Abstract
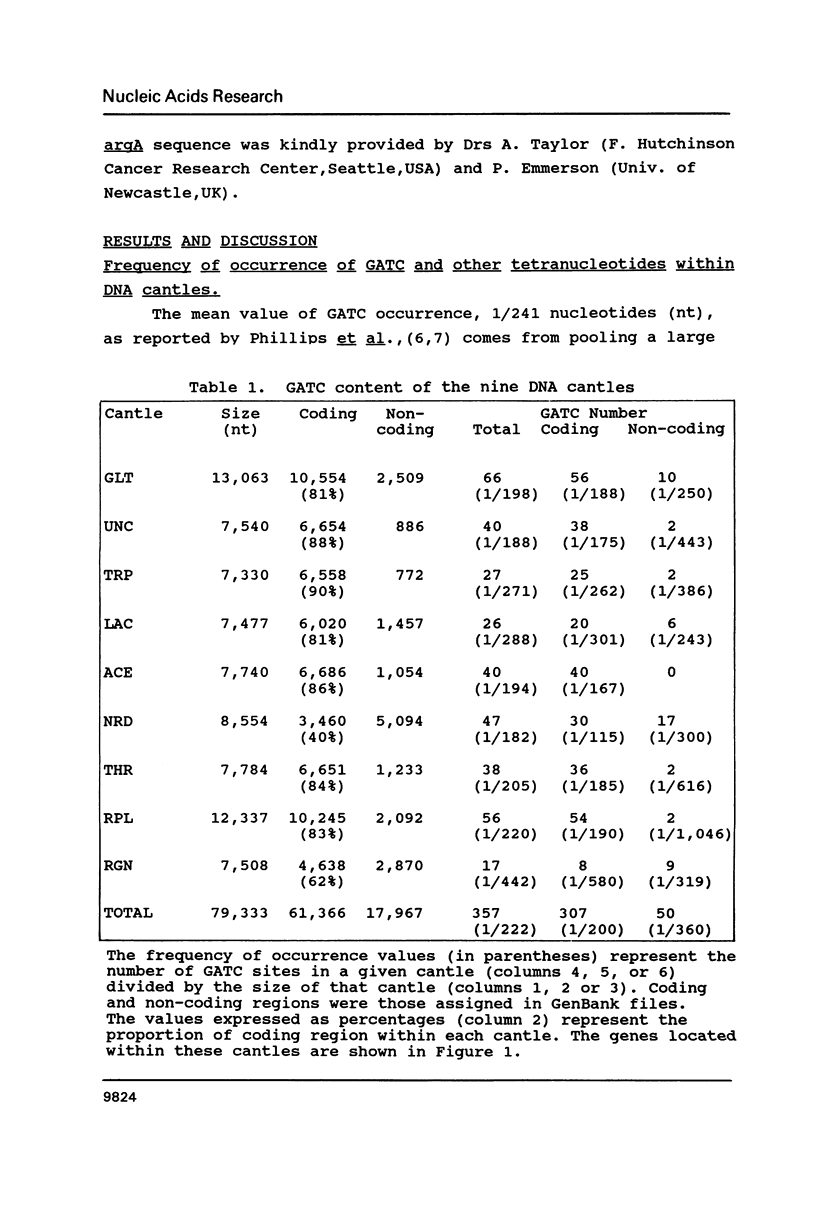
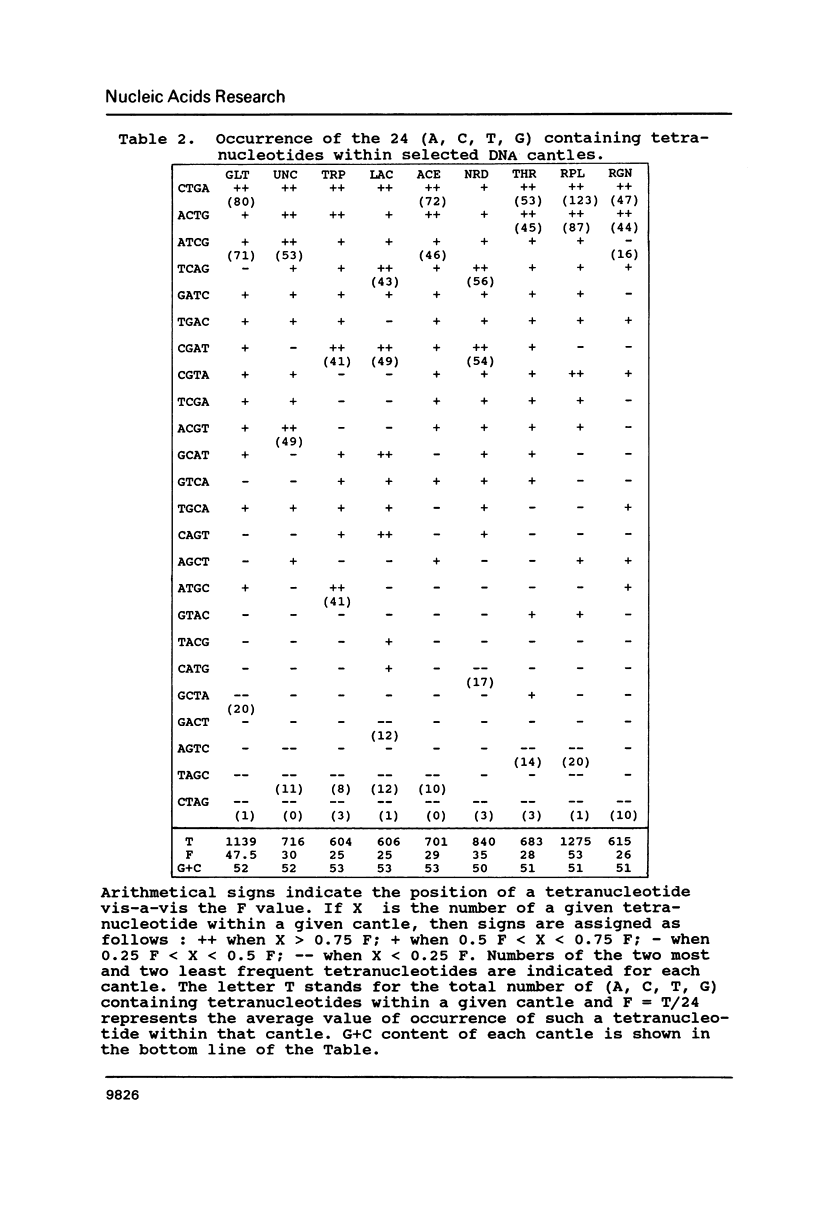
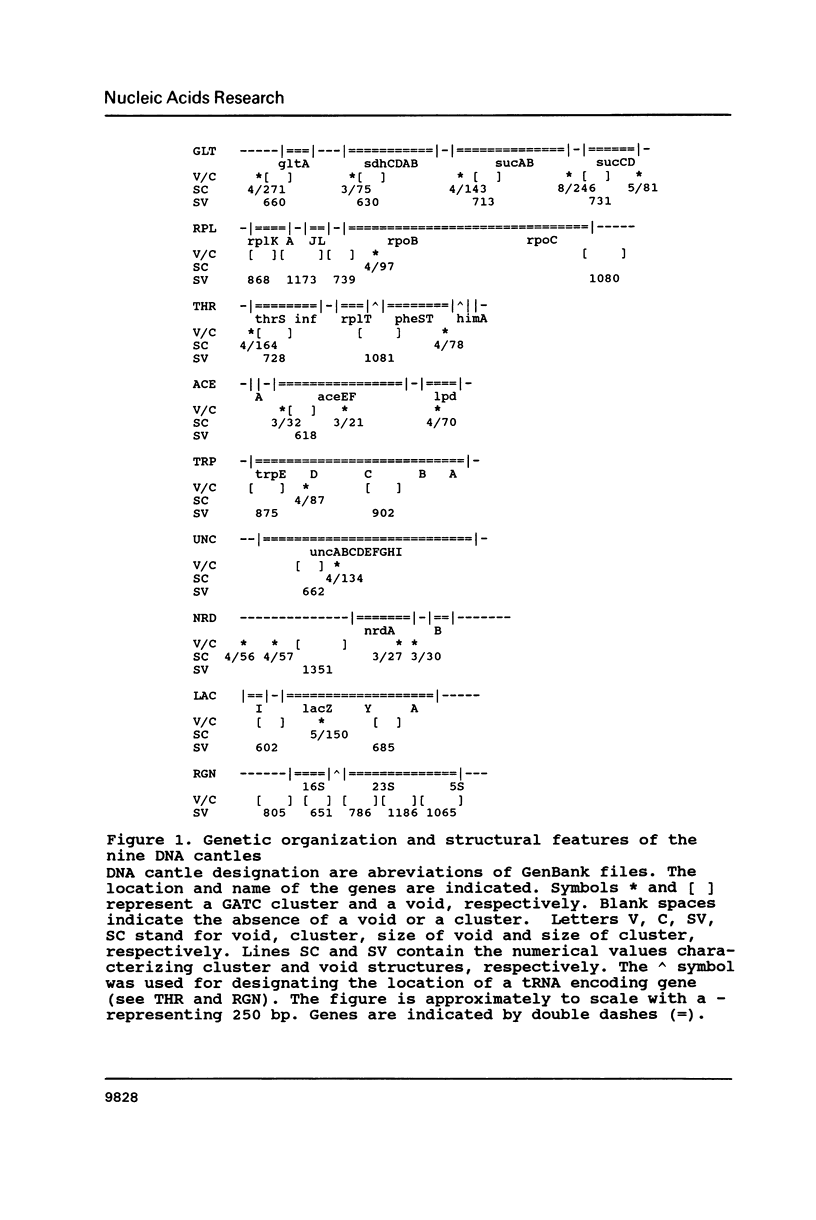
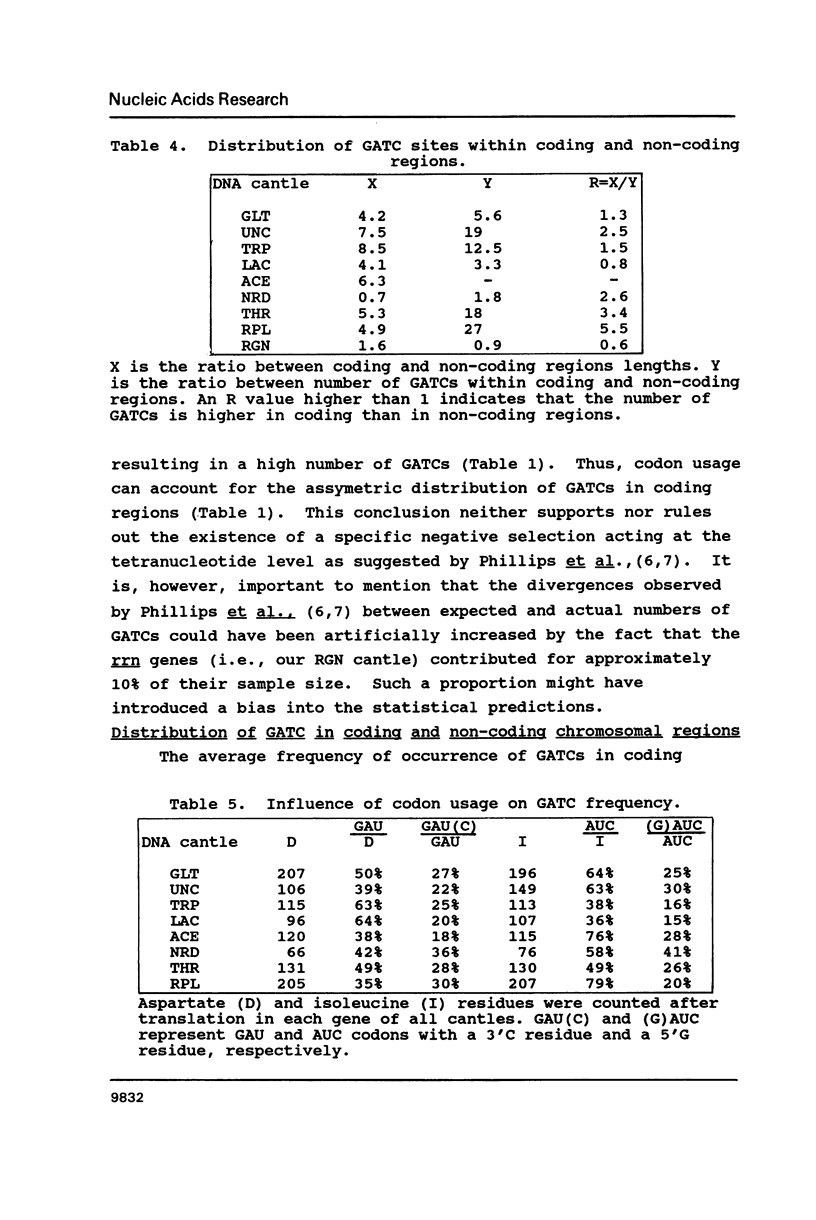
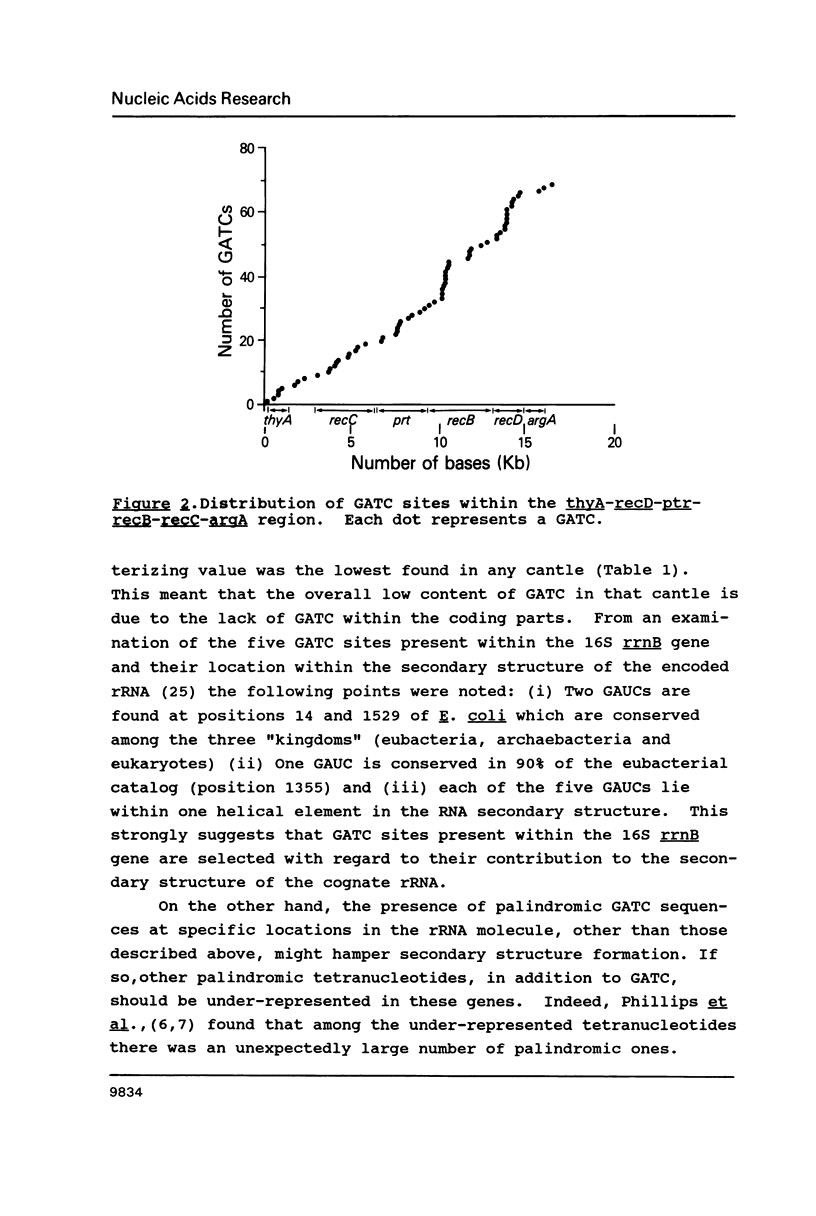
The occurrence of GATC (Dam-recognition) sites in available E. coli DNA sequences (representing about 2% of the chromosome) has been determined by a simple numerical analysis. Our approach was to analyze the nucleotide composition of nine large sequenced DNA stretches ("cantles") in order to identify patterns of GATC distribution and to rationalize such patterns in biological/structural terms. The following observations were made: (i) In addition to oriC, GATC-rich regions are present in numerous locations. (ii) There is a wide variation in GATC frequency both between and within DNA cantles which led to the identification of a void-cluster pattern of GATC arrangement. The distance between two GATCs was never greater than 2 kb. (iii) GATC sites are found more frequently in translated regions than (in decreasing order) non-coding or non-translated regions. In particular, rRNA and tRNA encoding genes exhibit the lowest GATC content.
