Abstract
Catalase, an antioxidant and hydroperoxidase enzyme protects the cellular environment from harmful effects of hydrogen peroxide by facilitating its degradation to oxygen and water. Molecular information on a cnidarian catalase and/or peroxidase is, however, limited. In this work an apparent full length cDNA sequence coding for a catalase (HvCatalase) was isolated from Hydra vulgaris using 3’- and 5’- (RLM) RACE approaches. The 1859 bp HvCatalase cDNA included an open reading frame of 1518 bp encoding a putative protein of 505 amino acids with a predicted molecular mass of 57.44 kDa. The deduced amino acid sequence of HvCatalase contained several highly conserved motifs including the heme-ligand signature sequence RLFSYGDTH and the active site signature FXRERIPERVVHAKGXGA. A comparative analysis showed the presence of conserved catalytic amino acids [His(71), Asn(145), and Tyr(354)] in HvCatalase as well. Homology modeling indicated the presence of the conserved features of mammalian catalase fold. Hydrae exposed to thermal, starvation, metal and oxidative stress responded by regulating its catalase mRNA transcription. These results indicated that the HvCatalase gene is involved in the cellular stress response and (anti)oxidative processes triggered by stressor and contaminant exposure.
Keywords: Hydra vulgaris, Catalase, Gene expression, Molecular biomarker
1. Introduction
Catalases (= Catalatic hydroperoxidases, CHPs) are ubiquitous enzymes and these proteins are placed into four main groups: (1) the classic heme-containing monofunctional catalases for which hydrogen peroxide is both electron donor and acceptor, (2) the heme-containing bifunctional catalatic peroxidases (CPXs) in which the catalatic activity is much higher than the peroxidatic activity, (3) the nonheme-containing catalases (Allgood and Perry, 1986), and (4) a miscellaneous group containing proteins with minor catalatic but no peroxidatic activities (Jones and Masters, 1978; Nadler et al., 1986). Most CHPs exist as tetramers of 60-65KD subunits (Nadler et al., 1986).
More than 300 CHPs sequences are now available, divided among monofunctional catalases (> 225), bifunctional catalase-peroxidases (> 50) and manganese-containing catalases (> 25) (Chelikani et al., 2004). Frequently, organisms use different isozymes, which are expressed simultaneously or under developmental-stage- and environment-specific conditions (Schrempf et al., 1999) to decompose hydrogen peroxides to ground-state O2. Catalases directly dismutate hydrogen peroxide to water and dioxygen by two-electron transfer redox reactions where as peroxidases remove the H2O2 by using it to oxidize another substrate (Schubert and Wilmer, 1991).
In an ultrastructural localization study (Hand, 1976) it is shown that catalase activity (diaminobenzidine reaction product) is present in small round or elongated bodies resembling microperoxisomes in the epitheliomuscular, digestive and gland cells of hydra. In the same study it is also demonstrated that microperoxisome-like bodies reactive for L-alpha-hydroxy acid oxidase is present in the epidermal cnidoblasts; however, catalase could not be demonstrated in them. This study (Hand, 1976) provided the first cytochemical evidence for the presence of an H2O2-producing oxidase in microperoxisomes. Peroxidase like activity has also been observed in the ectodermal foot mucous cells (Hoffmeister-Ullerich et al., 2002) and in lithium treated hydra (Jantzen et al., 1998). Hydroperoxides are also observed to play the role of second messengers in peroxidase activity. However peroxidase or catalase activity is not specifically attributed to any gene products in hydra, though few redox (and stress regulatory) proteins are reported in hydra (Gellner et al., 1992; Brennecke et al., 1998; van Dam et al., 2010). Our group have cloned and characterized two superoxide dismutases, manganese superoxide dismutase (HvMnSOD) and extracellular superoxide dismutase (HvEC-SOD) (Dash et al., 2007), and two phospholipid hydroperoxide glutathione peroxidases, mitochondrial (HvGPx41) and nuclear (HvGPx42) (Dash et al., 2006), from Hydra vulgaris.
Hydra, a fresh water cnidarian, is used as an ideal environmental toxicological model to study the acute and chronic toxicity effects of several environmental toxicants. Our laboratory (Mayura et al., 1991; Lum et al., 2003; Taylor et al., 2009) and other have used the changes in external gross morphology and anatomy, and physiology are useful as markers of toxicity or toxicity end points in the hydra bioassays (Johnson et al., 1982; Pollino and Holdway, 1999; Karntanut and Pascoe, 2000; Holdway et al., 2001; Pascoe et al., 2002; van Dam et al., 2010; Vernouillet et al., 2010; Ferreira et al., 2011; Trenfield et al., 2011). At molecular level, it may be postulated that, the detection of stress and/or redox sensitive messages in hydra can constitute an early-warning marker for the presence of potentially deleterious agents in water. Because H.vulgaris is sensitive to a variety of compounds, the detection stress protein messages such as catalase messages could be applied as a prescreening tool in determining the relative toxicity of many toxicants, and new compounds that are yet to be screened for toxicity.
In this work, a cDNA encoding a monofunctional catalase was identified and isolated from H. vulgaris. The expression of hydra monofunctional catalase (HvCatalase) mRNA is assayed with respect to both environmental contaminant challenge (i.e., arsenic, cadmium, zinc and copper) and stress (both oxidative and non-oxidative) in order to explore its possibility for use as biomarker of stress and toxicity.
2. Materials and methods
2.1. Hydra culture
Hydra vulgaris (formerly known as Hydra attenuata) were originally obtained from E. Marshall Johnson, Jefferson Medical College (Philadelphia, PA, USA). H. vulgaris were maintained in shallow glass dishes at 18 °C in a medium containing 1 mM CaCl2.2H2O, 0.012 mM EDTA, and 0.458 mM TES (N-tris(hydroxymethyl)-methyl-2-amino-ethanesulfonic acid, sodium salt) buffer (pH 7.0). Daily, hydrae were fed with brine shrimp (Artemia nauplii) hatched in a solution of 1% sodium chloride and treated with iodine (40 μg ml-1). Hydrae were maintained free from bacterial and fungal contamination and were not fed for 24 h before initiating the experiments. Deionized water was used throughout this portion of the study (Mayura et al., 1991).
2.2 RNA isolation and clean up
Total RNA was extracted from hydra by application of 2 ml of TRIzol® reagent (Invitrogen, USA) to approximately 20 mg of fresh tissue, using the manufacturer's instructions. The RNA was quantified by ultraviolet absorbance at 260 nm. Integrity of the total RNA was confirmed by 1 % formaldehyde agarose gel electrophoresis. The RNA isolated was cleaned up from contaminating DNA using RNeasy Mini Kit (Invitrogen, USA) following the manufacturer's instruction.
2.3 Identification of partial fragments of H. magnipapillata catalase cDNA
An EST was found from the hydra, H. magnipapillata, under the accession number gi|60408694. The expressed sequence tag coded for a catalase similar to the N-terminal end of Sus scrofa catalase. This EST sequence was used to design primers F2 (5’-ATGGTGTTGGATCGTAATCCTG-3’) and R3 (5’-CTTGAGGGCCATTAAAGCTG-3’) to clone a fragment of catalase from H. vulgaris.
2.4 Cloning and identification of partial fragments of H. vulgaris catalase cDNA
All oligonucleotides except as mentioned were procured from IDT Inc. (IA, USA). All polymerase chain reaction (RACE-PCR and RT-PCR) experiments were performed using Taq DNA polymerase (Invitrogen, USA) and a thermal cycler (MJ Research, USA).
