Abstract
Muricauda ruestringensis Bruns et al. 2001 is the type species of the genus Muricauda, which belongs to the family Flavobacteriaceae in the phylum Bacteroidetes. The species is of interest because of its isolated position in the genomically unexplored genus Muricauda, which is located in a part of the tree of life containing not many organisms with sequenced genomes. The genome, which consists of a circular chromosome of 3,842,422 bp length with a total of 3,478 protein-coding and 47 RNA genes, is a part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: facultatively anaerobic, non-motile, Gram-negative, mesophilic, marine, chemoheterotrophic, Flavobacteriaceae, GEBA
Introduction
Strain B1T (= DSM 13258 = LMG 19739 = KCTC 12928) is the type strain of the species Muricauda ruestringensis, which is the type species of the currently six species containing genus Muricauda [1,2]. The genus name was derived from the Latin words muris, of the mouse, and cauda, the tail; Muricauda, tail of the mouse, referring to the cellular appendages observed on some cells [1]. The species epithet is derived from the Neo-Latin word ruestringensis, pertaining to the former village of Rüstringen, which was destroyed by a tidal wave in 1362 [1]. Stain B1T was isolated from a seawater sediment suspension from intertidal sediment at the German North Sea coast, which contained hexadecane as the sole carbon source during the initial cultivation. Later, the organism either turned out to be unable to degrade hexadecane or lost its ability to do so [1]. Other isolates belonging to the species are not known, nor was strain B1T used for scientific work other than the description of the species M. ruestringensis. Here we present a summary classification and a set of features for M. ruestringensis strain B1T, together with the description of the complete genomic sequencing and annotation.
Classification and features
A representative genomic 16S rRNA sequence of M. ruestringensis B1T was compared using NCBI BLAST [3,4] under default settings (e.g., considering only the high-scoring segment pairs (HSPs) from the best 250 hits) with the most recent release of the Greengenes database [5] and the relative frequencies of taxa and keywords (reduced to their stem [6]) were determined, weighted by BLAST scores. The most frequently occurring genera were Muricauda (24.7%), Maribacter (24.0%), Cytophaga (12.3%), Zobellia (9.6%) and Flavobacterium (7.1%) (118 hits in total). Regarding the two hits to sequences from members of the species, the average identity within HSPs was 99.7%, whereas the average coverage by HSPs was 93.8%. Regarding the six hits to sequences from other members of the genus, the average identity within HSPs was 97.9%, whereas the average coverage by HSPs was 97.9%. Among all other species, the one yielding the highest score was Muricauda aquimarina (EU440979), which corresponded to an identity of 98.7% and an HSP coverage of 98.4%. (Note that the Greengenes database uses the INSDC (= EMBL/NCBI/DDBJ) annotation, which is not an authoritative source for nomenclature or classification.) The highest-scoring environmental sequence was HQ326265 ('Microbial structure biofilm on SWRO membranes clone SBS-FW-047'), which showed an identity of 98.5% and a HSP coverage of 98.0%. The most frequently occurring keywords within the labels of all environmental samples which yielded hits were 'microbi' (4.7%), 'sediment' (4.1%), 'sea' (2.9%), 'marin' (2.4%) and 'biofilm' (2.4%), (132 hits in total). Environmental samples which yielded hits of a higher score than the highest scoring species were not found.
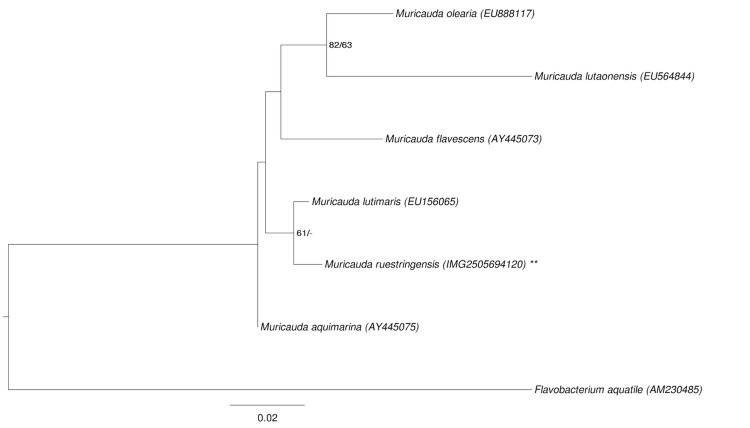
Figure 1 shows the phylogenetic neighborhood of M. ruestringensis in a 16S rRNA based tree. The sequences of the two identical 16S rRNA gene copies in the genome differ by one nucleotide from the previously published 16S rRNA sequence (AF218782).
