Abstract
This review examines why a knowledge of plant protein phosphorylation events is important in devising strategies to protect crops from both biotic and abiotic stresses, and why proteomics should be included when studying stress pathways. Most of the achievements in elucidating phospho-signaling pathways in biotic and abiotic stress are reported from model systems: while these are discussed, this review attempts mainly to focus on work done with crops, with examples of achievements reported from rice, maize, wheat, grape, Brassica, tomato, and soy bean after cold acclimation, hormonal and oxidative hydrogen peroxide treatment, salt stress, mechanical wounding, or pathogen challenge. The challenges that remain to transfer this information into a format that can be used to protect crops against biotic and abiotic stresses are enormous. The tremendous increase in the speed and ease of DNA sequencing is poised to reveal the whole genomes of many crop species in the near future, which will facilitate phosphoproteomics and phosphogenomics research.
Keywords: abiotic stress, biotic stress, phosphoproteomics, signaling
INTRODUCTION
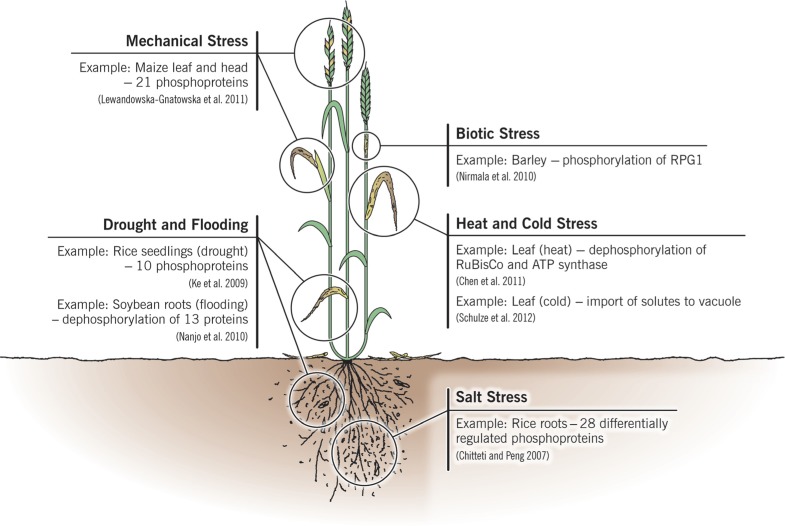
Agricultural plants have all been domesticated from their wild relatives, and as such have evolved and been selected to withstand many typical environmental stresses. These may be abiotic, such as drought, heat, cold, and soil salinity, or they may be biotic, principally from pathogenic fungi, oomycetes, bacteria, and viruses. The major types of stresses which crops face routinely, along with specific examples which will be discussed in the text, are indicated in Figure 1. Their ability to overcome these stresses and still be able to propagate are necessary adaptations for a sessile organism, and desirable traits for agricultural crops. With the advent of proteomics it has become evident that when plants are stressed there are changes in their proteome as they attempt to overcome the stress and maintain homeostasis (Kosová et al., 2011; Rampitsch and Bykova, 2012). Furthermore, some crop cultivars are genetically better equipped to withstand stress and although this is also reflected at the proteome level, the biochemical mechanisms resulting in the improved phenotype may not be understood at all. For example, the Australian bread wheat cultivar “Excalibur” is better able to withstand drought than “Kukri,” and quantitative changes have been documented at the proteome level during drought stress (Ford et al., 2011). Turning this knowledge into a tool to create improved crops (i.e., a selectable marker) is the ultimate goal. Drought resistance highlights many of the challenges encountered when attempting to breed for a quantitative trait whose successful deployment may also require resistance to other abiotic stresses that accompany drought: heat, intense sunlight, and possibly nutrient starvation (Fleury et al., 2010).
FIGURE 1.
A summary of the major categories of stresses which crops face routinely, showing their principal target organs and specific examples which are discussed in more detail in the text and in Table 1.
One biochemical manifestation common to all stresses is specific, regulated protein phosphorylation. It is universally accepted that a major part of the signal linking the environmental perception of the stress at the cell surface to the nucleus, where response proteins can be translated, is protein phosphorylation generally transmitted by protein kinase cascades (Kersten et al., 2009). A few kinase-mediated signaling pathways have been elucidated (e.g., Asai et al., 2002) and in the model plants Arabidopsis thaliana (van Bentem and Hirt, 2007; Pitzschke et al., 2009) and rice (Chen and Ronald, 2011) a picture of the complexity of these signaling pathways, with all their cross-talk and branch points, is beginning to emerge. Since these pathways rely principally on post-translational modification to transmit their signal, their elucidation is well served by a proteomic approach. In spite of this relatively simple problem statement, the challenges to elucidating stress pathways are significant. Kinase pathways have been demonstrated to become active within minutes – in some cases seconds – of a stimulus, even in crop plants (Nirmala et al., 2010; Schulze et al., 2010) and furthermore, in most cases the signal is transient and weak, presenting challenges to detection and quantification. Furthermore, phosphorylation cross-talk with other PTMs and with hormone signals also occurs, which further increases the complexity of understanding and ultimately manipulating this signal. Integration with other cellular signaling pathways is only starting to come under systematic investigation.
Plant responses to abiotic stresses are dynamic and complex, and quite different depending on the type, level, and duration of the stress involved, and on the type of tissue under stress (for review, see Cramer et al., 2011). Several research groups are deciphering the involvement of different kinases such as CDPKs (Wan et al., 2007), CIPKs (Batistic and Kudla, 2009; Luan, 2009), and MAPKs (Lee et al., 2008; Popescu et al., 2009) in abiotic and biotic stress signaling networks, in both A. thaliana and Oryza sativa (rice). The whole complement of protein phosphatases in A. thaliana (Kerk et al., 2002) and rice (Singh et al., 2010) genomes have recently been identified based on the availability of genome sequences, online databases, and in silico search tools. The latter revealed the presence of 112 phosphatase-coding genes in A. thaliana (Kerk et al., 2002) and 132 in rice (Singh et al., 2010).
Phosphorylation sites can be affected in diverse ways in plant species, and one such way is through single-nucleotide polymorphisms (SNPs). The effect of non-synonymous SNPs (nsSNPs) on phosphorylation sites was analyzed using a large number of experimentally verified phosphorylation sites (7,178 unique sites assigned to 4,252 protein-coding loci), combined with predicted phosphorylation sites to achieve better proteome coverage (80% of all protein-coding loci), and natural variation datasets in A. thaliana (Riaño-Pachón et al., 2010). It was found that phosphorylation hotspots are preferentially located outside conserved domains, and nsSNPs affected experimental phosphorylation sites as much as the corresponding non-phosphorylated amino-acid residues. Interestingly, proteins in which predicted phosphorylation sites were affected by nsSNPs (loss and gain), were determined to be mainly receptor proteins, stress response proteins, and proteins involved in nucleotide and protein binding. In contrast, proteins involved in metabolism, catalytic activity, and biosynthesis were less affected. Elucidating the effect of phosphorylation sites affected by nsSNPs on adaptive responses represents an exciting research goal.
A. thaliana represents the most successful system for making progress in understanding plant protein phosphorylation. All of the advantages of using A. thaliana for genomics research also hold for proteomics and phosphoproteomics: a small, fully sequenced, and well-annotated genome; the ability to be grown and manipulated in cell culture; easy access to a comprehensive collection of mutants and genomic resources. No agricultural crop is backed by these resources, and within the major monocots only rice can be in any way considered as a model suitable for high-throughput phosphoproteomics, for which a fully sequenced genome is all but essential. Without this prerequisite, phosphoprotein identification from enriched phosphopeptides through homology-based matching becomes much less certain. Since modern mass spectrometry relies extensively on this type of search, it is important that the database is complete to help reduce the impact of many of the assumptions that are necessarily made when interpreting tandem mass spectra produced by collision-induced dissociation (CID). The complex task of phosphorylation site identification by MS has been reviewed recently (St-Denis and Gingras, 2012).
Here, we review the latest research in phosphoproteomics, designed first to discover novel phosphoproteins, and second to begin to understand their role in transmitting stress signals from the cell surface to the nucleus, so that an appropriate response is elicited. Although A. thaliana is not a crop, its extensive use in research necessitates its inclusion here.
BIOTIC STRESS
Implied within the term proteomics is a high-throughput, global approach, and high-throughput phosphopeptide discovery has been applied to many biological systems including plants (e.g, Nakagami et al., 2010). The most productive approach is based on the enrichment of tryptic phospho-serine, -threonine, and -tyrosine peptides using either metal affinity or ion exchange chromatography, or a combination of the two. This is followed by LC-MS analysis using CID and/or electron-transfer dissociation (ETD) to fragment peptides, usually in a high-resolution mass spectrometer. A typical approach is described in detail by Villén and Gygi (2008). CID most often produces MS2 spectra containing a prominent neutral-loss ion (m/z = [M + H] - 98.0), representing the loss of H3PO4 from the precursor ion, and only a few other product ions. Such spectra can be difficult to interpret because of the lack of y- and b-product ions and its localization is often ambiguous because the phosphate group is lost in most phospho-serine and -threonine peptides. Phospho-tyrosine is stable in CID, but is rare, present at typically 1% of phospho-serine and -threonine combined. During ETD, there is no neutral loss of H3PO4, however, this type of dissociation reduces the charge of the product ions, and so is best suited for precursor ions whose charge is >3+ (Goods et al., 2007); most tryptic peptides carry a charge of 2+. The phosphoproteomics study described by Nakagami et al. (2010) is a good example of the high-throughput phosphoproteomics approach applied to a plant system, in this case rice. They identified 6,919 phosphopeptides from 3,393 proteins from Oryza sativa japonica “Nipponbare” and went on to demonstrate a high degree of conservation of phosphorylation sites between rice and other plants. Even though the plant material studied was not stressed, this result indicates that findings from rice phosphoproteomics may indeed be applicable to other crop plants. Reports of knowledge transfer from A. thaliana to benefit crop improvement are still surprisingly rare (Spannagl et al., 2011).