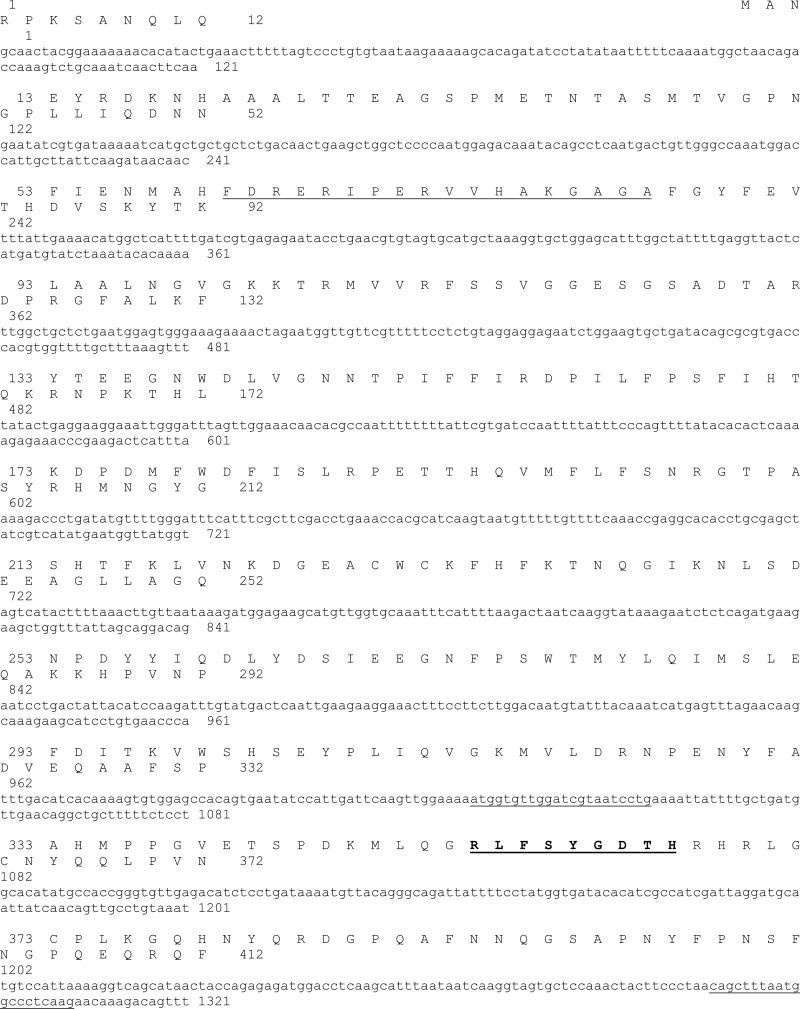
RNA (5 μg) was reverse-transcribed to cDNA at 37 °C for 60 min using the oligo(dT) bifunctional primer N (5’-AACTGGAAGAATTCGCGGCCGCAGGAAd(T)18-3’) and the AMV RT supplied in the cDNA synthesis kit (Amersham Biosciences, USA). The first-strand cDNA was amplified using the primer pairs: F2 and R3 for cloning and identifying partial fragments of H. vulgaris catalase cDNA. The PCR was performed for 30 cycles, consisting of 94 °C for 30 s, 50 °C for 30 s and 72 °C for 1 min and a final extension at 72 °C for 10 min. The resultant PCR products was subcloned into the pCR®II-TOPO® vector using a TA cloning kit (Invitrogen, USA). Multiple independent clones were sequenced using automated methods (DNA Technologies Lab, Department of Veterinary Pathobiology, Texas A&M University) on an ABI PRISM™ 310 Genetic Analyzer (Applied Biosystems, USA) using a Big-Dye sequencing kit (Applied Biosystems, USA) and M13 primers. The identity of the clones was evaluated by matching the sequences to the nucleotide/protein sequences available at the GenBank. The cloned sequence constituted residues 1019 to 1247 in the catalase nucleotide sequence as shown in Fig.1.
Fig.1.
Nucleotide and deduced amino acid sequences of H. vulgaris monofunctional catalase cDNA. The deduced amino acid sequence is shown in single-letter code above the nucleotide sequence. The nucleotide and amino acid sequences are numbered from the 5’-end of the 1859 bp cDNA sequence, and from the N-terminal start codon methionine, respectively. The asterisk denotes the translation stop signal. Underlined nucleotide sequences indicate primer positions and/sequences for the initial cloning of an HvCatalase cDNA fragment. Putative proximal active site signature is underlined and the proximal heme ligand signature is bold and underlined. Putative amino acid residues in bold letter with light gray shade indicate putative NADPH binding residues in reference to H. sapiens catalase (Putnam et al., 2000).
2.5 3’-RACE of the HvCatalase cDNA
In order to clone the 3’-end of the HvCatalase cDNA, the first-strand cDNA prepared above was amplified using the oligo(dT) bifunctional primer N and a gene-specific primer F2 (complementary to nucleotide (nt) 1019 to 1040, Fig. 1) for 35 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 3 min. The first-round PCR products were reamplified using the primer F3 (5’-CGGGTGTTGAGACATCTCCT-3’) (complementary to nucleotide (nt) 1096 to 1115, Fig. 1) and oligo(dT) bifunctional primer N using the same temperature parameters. The PCR products were subcloned and sequenced as described above. The identity of the clones was evaluated by matching the sequences to the nucleotide/protein sequences available at the GenBank.
2.6 5’-RACE of the HvCatalase cDNA
FirstChoice ®RLM-RACE (RNA Ligase Mediated Rapid Amplification of cDNA Ends) (Ambion Inc., USA) was employed to clone the 5’-end of the HvCatalase cDNA. In brief total RNA (10 μg) was treated with Calf Intestine Alkaline Phosphatase (CIP) to remove free 5'-phosphates from molecules such as ribosomal RNA, fragmented mRNA, tRNA, and contaminating genomic DNA. The cap structure found on intact 5' ends of mRNA is not affected by CIP. The RNA was then treated with Tobacco Acid Pyrophosphatase (TAP) to remove the cap structure from full-length mRNA, leaving a 5'-monophosphate. A 45 base RNA adapter oligonucleotide (5'-GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA-3') was ligated to the RNA population using T4 RNA ligase. The adapter cannot ligate to dephosphorylated RNA because these molecules lack the 5'-phosphate necessary for ligation. During the ligation reaction, the majority of the full length decapped mRNA acquires the adapter sequence as its 5' end. A random-primed reverse transcription reaction using MMLV reverse transcriptase and nested PCR then amplified the 5' end of the catalase transcript. First round of PCR was performed using 5’RACE outer primer (O1) (5'-GCTGATGGCGATGAATGAACACTG-3’) and a 5’RACE gene specific outer primer (R3). Second round of PCR used 5’RACE inner primer (O2) (5'-CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG-3') and 5’RACE gene specific inner primer (R2) (5'-AGGAGATGTCTCAACACCCG-3’).
2.7 GenBank accession number
The nucleotide sequence of the HvCatalase mRNA is available in the GenBank databases under the accession number JN580276.
2.8 Bioinformatic analyses and homology modelling
Conceptual translation of the cDNA sequence was performed using the program SIXFRAME (http://biologyworkbench.ucsd.edu). Homology to other catalase genes and proteins were identified by using the Blast program with default settings. Several catalase domain-containing sequences retrieved from the National Center for Biotechnology Information (NCBI) Entrez Web service were aligned to each other using the web program ClustalW (Fig. 2).
Fig. 2.
Multiple sequence alignment of catalse proteins using the program ClustalW. Accession numbers of the proteins as extracted using the Entrez web service are: NP_036652 (Rattus norvegicus), (Mus musculus), (Sus scrofa), (Bombyx mori), CAB45236 (Homo sapiens), AAH54964 (Xenopus laevis), AAH51626 (Danio rerio), CAA36529 (Drosophila melanogaster), and JN580276 (Hydra vulgaris). The properties of amino acids are identified by shading as follows: No shading indicates fully conserved amino acids, light gray shading indicates strongly conserved residues, and dark gray shading indicates weakly conserved residues.
