Abstract
Thermovirga lienii Dahle and Birkeland 2006 is a member of the genus Thermovirga in the genomically moderately well characterized phylum 'Synergistetes'. Members of this relatively recently proposed phylum ‘Synergistetes’ are of interest because of their isolated phylogenetic position and their diverse habitats, e.g. from humans to oil wells. The genome of T. lienii Cas60314T is the fifth genome sequence (third completed) from this phylum to be published. Here we describe the features of this organism, together with the complete genome sequence and annotation. The 1,999,646 bp long genome (including one plasmid) with its 1,914 protein-coding and 59 RNA genes is a part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: anaerobic, chemoorganotrophic, Gram-negative, motile, thermophilic, marine oil well, Synergistaceae, GEBA
Introduction
Strain Cas60314T (= DSM 17291 = ATCC BA-1197) is the type strain of the species Thermovirga lienii [1] of the monospecific genus Thermovirga [1]. The strain was originally isolated from 68°C hot oil-well production water from an oil reservoir in the North Sea (Norway) [1]. The genus name Thermovirga was derived from the Greek word for thermê, heat, and the Latin word virga, rod, meaning the hot rod [1]; the species epithet was derived of Lien, in honor of the Norwegian microbiologist Professor Torleiv Lien, for his important contribution in the study of anaerobes from petroleum reservoirs [1]. Whether or not strain Cas60314T occurs naturally in the oil reservoir is not clear, but is likely because the oil well was not injected with seawater (which eliminates a common source of contamination) [1]. Here we present a summary classification and a set of features for T. lienii Cas60314T, together with the description of the genomic sequencing and annotation.
Classification and features
A representative genomic 16S rRNA sequence of T. lienii Cas60314T was compared using NCBI BLAST [2,3] under default settings (e.g., considering only the high-scoring segment pairs (HSPs) from the best 250 hits) with the most recent release of the Greengenes database [4] and the relative frequencies of taxa and keywords (reduced to their stem [5]) were determined, weighted by BLAST scores. The most frequently occurring genera were Dethiosulfovibrio (45.6%), Thermanaerovibrio (30.2%), Aminobacterium (12.5%) and Thermovirga (11.7%) (16 hits in total). Regarding the single hit to sequences from members of the species, the average identity within HSPs was 100.0%, whereas the average coverage by HSPs was 92.2%. Among all other species, the one yielding the highest score was Thermanaerovibrio velox (FR733707), which corresponded to an identity of 90.1% and an HSP coverage of 85.5%. (Note that the Greengenes database uses the INSDC (= EMBL/NCBI/DDBJ) annotation, which is not an authoritative source for nomenclature or classification.) The highest-scoring environmental sequence was HM041945 ('aggregate-forming and crude-oil-adhering microbial biodegraded -temperature petroleum reservoir produced fluid Niiboli oilfield clone NRB28'), which showed an identity of 99.7% and an HSP coverage of 98.3%. The most frequently occurring keywords within the labels of all environmental samples which yielded hits were 'digest' (10.9%), 'anaerob' (7.4%), 'sludg' (5.8%), 'mesophil' (5.6%) and 'wastewat' (5.6%) (234 hits in total). The most frequently occurring keywords within the labels of those environmental samples which yielded hits of a higher score than the highest scoring species were 'microbi' (6.3%), 'temperatur' (4.4%), 'oil' (3.6%), 'water' (3.6%) and 'anaerob' (3.2%) (43 hits in total). Although these keywords are not in conflict with the habitat from which strain Cas60314T was isolated, they do not reveal new insights into the largely unknown natural habitat for members of the species.