High-throughput phosphoproteomic analyses performed on plants challenged with a pathogen (or pathogen elicitor) have only been reported from the A. thaliana–Pseudomonas syringae pv. tomato interactions, as this is the simplest and most intensely studied plant-biotic stress interaction system (Boller and Felix, 2009). Benschop et al. (2007) described early changes in the phosphoproteome of A. thaliana proteins from cells treated with either the fungal elicitor xylanase or the bacterial elicitor Flg22. The latter is a 22 amino-acid peptide from the bacterial protein flagellin and is ultimately derived from P. syringae pv. tabaci; it elicits many of the same downstream signals as the pathogen itself (Felix et al., 1999). Benschop et al. (2007) found 1,186 phosphopeptides in their membrane-enriched fraction, with 76 membrane-associated proteins being differentially phosphorylated in response to elicitors (Table 1). Although they were able to identify several kinases known to be involved in the plant defense response, most proteins that were identified had no known role in plant immunity. These remain to be validated. Their approach included 14N/15N labelling in culture to permit relative quantification of phosphopeptides from treated vs. untreated cells – an important piece of information that permits identification of proteins truly responding to the stimulus. Jones et al. (2006) performed a similar experiment with whole plants inoculated with P. syringae pv. tomato DC3000, except that they used iTRAQ for relative quantification of peptides. They identified five proteins which were phosphorylated reproducibly and rapidly in response to pathogen challenge. Their findings (i.e., identified proteins) did not overlap with those of the previous study (Benschop et al., 2007), possibly highlighting slight differences in their experimental approach, but more likely indicating that many more phosphoproteins remain to be discovered before a complete picture can emerge. Significantly, no high-throughput proteomics approach has been applied to any crop plants because the task is still technically very demanding, most especially in the area of phosphopeptide identification and site placement in crops lacking a complete genomic sequence. Even in rice, phosphoproteomics endeavors have been limited to studying development and abiotic stress.
Table 1.
Survey of stress-related phosphoproteomics studies.
| Condition; Type of stress | Proteomic approach; Subcellular fractions | # Phosphoproteins # Sites identified | Significant findings Phosphoproteomic response | Reference |
|---|---|---|---|---|
| Biotic: A. thaliana cells treated with elicitors Flg22 and xylanase | Comparative phosphoproteome of 14 N/15 N labeled proteins in culture for relative quantification of phosphopeptides; LC-MS with FT-ICR | 1,186 phosphopeptides identified, 76 differentially phosphorylated membrane-associated proteins identified | Data indicate a high degree of complexity at the level of post-translational phosphorylation, and suggest that protein translocation and vesicle traffic are important aspects of early signaling and defense in response to general elicitors | Benschop et al. (2007) |
| Biotic: A. thaliana with P. syringae pv. tomato DC3000 | Comparative phosphoproteome with iTRAQ quantification; LC-MS with linear IT (Q-Trap) | 5 proteins identified that were phosphorylated reproducibly and rapidly in response to pathogen challenge | Identification of proteins potentially phosphorylated as part of a plant basal defence response in A. thaliana | Jones et al. (2006) |
| Heat 42°C treatment of rice seedlings | Total protein, IEF 2D-PAGE; staining with Pro-Q Diamond and Coomassie R-250, MALDI-TOF/TOF MS and MS/MS, immunoblotting of ATP-β | 10 differentially phosphorylated proteins | Dephosphorylation of RuBisCo and phosphorylation of ATP-β decreasing the activities of these proteins; dephosphorylation of the mRNA binding protein and 2-Cys peroxiredoxin BAS1 | Chen et al. (2011) |
| Cold acclimation of A. thaliana plants at 4°C for 5 days | Tonoplast proteins; LC-MS/MS using an LTQ-Orbitrap; Label-free quantification; AQUA SRM using a Triple Quadrupole MS | The phosphorylated and non-phosphorylated peptides QTT(pS)MDK (fromTMT1), QTT(pS)MEK (fromTMT2), and T(pS)GLAEVLSK (fromTDT) were targets of analysis | Import of solutes into the vacuole is enhanced by an increased abundance of transporters (e.g., TDT) or by post-translational activation (e.g.,TMT phosphorylation) | Schulze et al. (2012) |
| Drought treatment of rice (Oryza sativa L. cv. Nipponbare) seedlings | Total protein, IEF 2D-PAGE; immunodetection of phospho-Ser and phospho-Thr; MALDI-TOF/TOF MS and MS/MS | 10 phosphoproteins | Responsive phosphoproteins: NAD-malate dehydrogenase, ABA- and stress-inducible protein, ribosomal protein, drought-induced S-like ribonuclease, OSJNBa0084K20.14, ethylene-inducible protein, GTP-binding protein beta subunit-like; dephosphorylation of OSJNBb0039L24.13 protein, r40c1 protein, and germin-like protein 1 | Ke et al. (2009) |
| ABA treatment of rice (Oryza sativa L. cv. Nipponbare) seedlings | Total protein, IEF 2D-PAGE; immunodetection of phospho-Ser and phospho-Thr; MALDI-TOF/TOF MS and MS/MS | 6 responding phosphoproteins out of 22 phosphoserine- and 6 phosphothreonine-containing proteins | Identified G protein beta subunit-like, ascorbate peroxidase, manganese superoxide dismutase, triosephosphate isomerase, putative Ca2+/H+ antiporter regulator protein, and glyoxysomal malate dehydrogenase | He and Li (2008) |
| ABA treatment of A. thaliana | 14 N/15 N reciprocal metabolic labelling, total protein extracts, TiO2 enrichment, LC-MS/MS on an LTQ-Orbitrap with CID or ETD, quantitative untargeted analysis | 50 ABA-responding phosphopeptides out of 4,675 total; over 20 different phosphoproteins out of 2,185 detected | Increases in phosphorylation of subfamily 2 SNF1-related kinases and ABA-responsive basic leucine zipper transcription factors; four members of the aquaporin family showed decreased phosphorylation at a C-terminal Ser | Kline et al. (2010) |
| Flooding of early-stage soybean seedling, effect on roots and hypocotyls | Total protein extracts; multiplexed 2D-DIGE analysis and 2D-gel staining with Pro-Q Diamond; nanoLC-MS/MS on LTQ XL Orbitrap MS | 3 protein spots were phosphorylated, while 13 were dephosphorylated in flooded seedlings; 2 phosphopeptides | Most of the identified proteins were dephosphorylated in response to flooding stress and included stem glycoprotein, class I small heat shock protein, eukaryotic translation initiation factor 5A2 and cyclophilin; two phosphopeptides verified in malate dehydrogenase | Nanjo et al. (2010) |
| Flooding of early-stage soybean seedlings, effect on root tips | Total protein extracts; phosphoprotein enrichment with Qiagen affinity column; nanoLC-MS/MS analysis on LTQ-Orbitrap; label-free SIEVE quantification | 3 phosphorylated and 3 dephosphorylated proteins affected by flooding; while a total of 40 phosphoproteins and 18 phosphopeptides were identified | The lipoxygenase and two of the pyruvate kinases were both phosphorylated and increased in abundance; heat shock protein 70 and two alpha-tubulins were found to be decreased in abundance and dephosphorylated; identified proteins involved in energy generation, protein synthesis, and cell structure maintenance | Nanjo et al. (2012) |
| Salt treatment of rice roots | Total protein extracts; IEF 2D-PAGE; Pro-Q Diamond; MALDI-TOF/TOF PMF and MS/MS | 17 differentially upregulated and 11 differentially downregulated putative phosphoproteins | Upregulated: salt-induced protein; mannose-binding lectin; glutathione S-transferase; small GTP binding; transposase, dnaK-type Hsp70, disease resistant protein, dimethylaniline monooxygenase, six hypothetical and unknown function proteins. Downregulated: glucan endo-1,3-beta-d-glucosidase, protein kinase, retroelement, glyceraldehyde-3-phosphate dehydrogenase, and unknown function | Chitteti and Peng (2007) |
| Salt and H2O2 treatment of A. thaliana plants | Plasma membrane vesicles; enrichment with TiO2columns; AQUA quantification; MALDI-TOF MS and MS/MS; mutation; GFP fusion and confocal microscopy | Adjacent sites in the C-terminal tail of several AtPIPs: two sites in AtPIP2;1/2;2/2;3; three sites in AtPIP2;4; three sites in AtPIP2;7; two sites in SoPIP2;1 | Quantitative changes in aquaporin phosphorylation were linked to plant AtPIP2;1 aquaporin subcellular localization; identified six novel phosphorylation sites in AtPIPs | Prak et al. (2008) |
| High salinity treatment of A. thaliana seedlings | Membrane fractions, protease shaving, enrichment using Zr(IV)-IMAC magnetic beads; nanoLC-MS/MS; dimethyl D/H stable-isotope labeling; label-free quantification | 200 mM salt treatment – 18 phosphorylation sites from 15 proteins were identified; 400 mM salt treatment – a novel phosphopeptide in AtMKK2 | The level of phosphopeptides on 14-3-3-interacting ATPase AHA1, sugar transporter STP1, Patellin-2, probable inactive receptor kinase (At3g02880), and probable purine permease 18 showed at least twofold increase; two novel phosphorylation sites found in aquaporin PIP2;4, and salt-induced phosphorylation site in AtMKK2 | Hsu et al. (2009) |
| Moderate short-term saline growth conditions of maize | Salt-resistant maize hybrid genotype SR12, 25 mM NaCl for 1 h treatment; root proteins; IEF 2D-PAGE; PhosTag stain; MALDI-TOF/TOF PMF | 16 total responsive: 10 proteins were phosphorylated; 6 proteins were dephosphorylated | Phosphorylation of fructokinase, UDP-glucosyl transferase BX9, and 2-Cys-peroxyredoxine was enhanced, whereas an isocitrate-dehydrogenase, calmodulin, maturase, and a 40-S-ribosomal protein were dephosphorylated | Zörb et al. (2010) |