Using web-available threading methods, several templates for the HvCatalase protein sequence were found. These templates were further screened using the SWISS-MODEL Protein Modeling Server (Guex and Peitsch, 1997) on the Web. Protein 1f4j (1f4j.pdb) (Safo et al., 2001) was selected as the structure template for the query sequence HvCatalase. Hence the 3D model of the HvCatalase protein was built on the coordinates of human erythrocyte catalase (1f4j.pdb) protein. The overall stereo-chemical quality of the final model was assessed by the program PROCHECK (Laskowski et al., 1996). The structural quality of the model was also verified using the program Verify-3D (http://nihserver.mbi.ucla.edu/Verify_3D/) that measures the compatibility of a protein model with its sequence where the values are calculated using 3D profile (Bowie et al., 1991).
2.9 Stress treatment
Hydrae were subjected to various stress treatments to evaluate their relative levels of catalase mRNA expression. In each treatment nearly 500 hydrae were incubated in (i) 18 (control temperature), 30 or 37 (maximum induction temperature) °C for 1 or 6 h for heat shock treatments; (ii) 100 ppm 5 ml solution of CuSO4.5H2O [≡ 0.4 mM Cu (II)], ZnCl 2[≡ 0.73 mM Zn (II)], CdCl2.2.5H2O [≡ 0.43 mM Cd (II)] , K2Cr2O7 [≡ 0.33 mM Cr (VI)], As2O3 [≡ 0.506 mM As (III)], Na2HAsO4 [≡ 0.302 mM As (V)] or Na2SeO3 [≡ 0.57 mM Se (IV)] for 1 h for metal stress treatments; (iii) 30 (= 0.979 mM) or 300 (=9.79 mM) ppm 5 ml solution of H2O2 or 100 ppm 5ml solution of paraquat (= 0.38mM) or sodium azide (= 1.538 mM) for oxidative stress treatments. All stressor solutions were made in hydra media. For starvation stress hydrae were unfed for 5 days and as usual were maintained at 18 °C or incubated at 30 °C for 1 hr after 5days of starvation.
Hydrae were collected from dishes (maintained at 18 °C) and subjected to respective treatments or controls. All hydrae treatment groups including controls went through same handling practices. Experiments for each group of stressors were conducted in batches in the same day and included its own control. For temperature treatments hydra media was pre-incubated at the intended temperature (30 or 37 °C) before subjecting hydrae to heat stress for their duration of treatments. All metal or oxidative stress treatments were done at 18 °C. In all cases, treatments or controls were carried out in triplicate in 14 ml polypropylene round-bottom tubes (Becton-Dickinson, NJ, USA) containing 5 ml media or stressor solution. At the end of the treatment, hydrae were collected by centrifugation at 7,500 Xg for 5 min. The supernatants were discarded. Then hydrae were washed once in 5 ml of 0.05 M PBS for 5 min at 7,500 Xg. The supernatants were discarded as before and all the animals in control or treatment groups were homogenized immediately in 3 ml of TRIzol®.
2.10 Expression analysis of HvCatalase mRNA
Total RNA was extracted from TRIzol® (Invitrogen, USA) treated whole hydrae and cleaned as before using RNeasy Mini Kit (Invitrogen, USA) following the procedure described there in. For RT-PCR, 5 μg of total cleaned RNA was reverse transcribed using 500 ng of oligo(dT)12–18 primer (Invitrogen, USA) and 200 units of the Superscript II enzyme (Invitrogen, USA) for 50 min at 42 °C. The reaction was inactivated by heating the mixture at 70 °C for 15 min. PCR assays were designed to normalize HvCatalase gene expression levels to actin transcription rate. Two μl of first strand cDNA (from 25 μl of reverse transcription mix) was diluted 100 times prior to PCR amplification. PCR was carried out in 50 μl total volume of 1 × PCR buffer (20 mM Tris-HCl, pH 8.4, 50 mM KCl), 2 mM MgCl2, 0.1 mM each of dNTPs, 10 pmol each of primers, and 0.5 U Taq DNA polymerase (Invitrogen, USA) using 2 μl of (1:100) diluted RT-product. Actin mRNA was amplified using the AF (5’-AAGCTCTTCCCTCGAAGAATC-3’) and AR (5’-CCAAAATAGATCCTCCGATCC-3’) primers and catalase mRNA was amplified using F2 and R3 primers. Cycling profile after initial denaturation at 94 °C for 4 min was 30 cycles of amplification as follows: denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72°C for 45 s. These number of PCR cycles ensured quantification within the exponential phase of amplification. Equal amounts of RT-PCR reactions (9.5 μl) were loaded on standardized 2 % agarose gels containing 0.1 μg/ml ethidium bromide. The gel images (Fig. 3A) were digitalized by a gel documentation system (Kodak Laboratories, USA) and HvCatalase mRNA bands were quantified by NIH ImageJ software. Respective mRNA levels were normalized to the control (18 °C) after the normalization to actin mRNA levels.
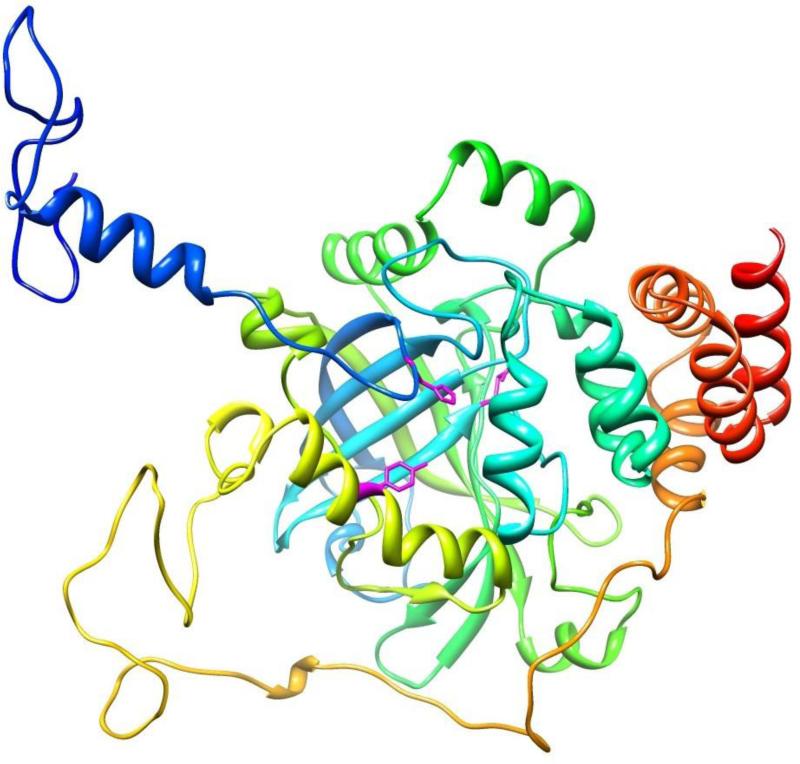
Fig. 3.
A 3D model of the H. vulgaris catalase. Catalase model was generated based on the crystal structure of human erythrocyte catalase (PDB code: 1f4j:A). An individual subunit/monomer of HvCatalase can be seen to possess an eight-stranded antiparallel β-barrel comprising of two four-stranded sheets. The active site heme is surrounded by the β-barrel and α-helices and loops. Conserved catalytic amino acids [His(71), Asn(145), and Tyr(354)] are identified in stick fashion. The figure displayed was drawn using the program Chimera.
2.11 Data analysis
Results are expressed as the mean of three replicates with standard deviation (mean±S.D; n=3). Error bars in the graphs (Fig.3B) indicate standard deviation (S.D.) of the mean. Student's t test was used to detect significant differences between the mean mRNA levels of control and metal treatments or starvation treatments or oxidative stress treatments of paraquat or sodium azide. One-way analyses of variance (ANOVA) followed by Tukey's multiple comparison tests was conducted to detect the significant differences between the mean mRNA levels of the control and experimental groups of heat treatment or H2O2 treatment. Statistically significant results were defined as those with a p-value of less than 0.05. The statistical package GraphPad Prism 4 (GraphPad Software, San Diego, CA) was used to analyze the data.