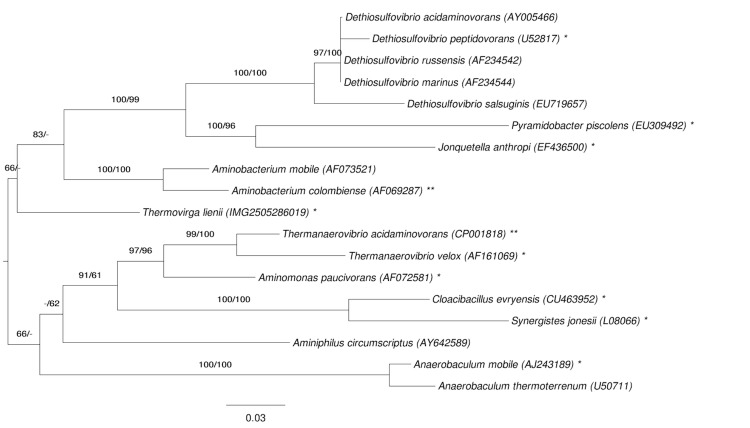
Figure 1 shows the phylogenetic neighborhood of T. lienii in a 16S rRNA based tree. The sequences of the three 16S rRNA gene copies in the genome differ from each other by up to three nucleotides, and differ by up to three nucleotides from the previously published 16S rRNA sequence (DQ071273)
Figure 1.
Phylogenetic tree highlighting the position of T. lienii relative to the type strains of the other species within the phylum Synergistetes. The tree was inferred from 1,385 aligned characters [6,7] of the 16S rRNA gene sequence under the maximum likelihood (ML) criterion [8]. Rooting was done initially using the midpoint method [9] and then checked for its agreement with the current classification (Table 1). The branches are scaled in terms of the expected number of substitutions per site. Numbers adjacent to the branches are support values from 1,000 ML bootstrap replicates [10] (left) and from 1,000 maximum parsimony bootstrap replicates [11] (right) if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [12] are labeled with one asterisk, those also listed as 'Complete and Published' with two asterisks [13,14]. Dethiosulfovibrio peptidovorans [15] and Aminomonas paucivorans [16] are without a second asterisk, because these publications were based on 'non-complete' permanent draft sequences.
Table 1. Classification and general features of T. lienii Cas60314T according to the MIGS recommendations [17].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [18] | |
| Phylum “Synergistetes” | TAS [19] | ||
| Class Synergistia | TAS [19] | ||
| Order Synergistales | TAS [19] | ||
| Family Synergistaceae | TAS [19] | ||
| Genus Thermovirga | TAS [1] | ||
| Species Thermovirga lienii | TAS [1] | ||
| Type strain Cas60314 | TAS [1] | ||
| Gram stain | negative | TAS [1] | |
| Cell shape | rod-shaped | TAS [1] | |
| Motility | motile | TAS [1] | |
| Sporulation | none | TAS [1] | |
| Temperature range | thermophilic | TAS [1] | |
| Optimum temperature | 58°C | TAS [1] | |
| Salinity | optimum 2-3% (w/v) NaCl | TAS [1] | |
| MIGS-22 | Oxygen requirement | obligate anaerobic | TAS [1] |
| Carbon source | amino acids, proteinous substrates, organic acids | TAS [1] | |
| Energy metabolism | chemoorganotrophic | TAS [1] | |
| MIGS-6 | Habitat | fresh water, oil fields | TAS [1] |
| MIGS-15 | Biotic relationship | free living | TAS [1] |
| MIGS-14 | Pathogenicity | none | NAS |
| Biosafety level | 1 | TAS [20] | |
| MIGS-23.1 | Isolation | production water from oil well | TAS [1] |
| MIGS-4 | Geographic location | Troll C Reservoir, North Sea, Norway | TAS [1] |
| MIGS-5 | Sample collection time | September 2003 | NAS |
| MIGS-4.1 | Latitude | 60.886 | TAS [1] |
| MIGS-4.2 | Longitude | 3.612 | TAS [1] |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | not reported |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [21]. If the evidence code is IDA, then the property was directly observed for a living isolate by one of the authors or an expert mentioned in the acknowledgements.