| High salinity treatment of A. thaliana and rice plants | Microsomal fractions, trypsin shaving, tube-gel assisted membrane protein digestion; enrichment using Zr(IV)-IMAC and TiO2 magnetic beads; Q-TOF nanoLC-MS/MS; label-free quantification on Q-TOF, LTQ-FT and Q-Trap | 400 mM salt treatment – 27 phosphorylation sites from 20 A. thaliana proteins; 200 mM salt treatment – 13 phosphorylation sites from 8 proteins | The phosphorylation sites of photosystem II reaction center protein H PsbH are regulated by salt stress in A. thaliana and rice and are evolutionarily conserved in plants | Chang et al. (2012) |
| Oxidative H2O2-treated A. thaliana root cell culture | Nuclear and cytosolic fractions, IMAC, neutral loss-triggered MS3 on LTQ; LTQ-FT; Peptide Chip analysis | 3 phosphopeptides from 3 proteins, and 4 phosphorylation sites | A peptide of the SR splicing factor SCL30 was most strongly affected and phosphorylated by SRPK4 constitutively and by MAPKs in response to oxidative stress; increased phosphorylation of a peptide from a putative chromatin remodeling protein (At5g07350) and a peptide from the PK At5g35980 | de la Fuente van Bentem et al. (2008) |
| In silico expression profiles of protein phosphatase genes in rice under salinity, cold and drought stress | Genome sequences, microarray gene expression analysis, online databases and in silico search tools, RT-PCR analysis | The presence of 132 protein phosphatase-coding genes in rice genome; 46 protein phosphatase genes were differentially expressed in 3 abiotic stresses | 31 phosphatase genes upregulated and 15 exhibited downregulation; 6 genes were upregulated in all stress conditions tested; 13 OsPP genes were upregulated in salt and drought stress together; 82 genes were differentially expressed in different developmental stages | Singh et al. (2010) |
| Differential hormonal GAs; BL; 2, 4-D, ABA, cold, or salt treatment of rice seedlings | Total protein extracts from leaf sheaths, leaf blades and roots, in vitro 32P labeling, IEF 2D-PAGE; ESI Q-TOF MS/MS; MALDI-TOF MS analysis | 42 phosphoproteins identified, among them 13 were stress/hormonal responsive; 8 sites of phosphorylation in 6 phosphoproteins | Phosphorylation of 13 proteins enhanced differentially by hormone and stress treatments; cytoplasmic malate dehydrogenase found to be phosphorylated in all tissues; a common enhanced phosphorylation of calreticulin by different stresses | Khan et al. (2005) |
| Sucrose resupply of A. thaliana seedlings | Plasma membrane vesicles, 4 time points 3–30 min after resupply, protease shaving, IMAC, nanoLC-MS/MS on LTQ, neutral loss-driven MS3, label-free quantitation | 40 out of total detected 67 phosphorylation sites showed sucrose-induced changes | Increased phosphorylation level at the C terminus of plasma membrane H+-ATPases; new site in AHA1 and/or AHA2 crucial for activity; novel sites with rapid increases for both receptor kinases and cytosolic kinases, and sucrose transporter SUC1 | Niittylä et al. (2007) |
| Mechanical wounding of maize leaves | Total protein extract at 5 time points 10–180 min after wounding, IEF 2D-PAGE; staining with Pro-Q Diamond and Sypro Ruby | 21 phosphoproteins | Phosphoproteins involved in either central metabolism, protein folding, or cellular signaling; the Gypsy retrotransposon-related protein and AtPUF43 | Lewandowska-Gnatowska et al. (2011) |
One of the most intriguing and challenging aspects of protein phosphorylation is how rapidly, and often transiently, proteins are phosphorylated. This makes it difficult to catch the participants in action. Peck et al. (2001) convincingly demonstrated using [32P]-ortho phosphate pulse labelling in cells and in planta that AtPhos43, and related proteins in rice and tomato, are phosphorylated within minutes of treatment with flagellin in A. thaliana suspension cells. More recent work has indicated that changes occur even much more rapidly than this. Phosphorylation of FLS2, the flagellin receptor, and the receptor-like kinase BAK1 (brassinosteroid receptor-associated receptor kinase 1), occurs within 15 s of treatment with Flg22 in A. thaliana suspension cells (Schulze et al., 2010). Such a rapid response is difficult to detect in whole plants because of the difficulty of obtaining a synchronous response, even under the most controlled conditions. Nevertheless, Nirmala et al. (2010) demonstrated in planta that the barley protein RPG1 (so named because it confers resistance to Puccinia graminis, the causal agent of stem rust in barley and wheat) is phosphorylated within 5 min of exposure to spores from avirulent (but not virulent) races of P. graminis. Furthermore, non-viable spores, or spores from other pathogens, e.g., the closely related stipe rust, P. striiformis. f. sp. hordei, did not trigger this response. The authors also showed that phosphorylation of RPG1 was correlated with resistance to stem rust, by using suitable loss-of-function barley mutants and kinase inhibitors. They hypothesized that an effector must be present on the surface of the spore, since rust spores do not actually germinate until at least 30 min have elapsed post-inoculation. Since RPG1 does not have a known receptor motif, it is possible that it associates with a receptor to perceive the presence of avirulent, viable rust spores. This causes autophosphorylation of RPG1 through its pseudokinase domain and presumably initiates phosphorylation cascades which help to coordinate the defense response, including the expression of suitable resistance genes.
The actual biochemical events occurring in this pathway are not yet know, and this is a recurring theme in phosphoproteomics. Although phosphopeptide discovery is challenging enough, it is much more challenging – and more time consuming – to validate downstream phosphorylation events, which must be done individually. A necessary prerequisite to this work is the discovery of candidates to be validated, an activity which has been restricted mainly to A. thaliana. Some of the findings from A. thaliana have been transferred to related crops, such as tomato. For example, Stulemeijer et al. (2007) demonstrated that transgenic tomato plants expressing both the Cf-4 resistance gene and the avirulence gene Avr4 activate at least three MAP kinases: LeMPK1, 2, and 3. The Cf-4 gene confers resistance to the phytopathogen Cladosporium fulvum, and the avirulence protein Avr4, when co-expressed in planta, presents a hypersensitive response only at the permissive temperature of 33°C or higher. These transgenic plants are convenient tools for studying the hypersensitive response, because it can be induced in a synchronous manner without the need for inoculation with a pathogen, simply by moving the plants to a warmer location. In spite of this advantage, there are no shortcuts in validating phosphorylation events. No analogous models currently exist for other agricultural crops, although Avr proteins have been cloned from pathogens of widely grown crop species, including Magnaporthe grisea (rice; Jia et al., 2000), Phytophthora sojae (soybean; Shan et al., 2004), Phytophthora infestans (potato; Armstrong et al., 2005), and Melampspora lini (flax; Catanzariti et al., 2006).
In spite of the lack of high-throughput phosphoproteomics studies on pathogen-challenged rice, some details of the phosphosignaling pathways are emerging (reviewed by Chen and Ronald, 2011). A good example is Xa21-mediated immunity (Park et al., 2010). The gene Xa21 confers resistance to Xanthomonas oryzae which is a serious bacterial pathogen of rice, causing bacterial blight. Its product, a receptor kinase, recognizesthe highly conserved Ax21 protein which is expressed in a wide range of Xanthomonas species, including X. oryzae (Song et al., 1995). Downstream phosphorylation events include activation of mitogen-activated protein kinases, transcription factors, and at least one protein phosphatase. Similarly, the effects of the fungal elicitor chitin have also been examined in rice, because the most serious disease of rice (rice blast) is caused by the fungus Magnaporthe grisea. Again, downstream protein discovery has been achieved individually rather than through a high-throughput approach (Chen and Ronald, 2011 and references therein).
ABIOTIC STRESS
TEMPERATURE: HEAT STRESS AND COLD ACCLIMATION
High temperature is one of the most important abiotic stresses that reduce crop yield and quality. The mean global temperature has steadily increased in recent decades due to increasing atmospheric greenhouse gases especially CO2 and CH4. Crop plant production is likely to be severely affected by the increasing temperature. Physiological mechanisms for heat tolerance of rice plants have been investigated (Lee et al., 2007). Rice plants were shown to remodel their proteomes and phosphoproteomes in response to high temperature stress (Scafaro et al., 2010; Chen et al., 2011). In a recent study, 10 differentially expressed phosphoproteins were identified in rice leaves in response to heat stress (Chen et al., 2011). Analysis of the biological processes revealed that three of the variable phosphoproteins are involved in the Calvin cycle, two are part of hydrogen peroxide (H2O2) catabolism, two participate in ATP synthesis-coupled proton transport, one is involved in microtubule-based movement, and one in cellular metabolic processes; the remainder have unknown functions (Table 1). Heat stress induced the dephosphorylation of ribulose bisphosphate carboxylase (RuBisCo) and the phosphorylation of ATP synthase subunit-β. This modification decreases the activities of these enzymes, but the functional significance of other phosphorylation events remains to be examined.