Figure 1.
Phylogenetic tree highlighting the position of M. ruestringensis relative to the type strains of the other species within the genus Muricauda. The tree was inferred from 1,481 aligned characters [7,8] of the 16S rRNA gene sequence under the maximum likelihood (ML) criterion [9]. Flavobacterium aquatile was included in the dataset for use as outgroup taxa. The branches are scaled in terms of the expected number of substitutions per site. Numbers adjacent to the branches are support values from 850 ML bootstrap replicates [10] (left) and from 1,000 Maximum-Parsimony bootstrap replicates [11] (right) if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [12] are labeled with one asterisk, those also listed as 'Complete and Published' with two asterisks.
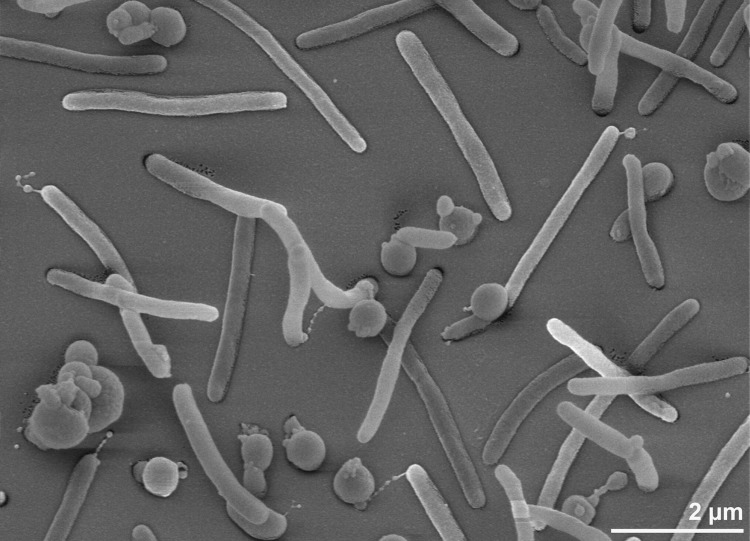
Cells of strain B1T are rod-shaped with rounded ends, 0.3 - 0.6 µm wide and 1.1 - 2.7 µm long (Figure 2 and Table 1) [1]. Cells of older cultures are characterized by mainly polar appendages with vesicle-like structures (blebs) at the end (Figure 2), which were discussed in detail by Bruns et al. in [1] and probably serve to contact cells to each other or for colonization of a substratum [1]. The non-motile cells (see missing genes in the motility category in COGs table) stain Gram-negative and grow as facultative anaerobes in seawater. The temperature range for growth is between 8°C and 40°C, with an optimum between 20 and 30°C [1]. The pH range for growth is 6.0-8.0, with an optimum at pH 6.5-7.5 [1]. Physiology and metabolism are discussed in detail in [1], with the surprising discovery that although strain B1T was isolated from a continuous-flow culture containing hexadecane as a sole carbon source, the strain was unable to degrade hexadecane (even if it was offered as cosubstrate along with other carbon sources); nor could it use acetate or pyruvate as sole carbon sources, but required a wide spectrum of amino acids as carbon and energy sources in addition to some carbohydrates [1].
Figure 2.