3. Results
3.1 Cloning and analysis of HvCatalase cDNA
Prior to cloning of the HvCatalase cDNA, GenBank was searched for the cnidaria/hydra catalase(s) and an EST was found from H. magnipapillata coding for a protein similar to the N-terminal end of Sus scrofa catalase. This cDNA sequence was used to design primers (F2 and R3, see materials and methods) to clone a partial fragment of Catalase cDNA from H. vulgaris based on the realization that the conserved proteins between H. magnipapillata and H. vulgaris share high sequence similarity. Primers F2 and R3 were used in RT-PCR experiments to amplify and clone a segment of HvCatalase cDNA. Sequence analysis showed that the cloned PCR fragment was 289 bp in length. Also sequence analysis indicated the presence of an open reading frame (ORF) encoding a polypeptide with a high degree of similarity to Catalases of many organisms.
The full-length cDNA sequence encoding the putative catalase was cloned using the 5’-/3’-RACE method using gene specific primers designed from the above cloned segment. To confirm that the cDNA and PCR cloning products are indeed from the same gene, the full-length ORFs are cloned by PCR and sequenced (data not shown). The putative transcription initiation site found by 5’-RACE is located at nucleotide (nt) position 1 (Fig. 1). The initiation site of translation was placed at nt 86, inferred by conceptual translation of the sequence in all three reading frames and alignment with the known sequences of catalase proteins available in the GenBank database. The putative catalase gene (Fig. 1) is shown to contain a 1515 bp ORF and an in-frame TGA stop codon at the 3’-end of the coding region. The ORF is flanked by a 235 bp 3’-untranslated region followed by the putative 21 bp poly(A) tail. The cDNA sequence also contained a splice leader (nt 4 to 49) in the 5′-untranslated region and belonged to the splice leader B (SL-B) category (Derelle et al., 2010). The H. vulgaris SL sequence was 100% identical to SL sequences of H. vulgaris mRNA for 5S ribosomal RNA gene and cAMPresponse element binding protein (X83872) (E<10-16) or H. littoralis Pax-AmRNA (U96193) (E<10-16).
The predicted amino acid sequence of the catalase cDNA is shown in Fig. 1. The deduced protein is composed of 505 amino acid residues. The theoretical isoelectric point (pI) and molecular weight (Mw) of the protein are calculated to be 6.59 and 57444.12 Da, respectively. As shown in Fig. 1, a variety of amino acid residues were conserved in HvCatalase including 36 residues interacting with a heme cofactor (Murthy et al., 1981), 77 consensus residues in distal and proximal sides of the prosthetic heme group (Zamocky et al., 2004), and 8 residues responsible for the NADPH binding (Putnam et al., 2000). The predicted HvCatalase amino acid sequence exhibited the characteristic catalase signature residues: RLFSYgDTH (residues 350-358).
The deduced HvCatalase amino acid sequence is aligned with representative catalases by the ClustalW method and is shown in Fig. 2. There were many absolutely conserved residues in all these sequences. The HvCatalase protein when compared to the proteins of GenBank database using the program BLAST had significant similarity scores with catalase of Sus scrofa (79 %), Homo sapiens (78 %), Canis familiaris (78 %), Mus musculus (77 %), Melopsittacus undulates (78 %), Rattus norvegicus (77 %), Xenopus laevis (77 %), Drosophila melanogaster (77 %), Danio rerio (75 %), Bombyx mori (77 %), Caenorhabditis elegans (75 %), Saccharomyces pombe (69 %); etc.
Hence identification of catalase signature RLFSYgDTH (residues 350-358) residues, presence of heme and NADPH binding residues and homology to other catalase proteins strongly suggest that this protein is a monofunctional catalase. Overall, these results suggested that the putative catalase of the H. vulgaris identified possessed the essential properties of a monofunctional catalase and can be classified accordingly.
3.2 The structural models of HvCatalase
Human erythrocyte catalase (1f4j.pdb, 2.40 Å) served as the best template for homologous modeling of the HvCatalase. HvCatalase and human erythrocyte catalase proteins shared a high degree of homology across their entire length [Sequence Identity (%):64.29, E-value: 0.00e-1]. The HvCatalase model displays the conserved features of mammalian catalase fold. For simplicity sake only a cartoon of a monomer (out of tetramer) is presented (Figs. 2 and 3). The extensive hydrophobic core of the monomer is generated by an eight-stranded antiparallel β-barrel. The β-barrel is comprised of two four-stranded sheets. Also structural characteristics necessary for binding of NADPH and heme cofactor, as seen in erythrocyte catalase proteins, is present (data not shown).
The comparative stereo-chemical analysis of the φ-ψ plots (Ramachandran diagram) of the model (vs. template 1f4j: A) is as follows: 84.8 (vs. 85.1) % of residues in the most favorable, 14 (vs. 14.7) % of residues in additional allowed regions, 0.7 (vs. 0) % of the residues in generously allowed regions, and 0.5(vs. 0.2) of the residues in disallowed regions. These results indicated that the molecular model presented here has good overall stereo-chemical qualities. AVerify-3D run on the model also showed a good stereo-chemical quality of the model (data not shown).
3.3 HvCatalase mRNA expression analysis
To examine the level of catalase transcripts in H. vulgaris before and after exposure of stressors, the expression patterns of the HvCatalase mRNA were investigated in whole organisms by RT-PCR experiments (Fig. 4). The results obtained demonstrate that there is considerable variation in the levels of HvCatalase mRNA expression following different stressor exposure.
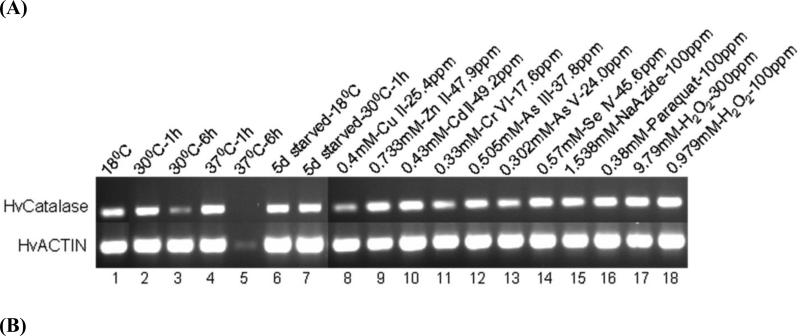
Fig. 4.