An altered phosphorylation of tonoplast monosaccharide transporters (TMTs) was found in response to cold acclimation in A. thaliana (Schulze et al., 2012). Single reaction monitoring of AtTMT1 and AtTMT2 phosphorylation in cold-treated and control plants was used for targeted quantitative analysis of specific peptides (Table 1). A general increase in the abundance of tonoplast disaccharide transporter indicated that although this protein was also present in a phosphorylated form under cold acclimation, the phosphorylation level at Ser409 was not greatly altered. A significantly higher phosphorylation of one serine residue in the central hydrophilic loop of TMT1 and TMT2 was observed upon incubation of the plants at 4°C. This loop distinguishes TMT proteins from all other plant sugar transporters, and possesses a number of putative and experimentally identified phosphorylation sites (Schulze et al., 2012). The transporters TMT1 and TMT2 have been proposed to function as the main proteins catalyzing the cold-induced import of glucose and fructose into the large central vacuole of mesophyll cells (Wormit et al., 2006). Phosphoproteomic analyses have been performed using tonoplast membrane proteins isolated from rice, barley, and A. thaliana (Whiteman et al., 2008a,b; Endler et al., 2009). Phosphorylation sites for TMT1 orthologs from A. thaliana and barley were identified. A possible direct or indirect regulation of the activity of TMT proteins by phosphorylation could explain the cold-induced redistribution of glucose and fructose toward the vacuole without alterations in TMT protein abundance. However, the precise phosphorylation stoichiometry of TMT proteins could not be deduced. A mitogen-activated triple kinase-like protein kinase has been identified recently that transfers phosphate groups to amino-acid residues within the hydrophilic loop of TMT1. Glucose uptake into isolated vacuoles was stimulated by this protein kinase, indicating that phosphorylated vacuolar glucose carriers exhibit higher transport activity. Furthermore, T-DNA insertion mutants of this kinase phenocopied TMT loss-of-function mutant plants when high external glucose was applied and when plants were exposed to cold temperatures (Wingenter et al., 2011). Thus, in the physiological context of cold acclimation, import of solutes into the vacuole was enhanced by an increased abundance of transporters (e.g., TDT) or by post-translational activation (e.g., TMT phosphorylation).
WATER SUPPLY: DROUGHT AND FLOODING
Drought-induced crop losses and their economic impact are predicted to increase with global climate change. In many parts of Asia, drought has become the largest constraint on rice production and efforts have been focused on identifying drought-resistant germplasm, and on discovering the genetic basis of drought-adaptive traits in rice (for review, see Zhang, 2007), with the long-term goal to develop rice cultivars referred to as Green Super Rice. It has proven difficult to identify genetic segments with clear, reproducible effects on yield under stress, and challenges remain before rice production can be increased sustainably under real agricultural field conditions. Proteomics and phosphoproteomics are recent additions to the molecular arsenal used to analyze plants affected by drought.
Differential regulation of proteins and phosphoproteins in rice under drought stress was recently investigated using 2-DE, phospho-specific antibody immunoblotting, and mass spectrometry (Ke et al., 2009). Among 10 proteins that exhibited altered phosphorylation, 3 were previously shown to be phosphorylated under various conditions of drought and/or by exogenous abscisic acid (ABA) in different species. These were NAD-malate dehydrogenase, ribosomal protein, and G protein beta subunit-like protein. Four other proteins detected by phospho-specific immunoblotting, ABA- and stress-inducible protein, ethylene-inducible protein, drought-induced S-like ribonuclease, and an unannotated protein, function in stress responses or metabolism (Table 1). On the other hand, three responding proteins possibly involved in signal transduction were greatly dephosphorylated during drought stress.
Abscisic acid is a central regulator of many plant responses to environmental stresses, particularly osmotic stresses. ABA initiates the water removal and overall dormancy program that plant embryos undergo during seed formation and also mediates the cold and drought stress responses that occur in vegetative tissues (Hubbard et al., 2010; Cramer et al., 2011). Recent studies showed that a pathway based on PYR/RCAR ABA receptors, protein phosphatase 2Cs (PP2Cs), and SNF1-related protein kinase 2s (SnRK2s) forms the primary basis of an early ABA signaling module (Ma et al., 2009; Park et al., 2009). This pathway interfaces with ion channels, transcription factors, and other targets, thus providing a mechanistic connection between the phytohormone and ABA-induced responses. In the absence of ABA, the PP2Cs are active and repress SnRK2 activity and downstream signaling. In the presence of ABA, PYR/RCARs interact with PP2Cs and inhibits phosphatase activity, allowing SnRK2 activation and phosphorylation of target proteins. Accumulating information on ABA-regulated phosphoproteins should reveal new and potentially key components in ABA signal transduction.
A recent study on the effect of ABA on rice leaf phosphoproteins gave insight into the regulatory mechanism for some ABA signaling proteins and implicated several proteins previously unrecognized in ABA action (He and Li, 2008). Phospho-specific antibodies, 2-DE and MS were used to detect and identify phosphoproteins and revealed the type of amino-acid phosphorylated. Six identified proteins exhibited altered phosphorylation in response to 3–6 h of ABA treatment (Table 1). Among these, phosphorylation of G protein beta subunit-like protein in rice is likely to be important in ABA signal transduction. ABA dramatically decreased serine phosphorylation of ascorbate peroxidase and Mn superoxide dismutase, which could enhance their catalytic activities. However, the functional importance of dephosphorylation of two other proteins, triosephosphate isomerase and the ChaC-like protein remains to be determined. In addition, ABA increased both serine and threonine phosphorylation of glyoxysomal malate dehydrogenase. In silico analysis revealed that five of the six ABA-regulated phosphoproteins contain the SnRK2 or MAPK substrate motif.
A quantitative untargeted mass spectrometric analysis with 14N/15N metabolic labeling of the A. thaliana phosphoproteome following ABA treatment was used to identify more broadly the phosphoproteins affected by this hormone in vivo (Kline et al., 2010). This study identified potential early protein targets of the phosphorylation/dephosphorylation machinery of the ABA response (Table 1). Using an LC-MS/MS approach with ETD, all three members of the SnRK2 subclass III family (SnRK2.2, SnRK2.3, and SnRK2.6), and a non-subclass III SnRK2 family member SnRK2.4 were identified with an increase in phosphorylation in their activation loops following 30-min ABA treatment. The bZIP transcription factor family proteins, known downstream substrates of the SnRK kinase family, showed increases in steady-state phosphorylation. Members of the aquaporin family showed decreased phosphorylation at a C-terminal serine, which is predicted to cause closure of the water-transporting aquaporin gate (Törnroth-Horsefield et al., 2006; Kline et al., 2010), consistent with ABA’s role in ameliorating the effect of drought. Multiple proteins not previously implicated in ABA action were found to have significantly altered phosphorylation status in many cases a phosphorylation decrease. Therefore, the interaction with and inhibition of protein phosphatases was not the only means of direct interaction of the ABA receptor family with regulatory proteins, and other currently unknown ABA signaling pathways that result in decreased phosphorylation of downstream targets must be involved. However, this was not an exhaustive survey because an untargeted quantitative phosphoproteomic analysis using a data-dependent acquisition is inherently biased toward the identification of higher abundance peptides.
Flooding impacts the productivity of arable farmland severely, and the vast majority of crops cannot grow under flooding stress. Roots are stressed from oxygen starvation and from the production of toxic substances in different soils (Setter and Waters, 2003). Flooding events are diverse and are defined through their timing, duration, severity, and complexity of waterlogging stress, suggesting that for each set of environmental conditions there could be different priorities for mechanisms of tolerance. The effect of flooding on soybean has been extensively studied because soybean is a flood-intolerant crop, whose growth and grain yield are significantly reduced under flooding stress (). Comparative gel-free proteomics and gel-based phosphoproteomics techniques were used to investigate early responses to flooding stress in the roots and hypocotyls of soybean seedlings (Nanjo et al., 2010). Dephosphorylation of proteins involved in protein folding and synthesis was found to be one of the early responses (Table 1). The authors suggested that the translational or post-translational control of proteins involved in protein folding and synthesis during flooding induces an imbalanced expression of proteins involved in several metabolic pathways, including carbohydrate metabolism, which may cause flooding-induced injury of the seedlings. Recently, gel-free mass spectrometry-based proteomics techniques were used to compare protein phosphorylation states in the root tips of flooded soybean seedlings (Nanjo et al., 2012). A comparison of the proteins identified in the phosphoproteomic and quantitative proteomic analyses revealed six proteins affected by flooding including those involved in energy generation, protein synthesis, and cell structure maintenance showed changes in both abundance and phosphorylation status (Table 1). Therefore, flooding-induced changes in protein phosphorylation possibly modulate energy production in flooded soybean root tips. On the other hand, the decreased expression and changes in the phosphorylation state of proteins involved in root elongation resulted in the suppression of this process during flooding.