Full text
PDF












Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Braun R. E., Wright A. DNA methylation differentially enhances the expression of one of the two E. coli dnaA promoters in vivo and in vitro. Mol Gen Genet. 1986 Feb;202(2):246–250. doi: 10.1007/BF00331644. [DOI] [PubMed] [Google Scholar]
- Brown K., Finch P. W., Hickson I. D., Emmerson P. T. Complete nucleotide sequence of the Escherichia coli argA gene. Nucleic Acids Res. 1987 Dec 23;15(24):10586–10586. doi: 10.1093/nar/15.24.10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruni R., Martin D., Jiricny J. d(GATC) sequences influence Escherichia coli mismatch repair in a distance-dependent manner from positions both upstream and downstream of the mismatch. Nucleic Acids Res. 1988 Jun 10;16(11):4875–4890. doi: 10.1093/nar/16.11.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad B., Mount D. W. Microcomputer programs for DNA sequence analysis. Nucleic Acids Res. 1982 Jan 11;10(1):31–38. doi: 10.1093/nar/10.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger M. A., Johnson D. F., Nierlich D. P., Zabin I. DNA sequence of the lactose operon: the lacA gene and the transcriptional termination region. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6414–6418. doi: 10.1073/pnas.82.19.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977 Jul;114(1):153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- Lu A. L. Influence of GATC sequences on Escherichia coli DNA mismatch repair in vitro. J Bacteriol. 1987 Mar;169(3):1254–1259. doi: 10.1128/jb.169.3.1254-1259.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G. DNA methylation in Escherichia coli. Annu Rev Genet. 1987;21:113–131. doi: 10.1146/annurev.ge.21.120187.000553. [DOI] [PubMed] [Google Scholar]
- McClelland M., Jones R., Patel Y., Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987 Aug 11;15(15):5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. Selection against dam methylation sites in the genomes of DNA of enterobacteriophages. J Mol Evol. 1984;21(4):317–322. doi: 10.1007/BF02115649. [DOI] [PubMed] [Google Scholar]
- Messer W., Bellekes U., Lother H. Effect of dam methylation on the activity of the E. coli replication origin, oriC. EMBO J. 1985 May;4(5):1327–1332. doi: 10.1002/j.1460-2075.1985.tb03780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R. Doublet frequencies in evolutionary distinct groups. Nucleic Acids Res. 1984 Feb 10;12(3):1749–1763. doi: 10.1093/nar/12.3.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R. Theoretical molecular biology: prospectives and perspectives. J Theor Biol. 1987 Mar 21;125(2):219–235. doi: 10.1016/s0022-5193(87)80043-7. [DOI] [PubMed] [Google Scholar]
- Ogden G. B., Pratt M. J., Schaechter M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell. 1988 Jul 1;54(1):127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- Phillips G. J., Arnold J., Ivarie R. Mono- through hexanucleotide composition of the Escherichia coli genome: a Markov chain analysis. Nucleic Acids Res. 1987 Mar 25;15(6):2611–2626. doi: 10.1093/nar/15.6.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips G. J., Arnold J., Ivarie R. The effect of codon usage on the oligonucleotide composition of the E. coli genome and identification of over- and underrepresented sequences by Markov chain analysis. Nucleic Acids Res. 1987 Mar 25;15(6):2627–2638. doi: 10.1093/nar/15.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila P. J., Peterson J., Herman G., Modrich P., Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983 Aug;104(4):571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D. W., Zinder N. D. Hemimethylation prevents DNA replication in E. coli. Cell. 1987 Sep 25;50(7):1071–1079. doi: 10.1016/0092-8674(87)90173-5. [DOI] [PubMed] [Google Scholar]
- Schmid K., Thomm M., Laminet A., Laue F. G., Kessler C., Stetter K. O., Schmitt R. Three new restriction endonucleases MaeI, MaeII and MaeIII from Methanococcus aeolicus. Nucleic Acids Res. 1984 Mar 26;12(6):2619–2628. doi: 10.1093/nar/12.6.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. W., Garland A. M., Herman G., Enns R. E., Baker T. A., Zyskind J. W. Importance of state of methylation of oriC GATC sites in initiation of DNA replication in Escherichia coli. EMBO J. 1985 May;4(5):1319–1326. doi: 10.1002/j.1460-2075.1985.tb03779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N. Evidence that adenine methylation influences DNA-protein interactions in Escherichia coli. J Bacteriol. 1985 Oct;164(1):490–493. doi: 10.1128/jb.164.1.490-493.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M., Gruenbaum Y., Urieli-Shoval S., Razin A. Studies on the biological role of DNA methylation: V. The pattern of E.coli DNA methylation. Nucleic Acids Res. 1982 Nov 25;10(22):7247–7259. doi: 10.1093/nar/10.22.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh K. M., Lu A. L., Clark S., Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem. 1987 Nov 15;262(32):15624–15629. [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyskind J. W., Smith D. W. The bacterial origin of replication, oriC. Cell. 1986 Aug 15;46(4):489–490. doi: 10.1016/0092-8674(86)90873-1. [DOI] [PubMed] [Google Scholar]
- de Massy B., Fayet O., Kogoma T. Multiple origin usage for DNA replication in sdrA(rnh) mutants of Escherichia coli K-12. Initiation in the absence of oriC. J Mol Biol. 1984 Sep 15;178(2):227–236. doi: 10.1016/0022-2836(84)90141-4. [DOI] [PubMed] [Google Scholar]