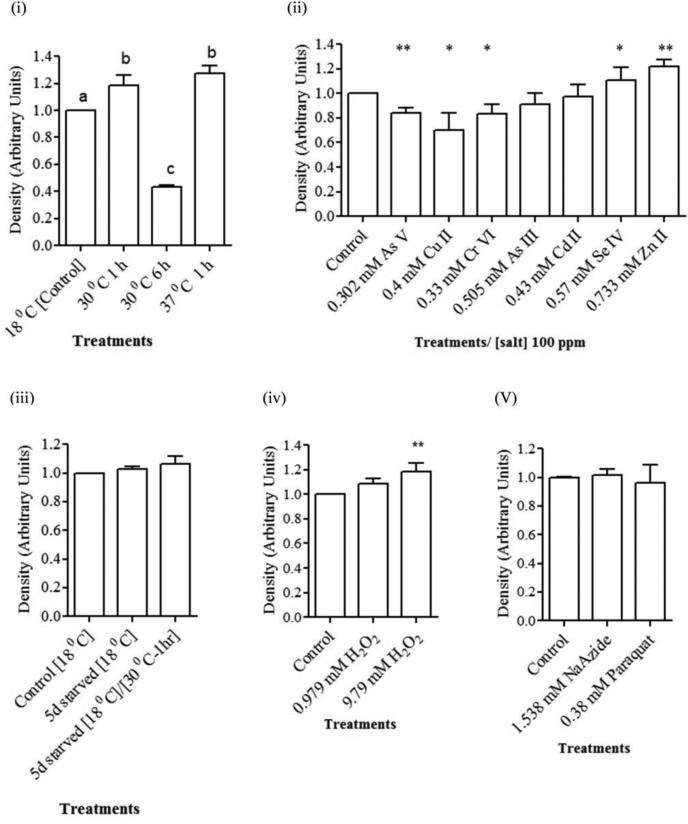
Expression analysis of HvCatalase mRNA from H. vulgaris exposed to thermal, starvation, metal and oxidative stress. (A) A representative agarose gel image of the expression of HvCatalase mRNA due to different stress conditions: Thermal stress (lanes 2-5), starvation stress (lanes 6-7), metal stress for 1 h (lanes 2-14), and oxidative stress for 1 h (lanes 15-18). The expression HvCatalase mRNA is compared to that of actin. (B) Detected HvCatalase mRNA bands were quantified by NIH ImageJ software. Mean levels of HvCatalase mRNA were normalized to that of the control (18 °C) after normalization to the mean levels of actin mRNA (i.e., double normalization). Hence there are no error bars in the control group in any of the graphs. Relative levels of catalase expression are presented for (i) thermal stress: HvCatalase mRNA levels were increased (p<0.01) in hydra groups exposed to 30 and 37 °C temperature for 1 h and decreased (p<0.01) in hydra groups exposed to 30 °C temperature for 6 h; (ii) metal stress: HvCatalase mRNA levels were increased (p<0.05) in hydra groups exposed to Se (IV) and Zn (II) and decreased (p<0.05) in hydra groups exposed to As (V), Cu (II) and Cr (VI) at the tested concentrations relative to control; (iii) starvation stress; (iv) oxidative stress involving H2O2 and (v) oxidative stress involving paraquat and sodium azide. Error bars in the graphs indicate standard deviation (S.D.) of the mean of three independent experiments. Bars without the same letters are statistically different (p<0.01). Asterisks indicate statistical significance of differences between values of control and those obtained for the treatments: *, p<0.05; and **, p<0.01.
Transcription of actin DNA was almost constant before and after exposure to stressors with the exception that the level of actin expression was drastically suppressed following incubation at 37 °C for 6 h (Fig. 4A, lane 5). Also expression of catalase mRNA is almost abolished. This in turn limited our ability to quantify expressed level of catalase mRNA for this treatment. This treatment or 30 °C exposure for 6 h (Fig. 4A, lane 3) was envisioned to serve as a negative control for the transcription of actin or HvCatalase mRNA as it would be expected that a higher temperature exposure for longer duration would impair the transcriptional machinery of the cell thereby negatively affecting transcription of actin or HvCatalase mRNA.
On the other hand heat treatments for 1 h at 30 or 37 °C was envisioned to see the impact of temperature on catalase transcription as literature indicates varied effect of temperature on catalase mRNA expression and enzyme activity (Wieser et al., 1991; Ozmen et al., 2007). One way ANOVA demonstrated significant differences in the mean level of HvCatalase mRNA expression in hydrae groups receiving different temperature treatments (F3,8=190, p<0.0001). Thermal stress for shorter time period of time (1 h) enhanced the HvCatalase mRNA expression. Heat treatments for 1 h at 30 °C (p<0.01) or 37 °C (p<0.001) induced the expression of HvCatalase mRNA (Fig. 4A, lanes 2 and 4 and 3B (i)). In contrast expression of the HvCatalase mRNA was drastically reduced when hydrae were exposed to 30 °C for 6 h (p<0.001). Also the mean level of HvCatalase mRNA expression was reduced (p<.0.001) in hydra groups exposed 30 °C for 6 h compared to the groups that received heat treatments for 1 h at 30 °C (p<0.01) or 37 °C (p<0.001).
Hydrae starved for 5 days at 18 °C (p=0.11) or incubated at 30 °C for 1 h following 5 days of starvation at 18 °C (p=0.12) didn't show a change in expression of HvCatalase mRNA compared to the control maintained at 18 °C (Fig. 4, lanes 6-7 and 3B(iii)).
When hydrae were exposed to metal toxicants Cd (II) (p=0.69) and As (III) (p=0.17) for 1 h, there wasn't a change in the expression level of HvCatalase mRNA at the treated concentrations (Fig. 4A & B (ii)) compared to control. However, hydrae exposed to Cu (II) (p<0.02), Cr (VI) (p<0.02) and As (V) (p<0.002) reduced their level of catalase mRNA expression compared to control. Exposure of hydrae to Zn (II) (p<0.002) and Se (IV) (p<0.02) at the tested concentrations for 1 h enhanced the expression of HvCatalase mRNA compared to control treatment (Fig. 4A & B (ii)).
One way ANOVA demonstrated significant differences in the mean level of HvCatalase mRNA expression in hydra groups exposed to different concentrations of H2O2 treatments (F2,6=12, p=0.008). Exposure of hydrae to 9.79 mM H2O2 (1 h) but not 0.979 mM H2O2 (Fig. 4A, lanes 17-18 and 3B) enhanced (p<0.05) the expression of HvCatalase mRNA.
Two other toxicants and oxidants sodium azide (p=0.51) and paraquat (p=0.6) didn't have any effect on HvCatalase mRNA expression.
4. Discussion
In this study, a full length cDNA encoding a catalase protein (HvCatalase) from H. vulgaris is cloned. Messenger RNA expression levels of HvCatalase are analyzed following exposure of hydrae to several commonly occurring stressors including some well known oxidative stressors that could be found in aquatic environments naturally or released to aquatic environments due to anthropogenic activities. Results indicate possible detection of a compensatory effect in catalase expression which is implicated in the detoxification of reactive oxygen species (ROS) in various prior studies.
The deduced protein sequence is homologous to the catalase proteins from many organisms including that of B. mori, D. melanogaster, C. elegans; etc. (Furuta et al., 1986; Lin et al., 1997). HvCatalase has catalase signature residues (residues 350-358: RLFSYgDTH) and also contains 36 amino acid residues typical of many catalases that interact with a heme cofactor (Murthy et al., 1981). It also contains consensus sequence containing 77 residues that is highly conserved in many catalases (Zamocky et al., 2004). All of these residues are present in distal and proximal sides of the prosthetic heme group. Also 8 amino acid residues out of a total of 12 amino acid residues that are identified to be responsible for the binding of dinucleotide NADPH (Putnam et al., 2000) in human erythrocyte catalase is present in HvCatalase. The calculated molecular size of HvCatalase, ~57 kDa, is similar to other catalases isolated so far (Switala and Loewen, 2002). Molecular modeling reveals the presence of a conserved mammalian catalase fold in hydra catalase as well. Hence, based on the molecular features described here it is almost certain that the putative cDNA codes for a hydra catalase.
Messenger RNAs have been identified in four metazoan phyla (Nematoda, Platyhelminthes, Chordata and Cnidaria) and in one unicellular eukaryotic phylum (Sarcomastigophora) that can receive either splice leader A or B (SL-A or -B), although the impact of the two different SLs on the function of the mRNA is not known (Stover and Steele, 2001). So the presence of SL-B in the HvCatalase mRNA is another addition to the repertoire of mRNAs that receive splice leaders in cnidaria.
Organisms have antioxidant systems to protect oxidative damage to key molecules such as lipids, DNA; etc. from ROS mediated damage (Halliwell, 2007). In antioxidant systems, superoxide dismutases (SOD) are the first line of defense as they catalyze the dismutation of oxygen radical (O2-) into molecular oxygen and H2O2. Thereafter peroxiredoxins (Prxs), catalases and glutathione peroxidases (GPxs) scavenge H2O2 from their cellular environments. Also these genes that constitute the antioxidant defense system have been proposed as a possible tools for biomonitoring environmental toxicants and stress (Niyogi et al., 2001; Sayeed et al., 2003; Zhang et al., 2004) as their normal expression pattern will be altered in the presence of the stressors. Therefore in this study we have tried to see if such an alteration of hydra catalase expression would occur when hydrae are challenged with various stimuli (i.e., heat, heavy metals, starvation, oxidants like H2O2, paraquat; etc.) that are generally known to cause and/or induce oxidative stress and damage.