SALT STRESS
Salinity imposes both osmotic stress and ion toxicity to plants. To survive, a sophisticated signal perception, transduction, and cellular and morphological response network has evolved to combat this stress. Plants respond to salt stress by triggering phosphorylation cascades to turn on the salt overly sensitive (SOS) signaling pathway (Gong et al., 2004). Protein kinase activity assays, yeast two-hybrid analyses, and studies with mutants have identified an MAPK signaling pathway, consisting of AtMEKK1, AtMEK1/AtMKK2, and AtMPK4/AtMPK6, that transduces salt and other abiotic stress signals in A. thaliana. Overexpression of AtMKK2 in A. thaliana resulted in constitutive activation of AtMPK4 and AtMPK6 and enhanced salt and freezing tolerance. Meanwhile, mkk2 mutants exhibited hypersensitivity to salt and cold stress (Teige et al., 2004). In rice, OsMAPK5 kinase activity and gene expression were shown to be regulated by salt, drought, wounding, cold, and ABA. Overexpression of OsMAPK5 resulted in an increase in tolerance to several abiotic stresses, including salt stress (Xiong and Yang, 2003). Protein phosphorylation also plays a critical role in ion homeostasis under salinity stress (Zhu, 2003).
Many of the soil-stress (i.e., water supply and salinity) studies focus on the root proteome, because this is the organ that faces the stress directly. Phosphoproteome differential expression analysis under salinity stress in rice roots used Pro-Q Diamond phosphoprotein stain (Schulenberg et al., 2003) and SYPRO Ruby total protein stain following 2-DE gel separation (Chitteti and Peng, 2007). The Pro-Q Diamond in-gel stain was useful for a direct visualization of the putative phosphoprotein spots. Seventeen of the twenty upregulated and eleven of the eighteen downregulated putative phosphoproteins were identified in this study (Table 1). Among the upregulated proteins, two were typical salt-stress response proteins, the salt-induced protein and mannose-binding rice lectin. Other responding phosphoproteins, glutathione S-transferase, a small GTP binding protein, transposase, dnaK-type Hsp70, and dimethylaniline monooxygenase may also be involved in the salt-stress response. Experimental evidence indicated that the Pro-Q Diamond stain intensity was not proportional to protein concentration but was likely proportional to the level of phosphorylation of each phosphoprotein. However, the limited protein quantity from an excised protein spot of 2-DE gel made mapping the phosphorylated residues extremely challenging. In addition, an appropriate concentration of Pro-Q Diamond dye was critical for reducing non-specific background.
In a recent study, the role of phosphorylation of plasma membrane intrinsic protein (PIP) aquaporins from A. thaliana roots in regulating the water permeability in response to NaCl and H2O2 treatments was investigated by quantitative mass spectrometry and cellular biology approaches (Prak et al., 2008). In plants, aquaporins play an important role in the regulation of root water transport in response to abiotic stresses. Phosphorylation sites in aquaporins were well characterized and generally found at the C terminus of the protein (Maurel et al., 2008). A systematic inventory of phosphorylation sites in the C terminus of AtPIP aquaporins was performed and novel phosphoresidues were discovered (Table 1). Quantitative analysis using stable-isotope labeling detected reduced phosphorylation level of PIP2;1 in salt-stressed A. thaliana (Prak et al., 2008). The emerging role of stimulus-dependent trafficking of plant aquaporins between the plasma membrane and intracellular compartments was explored using AtPIPs fused to GFP, the roots of transgenic lines carrying Ser to Ala mutations to abolish phosphorylation or Ser to Asp mutations to mimic a constitutive phosphorylation, and confocal microscopy, in conjunction with phosphoproteomic analysis. It was demonstrated that in resting conditions one of the phosphorylation sites was necessary to target AtPIP2;1 to the plasma membrane. The salt treatment induced an intracellular accumulation of AtPIP2;1 in distinct intracellular structures by exerting specific actions onto differentially phosphorylated forms.
The initial effects on the maize root proteome and phosphoproteome in the first hour after the adjustment to saline conditions were monitored to elucidate initial responses (Zörb et al., 2010). The subsequent proteome change was monitored using a 2-DE approach. The use of a resistant genotype helped to detect proteins that were affected by short-term salt stress and compensated for the fact that maize is a salt-sensitive crop plant. A set of phosphoproteins in maize was detected but only 10 proteins were phosphorylated and 6 proteins were dephosphorylated after the application of short-term salt treatment (Table 1). The authors suggested that changes in phosphorylation status of fructokinase 2 and sucrose synthase from maize roots may serve as markers for early signals of salt stress. This study revealed early members of sugar signaling and cell signaling pathways such as calmodulin (Zörb et al., 2010). The increase in the phosphorylation of 14-3-3 proteins immediately after the adjustment to salt stress may change plasma membrane ATPase activity and lead to a change of apoplastic pH, and subsequently a phosphorylation state of cell wall growth regulators such as XET hydrolase.
Phosphorylation sites in the membrane proteins of 200 mM salt-stressed A. thaliana plants were profiled using Zr(IV)-IMAC magnetic beads for phosphopeptide enrichment (Hsu et al., 2009). Nine out of eighteen total identified phosphorylation sites were from membrane proteins (Table 1). The combination of label-free and labeling quantitative analyses provided evidence for at least twofold differential expression of phosphorylated peptides in five membrane proteins. In addition, novel phosphorylation sites in aquaporin PIP2;4, a member of an MAPK signaling pathway AtMKK2, and a calcium-binding protein were identified after high-salt treatment (Hsu et al., 2009). Another putative phosphorylation site in AtMKK2 has previously been reported (Cardinale et al., 2002). The MAPKK (SIMK kinase) of a salt-induced MAPK (Munnik et al., 1999) has also been identified in alfalfa (Kiegerl et al., 2000). The involvement of the MAPK signaling pathway in the osmotic stress response has previously been reported (Zhu, 2002). In A. thaliana, AtMKK2 was shown to be activated by MEKK1 and involved in salt-stress signaling (Teige et al., 2004). The same kinase also plays a role in disease resistance (Brader et al., 2007).
The function of aquaporin phosphorylation under salinity stress has recently been studied in barley roots (Horie et al., 2011). Water transport activity in intact roots was shown to require phosphorylation by a staurosporine-sensitive kinase. These findings suggest that HvPIP2 channels predominantly function via an activation process involving phosphorylation and severe salinity stress causes reductions in the root hydraulic conductivity (Lpr), similar to findings in other plant species.
A large-scale phosphoproteome analysis in rice identified conserved phosphorylation sites of membrane proteins in plants (Nakagami et al., 2010). Further comparative analyses against the A. thaliana and Medicago truncatula phosphoproteomes showed that nearly half of the phosphorylated orthologous proteins were phosphorylated at equivalent sites. This study provided more direct evidence for conserved regulatory mechanisms based on phosphorylation in plants.
Phosphopeptide enrichment is a common strategy to uncover phosphopeptides of low abundance, often those with key roles in signaling. Chang et al. (2012) used Zr(IV)-IMAC and TiO2 magnetic beads to enrich phosphopeptides shaved from the microsomal fractions of 400 mM salt-stressed A. thaliana seedlings. A total of 27 phosphorylation sites were identified of which 10 were membrane proteins (Table 1). Label-free quantitative analyses provided evidence to support the identification of upregulation or downregulation of the phosphopeptide abundance from membrane proteins. A cluster analysis revealed differential abundance of each phosphopeptide under 200 mM (Hsu et al., 2009) or 400 mM salt stress (Chang et al., 2012). In addition to A. thaliana, microsomal fractions of 200 mM salt-stressed rice were analyzed by either protease shaving or tube-gel methods, which identified seven phosphorylation sites of rice aquaporins. Phosphorylation motifs were predicted that may be important in salt-stress responses in plants (Chang et al., 2012). Label-free quantitative analysis revealed salt-regulated changes in the abundance of two phosphorylation sites-containing phosphopeptide in the photosystem II reaction center protein H PsbH from two independent plant systems. However, the functional significance of these sites in relation to salt-stress responses has not been elucidated. The label-free ratio indicated that the abundance of phosphopeptides of rice aquaporin PIPs was downregulated, suggesting that OsPIPs negatively mediate water flux in response to salt stress. In addition, consensus phosphorylation motifs that may be important in high-salt stress responses in plants were predicted.
OXIDATIVE STRESS
A common plant response to different abiotic and biotic stresses is the accelerated generation and accumulation of reactive oxygen species, including H2O2, superoxide anions (O2.-), and hydroxyl radicals (OH.) (Mittler et al., 2011). In many eukaryotes, the transduction of oxidative signals is controlled by protein phosphorylation involving MAPKs (Kovtun et al., 2000). Using epitope tagging and a protoplast transient expression assay, Kovtun et al. (2000) showed that H2O2 can activate a specific A. thaliana mitogen-activated protein kinase kinase kinase, ANP1, which initiates a phosphorylation cascade involving two stress MAPKs, AtMPK3 and AtMPK6. Furthermore, evidence was provided for a molecular link between oxidative stress and auxin signal transduction. Constitutively, active ANP-like MAPKKKs mimicked oxidative stress signal by inducing stress MAPKs and protective gene expression, as well as by repressing an auxin-responsive promoter.