Scanning electron micrograph of M. ruestringensis B1T
Table 1. Classification and general features of M. ruestringensis B1T according to the MIGS recommendations [13].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [14] | |
| Phylum Bacteroidetes | TAS [15,16] | ||
| Class Flavobacteria | TAS [17,18] | ||
| Order Flavobacteriales | TAS [19,20] | ||
| Family Flavobacteriaceae | TAS [21-24] | ||
| Genus Muricauda | TAS [1,25,26] | ||
| Species Muricauda ruestringensis | TAS [1] | ||
| Type strain B1 | TAS [1] | ||
| Gram stain | negative | TAS [1] | |
| Cell shape | rod-shaped | TAS [1] | |
| Motility | non-motile | TAS [1] | |
| Sporulation | not reported | ||
| Temperature range | mesophile, 20°C–30°C | TAS [1] | |
| Optimum temperature | 30°C | TAS [1] | |
| Salinity | slightly halophilic, optimum 3% NaCl (w/v) | TAS [1] | |
| MIGS-22 | Oxygen requirement | facultatively anaerobic | TAS [1] |
| Carbon source | various sugars and amino acids | TAS [1] | |
| Energy metabolism | chemoheterotroph | TAS [1] | |
| MIGS-6 | Habitat | marine | TAS [1] |
| MIGS-15 | Biotic relationship | free-living | TAS [1] |
| MIGS-14 | Pathogenicity | none | NAS |
| Biosafety level | 1 | TAS [27] | |
| Isolation | seawater sediment suspension | TAS [1] | |
| MIGS-4 | Geographic location | Jadebusen Bay, coast of North Sea, Germany | TAS [1] |
| MIGS-5 | Sample collection time | 1998 or earlier | NAS |
| MIGS-4.1 | Latitude | 53.45 | NAS |
| MIGS-4.2 | Longitude | 8.20 | NAS |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | about 0 m, sea level | NAS |
Evidence codes - NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [28].
Chemotaxonomy
The spectrum of whole-cell fatty acids represents the only chemotaxonomical data published thus far for strain B1T. The spectrum of fatty acids was dominated by branched-chain acids (72%): iso-C17:0 3OH (28.7%), iso-C15:1 (16.3%), iso-C15:0 (15.5%), iso-C15:0 3OH (4.9%), iso-C16:0 3OH (2.9%), iso-C17:0 2OH (2.8%), iso -C15:0 2OH (2.5%), C16:1 ω7c (2.5%), anteiso-C15:0 (2.4%), other acids below 2% [1].
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position [29], and is part of the Genomic Encyclopedia of Bacteria and Archaea project [30]. The genome project is deposited in the Genomes On Line Database [12] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Four genomic libraries: one 454 pyrosequence standard library, two 454 PE libraries (4 kb and 8 kb insert size), one Illumina library |
| MIGS-29 | Sequencing platforms | Illumina GAii, 454 GS FLX Titanium |
| MIGS-31.2 | Sequencing coverage | 996.4 × Illumina; 36.4 × pyrosequence |
| MIGS-30 | Assemblers | Newbler version 2.3, Velvet version 0.7.63, phrap version SPS - 4.24 |
| MIGS-32 | Gene calling method | Prodigal 1.4, GenePRIMP |
| INSDC ID | CP002999 | |
| Genbank Date of Release | August 19, 2011 | |
| GOLD ID | Gc01927 | |
| NCBI project ID | 52467 | |
| Database: IMG-GEBA | 2505679007 | |
| MIGS-13 | Source material identifier | DSM 13258 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
M. ruestringensis strain B1T, DSM 13258, was grown in DSMZ medium 917 (Modified Sea Water Agar) [31] at 30°C. DNA was isolated from 0.5-1 g of cell paste using Jetflex Genomic DNA Purification Kit (GENOMED 600100) following the manufacturer’s instructions, with a modified procedure for cell lysis: incubation with 40 μl proteinase K for 40 minutes at 58°C. DNA is available through the DNA Bank Network [32].
Genome sequencing and assembly
The genome was sequenced using a combination of Illumina and 454 sequencing platforms. All general aspects of library construction and sequencing can be found at the JGI website [33]. Pyrosequencing reads were assembled using the Newbler assembler (Roche). The initial Newbler assembly consisting of 26 contigs in one scaffold was converted into a phrap [34] assembly by making fake reads from the consensus, to collect the read pairs in the 454 paired end library. Illumina GAii sequencing data (3,847 Mb) was assembled with Velvet [35] and the consensus sequences were shredded into 1.5 kb overlapped fake reads and assembled together with the 454 data. The 454 draft assembly was based on 268.3 Mb 454 draft data and all of the 454 paired end data. Newbler parameters are -consed -a 50 -l 350 -g -m -ml 20. The Phred/Phrap/Consed software package [34] was used for sequence assembly and quality assessment in the subsequent finishing process. After the shotgun stage, reads were assembled with parallel phrap (High Performance Software, LLC). Possible mis-assemblies were corrected with gapResolution [33], Dupfinisher [36], or sequencing cloned bridging PCR fragments with subcloning. Gaps between contigs were closed by editing in Consed, by PCR and by Bubble PCR primer walks (J.-F. Chang, unpublished). A total of 46 additional reactions were necessary to close gaps and to raise the quality of the finished sequence. Illumina reads were also used to correct potential base errors and increase consensus quality using a software Polisher developed at JGI [37]. The error rate of the completed genome sequence is less than 1 in 100,000. Together, the combination of the Illumina and 454 sequencing platforms provided 1,032.9 × coverage of the genome. The final assembly contained 422,407 pyrosequence and 49,819,141 Illumina reads.