Induction of HvCatalase mRNA was detectable over a temperature range of 30 - 37 °C, but the level of synthesis depended on the stress temperature and exposure duration, with the highest level of expression at the highest temperature 37 °C. The observation that heat shock enhances catalase mRNA expression in hydra is consistent with the earlier finding that yeast CTT1 (cytosolic catalase T) transcription is induced by heat shock (Wieser et al., 1991). Also catalase activities in the erythrocyte of rainbow trout (Oncorhynchus mykiss) experiencing high temperature was significantly higher than in the control group experiencing no such treatments (Ozmen et al., 2007).
Catalase is extensively regulated in the responses of cells to extracellular H2O2. Upon exposure to H2O2 catalase mRNA levels are increased through mRNA expression and/or stabilization (Gutierrez-Uzquiza et al., 2012) and catalase activity is regulated through protein stabilization (Gutierrez-Uzquiza et al., 2012) and posttranslational modifications (Cao et al., 2003; Rhee et al., 2005). In one of the very early experiments it is shown that stresses which induce the catalase activity also induce the transcription of the catalase gene (Nakagawa et al., 1995; Nakagawa et al., 1999). Concurrent with previous observations about catalase expression in other organisms, in the current study exposure of hydrae to oxidant H2O2 (0.979 or 9.79 mM) for 1 h induced the expression of catalase mRNA, a larger and significant induction at 9.79 mM H2O 2than at 0.979 mM H2O2. Because the primary role of catalase is to rid the cell of H2O2 before it causes unwanted reactions or gives rise to even more reactive hydroxyl radical, the increase in catalase expression in response to H2O2 at the transcriptional level could be understood as a protective response to an oxidative stress.
Two other compounds: paraquat, a bipyridinium salt and a potent herbicide; and sodium azide, an ionic industrial effluent and peroxidase inhibitor, known to generate ROS and inhibit antioxidant systems were also assayed in the current study to assess their effect on expression of hydra catalase mRNA. Paraquat is shown to induce several drug metabolizing and antioxidant defense enzymes (Lee et al., 2003; Olesen et al., 2008). However in this we didn't observe a detectable change in catalase mRNA levels when hydrae are treated with either paraquat or sodium azide.
Heavy metals are known to produce ROS and induce oxidative damage to cells. In order to protect from and/or cope with ROS and oxidative damage various organisms regulate their transcriptional and translational machinery that may result in transcriptional upregulation and/or downregulation of various genes; and also increased and/or decreased activity of various enzymes and proteins. In this study several metals are tested to assess their effect on hydra catalase expression. Our results indicate that the HvCatalase expression is either increased or decreased or remain unaltered compared to control. If a positive relationship between catalase mRNA expression and its activity (Liu et al., 2007) can be assumed notwithstanding the fact that catalase activity can be modulated independent of catalase expression (Pasquali et al., 2008), then most of our HvCatalase expression results are in agreement with the effect of various stressors on catalase expression. Algae Scenedesmus sp. exposed to elevated levels of heavy metals [Cu2+ (2.5 μM) and Zn2+ (25 μM )] for 6 h (short-term ) and 7 d (long-term) resulted in increased catalase activity, but were inhibited at 10 μM Cu2+ under intense oxidative stress (Tripathi et al., 2006). However, catalase activity is increased in the marine microalga (Pavlova viridis) when algal cells are grown in copper or zinc solutions that includes a maximum of 47 μM of Cu2+ or 99.3 μM Zn2+ (Li et al., 2006). Also in another species of cnidaria, symbiotic sea anemone, Aiptasia pallida, exposure to sublethal copper concentrations (0, 5, 15, and 50 microg/L) for 7 d increased catalase activity only in response to the highest two copper concentrations (i.e., 0.23 and 0.79 μM) (Main et al., 2010). Our results suggested that catalase gene expression is inhibited when hydrae are exposed to 400 μM Cu2+ and this probably is due to induction of excessive oxidative stress. In contrary, hydrae exposed to Zn2+ have enhanced the expression of catalase mRNA probably to counteract or overcome the oxidative stress imposed by Zn2+. In algivorous marine gastropods catalase mRNA expression is increased and has reached the maximum at the dietary zinc level of 33.8 mg/kg, and then dropped progressively (Wu et al., 2011). In recent studies it is shown that green alga (Ulva lactuca) exposed to 0.4 mM CdCl2 for 4 days or zebrafish (Danio rerio) exposed to CdCl2 (0.4 mg/l) for 3 weeks showed diminished catalase activity (Kumar et al., 2010; Banni et al., 2011). However, in this study the transcriptional activity of catalase remains unaltered when hydrae are exposed to 0.34 mM CdCl2 for 1 h. Earlier studies have shown that selenium supplementation or exposure play an important role in protection of aquatic organisms against oxidative stress by induction of key antioxidant defenses such as glutathione and selenium-dependent glutathione peroxidase (Trevisan et al., 2011; Kumar et al., 2012). Liver catalase expression is decreased in the liver of chickens when fed with high Se containing diet at 4th week of supplementation but is not significantly affected at 6th week of supplementation (Zoidis et al., 2010). However, our results indicate that Se (IV) increases HvCatalase expression. As (III) and As (V), two forms of inorganic arsenic found in the aquatic environment. As (III) (0.505 mM) treatment for 1 hr didn't affect the expression of HvCatalase while 0.302 mM As (V) treatment for 1 h increased the expression HvCatalase. However, CAT activity, mRNA expression and protein levels were decreased in established human cell lines of keratinocytes (HaCaT) when cells are exposed to 5-20 μmol/l of sodium arsenite (AsIII) (Sun et al., 2006). Hexavalent Chromium Cr (VI) is the predominant chemical form of the metal in the aquatic ecosystems and exposure to Cr (VI) in the low ppb range did not result in change in the catalase activity in the digestive gland of the mussel (Mytilus galloprovincialis) (Barmo et al., 2011). In this study we observe that 0.33 mM Cr (VI) treatment for 1 h decreased HvCatalase expression.
It has been demonstrated that starvation can cause several physiological, metabolic and behavioral changes in aquatic organisms including oxidative stress (Pascual et al., 2003; Sanchez-Paz et al., 2007; Matozzo et al., 2011). For example, catalase activity is decreased in the liver of fish (Sparus aurata) fasting for 39 days (Pascual et al., 2003) or unaltered significantly in the gills and digestive gland from 7 d starved crabs (Carcinus aestuarii) (Matozzo et al., 2011) compared to their fully fed counterparts respectively. Similar other studies indicated that aquatic organism can suffer from reductions in antioxidant status after prolonged starvation only (Dissanayake et al., 2008). However we didn't observe a significant change in catalase expression in hydra following 5 d of starvation. It probably indicates that 5 d of starvation may not be long enough to induce compensatory catalase expression in hydra.
The concentration of some of the toxicants chosen was based on previous scientific reports on whole organism based bioassay reports in hydra (Pollino and Holdway, 1999; Karntanut and Pascoe, 2000; Holdway et al., 2001). In the context of known inducers (metals and oxidative stressors) not inducing a significant change in HvCatalase expression at the tested concentration offers few possibilities. It may be due to that the duration of exposure is not sufficient or doses tested couldn't provoke a response or the doses tested cause toxicity impairing the transcriptional ability of H. vulgaris.
The gene expression data presented here gives further basis to conduct dose-response and dose-equivalent studies to elucidate contaminant specific gene expression and mechanistic basis of catalase expression in hydra. Thus the catalase mRNA expression system in hydra could be a useful tool for testing involvement of H2O2 in toxicological responses and processes associated with reactive oxygen species like heavy metal exposure. Thus the evaluation of catalase expression levels could be considered as a potential general, if not selective, biomarker of toxicity associated with contaminant exposure in hydra.