An original approach was used for differential screening for oxidative stress-regulated phosphorylation events using peptide chips and identification of responsible protein kinases (de la Fuente van Bentem et al., 2008). To perform quantitative phosphorylation analysis, peptide arrays were used with peptides corresponding to in vivo phosphorylation sites. These peptide chips were used for kinome profiling of subcellular cytoplasmic and nuclear fractions as well as H2O2-treated A. thaliana cells. An oxidative stress-induced protein kinase activity was detected that phosphorylated a peptide of the mRNA splicing factor SCL30. Further analysis suggested that SRPK4 and MAPKs act together to mediate constitutive and oxidative stress-induced phosphorylation of SCL30. SR proteins control both splice site selection and spliceosome assembly, and interactions between SR proteins and other spliceosome components are regulated by phosphorylation (Lorkovic et al., 2004). Therefore, MAPKs might connect stress signals to the mRNA splicing machinery via SR protein phosphorylation, potentially leading to differential splicing of pre-mRNAs (Iida et al., 2004). In addition, an increased phosphorylation of a peptide derived from a putative chromatin remodeling protein (At5g07350) and a peptide from the activation loop of the PK At5g35980 phosphorylated in vivo on a tyrosine residue was observed. However, the substrate specificities of most plant kinases are unknown, and more research is needed for the high-throughput discovery of kinases-substrate links.
HORMONAL EFFECT
In an early study, a phosphoproteome analysis in different tissues of rice used an in vitro protein phosphorylation approach followed by MS analysis (Khan et al., 2005). Changes in protein phosphorylation were investigated in response to different hormones and stresses (Table 1). It was shown that the phosphorylation of glyceraldehyde-3-phosphate dehydrogenase, cytoplasmic malate dehydrogenase, putative zinc finger protein, and glyoxalase-I was increased by both gibberellin (GA3) and 2, 4-dichlorophenoxyacetic (2, 4-D) treatment. In addition, the hormone-specific phosphorylation of aldo/keto reductase family protein was increased by Brassinolide (BL) alone, while the phosphorylation of calmodulin-related protein was increased by 2, 4-D alone. The phosphorylation of two proteins, calcium binding and an unknown protein, was altered with GA3 treatment. In contrast, the phosphorylation of glyceraldehyde-3-phosphate dehydrogenase and cytoplasmic malate dehydrogenase was enhanced by all of the hormones used (Khan et al., 2005), indicating hormonal modulation of energy metabolism. Interestingly, phosphorylation of calreticulin was regulated by ABA, NaCl, and cold treatments, and the phosphorylated form was only detected under stress conditions but not during normal plant growth. Furthermore, evidence was presented for the possible promotion of cold tolerance and GA-dependent elongation through distinct signaling pathways that cross-talk at the level of OsCDPK13 (Abbasi et al., 2004; Khan et al., 2005). Therefore, glycolytic metabolism and Ca2+-signaling processes were found to be important targets of the phosphorylation cascades in rice under hormone and stress conditions, and the identification of a common signaling molecule indicated a coordinated response of the plant to various stimuli.
METABOLITE SIGNALING
Many abiotic stresses directly or indirectly affect the synthesis, concentration, metabolism, transport, and storage of sugars (Cramer et al., 2011). Sugars, as well as hormones, can act as primary messengers and may regulate signals that control the expression of various genes under different environmental influences. Sugar signaling interacts with stress pathways to modulate metabolism (Osuna et al., 2007). Sugar sensing and signaling is involved in the control of growth and development during the entire plant life cycle (Esatmond and Graham, 2001; Gazzarini and McCourt, 2001).
A label-free quantitative correlation profiling phosphoproteomics approach was used to identify plasma membrane proteins that are dynamically phosphorylated in response to sucrose resupply after depletion to A. thaliana seedlings (Niittylä et al., 2007). A novel phosphorylation site was identified that determines the general activity state of the plasma membrane H+-ATPase (Table 1), a protein providing the proton motive force that drives sucrose transport (Palmgren, 2001). Moreover, a conserved phosphorylation site in the C terminus of PIP2e had increased phosphorylation, suggesting that addition of sucrose leads to activation potentially via a trans-regulation within the complex (Niittylä et al., 2007). A novel sucrose transporter phosphorylation site was found that could play an important role in acclimating sucrose uptake to the extracellular supply. In this study, three membrane-bound kinases were identified that showed a rapid transient change in relative phosphorylation, whereas the identified soluble kinases showed peaks in relative phosphorylation at later time points. These findings on dynamics of phosphorylation under sucrose depletion and resupply indicate that protein phosphorylation provides another level of regulation that may not affect transcription (Niittylä et al., 2007; Osuna et al., 2007), especially with respect to very short-term changes in environmental conditions for the plant, whereas in some cases specific proteins/genes may be affected on both regulatory levels.
MECHANICAL WOUNDING
Mechanical wounding can result from either abiotic stress (wind, rain, hail, etc.) or biotic stress (herbivore feeding). Wounding can provide a gate for pathogen invasion and triggers specific defense responses, which in part serve to activate developmental programs that prepare the host to resist further attack. Wound signaling pathways are regulated by reversible protein phosphorylation (Seo et al., 2007). In plants two classes of protein kinases, MAPKs and CPKs, are activated by wounding. The mechanical wounding of maize leaves induced accumulation of transcript encoding the CDPK, ZmCPK11, and stimulated kinase activity both locally at the injury site and systemically in distant non-wounded leaves as monitored using in-gel kinase assays (Szczegielniak et al., 2005).
Changes in the maize leaf phosphoproteome in response to mechanical injury were analyzed using multiplex staining of high-resolution 2D gels for protein phosphorylation together with a strategy for quantifying changes in the stoichiometry of phosphorylation after wounding (Lewandowska-Gnatowska et al., 2011). Nearly all of the presumptive maize leaf phosphoproteins were involved in central metabolism, protein folding, or cellular signaling, and have previously been identified in phosphoproteomics profiling studies. Further elaboration of this descriptive platform will be needed to develop a system understanding of the interactive processes involved to address the complex dynamics of information flow through the wound-induced signaling pathways.
CONCLUDING REMARKS
In spite of the progress reviewed here, biotic and abiotic stress will continue to be a problem until stress resistance is better understood at the molecular level. Thus, breeding crops better adapted to specific stresses, such as the Australian bread wheat cultivar “Excalibur” (see Introduction), will remain challenging. Fortunately, many stresses share similar response pathways and an understanding of one can be helpful to understand another. It is clear that most of the progress in plant stress-related phosphoproteomics is being reported in A. thaliana. Being the first plant with a fully sequenced genome rapidly led to its wide adoption as a model system and led to the development of numerous molecular research tools for this system, many of which facilitate phosphoproteomics. Sequencing the genomes of crop plants is now actively being pursued, and even wheat, a complex hexaploid with a large genome containing numerous repeat sequences, is expected to be sequenced within a few years. This will support crop phosphoproteomics research because phosphopeptide identification and site localization depend largely on homology-based matching of mass spectra to databases. The success of this is enormously increased if the databases contain complete genomic sequences.
One fruitful approach to understanding the roles of critical participants of signaling cascades (especially kinases) is through T-DNA insertion mutagenesis. This has been used to study, for example, OsABC1-2 a kinase conferring enhanced tolerance to dark-induced stress in rice (Gao et al., 2012) and to investigate, in a high-throughput manner, the effects of T-DNA mutagenesis of 69 root-specific RLKs on root phenotypes and alterations in the response to environmental stresses (ten Hove et al., 2011). Pitorre et al. (2010) used molecular genetics, phenomics, and proteomics to uncover biological processes orchestrated by a leucine-rich repeat receptor-like kinase in a T-DNA insertion mutant of A. thaliana. Although none of these studies employed phosphoproteomics, they demonstrate the utility of the approach.
Applying knowledge learned through phosphoproteomics, for example, the discovery of a phosphorylation event associated with resistance to a biotic or abiotic stress, to crop improvement has not yet been reported and remains a significant challenge. The task of finding a selectable marker segregating with a regulatory gene is formidable and only beginning to be explored (Lee et al., 2011), and it is likely going to be many more years before robust strategies that can be used by molecular breeders to improve crop resistance to stress can be implemented routinely.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge AAFC for funding support and Michael Shillinglaw for preparing the figure.