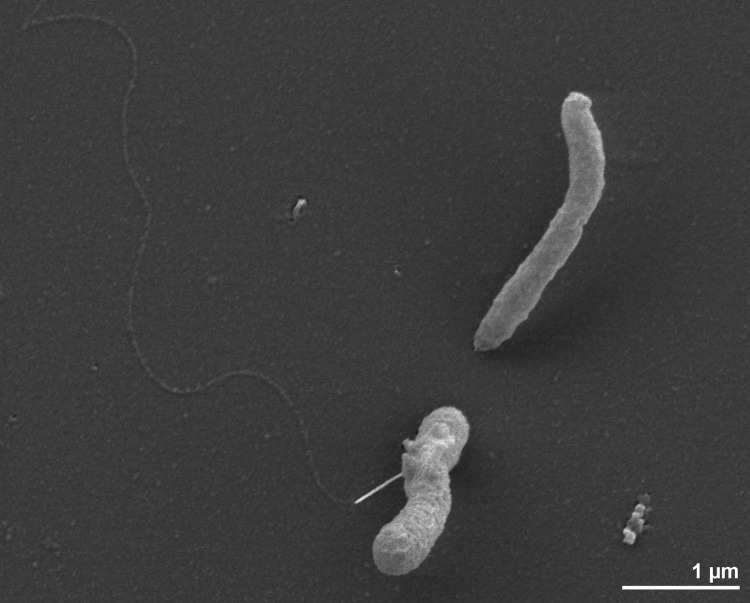
Cells of T. lienii Cas60314T are Gram-negative staining, motile (only in the early exponential growth phase), straight rods of 0.4 to 0.8 μm diameter and 2 to 3 μm length (Figure 2) [1]. Cells appeared to be singly, or in chains of 2 to 5 cells, but can also form aggregates of several hundred cells [1]. The growth range of strain Cas60314T spans from 37-68°C, with an optimum at 58°C, and pH 6.2-8.5, with an optimum at 6.5-7 [1]. Strain Cas60314T grow best in medium containing 2-3% NaCl [1]. Physiological characteristics such as growth substrates and products formed are described in great detail by Dahle and Birkeland [1]; the organism has a fermentative metabolism and utilizes amino acids, proteinaceous substrates and selected organic acids, but no sugars, fatty acids or alcohols [1]. Strain Cas60314T reduces cysteine and elemental sulfur to sulfide, but not thiosulfate [1]. The strain could not grow in unreduced medium under an N2/air (20:1, v/v) gas phase, but was able to grow with 100% H2 in the headspace, with peptone as substrate [1].
Figure 2.
Scanning electron micrograph of T. lienii Cas60314T
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position [22], and is part of the Genomic Encyclopedia of Bacteria and Archaea project [23]. The genome project is deposited in the Genomes On Line Database [12] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Three genomic libraries: one 454 pyrosequence standard library, one 454 PE library (8 kb insert size), one Illumina library |
| MIGS-29 | Sequencing platforms | Illumina GAii, 454 GS FLX Titanium |
| MIGS-31.2 | Sequencing coverage | 223.9 × Illumina; 78.8 × pyrosequence |
| MIGS-30 | Assemblers | Newbler version 2.0.00.20-PostRelease-11-05-2008, Velvet version 1.0.13, phrap version SPS - 4.24 |
| MIGS-32 | Gene calling method | Prodigal |
| INSDC ID | CP003096 (chromosome) CP003097 (plasmid) |
|
| GenBank Date of Release | November 03, 2011 | |
| GOLD ID | Gc02016 | |
| NCBI project ID | 33163 | |
| Database: IMG | 2505119043 | |
| MIGS-13 | Source material identifier | DSM 17291 |
| Project relevance | Bioenergy and phylogenetic diversity |
Growth conditions and DNA isolation
T. lienii strain Cas60314T, DSM 17291, was grown anaerobically in DSMZ medium 383 (Desulfobacterium medium) [24] at 55°C. DNA was isolated from 0.5-1 g of cell paste using MasterPure Gram-positive DNA purification kit (Epicentre MGP04100) following the standard protocol as recommended by the manufacturer, with the following modification for cell lysis: 1µl lysozyme and 5 µl mutanolysin added to the standard lysis solution for 40 min at 37°C; after the MPC-step the lysis solution was incubated for one hour on ice. DNA is available through the DNA Bank Network [25].
