Abstract
The thiocillins from Bacillus cereus ATCC 14579 are natural products from the broader class of thiazolyl peptides. Their biosynthesis proceeds via extensive post-translational modification of a ribosomally encoded precursor peptide. This post-translational tailoring involves a key step formal cycloaddition between two distal serine residues. In the wild type structure this cycloaddition forms a major macrocycle circumscribed by 26-atoms (shortest path). Results presented herein demonstrate the promiscuity of this last step by means of a set of “competition” experiments. Cyclization proceeds in many cases to provide altered ring sizes, giving access to several variant rings sizes that have not previously been observed in nature.
The vast majority of peptide bonds within known natural product scaffolds are installed by action of non-ribosomal peptide synthetases (NRPSs), multi-modular complexes capable of creating a wide range of functionality around the nascent peptide.1 However, an increasing number of natural products have been found to derive from ribosomally-encoded peptides, which are subsequently post-translationally modified to provide active compounds.2 Structurally, ribosomal peptide natural products can be limited: of necessity, frameworks are defined by a peptidic backbone, comprised of the twenty proteinogenic amino acids. Never the less, the array of observed post-translational modifications has proven to be both vast and profound, leading to tremendous structural diversity. Additionally, the ribosmal-encodement of biosynthetic starting materials provides great advantage in terms of combinatorial biosynthesis: simple modifications to sequences of the peptide starting materials can render profound changes to the natural product structure.3
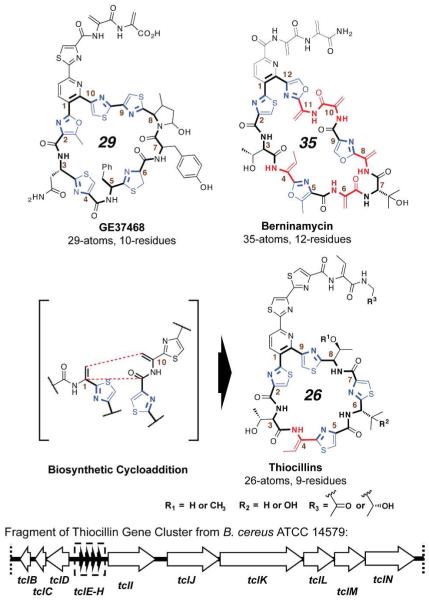
The thiazolyl peptide antibiotics are a class of ribosomally encoded peptide-derived natural products.4 The class is defined by a core complement of “NRPS-like” post-translational modifications, including dehydroalanines (Dhas), dehydrobutyrines (Dhbs), oxazolines/oxazoles, and thiazolines/thiazoles. These varied motifs contribute to the three-dimensional architecture of a constrained macrocycle, which is 26-35 atoms (9-12 residues) in circumference and closed by a central nitrogenous heterocycle. The different macrocycles all have antibiotic activity against gram-positive bacteria but can have different targets, depending on ring size. The thiocillin and thiostrepton subclasses (26 atoms, 9 residues in the major macrocycle) target a crevice in the 50S ribosomal subunit between the 23S rRNA and protein L11.5 The 29-atom GE2270A, GE37468, and thiomuracin instead target the aminoacyl tRNA chaperone EF-Tu.6 The target of the 35-atom members, such as berninamycin, has yet to be determined.7
In prior efforts we have identified the thiocillin biosynthetic gene cluster in a strain of Bacillus cereus (ATCC 14579) and demonstrated that compound maturation proceeds via a cascade of fourteen enzymatic transformations about a 52 residue prepeptide.8 We have probed much of the process by a site-directed mutagenesis gene-replacement strategy, demonstrating profound substrate promiscuity at each step.9 That promiscuity contrasts with the highly defined architecture necessary for antibiotic activity; the cluster is capable of producing large quantities of apparently inactive analogs. More recently, this strategy has uncovered a role for protein TclM in pyridine-ring formation.10
TclM appears to be instrumental in combination of Dhas 1 and 10 in a putative aza-Diels-Alder reaction, which proceeds to form the central pyridine ring, while at the same time closing the 26 atom (9 residue) major macrocycle (figure 1).11 Herein we present a compact set of mutants aimed at interrogating the ability of the B. cereus biosynthetic machinery (in particular the putative cycloaddition) to construct macrocycles of differing ring size. These efforts represent a prelude to examination of the structural basis for ribosome/EF-Tu target switching within this antibiotic class.