Genome annotation
Genes were identified using Prodigal [38] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [39]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGR-Fam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes – Expert Review (IMG-ER) platform [40].
Genome properties
The genome consists of a 3,842,422 bp long circular chromosome with a G+C content of 41.4% (Table 3 and Figure 3). Of the 3,525 genes predicted, 3,478 were protein-coding genes, and 47 RNAs; 46 pseudogenes were also identified. The majority of the protein-coding genes (66.6%) were assigned with a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 3,842,422 | 100.00% |
| DNA coding region (bp) | 3,479,569 | 90.56% |
| DNA G+C content (bp) | 1,589,148 | 41.36% |
| Number of replicons | 1 | |
| Extrachromosomal elements | 0 | |
| Total genes | 3,525 | 100.00% |
| RNA genes | 47 | 1.33% |
| rRNA operons | 2 | |
| tRNA genes | 38 | 1.08% |
| Protein-coding genes | 3,478 | 98.67% |
| Pseudo genes | 46 | 1.30% |
| Genes with function prediction | 2,349 | 66.64% |
| Genes in paralog clusters | 1,644 | 46.64% |
| Genes assigned to COGs | 2,433 | 69.02% |
| Genes assigned Pfam domains | 2,500 | 70.92% |
| Genes with signal peptides | 970 | 27.52% |
| Genes with transmembrane helices | 809 | 22.95% |
| CRISPR repeats | 0 |
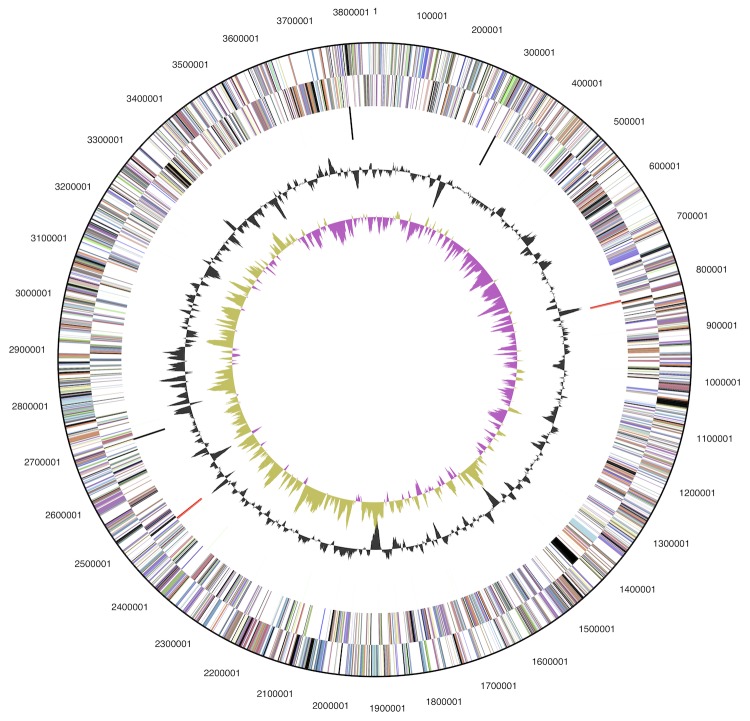
Figure 3.
