cycle. Citrate can then be shuttled out of the mitochondria and cleaved in the cytosol so that acetyl-CoA can be used as a carbon supply for fatty acid synthesis (Fig. 6). The remaining cytosolic oxaloacetate can be converted to pyruvate through malic enzyme, during which nicotinamide adenine dinucleotide phosphate (NADPH) is also produced for use in long chain fatty acid synthesis (Fig. 6). Alternatively, cytosolic oxaloacetate can re-enter the mitochondria and TCA cycle as malate, where it can then be converted to oxaloacetate by malate dehydrogenase (MDH). Increased PDHC and MDH activity for fatty acid synthesis would produce an increase in mitochondrial NADH (Fig. 6). Metabolic flux analysis of 3T3-L1 differentiating preadipocytes in previous work has demonstrated increases in PDHC activity coinciding with an increase in mitochondrial NADH [27]. In fact, a lower baseline NAD+/NADH ratio in the mitochondria may be needed to help facilitate citrate transport out of the mitochondria rather than continued catabolism through the TCA cycle [28–29]. Collectively, an increase in flux through anabolic pathways in the recently expanded mitochondrial networks of differentiating cells without a proportional increase in ATP demand, likely combine to produce a lower optical redox ratio during the initial stages of differentiation.
Figure 6. Schematic of the fatty acid synthesis and oxidative phosphorylation pathways with an emphasis on the roles of NADH and FAD.
An increase in the demand for fatty acid synthesis relative to oxidative phosphorylation may produce an accumulation of mitochondrially-bound NADH and a decrease in NAD+. A lack of available NAD+ in the mitochondria would produce a lower ratio of FAD/FADH2 from enzyme complexes containing lipoamide dehydrogenase (LipDH), such as pyruvate dehydrogenase (PDH). ETC= electron transport chain; PC= pyruvate carboxylase; MDH = malate dehydrogenase; TCA= tricarboxylic acid.
The lack of cell proliferation in the hASC group in this study further suggests that cells are undergoing differentiation. The gradual increase in cell numbers in the co-culture group suggests HMVECs may continue to either proliferate or promote some hASC proliferation throughout the culture duration (Fig. 3). However, the overlapping redox ratio distributions of HMVECs and hASCs present a challenge in discriminating cell types from the acquired TPEF image volumes. The increased cell numbers deeper within the co-culture tissue relative to hASC mono-culture tissue suggest that HMVECs may help support hASC viability (Fig. 4). Although, the identification of HMVEC lumen formation in this (Fig. 5b) and previous work [22] suggests oxygen or nutrient delivery may be improved deeper within co-culture tissues (Fig. 5b), CD-31 staining also indicates that the mere presence of HMVEC clusters may improve hASC viability deeper within the tissue. Indeed, both in vitro work and studies of embryo development suggest endothelial cells guide stem cell migration and promote adipogenesis through paracrine signaling [30–31].
Although the decrease in optical redox ratio observed in this study is likely driven by sensitivity to NADH bound to mitochondrial proteins, additional molecules may contribute to the detected fluorescence. Concentrations of cytosolic NADH may change as it is reduced through glycolysis and oxidized by cytosolic MDH during adipogenesis. Additionally, NADPH, which cannot be distinguished from NADH using standard TPEF imaging, is produced in the cytosol so that it can be oxidized during lipogenesis [32–34]. However, previous work suggests the majority of fluorescence emitted in the blue range is produced by protein-bound NADH in mitochondria, rather than by cytosolic NADH or NADPH [35–36]. In addition, it has been shown that FAD fluorescence is quenched when bound to most proteins, and the majority of FAD fluorescence is produced by lipoamide dehydrogenase-containing enzyme complexes such as PDHC and oxoglutarate dehydrogenase complex [12, 37–38]. A decrease in NAD+ availability during lipogenesis would produce a decrease in the FAD/FADH2 ratio of enzyme complexes such as PDHC, which donate electrons to NAD+. As a result, one would expect the optical redox ratio of FAD/(NADH+FAD) measured in this study to be proportional to a ratio of NAD+ and NADH. Direct correlations of normalized fluorescence intensities and actual cofactor concentrations measured through mass spectroscopy techniques will be necessary to provide context for any studies aiming to quantify absolute concentration levels. Associating absolute cofactor concentrations with fluorescence can be further complicated by potential differences in excitation or collection efficiency with respect to depth and/or time. In fact, while the increased cell fluorescence intensity over 183 days certainly supports previous findings of mitochondrial biogenesis, significant variability was detected in those data. By measuring relative changes in the ratio of FAD and NADH fluorescence intensities, this study provides a much more robust biomarker of differentiation that accounts for variable excitation and collection efficiency.