Highlights.
BACKGROUND:
Little is known about presence and role of antioxidant enzyme catalase in hydra.
PRINCIPAL FINDINGS:
Hydra Catalase is homologous to bilaterian catalases and displays catalase fold.
Its transcription is regulated by various environmental stressors.
CONCLUSIONS:
This is first report of a full length Catalase cDNA from Cnidaria.
It could be of use as a molecular biomarker for stress response.
Acknowledgement
This work is funded by USAID TAM50 and NIEHS P42-ES04917.
Abbreviations
- H.
Hydra
- Hv
Hydra vulgaris
- cDNA
DNA complementary to RNA
- PCR
polymerase chain reaction
- RT-PCR
reverse transcription-polymerase chain reaction
- RACE
rapid amplification of cDNA ends
- ORF
open reading frame
- UTR
untranslated region
- nt
nucleotide(s)
- aa
amino acid(s)
- bp
base pair(s)
- kDa
kilodalton(s)
- dNTP
deoxyribonucleoside triphosphate
- EDTA
ethylenediaminetetraacetic acid
- ppm
parts per million
- Cu
copper
- Zn
zinc
- Cd
cadmium
- Cr
chromium
- As
arsenic
- Se
selenium
- Na
sodium
- H2O2
hydrogen peroxide
- ROS
reactive oxygen species
- EST
expressed sequence tag
- catalase
catalase protein
- catalase
gene, cDNA or mRNA encoding catalase
- HvCatalase
gene, cDNA or mRNA encoding HvCatalase protein
- NADPH
nicotinamide adeninedinucleotide phosphate reduced form
- CAT
catalase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allgood GS, Perry JJ. Characterization of a manganese-containing catalase from the obligate thermophile Thermoleophilum album. J Bacteriol. 1986;168:563–7. doi: 10.1128/jb.168.2.563-567.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banni M, Chouchene L, Said K, Kerkeni A, Messaoudi I. Mechanisms underlying the protective effect of zinc and selenium against cadmium-induced oxidative stress in zebrafish Danio rerio. Biometals. 2011;24:981–92. doi: 10.1007/s10534-011-9456-z. [DOI] [PubMed] [Google Scholar]
- Barmo C, Ciacci C, Fabbri R, Olivieri S, Bianchi N, Gallo G, Canesi L. Pleiotropic effects of hexavalent chromium (CrVI) in Mytilus galloprovincialis digestive gland. Chemosphere. 2011;83:1087–95. doi: 10.1016/j.chemosphere.2011.01.037. [DOI] [PubMed] [Google Scholar]
- Bowie JU, Luthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–70. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- Brennecke T, Gellner K, Bosch TC. The lack of a stress response in Hydra oligactis is due to reduced hsp70 mRNA stability. Eur J Biochem. 1998;255:703–9. doi: 10.1046/j.1432-1327.1998.2550703.x. [DOI] [PubMed] [Google Scholar]
- Cao C, Leng Y, Liu X, Yi Y, Li P, Kufe D. Catalase is regulated by ubiquitination and proteosomal degradation. Role of the c-Abl and Arg tyrosine kinases. Biochemistry. 2003;42:10348–53. doi: 10.1021/bi035023f. [DOI] [PubMed] [Google Scholar]
- Chelikani P, Fita I, Loewen PC. Diversity of structures and properties among catalases. Cell Mol Life Sci. 2004;61:192–208. doi: 10.1007/s00018-003-3206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash B, Metz R, Huebner HJ, Porter W, Phillips TD. Molecular characterization of phospholipid hydroperoxide glutathione peroxidases from Hydra vulgaris. Gene. 2006;381:1–12. doi: 10.1016/j.gene.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Dash B, Metz R, Huebner HJ, Porter W, Phillips TD. Molecular characterization of two superoxide dismutases from Hydra vulgaris. Gene. 2007;387:93–108. doi: 10.1016/j.gene.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle R, Momose T, Manuel M, Da Silva C, Wincker P, Houliston E. Convergent origins and rapid evolution of spliced leader trans-splicing in metazoa: insights from the ctenophora and hydrozoa. RNA. 2010;16:696–707. doi: 10.1261/rna.1975210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissanayake A, Galloway TS, Jones MB. Nutritional status of Carcinus maenas (Crustacea: Decapoda) influences susceptibility to contaminant exposure. Aquat Toxicol. 2008;89:40–6. doi: 10.1016/j.aquatox.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Ferreira LF, Aguiar MM, Messias TG, Pompeu GB, Lopez AM, Silva DP, Monteiro RT. Evaluation of sugar-cane vinasse treated with Pleurotus sajor-caju utilizing aquatic organisms as toxicological indicators. Ecotoxicol Environ Saf. 2011;74:132–7. doi: 10.1016/j.ecoenv.2010.08.042. [DOI] [PubMed] [Google Scholar]
- Furuta S, Hayashi H, Hijikata M, Miyazawa S, Osumi T, Hashimoto T. Complete nucleotide sequence of cDNA and deduced amino acid sequence of rat liver catalase. Proc Natl Acad Sci U S A. 1986;83:313–7. doi: 10.1073/pnas.83.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellner K, Praetzel G, Bosch TC. Cloning and expression of a heat-inducible hsp70 gene in two species of Hydra which differ in their stress response. Eur J Biochem. 1992;210:683–91. doi: 10.1111/j.1432-1033.1992.tb17469.x. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Uzquiza A, Arechederra M, Bragado P, Aguirre-Ghiso JA, Porras A. p38alpha mediates cell survival in response to oxidative stress via induction of antioxidant genes: effect on the p70S6K pathway. J Biol Chem. 2012;287:2632–42. doi: 10.1074/jbc.M111.323709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B. Biochemistry of oxidative stress. Biochemical Society Transactions. 2007;35:1147–1150. doi: 10.1042/BST0351147. [DOI] [PubMed] [Google Scholar]
- Hand AR. Ultrastructural localization of catalase and L-alpha-hydroxy acid oxidase in microperoxisomes of Hydra. J Histochem Cytochem. 1976;24:915–25. doi: 10.1177/24.8.956644. [DOI] [PubMed] [Google Scholar]
- Hoffmeister-Ullerich SA, Herrmann D, Kielholz J, Schweizer M, Schaller HC. Isolation of a putative peroxidase, a target for factors controlling foot-formation in the coelenterate hydra. Eur J Biochem. 2002;269:4597–606. doi: 10.1046/j.1432-1033.2002.03159.x. [DOI] [PubMed] [Google Scholar]
- Holdway DA, Lok K, Semaan M. The acute and chronic toxicity of cadmium and zinc to two hydra species. Environ Toxicol. 2001;16:557–65. [PubMed] [Google Scholar]
- Jantzen H, Hassel M, Schulze I. Hydroperoxides mediate lithium effects on regeneration in Hydra. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1998;119:165–75. doi: 10.1016/s0742-8413(97)00204-1. [DOI] [PubMed] [Google Scholar]
- Johnson EM, Gorman RM, Gabel BE, George ME. The Hydra attenuata system for detection of teratogenic hazards. Teratog Carcinog Mutagen. 1982;2:263–76. doi: 10.1002/1520-6866(1990)2:3/4<263::aid-tcm1770020308>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Jones GL, Masters CJ. On the synthesis and degradation of the multiple forms of catalase in mouse liver: effects of aminotriazole and p-chlorophenoxyisobutric acid ethyl ester. Arch Biochem Biophys. 1978;187:431–40. doi: 10.1016/0003-9861(78)90054-1. [DOI] [PubMed] [Google Scholar]