Graphical map of the chromosome. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | %age | Description |
|---|---|---|---|
| J | 151 | 5.8 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.0 | RNA processing and modification |
| K | 206 | 7.9 | Transcription |
| L | 130 | 5.0 | Replication, recombination and repair |
| B | 2 | 0.1 | Chromatin structure and dynamics |
| D | 23 | 0.9 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | 0.0 | Nuclear structure |
| V | 77 | 2.9 | Defense mechanisms |
| T | 145 | 5.5 | Signal transduction mechanisms |
| M | 186 | 7.1 | Cell wall/membrane/envelope biogenesis |
| N | 7 | 0.3 | Cell motility |
| Z | 1 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 50 | 1.9 | Intracellular trafficking, secretion, and vesicular transport |
| O | 106 | 4.0 | Posttranslational modification, protein turnover, chaperones |
| C | 129 | 4.9 | Energy production and conversion |
| G | 136 | 5.2 | Carbohydrate transport and metabolism |
| E | 220 | 8.4 | Amino acid transport and metabolism |
| F | 65 | 2.5 | Nucleotide transport and metabolism |
| H | 138 | 5.3 | Coenzyme transport and metabolism |
| I | 86 | 3.3 | Lipid transport and metabolism |
| P | 141 | 5.4 | Inorganic ion transport and metabolism |
| Q | 49 | 1.9 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 339 | 12.9 | General function prediction only |
| S | 236 | 9.0 | Function unknown |
| - | 1,092 | 31.0 | Not in COGs |
Acknowledgements
We would like to gratefully acknowledge the help of Maren Schröder (DSMZ) for growing M. ruestringensis cultures. This work was performed under the auspices of the US Department of Energy Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, UT-Battelle and Oak Ridge National Laboratory under contract DE-AC05-00OR22725, as well as German Research Foundation (DFG) INST 599/1-2.
References
- 1.Bruns A, Rohde M, Berthe-Corti L. Muricauda ruestringensis gen.nov., sp. nov., a facultatively anaerobic, appendaged bacterium from German North Sea intertidal sediment. Int J Syst Evol Microbiol 2001; 51:1997-2006 10.1099/00207713-51-6-1997 [DOI] [PubMed] [Google Scholar]
- 2.Garrity G. NamesforLife. BrowserTool takes expertise out of the database and puts it right in the browser. Microbiol Today 2010; 37:9 [Google Scholar]
- 3.Altschul SF, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol 1990; 215:403-410 [DOI] [PubMed] [Google Scholar]
- 4.Korf I, Yandell M, Bedell J. BLAST, O'Reilly, Sebastopol, 2003. [Google Scholar]
- 5.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069-5072 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porter MF. An algorithm for suffix stripping. Program: electronic library and information systems 1980; 14:130-137.
- 7.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 8.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- 9.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 10.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? Lect Notes Comput Sci 2009; 5541:184-200 10.1007/978-3-642-02008-7_13 [DOI] [PubMed] [Google Scholar]
- 11.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0 b10. Sinauer Associates, Sunderland, 2002. [Google Scholar]
- 12.Pagani I, Liolios K, Jansson J, Chen IM, Smirnova T, Nosrat B, Markowitz VM, Kyrpides NC. The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2012; 40:D571-D579 10.1093/nar/gkr1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Validation List No 143. Int J Syst Evol Microbiol 2012; 62:1-4 10.1099/ijs.0.039487-0 [DOI] [Google Scholar]
- 16.Krieg NR, Ludwig W, Euzéby J, Whitman WB. Phylum XIV. Bacteroidetes phyl. nov. In: Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, Ludwig W, Whitman WB (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 4, Springer, New York, 2010, p. 25. [Google Scholar]
- 17.Ludwig W, Euzeby J, Whitman WG. Draft taxonomic outline of the Bacteroidetes, Planctomycetes, Chlamydiae, Spirochaetes, Fibrobacteres, Fusobacteria, Acidobacteria, Verrucomicrobia, Dictyoglomi, and Gemmatimonadetes http://www.bergeys.org/outlines/Bergeys_Vol_4_Outline.pdf Taxonomic Outline 2008.