Figure 1.
Structures of naturally occurring thiazolyl peptide ring size variants and fragment of thiocillin gene cluster from B. cere- us ATCC 14579.
The first set of alterations is a direct test of ring size promiscuity. Site-directed mutagenesis was employed to sequentially incorporate up to three glycine residues between threonines 3 and 4 of the precursor peptide. In this instance, glycine was chosen for incorporation due to its lower conformational rigidity relative to the set of substituted amino acids. A threonine-3 deletion mutant was also engineered to examine the possibility for ring contraction. Compound production in all four mutants was confirmed by LC/MS and signature MS/MS of isolates (see Supporting Information).12
This set of mutants confirmed the ability of the thiocillin biosynthetic machinery to accommodate ring size formation from a contracted 23-mer stepwise up to the berninamycin-like 35-mer. The 32-mer, resulting from introduction of two glycines, represents a previously unknown thiazolyl peptide ring size variant; to the best of our knowledge, all naturally occurring thiazolyl peptides fall into the three sub-classes described above (26-, 29-, and 35-membered macrocycles) and there have been no 32-mers observed to date. Similarly, the 23-atom ring size variant resulting from the deletion mutant represents the first such thiazolyl peptide of its kind.13
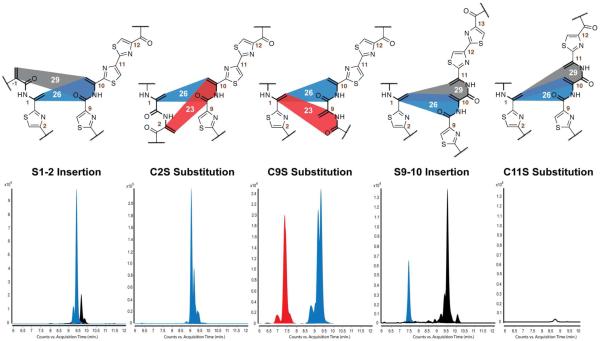
Given the now proven ability of the B. cereus system to produce macrocycles of at least five different sizes, 23- (8 residues), 26- (9 residues), 29- (10 residues), 32- (11 residues), and 35-atoms (12 residues) a second set of mutants was examined to determine any degree of preference within the system for a given ring size. The five mutants, three serine for cysteine substitutions (C2S, C9S, and C11S) and two serine insertions (S1-2 and S9-10) are illustrated in figure 2, together with the LC/MS traces for compound isolates. By selective introduction of a serine residue, each of these mutants presents the enzymatic machinery with at least the theoretical possibility of two different cyclization routes and, thereby, two different ring size variant products. The first two in the series, the S1-2 insertion and C2S substitution present this choice by means of two adjacent N-terminal 2-π components (in the terminology of cycloaddition-type mechanism) and a single C-terminal 4-π component. In both cases the overwhelming preference is for the “natural”, 26-atom ring size. In the S1-2 insertion mutant, both ring sizes, the 26-membered and, to a much lesser extent, the 29-membered, are observed. Interestingly, the pattern of thiazole and dehydroamino acid modifications does not appear to have been perturbed: the resultant 26-member compounds match wild type, as would be expected for standard processing of the mutant precursor peptide. Formation of the 29-member compound demonstrates that the two successive serines are capable of undergoing enzymatic dehydration as well as cycloaddition.
Figure 2.
Overlayed extracted ion chromatograms (EICs) for expected thiazolyl peptide masses in isolates from ring size competition mutants; peaks of compounds with 26-membered rings in blue, 23-memebered rings in red, and 29-membered in black. C11S depicts the total ion count (TIC) as no expected products were detected. Multiple peaks for each ring size were detected due to stochastic modification at Val6 and Thr8 and combined to create a single chromatogram. A detailed breakdown of ions present in the composite peaks above is given in the supporting information.