The non-invasive optical methods employed in this study offer a complimentary role to traditional histological and biochemical assays for evaluating engineered tissue development. Although IHC, qPCR, western blot, and MS techniques offer sensitivity to a vast array of biomolecules to evaluate the differentiation status of cells, these destructive techniques cannot monitor the development of a single tissue. Rather, large sample sizes are required in tissue engineering studies to evaluate development at multiple discrete time points. The nondestructive nature of TPEF imaging in this study allowed for the identification of dynamic increases in redox ratio occurring at later time points between Days 57 through 183 (Fig. 1), suggesting a reduction in fatty acid synthesis relative to oxidative phosphorylation. This finding was also confirmed by the similar lipid droplet density and size observed between Days 57 and 183 in Oil Red O staining (Fig. 2). Future work utilizing TPEF imaging to pinpoint the cessation of lipogenesis at these later time points can be used to guide additional biochemical assays to elucidate the underlying mechanism of this phenomenon.
5. CONCLUSION
The methodology presented in this study describes the ability to quantify individual cell redox ratios within a 3D tissue culture undergoing adipogenic differentiation. Through this approach, we were able to demonstrate, for the first time, repeated assessments of the differentiation status of individual tissues over 6 months. A decrease in the optical redox ratio of metabolic cofactors was observed over the first two months of culture, which coincided with stem cell differentiation and the accumulation of lipid droplets. During the final four months, additional lipogenesis was not observed in Oil Red O staining, and the average cell redox ratio increased over time. The sensitivity of our non-invasive redox ratio measurements to adipogenic differentiation is likely related to mitochondrial biogenesis observed during differentiation in this study and others, as well as the anabolic requirements of adipogenesis. Through the automated segmentation and identification of cell objects within the 3D image volumes, we also found that the presence of endothelial cells facilitated greater cell numbers deeper within the engineered adipose tissue. As tissue engineering research progresses toward evaluations of tissue function at later in vitro time points and assessments of implant integration in vivo, these methods to noninvasively localize cell differentiation and proliferation within a 3D volume will become increasingly advantageous.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by Grant Numbers R01EB007542 from NIBIB/NIH and F32AR061933 from NIAMS/NIH, as well as the Tissue Engineering Resource Center (TERC) through Grant Number P41EB002520 from NIBIB/NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Coleman SR. Structural fat grafting: more than a permanent filler. Plast Reconstr Surg. 2006;118:108S–120S. doi: 10.1097/01.prs.0000234610.81672.e7. [DOI] [PubMed] [Google Scholar]
- 2.Soto-Gutierrez A, Yagi H, Uygun BE, Navarro-Alvarez N, Uygun K, Kobayashi N, et al. Cell delivery: from cell transplantation to organ engineering. Cell Transplant. 2010;19:655–665. doi: 10.3727/096368910X508753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dvir T, Timko BP, Kohane DS, Langer R. Nanotechnological strategies for engineering complex tissues. Nat Nanotechnol. 2011;6:13–22. doi: 10.1038/nnano.2010.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li WJ, Tuli R, Okafor C, Derfoul A, Danielson KG, Hall DJ, et al. A three-dimensional nanofibrous scaffold for cartilage tissue engineering using human mesenchymal stem cells. Biomaterials. 2005;26:599–609. doi: 10.1016/j.biomaterials.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Kim HJ, Kim UJ, Vunjak-Novakovic G, Min BH, Kaplan DL. Influence of macroporous protein scaffolds on bone tissue engineering from bone marrow stem cells. Biomaterials. 2005;26:4442–4452. doi: 10.1016/j.biomaterials.