REFERENCES
- Abbasi F., Onodera H., Toki S., Tanaka H., Komatsu S. (2004). OsCDPK13, a calcium-dependent protein kinase gene from rice, is induced by cold and gibberellin in rice leaf sheath. Plant Mol. Biol. 55 541–552 [DOI] [PubMed] [Google Scholar]
- Armstrong M. A., Whisson S. C., Pritchard L., Bos J. I., Venter E., Avrova A. O., Rehmany A., Böhme U., Brooks K., Cherevach I., Hamlin N., White B., Fraser A., Lord A., Quail M. A., Churcher C., Hall N., Berriman M., Huang S., Kamoun S., Beynon J. L., Birch P. R. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 102 7766–7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M. R., Gomez-Gomez L., Boller T., Ausubel F. M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983 [DOI] [PubMed] [Google Scholar]
- Batistic O., Kudla J. (2009). Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta 1793 985–992 [DOI] [PubMed] [Google Scholar]
- Benschop J. J., Mohammed S., O’Flaherty M., Heck A. J. A., Slijper M, Menke F. L. H. (2007). Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6 1198–1214 [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60 379–406 [DOI] [PubMed] [Google Scholar]
- Brader G., Djamei A., Teige M., Palva E. T., Hirt H. (2007). The MAP kinase kinase MKK2 affects disease resistance in Arabidopsis. Mol. Plant Microbe Interact. 20 589–596 [DOI] [PubMed] [Google Scholar]
- Cardinale F., Meskiene I., Ouaked F., Hirt H. (2002). Convergence and divergence of stress-induced mitogen-activated protein kinase signalling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell 14 703–711 [PMC free article] [PubMed] [Google Scholar]
- Catanzariti A., Dodds P. N., Lawrence G. J., Ayliffe M. A., Ellis J. G. (2006). Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang I. F., Hsu J. L., Hsu P. H., Sheng W. A., Lai S. J., Lee C., Chen C. W., Hsu J. C., Wang S. Y., Wang L. Y., Chen C. C. (2012). Comparative phosphoproteomic analysis of microsomal fractions of Arabidopsis thaliana and Oryza sativa subjected to high salinity. Plant Sci. 185–186 131–142 [DOI] [PubMed] [Google Scholar]
- Chen X., Ronald P. C. (2011). Innate immunity in rice. Trends Plant Sci. 16 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhang W., Zhang B., Zhou J., Wang Y., Yang Q., Ke Y., He H. (2011). Phosphoproteins regulated by heat stress in rice leaves. Proteome Sci. 9 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitteti B. R., Peng Z. (2007). Proteome and phosphoproteome differential expression under salinity stress in rice (Oryza sativa) roots. J. Proteome Res. 6 1718–1727 [DOI] [PubMed] [Google Scholar]
- Cramer G. R., Urano K., Delrot S., Pezzotti M., Shinozaki K. (2011). Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 11 163 10.1186/1471-2229-11-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente van Bentem S. D., Anrather D., Dohnal I., Roitinger E., Csaszar E., Joore J., Buijnink J., Carreri A., Forzani C., Lorkovic Z. J., Barta A., Lecourieux D., Verhounig A., Jonak C., Hirt H. (2008). Site-specific phosphorylation profiling of Arabidopsis proteins by mass spectrometry and peptide chip analysis. J. Proteome Res. 7 2458–2470 [DOI] [PubMed] [Google Scholar]
- Endler A., Reiland S., Gerrits B., Schmidt U. G., Baginsky S., Martinoia E. (2009). In vivo phosphorylation sites of barley tonoplast proteins identified by a phosphoproteomic approach. Proteomics 9 310–321 [DOI] [PubMed] [Google Scholar]
- Esatmond P. J., Graham I. A. (2001). Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 6 72–78 [DOI] [PubMed] [Google Scholar]
- Felix G., Duran J. D., Volko S., Boller T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18 265–276 [DOI] [PubMed] [Google Scholar]
- Fleury D., Jefferies S., Kuchel H., Langridge P. (2010). Genetic and genomic tools to improve drought tolerance in wheat. J. Exp. Bot. 61 3211–3222 [DOI] [PubMed] [Google Scholar]
- Ford K. L., Cassin A., Bacic A. (2011). Quantitative proteomic analysis of wheat cultivars with differing drought stress tolerance. Front. Plant Sci. 2: 44 10.3389/fpls.2011.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q. S., Yang Z. F., Zhou Y., Yin Z. T., Qiu J., Liang G. H., Xu C. W. (2012). Characterization of an Abc1 kinase family gene OsABC1-2 conferring enhanced tolerance to dark-induced stress in rice. Gene 498 155–163 [DOI] [PubMed] [Google Scholar]
- Gazzarini S., McCourt P. (2001). Genetic interactions between ABA, ethylene and sugar signalling pathways. Curr. Opin. Plant Biol. 4 387–391 [DOI] [PubMed] [Google Scholar]
- Gong D., Guo Y., Schumaker K. S., Zhu J. K. (2004). The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiol. 134 919–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goods D. M., Wirtala M., McAlister G. C., Coon J. J. (2007). Performance characteristics of electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics 6 1942–1950 [DOI] [PubMed] [Google Scholar]
- He H., Li J. (2008). Proteomic analysis of phosphoproteins regulated by abscisic acid in rice leaves. Biochem. Biophys. Res. Commun. 371 883–888 [DOI] [PubMed] [Google Scholar]
- Horie T., Kaneko T., Sugimoto G., Sasano S., Panda S. K., Shibasaka M., Katsuhara M. (2011). Mechanisms of water transport mediated by PIP aquaporins and their regulation via phosphorylation events under salinity stress in barley roots. Plant Cell 52 663–675 [DOI] [PubMed] [Google Scholar]
- Hsu J. L., Wang L. Y., Wang S. Y., Lin C. H., Ho K. C., Shi F. K., Chang I. F. (2009). Functional phosphoproteomic profiling of phosphorylation sites in membrane fractions of salt-stressed Arabidopsis thaliana. Proteome Sci. 7 42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard K. E., Nishimura N., Hitomi K., Getzoff E. D., Schroeder J. I. (2010). Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 24 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K., Seki M., Sakurai T., Satou M., Akiyama K., Toyoda T., Konagaya A., Shinozaki K. (2004). Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res. 32 5096–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., McAdams S. A., Bryan G. T., Hershey H. P., Valent B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast . EMBO J. 19 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. M. E., Bennett M. H., Mansfield J. W., Grant M. (2006). Analysis of the defence phosphoproteome of Arabidopsis thaliana using differential mass tagging. Proteomics 6 4155–4165 [DOI] [PubMed] [Google Scholar]
- Khan M., Takasaki H., Komatsu S. (2005). Comprehensive phosphoproteome analysis in rice and identification of phosphoproteins responsive to different hormones/stresses. J. Proteome Res. 4 1592–1599 [DOI] [PubMed] [Google Scholar]
- Ke Y., Han G., He H., Li J. (2009). Differential regulation of proteins and phosphoproteins in rice under drought stress. Biochem. Biophys. Res. Commun. 379 133–138 [DOI] [PubMed] [Google Scholar]
- Kerk D., Bulgrien J., Smith D. W., Barsam B., Veretnik S., Gribskov M. (2002). The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 129 908–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten B., Agrawal G., Durek P., Neigenfind J., Schulze W., Walther D., Rakwal R. (2009). Plant phosphoproteomics: an update. Proteomics 9 964–988 [DOI] [PubMed] [Google Scholar]
- Kiegerl S., Cardinale F., Siligan C., Gross A., Baudouin E., Liwosz A., Eklöf S., Till S., Bögre L., Hirt H., Meskiene I. (2000). SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell 12 2247–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline K. G., Barrett-Wilt G. A., Sussman M. R. (2010). In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107 15986–15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosová K., Vitámvás P., Prášil I. T., Renaut J. (2011). Plant proteome changes under abiotic stress – contribution of proteomics studies to understanding plant stress response. J. Proteomics 12 1301–22 [DOI] [PubMed] [Google Scholar]
- Kovtun Y., Chiu W. L., Tena G., Sheen J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. U.S.A. 97 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Ahsan N. B., Lee S., Kang K., Bahk J., Lee I., Lee B. (2007). A proteomic approach in analyzing heat-responsive proteins in rice leaves. Proteomics 7 3369–3383 [DOI] [PubMed] [Google Scholar]
- Lee M. O., Cho K., Kim S. H., Jeong S. H., Kim J. A., Jung Y. H., Shim J., Shibato J., Rakwal R., Tamogami S. (2008). Novel rice OsSIPK is a multiple stress responsive MAPK family member showing rhythmic expression at mRNA level. Planta 227 981–990 [DOI] [PubMed] [Google Scholar]
- Lee E., Kim K., Park T. (2011). Genome-wide search for genetic modulators in gene regulatory pathways: weighted window-based peak identification algorithm. Genomics 97 386–393 [DOI] [PubMed] [Google Scholar]
- Lewandowska-Gnatowska E., Johnston M. L., Antoine W., Szczegielniak J., Muszyń G., Miernyk J. A. (2011). Using multiplex-staining to study changes in the maize leaf phosphoproteome in response to mechanical wounding. Phytochemistry 72 1285–1292 [DOI] [PubMed] [Google Scholar]
- Lorkovic Z. J., Lopato S., Pexa M., Lehner R., Barta A. (2004). Interactions of Arabidopsis RS domain containing cyclophilins with SR proteins and U1 and U11 small nuclear ribonucleoprotein-specific proteins suggest their involvement in pre-mRNA Splicing. J. Biol. Chem. 279 33890–33898 [DOI] [PubMed] [Google Scholar]
- Luan S. (2009). The CBL-CIPK network in plant calcium signalling. Trends Plant Sci. 14 37–42 [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324 1064–1068 [DOI] [PubMed] [Google Scholar]