In contrast, the biosynthetic machinery in the C2S mutant appeared to ignore the newly introduced pathway in of the favor of the natural 26-membered ring alone. Two compounds were obtained, one bearing a C2S-hydroxyl and the other a C2S oxazoline; in this mutant, the newly introduced serine did not undergo dehydration (we have previously characterized products of the C2S mutant by NMR and a solution phase model is published in reference 9b). As a result, the dominant cyclization product derives from the only available pathway, the 26-member pathway. A different cyclization pattern is observed in both the C9S substitution and S9-10 insertion mutants. These mutants present a single 2-π component with two different choices of C-terminal 4-π component. Isolates exhibit two “groups” of compounds, distinguishable by differences in LC/MS retention times, yet containing several identical masses. Extensive MS/MS on the subsets of compounds demonstrates that the two “groupings” result from the two different ring size products: 23- and 26-member in case of the C9S mutant and 26- and 29-member for the S9-10 insertion.
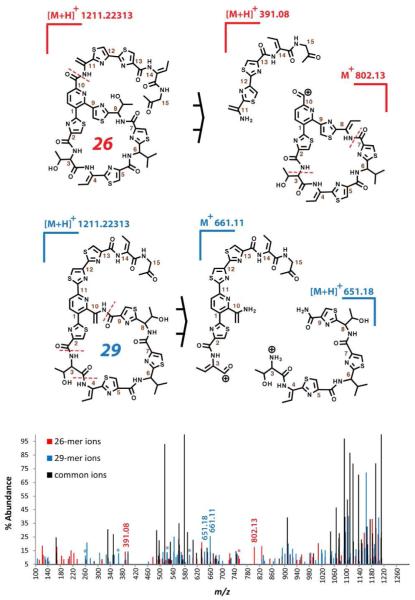
Due to the small amounts of compound produced in these cases and the complexity of the mixtures, MS/MS was the only available technique capable of distinguishing the ring size variants. Characterizations were aided by the familiarity of fragmentation pathways based on our previously reported thiocillin mutants, as well as the presence of a number of distinctive ions in these experiments. Overlayed MS/MS spectra for two compounds from the S9-10 mutant are presented in figure 3 as an example of the latter. The similar structures display many of the same ions, but ring size products could be distinguished by the presence of different northern and southern half fragments (spectra and fragmentation schema are presented in greater detail in the Supporting Information). In particular, ions for the C-term tail of the 26-membered ring compound exhibit an added dehydroalanine residue, not observed in previous mutants or the 29-memebred ring product. In general, fragmentation patterns were in accord with literature precedent.9 Through these experiments it was determined that compounds are present that possess dehydrations at the unreacted serines (serines not undergoing cycloaddition).
Figure 3.
MS/MS fragmentation pattern of 26-member and 29-member ring compounds from S9-10 insertion mutant. Two key fragmentations are indicated on structures above and according ions are labeled below in an overlayed spectra; derivatives of these ions are further labeled with asterisks. The same spectral data is also presented in the supporting information with a more detailed fragment assignment.
A last, and somewhat cryptic, result is presented in the form of a C11S mutant. This mutant provides a cyclization scenario similar to the S9-10 insertion mutant, wherein a single dienophile is again presented with an option of two partners, one leading to a 26-member and the other a 29-member macrocycle. The sole difference now is that the S9-10 mutant possesses two C-terminal thiazoles, where the C11S mutant has only one. In a number of different substitution mutations at cysteine 11, in addition to the C11S mutant, we observed no compound production, suggesting either that the system is incapable of cyclizing/producing these mutant variants, or that level is decreased to undetectable amounts.
In conclusion, the results presented herein clearly indicate that the B. cereus biosynthetic machinery is capable of transforming variant preprotein substrates to a broader range of macrocycle sizes than is observed naturally. The two components of ring closure, 2-π and 4-π, can be tethered at variable distance, at least 23- to 35-atoms apart, and still participate in the putative cycloaddition; tether length and rigidity does not appear necessary to preorganize the two components. The promiscuity of the B. cereus machinery contrasts with the highly modified structure of berninamycin from S. bernensis. Berninamycin possesses a 35-membered ring exhibiting Dhas (2-π alternatives) at positions capable of rendering a 26- or 29-membered ring. The S. bernensis system is currently under research in our laboratories.