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Rice WL, Kaplan DL, Georgakoudi I. Two-photon microscopy for non-invasive, quantitative monitoring of stem cell differentiation. PLoS One. 2010;5:e10075. doi: 10.1371/journal.pone.0010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi JH, Bellas E, Vunjak-Novakovic G, Kaplan DL. Adipogenic differentiation of human adipose-derived stem cells on 3D silk scaffolds. Methods Mol Biol. 2011;702:319–330. doi: 10.1007/978-1-61737-960-4_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 9.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 10.Chance B, Estabrook RW, Ghosh A. Damped sinusoidal oscillations of cytoplasmic reduced pyridine nucleotide in yeast cells. Proc Natl Acad Sci U S A. 1964;51:1244–1251. doi: 10.1073/pnas.51.6.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scholz R, Thurman RG, Williamson JR, Chance B, Bucher T. Flavin and pyridine nucleotide oxidation-reduction changes in perfused rat liver. I. Anoxia and subcellular localization of fluorescent flavoproteins. J Biol Chem. 1969;244:2317–2324. [PubMed] [Google Scholar]
- 12.Huang S, Heikal AA, Webb WW. Two-photon fluorescence spectroscopy and microscopy of NAD(P)H and flavoprotein. Biophys J. 2002;82:2811–2825. doi: 10.1016/S0006-3495(02)75621-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skala MC, Riching KM, Gendron-Fitzpatrick A, Eickhoff J, Eliceiri KW, White JG, et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc Natl Acad Sci U S A. 2007;104:19494–19499. doi: 10.1073/pnas.0708425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayevsky A, Chance B. Intracellular oxidation-reduction state measured in situ by a multichannel fiber-optic surface fluorometer. Science. 1982;217:537–540. doi: 10.1126/science.7201167. [DOI] [PubMed] [Google Scholar]
- 15.Bird DK, Yan L, Vrotsos KM, Eliceiri KW, Vaughan EM, Keely PJ, et al. Metabolic mapping of MCF10A human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme NADH. Cancer Res. 2005;65:8766–8773. doi: 10.1158/0008-5472.CAN-04-3922. [DOI] [PubMed] [Google Scholar]
- 16.Levitt JM, McLaughlin-Drubin ME, Munger K, Georgakoudi I. Automated biochemical, morphological, and organizational assessment of precancerous changes from endogenous two-photon fluorescence images. PLoS One. 2011;6:e24765. doi: 10.1371/journal.pone.0024765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostrander JH, McMahon CM, Lem S, Millon SR, Brown JQ, Seewaldt VL, et al. Optical redox ratio differentiates breast cancer cell lines based on estrogen receptor status. Cancer Res. 2010;70:4759–4766. doi: 10.1158/0008-5472.CAN-09-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schenke-Layland K, Riemann I, Damour O, Stock UA, Konig K. Two-photon microscopes and in vivo multiphoton tomographs--powerful diagnostic tools for tissue engineering and drug delivery. Adv Drug Deliv Rev. 2006;58:878–896. doi: 10.1016/j.addr.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Xu M, Wang X, Yan Y, Yao R, Ge Y. An cell-assembly derived physiological 3D model of the metabolic syndrome, based on adipose-derived stromal cells and a gelatin/alginate/fibrinogen matrix. Biomaterials. 2010;31:3868–3877. doi: 10.1016/j.biomaterials.2010.01.111. [DOI] [PubMed] [Google Scholar]
- 20.Gerlach JC, Lin YC, Brayfield CA, Minteer DM, Li H, Rubin JP, et al. Adipogenesis of human adipose-derived stem cells within three-dimensional hollow fiber-based bioreactors. Tissue Eng Part C Methods. 2012;18:54–61. doi: 10.1089/ten.tec.2011.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nazarov R, Jin HJ, Kaplan DL. Porous 3-D scaffolds from regenerated silk fibroin. Biomacromolecules. 2004;5:718–726. doi: 10.1021/bm034327e. [DOI] [PubMed] [Google Scholar]