- Maurel C., Verdoucq L., Luu D. T., Santoni V. (2008). Plant aquaporins: membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 59 595–624 [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Suzuki N., Miller G., Tognetti V. B., Vandepoele K., Gollery M., Shulaev V., Van B. F. (2011). ROS signalling: the new wave? Trends Plant Sci. 16 300–309 [DOI] [PubMed] [Google Scholar]
- Munnik T., Ligterink W., Meskiene I. I., Calderini O., Beyerly J., Musgrave A., Hirt H. (1999). Distinct osmo-sensing protein kinase pathways are involved in signalling moderate and severe hyper-osmotic stress. Plant J. 20 381–388 [DOI] [PubMed] [Google Scholar]
- Nakagami H., Sugiyama N., Mochida K., Daudi A., Yoshida Y., Toyoda T., Tomita M., Ishihama Y., Shirasu K. (2010). Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjo Y., Skultety L., Ashraf Y., Komatsu S. (2010). Comparative proteome analysis of early-stage soybean seedlings responses to flooding by using gel and gel-free techniques. J. Proteome Res. 9 3989–4002 [DOI] [PubMed] [Google Scholar]
- Nanjo Y., Skultety L., Uváková L., Klubicová K., Hajduch M., Komatsu S. (2012). Mass spectrometry-based analysis of proteomic changes in the root tips of flooded soybean seedlings. J. Proteome Res. 11 372–385 [DOI] [PubMed] [Google Scholar]
- Niittylä T., Fuglsang A. T., Palmgren M. G., Frommer W. B., Schulze W. X. (2007). Temporal analysis of sucrose induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol. Cell. Proteomics 6 1711–1726 [DOI] [PubMed] [Google Scholar]
- Nirmala J., Drader T., Chen X., Steffenson B., Kleinhofs A. (2010). Stem rust spores elicit rapid RPG1 phosphorylation. Mol. Plant Microbe Interact. 23 1635–1642 [DOI] [PubMed] [Google Scholar]
- Osuna D., Usadel B., Morcuende R., Gibon Y., Bläsing O. E., Höhne M., Günter M., Kamlage B., Trethewey R., Scheible W. R., Stitt M. (2007). Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J. 49 463–491 [DOI] [PubMed] [Google Scholar]
- Palmgren M. G. (2001). Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 817–845 [DOI] [PubMed] [Google Scholar]
- Park S. Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T. F., Alfred S. E., Bonetta D., Finkelstein R., Provart N. J., Desveaux D., Rodriguez P. L., McCourt P., Zhu J. K., Schroeder J. I., Volkman B. F., Cutler S. R. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Han S., Chen X., Ronald P. C. (2010). Elucidation of XA21-mediated innate immunity. Cell. Microbiol. 12 1017–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck S. C., Nühse T., Hess D., Iglesisa A., Meins F., Boller T. (2001). Directed proteomics identifies a plant-specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell 13 1467–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitorre D., Llauro C., Jobet E., Guilleminot J., Brizard J.-P., Delseny M., Lasserre E. (2010). A leucine-rich repeat receptor-like kinase, is required for proper germination speed and tolerance to oxidative stress in Arabidopsis thaliana. Planta 232 1339–1353 [DOI] [PubMed] [Google Scholar]
- Pitzschke A., Schikora A., Hirt H. (2009). MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12 421–426 [DOI] [PubMed] [Google Scholar]
- Popescu S. C., Popescu G. V., Bachan S., Zhang Z., Gerstein M., Snyder M., Dinesh-Kumar S. P. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prak S., Hem S., Boudet J., Viennois G., Sommerer N., Rossignol M., Maurel C., Santoni V. (2008). Multiple phosphorylations in the C-terminal tail of plant plasma membrane aquaporins: role in subcellular trafficking of AtPIP2;1 in response to salt stress. Mol. Cell. Proteomics 7 1019–1030 [DOI] [PubMed] [Google Scholar]
- Rampitsch C., Bykova N. V. (2012). Proteomics and plant disease: advances in combating a major threat to the global food supply. Proteomics 12: 673–690. [DOI] [PubMed] [Google Scholar]
- Riaño-Pachón D. M., Kleessen S., Neigenfind J., Durek P., Weber E., Engelsberger W. R., Walther D., Selbig J., Schulze W. X., Kersten B. (2010). Proteome-wide survey of phosphorylation patterns affected by nuclear DNA polymorphisms in Arabidopsis thaliana. BMC Genomics 11 411 10.1186/1471-2164-11-411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafaro A. P., Haynes P. A., Atwell B. J. (2010). Physiological and molecular changes in Oryza meridionalis Ng., a heat-tolerant species of wild rice. J. Exp. Bot. 61 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenberg B., Aggeler B., Beechem J., Capaldi R., Patton W. (2003). Analysis of steady-state protein phosphorylation in mitochondria using a novel fluorescent phosphosensor dye. J. Biol. Chem. 278 27251–27255 [DOI] [PubMed] [Google Scholar]
- Schulze B., Menzel T., Jehle A. K., Müller K., Beeler S., Boller T., Felix G., Chinchilla D. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285 9444–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze W. X., Schneider T., Starck S., Martinoia E., Trentman O. (2012). Cold acclimation induces changes in Arabidopsis tonoplast protein abundance and activity and alters phosphorylation of tonoplast monosaccharide transporters. Plant J. 69 529–541 [DOI] [PubMed] [Google Scholar]
- Seo S., Katou S., Seto H., Gomi K., Ohashi Y. (2007). The mitogen-activated protein kinases WIPK and SIPK regulate the levels of jasmonic acid and salicylic acid in wounded tobacco plants. Plant J. 49 899–909 [DOI] [PubMed] [Google Scholar]
- Setter T. L., Waters I. (2003) Review of prospect for germplasm improvement for waterlogging tolerance in wheat, barley and oats. Plant Soil 253 1–34. [Google Scholar]
- Shan W., Cao M., Leung D., Tyler B. M. (2004). The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant Microbe Interact. 17 394–403 [DOI] [PubMed] [Google Scholar]
- Singh A., Giri J., Kapoor S., Tyagi A., Pandey G. (2010). Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics 11 435 10.1186/1471-2164-11-435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W. Y., Wang G. L., Chen L. L., Kim H. S., Pi L. Y., Holsten T., Gardner J., Wang B, Zhai W. X., Zhu L. H., Fauquet C., Ronald P. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270 1804–1806 [DOI] [PubMed] [Google Scholar]
- Spannagl M., Mayer K., Durner K., Haberer G, Fröhlich A. (2011). Exploring the genomes: from Arabidopsis to crops. J. Plant Physiol. 168 3–8 [DOI] [PubMed] [Google Scholar]
- Stulemeijer I. J., Stratmann J. W., Joosten M. H. (2007). Tomato mitogen-activated protein kinases LeMPK1, LeMPK2, and LeMPK3 are activated during the Cf-4/Avr4-induced hypersensitive response and have distinct phosphorylation specificities. Plant Physiol. 144 1481–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Denis N., Gingras A.-C. (2012). Mass spectrometric tools for systematic analysis of protein phosphorylation. Prog. Mol. Biol. Transl. Sci. 106 3–32 [DOI] [PubMed] [Google Scholar]
- Szczegielniak J., Klimecka M., Liwosz A., Ciesielski A., Kaczanowski S., Dobrowolska G., Harmon A. C, Muszyńska G. (2005). A wound-responsive and phospholipid-regulated maize calcium-dependent protein kinase. Plant Physiol. 139 1970–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M., Scheikl E., Eulgem T., Dóczi R., Ichimura K., Shinozaki K., Dangl J. L., Hirt H. (2004). The MKK2 pathway mediates cold and salt stress signalling in Arabidopsis. Mol. Cell 15 141–152 [DOI] [PubMed] [Google Scholar]
- ten Hove C. A., Bochdanovits Z., Jansweijer V. M. A., Koning F. G., Berke L., Sanchez-Perez G. F., Scheres B., Heidstra R. (2011). Probing the roles of LRR RLK genes in Arabidopsis thaliana roots using a custom T-DNA insertion set. Plant Mol. Biol. 76 69–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Törnroth-Horsefield S., Wang Y., Hedfalk K., Johanson U., Karlsson M., Tajkhorshid E., Neutze R., Kjellbom P. (2006). Structural mechanism of plant aquaporin gating. Nature 439 688–694 [DOI] [PubMed] [Google Scholar]
- van Bentem S., Hirt H. (2007). Using phosphoproteomics to reveal signalling dynamics in plants. Trends Plant Sci. 12 404–411 [DOI] [PubMed] [Google Scholar]
- Villén J., Gygi S. (2008). The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan B., Lin Y., Mou T. (2007). Expression of rice Ca2+-dependent protein kinases (CDPKs) genes under different environmental stresses. FEBS Lett. 581 1179–1189 [DOI] [PubMed] [Google Scholar]
- Whiteman S. A., Nuhse T. S., Ashford D. A., Sanders D., Maathuis F. J. (2008a). A proteomic and phosphoproteomic analysis of Oryza sativa plasma membrane and vacuolar membrane. Plant J. 56 146–156 [DOI] [PubMed] [Google Scholar]
- Whiteman S. A., Serazetdinova L., Jones A. M., Sanders D., Rathjen J., Peck S. C., Maathuis F. J. (2008b). Identification of novel proteins and phosphorylation sites in a tonoplast enriched membrane fraction of Arabidopsis thaliana. Proteomics 8 3536–3547 [DOI] [PubMed] [Google Scholar]
- Wingenter K., Trentmann O., Winschuh I., Hoermiller I. I., Heyer A. G., Reinders J., Schulz A., Geiger D., Hedrich R., Neuhaus H. E. (2011). A member of the mitogen-activated protein 3-kinase family is involved in the regulation of plant vacuolar glucose uptake. Plant J. 68 890–900 [DOI] [PubMed] [Google Scholar]
- Wormit A., Trentmann O., Feifer I., Lohr C., Tjaden J., Meyer S., Schmidt U., Martinoia E., Neuhaus H. E. (2006). Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. Plant Cell 18 3476–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Yang Y. (2003). Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. (2007). Strategies for developing Green Super Rice. Proc. Natl. Acad. Sci. U.S.A. 104 16402–16409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. (2003). Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6 441–445 [DOI] [PubMed] [Google Scholar]
- Zörb C., Schmitt S, Mühling K. H. (2010). Proteomic changes in maize roots after short-term adjustment to saline growth conditions. Proteomics 10 4441–4449 [DOI] [PubMed] [Google Scholar]