This promiscuity bears promise in regards to redesigning and even repurposing cyclic thiopeptides. Initial assays have indicated that the new ring size variants from B. cereus are all but devoid of antibiotic activity, in-keeping with our previous results on the importance of ring constraint. However, new eukaryotic targets for thiazolyl peptides are emerging, recently among them the apicoplast ribosomes and 20S proteasomes of P. falciparum.14 The ability to more broadly manipulate thiopeptide structure and now ring size is also being applied by us in these new target areas.
Supplementary Material
ACKNOWLEDGMENTS
We thank Amber Janasch and Bruce Cooper (Purdue Metabolite Profiling Center) for help with mass spectrometry. This work was supported by NIH NIAID Grant Nos. AI057159 and GM20011 and the New England Regional Center of Excellence (NIAID 057159). Support for instrumentation was provided by the Taplin Funds for Discovery Program (Suzanne Walker, PI) and the Purdue Research Foundation.
Footnotes
Supporting Information Available: Supporting figures and tables, experimental procedures, and spectral data. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Nolan EM, Walsh CT. Chembiochem. 2009;10:34. doi: 10.1002/cbic.200800438. Meier JL, Burkart MD. Chem. Soc. Rev. 2009;38:2012. doi: 10.1039/b805115c. Challis GL, Naismith JH. Curr. Opin. Struct. Biol. 2004;14:748. doi: 10.1016/j.sbi.2004.10.005. Finking R, Marahiel MA. Annu. Rev. Microbiol. 2004;58:453. doi: 10.1146/annurev.micro.58.030603.123615. For an example of NRPS/RPS-independent peptide synthesis see: Oves-Costales D, Kadi N, Challis GL. Chem. Comm. 2009;21:6530. doi: 10.1039/b913092f.
- 2.(a) McIntosh JA, Donia MS, Schmidt EW. Nat. Prod. Rep. 2009;26:537. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Oman TJ, van der Donk WA. Nat. Chem. Bio. 2010;6:9. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Melby JO, Nard NJ, Mitchell DA. Curr. Opin. Chem. Biol. 2011;15:369. doi: 10.1016/j.cbpa.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Nat. Chem. Biol. 2006;2:729. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]; (b) Donia MS, Ravel J, Schmidt EW. Nat. Chem. Biol. 2008;4:341. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Widdick DA, Dodd HM, Barraille P, White J, Stein TH, Chater KF, Gasson MJ, Bibb MJ. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4316. doi: 10.1073/pnas.0230516100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Kelleher NL, Hendrickson CL, Walsh CT. Biochemistry. 1999;38:15623. doi: 10.1021/bi9913698. [DOI] [PubMed] [Google Scholar]; (e) Willey JM, van der Donk WA. Annu. Rev. Microbiol. 2007;61:477. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]; (f) Houssen WE, Wright SH, Kalverda AP, Thompson GS, Kelly SM, Jaspars M. ChemBioChem. 2010;11:1867. doi: 10.1002/cbic.201000305. [DOI] [PubMed] [Google Scholar]; (g) Pan SJ, Link AJ. J. Am. Chem. Soc. 2011;133:5016. doi: 10.1021/ja1109634. [DOI] [PubMed] [Google Scholar]; (h) Knappe TA, Manzenrieder F, Mas-Moruno C, Linne U, Sasse F, Kessler H, Xie X, Marahiel MA. Angew. Chem., Int. Ed. 2011;50:8714. doi: 10.1002/anie.201102190. [DOI] [PubMed] [Google Scholar]; (i) Li C, Zhang F, Kelly WL. Mol. Biosyst. 2011;7:82. doi: 10.1039/c0mb00129e. [DOI] [PubMed] [Google Scholar]