- 22.Kang JH, Gimble JM, Kaplan DL. In vitro 3D model for human vascularized adipose tissue. Tissue Eng Part A. 2009;15:2227–2236. doi: 10.1089/ten.tea.2008.0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zipfel WR, Williams RM, Christie R, Nikitin AY, Hyman BT, Webb WW. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc Natl Acad Sci U S A. 2003;100:7075–7080. doi: 10.1073/pnas.0832308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stringari C, Cinquin A, Cinquin O, Digman MA, Donovan PJ, Gratton E. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc Natl Acad Sci U S A. 2011;108:13582–13587. doi: 10.1073/pnas.1108161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeLany JP, Floyd ZE, Zvonic S, Smith A, Gravois A, Reiners E, et al. Proteomic analysis of primary cultures of human adipose-derived stem cells: modulation by Adipogenesis. Mol Cell Proteomics. 2005;4:731–740. doi: 10.1074/mcp.M400198-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Wilson-Fritch L, Burkart A, Bell G, Mendelson K, Leszyk J, Nicoloro S, et al. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol. 2003;23:1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Si Y, Yoon J, Lee K. Flux profile and modularity analysis of time-dependent metabolic changes of de novo adipocyte formation. Am J Physiol Endocrinol Metab. 2007;292:E1637–E1646. doi: 10.1152/ajpendo.00670.2006. [DOI] [PubMed] [Google Scholar]
- 28.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgunov IG, Solodovnikova NY, Sharyshev AA, Kamzolova SV, Finogenova TV. Regulation of NAD(+)-dependent isocitrate dehydrogenase in the citrate producing yeast Yarrowia lipolytica. Biochemistry (Mosc) 2004;69:1391–1398. doi: 10.1007/s10541-005-0086-3. [DOI] [PubMed] [Google Scholar]
- 30.Cao Y. Angiogenesis modulates adipogenesis and obesity. J Clin Invest. 2007;117:2362–2368. doi: 10.1172/JCI32239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai N, Jayaraman A, Lee K. Enhanced proliferation of human umbilical vein endothelial cells and differentiation of 3T3-L1 adipocytes in coculture. Tissue Eng Part A. 2009;15:1053–1061. doi: 10.1089/ten.tea.2008.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal. 2008;10:179–206. doi: 10.1089/ars.2007.1672. [DOI] [PubMed] [Google Scholar]
- 33.Georgakoudi I, Jacobson BC, Muller MG, Sheets EE, Badizadegan K, Carr-Locke DL, et al. NAD(P)H and collagen as in vivo quantitative fluorescent biomarkers of epithelial precancerous changes. Cancer Res. 2002;62:682–687. [PubMed] [Google Scholar]
- 34.Rocheleau JV, Head WS, Piston DW. Quantitative NAD(P)H/flavoprotein autofluorescence imaging reveals metabolic mechanisms of pancreatic islet pyruvate response. J Biol Chem. 2004;279:31780–31787. doi: 10.1074/jbc.M314005200. [DOI] [PubMed] [Google Scholar]
- 35.Blinova K, Levine RL, Boja ES, Griffiths GL, Shi ZD, Ruddy B, et al. Mitochondrial NADH fluorescence is enhanced by complex I binding. Biochemistry. 2008;47:9636–9645. doi: 10.1021/bi800307y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eng J, Lynch RM, Balaban RS. Nicotinamide adenine dinucleotide fluorescence spectroscopy and imaging of isolated cardiac myocytes. Biophys J. 1989;55:621–630. doi: 10.1016/S0006-3495(89)82859-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunz WS, Kunz W. Contribution of different enzymes to flavoprotein fluorescence of isolated rat liver mitochondria. Biochim Biophys Acta. 1985;841:237–246. doi: 10.1016/0304-4165(85)90064-9. [DOI] [PubMed] [Google Scholar]
- 38.Saks VA, Veksler VI, Kuznetsov AV, Kay L, Sikk P, Tiivel T, et al. Permeabilized cell and skinned fiber techniques in studies of mitochondrial function in vivo. Mol Cell Biochem. 1998;184:81–100. [PubMed] [Google Scholar]
- 39.Rehman J. Empowering self-renewal and differentiation: the role of mitochondria in stem cells. J Mol Med (Berl) 2010;88:981–986. doi: 10.1007/s00109-010-0678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen CT, Shih YR, Kuo TK, Lee OK, Wei YH. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells. 2008;26:960–968. doi: 10.1634/stemcells.2007-0509. [DOI] [PubMed] [Google Scholar]
- 41.Moyes CD, Mathieu-Costello OA, Tsuchiya N, Filburn C, Hansford RG. Mitochondrial biogenesis during cellular differentiation. Am J Physiol. 1997;272:C1345–C1351. doi: 10.1152/ajpcell.1997.272.4.C1345. [DOI] [PubMed] [Google Scholar]
- 42.Duguez S, Feasson L, Denis C, Freyssenet D. Mitochondrial biogenesis during skeqletal muscle regeneration. Am J Physiol Endocrinol Metab. 2002;282:E802–E809. doi: 10.1152/ajpendo.00343.2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.