Genome sequencing and assembly
The genome was sequenced using a combination of Illumina and 454 sequencing platforms. All general aspects of library construction and sequencing can be found at the JGI website [26]. Pyrosequencing reads were assembled using the Newbler assembler (Roche). The initial Newbler assembly consisting of 127 contigs in one scaffold was converted into a phrap [27] assembly by making fake reads from the consensus, to collect the read pairs in the 454 paired end library. Illumina GAii sequencing data (379.0 Mb) was assembled with Velvet [28] and the consensus sequences were shredded into 1.5 kb overlapped fake reads and assembled together with the 454 data. The 454 draft assembly was based on 156.8 Mb 454 draft data and all of the 454 paired end data. Newbler parameters are -consed -a 50 -l 350 -g -m -ml 20. The Phred/Phrap/Consed software package [27] was used for sequence assembly and quality assessment in the subsequent finishing process. After the shotgun stage, reads were assembled with parallel phrap (High Performance Software, LLC). Possible mis-assemblies were corrected with gapResolution [26], Dupfinisher [29], or sequencing cloned bridging PCR fragments with subcloning. Gaps between contigs were closed by editing in Consed, by PCR and by Bubble PCR primer walks (J.-F. Chang, unpublished). A total of 811 additional reactions and 23 shatter libraries were necessary to close gaps and to raise the quality of the finished sequence. Illumina reads were also used to correct potential base errors and increase consensus quality using a software Polisher developed at JGI [30]. The error rate of the completed genome sequence is less than 1 in 100,000. Together, the combination of the Illumina and 454 sequencing platforms provided 302.7 × coverage of the genome. The final assembly contained 569,599 pyrosequence and 12,441,8 Illumina reads.
Genome annotation
Genes were identified using Prodigal [31] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [32]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) non-redundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. These data sources were combined to assert a product description for each predicted protein. Non-coding genes and miscellaneous features were predicted using tRNAscan-SE [33], RNAMMer [34], Rfam [35], TMHMM [36], and signalP [37].
Genome properties
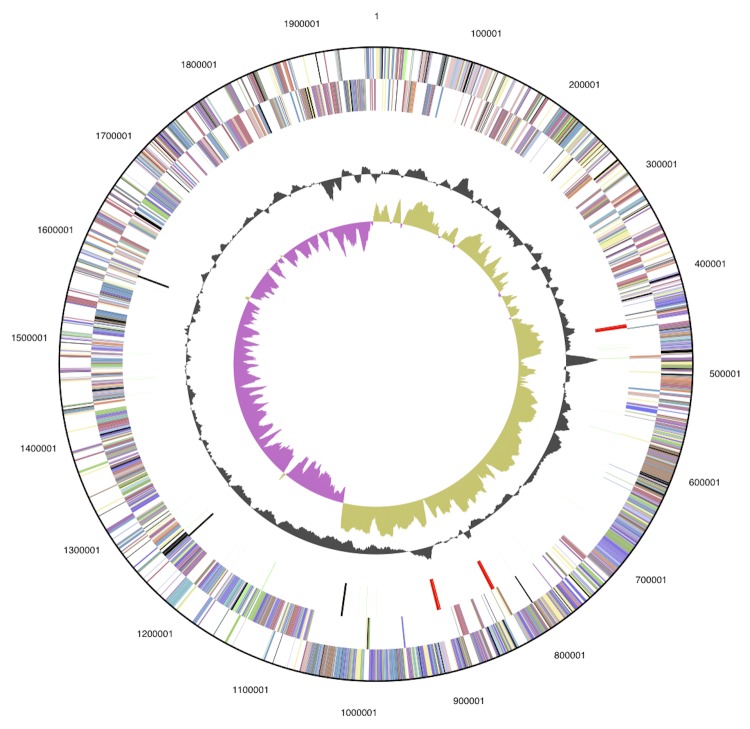
The genome consists of a 1,967,774 bp long chromosome and a 31,872 bp long circular plasmid with a 47.1% G+C content (Table 3 and Figure 3). Of the 1,973 genes predicted, 1,914 were protein-coding genes, and 59 RNAs; 38 pseudogenes were also identified. The majority of the protein-coding genes (79.0%) were assigned a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 1,999,646 | 100.00% |
| DNA coding region (bp) | 1,838,210 | 91.93% |
| DNA G+C content (bp) | 941,059 | 47.06% |
| Number of replicons | 2 | |
| Extrachromosomal elements | 1 | |
| Total genes | 1,973 | 100.00% |
| RNA genes | 59 | 2.99% |
| rRNA operons | 3 | |
| tRNA genes | 47 | 2.38% |
| Protein-coding genes | 1,914 | 97.01% |
| Pseudo genes | 38 | 1.93% |
| Genes with function prediction (proteins) | 1,558 | 78.97% |
| Genes in paralog clusters | 833 | 42.22% |
| Genes assigned to COGs | 1,685 | 85.40% |
| Genes assigned Pfam domains | 1,695 | 85.91% |
| Genes with signal peptides | 285 | 14.45% |
| Genes with transmembrane helices | 486 | 24.63% |
| CRISPR repeats | 0 |
Figure 3.