- 4.(a) Bagley MC, Dale JW, Merritt EA, Xiong X. Chem. Rev. 2005;105:685. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]; (b) Arndt HD, Schoof S, Lu JY. Angew. Chem. Int. Ed. 2009;48:6770. doi: 10.1002/anie.200901808. [DOI] [PubMed] [Google Scholar]; (c) Walsh CT, Acker MG, Bowers AA. J. Biol. Chem. 2010;285:27525. doi: 10.1074/jbc.R110.135970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CMT, Fucini P. Mol. Cell. 2008;30:26. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Parmeggiani A, Nissen P. FEBS Lett. 2006;580:4576. doi: 10.1016/j.febslet.2006.07.039. [DOI] [PubMed] [Google Scholar]
- 7.Thompson J, Cundliffe E, Stark MJ. J. Gen. Microbiol. 1982;128:875. doi: 10.1099/00221287-128-4-875. [DOI] [PubMed] [Google Scholar]
- 8.Wieland-Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA. Proc Natl Acad Sci U S A. 2009;106:2549. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Acker MG, Bowers AA, Walsh CT. J. Am. Chem. Soc. 2009;131:17563. doi: 10.1021/ja908777t. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bowers AA, Acker MG, Koglin A, Walsh CT. J. Am. Chem. Soc. 2010;132:7519. doi: 10.1021/ja102339q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowers AA, Walsh CT, Acker MG. J. Am. Chem. Soc. 2010;132:12182. doi: 10.1021/ja104524q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enzymatic Diels-Alder reactions: Kelly WL. Org. Biomol. Chem. 2008;6:4483. doi: 10.1039/b814552k. Kim HJ, Ruszczycky MW, Choi SH, Liu Y.-n., Liu H.-w. Nature. 2011;473:109–112. doi: 10.1038/nature09981. Auclair K, Sutherland A, Kennedy J, Witter DJ, Van den Heever JP, Hutchinson CR, Vederas JC. J. Am. Chem. Soc. 2000;122:11519. Watanabe K, Oikawa H, Yagi K, Ohashi S, Mie T, Ichihara A, Honma M. J. Biochem. 2000;127:467. doi: 10.1093/oxfordjournals.jbchem.a022629.
- 12.Observed fragmentation patterns correlate with our previously reported work on the thiazolyl peptides, as well as those of other groups, esp. Ketterning J, Colombo L, Ferrari P, Tavecchia P, Nebuloni M, Vekey K, Gallo G, Selva E. J. Antibiotics. 1991;44:702. doi: 10.7164/antibiotics.44.702. Kurz M, Sottani C, Bonfichi R, Lociuro S, Selva E. J. Antibiotics. 1994;47:1564. doi: 10.7164/antibiotics.47.1564. Reusser F. Biochemistry. 1969;8:3303. doi: 10.1021/bi00836a026. Favret ME, Boeck LD. J. Antibiot. 1992;45:1809. doi: 10.7164/antibiotics.45.1809. Debono M, Molloy RM, Occolowitz JL, Paschal JW, Hunt AH, Michel KH, Martin JW. J. Org. Chem. 1992;57:5200. Biskupiak JE, Meyers E, Gillum AM, Dean L, Trejo WH, Kirsch DR. J. Antibiot. 1988;41:684. doi: 10.7164/antibiotics.41.684. Yun B-S, Seto H. Biosci. Biotechnol. Biochem. 1995;59:876. doi: 10.1271/bbb.59.876. Colombo L, Tavecchia P, Selva E, Gallo GG, Zerilli LF. □Org. Mass Spectrom. 1992;27:219. Ferrari P, Colombo L, Stella S, Selva E, Zaerilli LF. J. Antibiotics. 1995;48:1304. doi: 10.7164/antibiotics.48.1304. McGibbon GA, Hrusak J, Lavorato DJ, Schwarz H, Terlouw JK. Chem. Eur. J. 1997;3:232. doi: 10.1002/chem.19970030211.
- 13.Changes in the tailoring state of the formerly valine-6 and threonine-8 residues can also be observed in these mutants. However, these modification states have proven batch dependent and do not as yet lend themselves to any helpful understanding of their ordering or relevance to the key ring-formation step.
- 14.(a) Aminake MN, Schoof S, Sologub L, Leubner M, Kirschner M, Arndt HD, Pradel G. Antimicrob. Agents Chemother. 2011;55:1338. doi: 10.1128/AAC.01096-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Clough B, Strath M, Preiser P, Denny P, Wilson I. FEBS Lett. 1997;406:123. doi: 10.1016/s0014-5793(97)00241-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.