- 18.Judicial Commission of the International Committee on Systematics of Prokaryotes The nomenclatural types of the orders Acholeplasmatales, Halanaerobiales, Halobacteriales, Methanobacteriales, Methanococcales, Methanomicrobiales, Planctomycetales, Prochlorales, Sulfolobales, Thermococcales, Thermoproteales and Verrucomicrobiales are the genera Acholeplasma, Halanaerobium, Halobacterium, Methanobacterium, Methanococcus, Methanomicrobium, Planctomyces, Prochloron, Sulfolobus, Thermococcus, Thermoproteus and Verrucomicrobium, respectively. Opinion 79. Int J Syst Evol Microbiol 2005; 55:517-518 10.1099/ijs.0.63548-0 [DOI] [PubMed] [Google Scholar]
- 19.Validation List No 143. Int J Syst Evol Microbiol 2012; 62:1-4 10.1099/ijs.0.039487-0 [DOI] [Google Scholar]
- 20.Bernardet JF. Order I. Flavobacteriales ord. nov. In: Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, Ludwig W, Whitman WB (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 4, Springer, New York, 2010, p. 105. [Google Scholar]
- 21.Reichenbach H. Order 1. Cytophagales Leadbetter 1974, 99AL. In: Holt JG (ed), Bergey's Manual of Systematic Bacteriology, First Edition, Volume 3, The Williams and Wilkins Co., Baltimore, 1989, p. 2011-2013. [Google Scholar]
- 22.Validation of the publication of new names and new combinations previously effectively published outside the IJSB. List No. 41. Int J Syst Bacteriol 1992; 42:327-328 10.1099/00207713-42-2-327 [DOI] [PubMed] [Google Scholar]
- 23.Bernardet JF, Segers P, Vancanneyt M, Berthe F, Kersters K, Vandamme P. Cutting a Gordian knot: emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriaceae, and proposal of Flavobacterium hydatis nom. nov. (Basonym, Cytophaga aquatilis Strohl and Tait 1978). Int J Syst Bacteriol 1996; 46:128-148 10.1099/00207713-46-1-128 [DOI] [Google Scholar]
- 24.Bernardet JF, Nakagawa Y, Holmes B. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae, and emended description of the family. Int J Syst Evol Microbiol 2002; 52:1049-1070 10.1099/ijs.0.02136-0 [DOI] [PubMed] [Google Scholar]
- 25.Yoon JH, Lee MH, Oh TK, Park YH. Muricauda flavescens sp. nov. and Muricauda aquimarina sp. nov., isolated from a salt lake near Hwajinpo Beach of the East Sea in Korea, and emended description of the genus Muricauda. Int J Syst Evol Microbiol 2005; 55:1015-1019 10.1099/ijs.0.03051-0 [DOI] [PubMed] [Google Scholar]
- 26.Hwang CY, Kim MH, Bae GD, Zhang GI, Kim YH, Cho BC. Muricauda olearia sp. nov., isolated from crude-oil-contaminated seawater, and emended description of the genus Muricauda. Int J Syst Evol Microbiol 2009; 59:1856-1861 10.1099/ijs.0.007708-0 [DOI] [PubMed] [Google Scholar]
- 27.BAuA. Classification of bacteria and archaea in risk groups. TRBA 466. p. 141. Bundesanstalt für Arbeitsschutz und Arbeitsmedizin, Germany. 2010. [Google Scholar]
- 28.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klenk HP, Göker M. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 2010; 33:175-182 10.1016/j.syapm.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 30.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 2009; 462:1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.List of growth media used at DSMZ: http//www.dsmz.de/catalogues/catalogue-microorganisms/culture-technology/list-of-media-for-microorganisms.html
- 32.Gemeinholzer B, Dröge G, Zetzsche H, Haszprunar G, Klenk HP, Güntsch A, Berendsohn WG, Wägele JW. The DNA Bank Network: the start from a German initiative. Biopreserv Biobank 2011; 9:51-55 10.1089/bio.2010.0029 [DOI] [PubMed] [Google Scholar]
- 33.JGI website. http://www.jgi.doe.gov
- 34.The Phred/Phrap/Consed software package. http://www.phrap.com
- 35.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821-829 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han C, Chain P. Finishing repeat regions automatically with Dupfinisher. In: Proceeding of the 2006 international conference on bioinformatics & computational biology. Arabnia HR, Valafar H (eds), CSREA Press. June 26-29, 2006: 141-146. [Google Scholar]
- 37.Lapidus A, LaButti K, Foster B, Lowry S, Trong S, Goltsman E. POLISHER: An effective tool for using ultra short reads in microbial genome assembly and finishing. AGBT, Marco Island, FL, 2008. [Google Scholar]
- 38.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods 2010; 7:455-457 10.1038/nmeth.1457 [DOI] [PubMed] [Google Scholar]
- 40.Markowitz VM, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]