Graphical map of the chromosome (plasmid not shown, but accessible through the img/er pages on the JGI web pages [26]). From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | %age | Description |
|---|---|---|---|
| J | 149 | 8.1 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.0 | RNA processing and modification |
| K | 98 | 5.3 | Transcription |
| L | 147 | 7.9 | Replication, recombination and repair |
| B | 2 | 0.1 | Chromatin structure and dynamics |
| D | 31 | 1.7 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | 0.0 | Nuclear structure |
| V | 14 | 0.8 | Defense mechanisms |
| T | 77 | 4.2 | Signal transduction mechanisms |
| M | 99 | 5.4 | Cell wall/membrane biogenesis |
| N | 61 | 3.3 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 50 | 2.7 | Intracellular trafficking and secretion, and vesicular transport |
| O | 60 | 3.2 | Posttranslational modification, protein turnover, chaperones |
| C | 160 | 8.6 | Energy production and conversion |
| G | 91 | 4.9 | Carbohydrate transport and metabolism |
| E | 229 | 12.4 | Amino acid transport and metabolism |
| F | 56 | 3.0 | Nucleotide transport and metabolism |
| H | 77 | 4.2 | Coenzyme transport and metabolism |
| I | 40 | 2.2 | Lipid transport and metabolism |
| P | 73 | 3.9 | Inorganic ion transport and metabolism |
| Q | 31 | 1.7 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 189 | 10.2 | General function prediction only |
| S | 117 | 6.3 | Function unknown |
| - | 288 | 14.6 | Not in COGs |
Acknowledgements
We would like to gratefully acknowledge the help of Maren Schröder (DSMZ) for growing T. lienii cultures. This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, UT-Battelle and Oak Ridge National Laboratory under contract DE-AC05-00OR22725, as well as German Research Foundation (DFG) INST 599/1-2.
References
- 1.Dahle H, Birkeland NK. Thermovirga lienii gen. nov., sp. nov., a novel moderately thermophilic, anaerobic, amino-acid-degrading bacterium isolated from a North Sea oil well. Int J Syst Evol Microbiol 2006; 56:1539-1545 10.1099/ijs.0.63894-0 [DOI] [PubMed] [Google Scholar]
- 2.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-410 [DOI] [PubMed] [Google Scholar]
- 3.Korf I, Yandell M, Bedell J. BLAST, O'Reilly, Sebastopol, 2003. [Google Scholar]
- 4.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069-5072 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porter MF. An algorithm for suffix stripping. Program: electronic library and information systems 1980; 14:130-137.
- 6.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 7.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- 8.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 9.Hess PN, De Moraes Russo CA. An empirical test of the midpoint rooting method. Biol J Linn Soc Lond 2007; 92:669-674 10.1111/j.1095-8312.2007.00864.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? Lect Notes Comput Sci 2009; 5541:184-200 10.1007/978-3-642-02008-7_13 [DOI] [PubMed] [Google Scholar]
- 11.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0 b10. Sinauer Associates, Sunderland, 2002. [Google Scholar]
- 12.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Kyrpides NC. The genomes on line database (GOLD) in 2009: Status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2010; 38:D346-D354 10.1093/nar/gkp848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chertkov O, Sikorski J, Brambilla E, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Lucas S, Tice H, Cheng JF, et al. Complete genome sequence of Aminobacterium colombiense type strain (ALA-1T). Stand Genomic Sci 2010; 2:280-289 10.4056/sigs.902116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chovatia M, Sikorski J, Schröder M, Lapidus A, Nolan M, Tice H, Glavina Del Rio T, Copeland A, Cheng JF, Lucas S, et al. Complete genome sequence of Thermanaerovibrio acidaminovorans type strain (SU883T). Stand Genomic Sci 2009; 1:254-261 10.4056/sigs.40645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaButti K, Mayilraj S, Clum A, Lucas S, Glavina Del Rio T, Nolan M, Tice H, Cheng JF, Pitluck S, Liolios K, et al. Permanent draft genome sequence of Dethiosulfovibrio peptidovorans type strain (SEBR 4207T). Stand Genomic Sci 2010; 3:85-92 10.4056/sigs.1092865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pitluck S, Yasawong M, Held B, Lapidus A, Nolan M, Copeland A, Lucas S, Glavina Del Rio T, Tice H, Cheng JF, et al. Non-contiguous finished genome sequence of Aminomonas paucivorans type strain (GLU-3T). Stand Genomic Sci 2010; 3:285-293 10.4056/sigs.1253298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms. Proposal for the domains Archaea and Bacteria. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jumas-Bilak E, Roudiére L, Marchandin H. Description of 'Synergistetes' phyl. nov. and emended description of the phylum 'Deferribacteres' and of the family Syntrophomonadaceae, phylum 'Firmicutes'. Int J Syst Evol Microbiol 2009; 59:1028-1035 10.1099/ijs.0.006718-0 [DOI] [PubMed] [Google Scholar]
- 20.BAuA. 2010, Classification of bacteria and archaea in risk groups. http://www.baua.de TRBA 466, p. 237.
- 21.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klenk HP, Göker M. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 2010; 33:175-182 10.1016/j.syapm.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 23.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven Genomic Encyclopaedia of Bacteria and Archaea. Nature 2009; 462:1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.List of growth media used at DSMZ. http://www.dsmz.de/catalogues/catalogue-microorganisms/culture-technology/list-of-media-for-microorganisms.html
- 25.Gemeinholzer B, Dröge G, Zetzsche H, Haszprunar G, Klenk HP, Güntsch A, Berendsohn WG, Wägele JW. The DNA Bank Network: the start from a German initiative. Biopreserv Biobank 2011; 9:51-55 10.1089/bio.2010.0029 [DOI] [PubMed] [Google Scholar]
- 26.The DOE Joint Genome Institute www.jgi.doe.gov
- 27.Phrap and Phred for Windows. MacOS, Linux, and Unix. www.phrap.com
- 28.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821-829 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han C, Chain P. Finishing repeat regions automatically with Dupfinisher. In: Proceeding of the 2006 international conference on bioinformatics & computational biology. Arabnia HR, Valafar H (eds), CSREA Press. June 26-29, 2006: 141-146. [Google Scholar]
- 30.Lapidus A, LaButti K, Foster B, Lowry S, Trong S, Goltsman E. POLISHER: An effective tool for using ultra short reads in microbial genome assembly and finishing. AGBT, Marco Island, FL, 2008. [Google Scholar]
- 31.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal Prokaryotic Dynamic Programming Genefinding Algorithm. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pati A, Ivanova N, Mikhailova N, Ovchinikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: A Gene Prediction Improvement Pipeline for microbial genomes. Nat Methods 2010; 7:455-457 10.1038/nmeth.1457 [DOI] [PubMed] [Google Scholar]
- 33.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955-964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagesen K, Hallin PF, Rødland E, Stærfeldt HH, Rognes T, Ussery DW. RNammer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res 2007; 35:3100-3108 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res 2003; 31:439-441 10.1093/nar/gkg006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol 2001; 305:567-580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 37.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 2004; 340:783-795 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]