- Karntanut W, Pascoe D. A comparison of methods for measuring acute toxicity to Hydra vulgaris. Chemosphere. 2000;41:1543–8. doi: 10.1016/s0045-6535(00)00068-0. [DOI] [PubMed] [Google Scholar]
- Kumar M, Bijo AJ, Baghel RS, Reddy CR, Jha B. Selenium and spermine alleviate cadmium induced toxicity in the red seaweed Gracilaria dura by regulating antioxidants and DNA methylation. Plant Physiol Biochem. 2012;51:129–38. doi: 10.1016/j.plaphy.2011.10.016. [DOI] [PubMed] [Google Scholar]
- Kumar M, Kumari P, Gupta V, Anisha PA, Reddy CR, Jha B. Differential responses to cadmium induced oxidative stress in marine macroalga Ulva lactuca (Ulvales, Chlorophyta). Biometals. 2010;23:315–25. doi: 10.1007/s10534-010-9290-8. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem. 2003;278:12029–38. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- Li M, Hu C, Zhu Q, Chen L, Kong Z, Liu Z. Copper and zinc induction of lipid peroxidation and effects on antioxidant enzyme activities in the microalga Pavlova viridis (Prymnesiophyceae). Chemosphere. 2006;62:565–72. doi: 10.1016/j.chemosphere.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Lin ZH, Wang YF, Sarai A, Yasue H. Swine catalase deduced from cDNA and localization of the catalase gene on swine chromosome 2p16-p15. Biochem Genet. 1997;35:297–302. doi: 10.1023/a:1021865603981. [DOI] [PubMed] [Google Scholar]
- Liu Y, Guo Y, Wang Z, Nie W. Effects of source and level of magnesium on catalase activity and its gene expression in livers of broiler chickens. Arch Anim Nutr. 2007;61:292–300. doi: 10.1080/17450390701432019. [DOI] [PubMed] [Google Scholar]
- Lum KT, Huebner HJ, Li Y, Phillips TD, Raushel FM. Organophosphate nerve agent toxicity in Hydra attenuata. Chem Res Toxicol. 2003;16:953–7. doi: 10.1021/tx034047k. [DOI] [PubMed] [Google Scholar]
- Main WP, Ross C, Bielmyer GK. Copper accumulation and oxidative stress in the sea anemone, Aiptasia pallida, after waterborne copper exposure. Comp Biochem Physiol C Toxicol Pharmacol. 2010;151:216–21. doi: 10.1016/j.cbpc.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Matozzo V, Gallo C, Marin MG. Can starvation influence cellular and biochemical parameters in the crab Carcinus aestuarii? Mar Environ Res. 2011;71:207–12. doi: 10.1016/j.marenvres.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Mayura K, Smith EE, Clement BA, Phillips TD. Evaluation of the developmental toxicity of chlorinated phenols utilizing Hydra attenuata and postimplantation rat embryos in culture. Toxicol Appl Pharmacol. 1991;108:253–66. doi: 10.1016/0041-008x(91)90116-v. [DOI] [PubMed] [Google Scholar]
- Murthy MR, Reid TJ, 3rd, Sicignano A, Tanaka N, Rossmann MG. Structure of beef liver catalase. J Mol Biol. 1981;152:465–99. doi: 10.1016/0022-2836(81)90254-0. [DOI] [PubMed] [Google Scholar]
- Nadler V, Goldberg I, Hochman A. Comparative-Study of Bacterial Catalases. Biochimica Et Biophysica Acta. 1986;882:234–241. [Google Scholar]
- Nakagawa CW, Mutoh N, Hayashi Y. Transcriptional regulation of catalase gene in the fission yeast Schizosaccharomyces pombe: molecular cloning of the catalase gene and northern blot analyses of the transcript. J Biochem. 1995;118:109–16. doi: 10.1093/oxfordjournals.jbchem.a124864. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Koide K, Watanabe K, Morita Y, Mizuguchi I, Akashi T. The expression of the pathogenic yeast Candida albicans catalase gene in response to hydrogen peroxide. Microbiol Immunol. 1999;43:645–51. doi: 10.1111/j.1348-0421.1999.tb02452.x. [DOI] [PubMed] [Google Scholar]
- Niyogi S, Biswas S, Sarker S, Datta AG. Seasonal variation of antioxidant and biotransformation enzymes in barnacle, Balanus balanoides, and their relation with polyaromatic hydrocarbons. Mar Environ Res. 2001;52:13–26. doi: 10.1016/s0141-1136(00)00257-9. [DOI] [PubMed] [Google Scholar]
- Olesen BT, Clausen J, Vang O. Characterization of the transcriptional profile in primary astrocytes after oxidative stress induced by Paraquat. Neurotoxicology. 2008;29:13–21. doi: 10.1016/j.neuro.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Ozmen I, Atamanalp M, Bayir A, Sirkecioglu AN, Cengiz M, Cengiz M. The effects of different stressors on antioxidant enzyme activities in the erythrocyte of rainbow trout (Oncorhynchus mykiss W., 1792). Fresenius Environmental Bulletin. 2007;16:922–927. [Google Scholar]
- Pascoe D, Carroll K, Karntanut W, Watts MM. Toxicity of 17alpha-ethinylestradiol and bisphenol A to the freshwater Cnidarian Hydra vulgaris. Arch Environ Contam Toxicol. 2002;43:56–63. doi: 10.1007/s00244-001-0016-3. [DOI] [PubMed] [Google Scholar]
- Pascual P, Pedrajas JR, Toribio F, Lopez-Barea J, Peinado J. Effect of food deprivation on oxidative stress biomarkers in fish (Sparus aurata). Chem Biol Interact. 2003;145:191–9. doi: 10.1016/s0009-2797(03)00002-4. [DOI] [PubMed] [Google Scholar]
- Pasquali MA, Gelain DP, Zanotto-Filho A, de Souza LF, de Oliveira RB, Klamt F, Moreira JC. Retinol and retinoic acid modulate catalase activity in Sertoli cells by distinct and gene expression-independent mechanisms. Toxicol In Vitro. 2008;22:1177–83. doi: 10.1016/j.tiv.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Pollino CA, Holdway DA. Potential of two hydra species as standard toxicity test animals. Ecotoxicol Environ Saf. 1999;43:309–16. doi: 10.1006/eesa.1999.1796. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Arvai AS, Bourne Y, Tainer JA. Active and inhibited human catalase structures: ligand and NADPH binding and catalytic mechanism. J Mol Biol. 2000;296:295–309. doi: 10.1006/jmbi.1999.3458. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Yang KS, Kang SW, Woo HA, Chang TS. Controlled elimination of intracellular H(2)O(2): regulation of peroxiredoxin, catalase, and glutathione peroxidase via post-translational modification. Antioxid Redox Signal. 2005;7:619–26. doi: 10.1089/ars.2005.7.619. [DOI] [PubMed] [Google Scholar]
- Safo MK, Musayev FN, Wu SH, Abraham DJ, Ko TP. Structure of tetragonal crystals of human erythrocyte catalase. Acta Crystallogr D Biol Crystallogr. 2001;57:1–7. doi: 10.1107/s0907444900013767. [DOI] [PubMed] [Google Scholar]
- Sanchez-Paz A, Garcia-Carreno F, Hernandez-Lopez J, Muhlia-Almazan A, Yepiz-Plascencia G. Itect of short-term starvation on hepatopancreas and plasma energy reserves of the Pacific white shrimp (Litopenaeus vannamei). Journal of Experimental Marine Biology and Ecology. 2007;340:184–193. [Google Scholar]
- Sayeed I, Parvez S, Pandey S, Bin-Hafeez B, Haque R, Raisuddin S. Oxidative stress biomarkers of exposure to deltamethrin in freshwater fish, Channa punctatus Bloch. Ecotoxicol Environ Saf. 2003;56:295–301. doi: 10.1016/s0147-6513(03)00009-5. [DOI] [PubMed] [Google Scholar]
- Schrempf H, Zou PJ, Borovok I, Lucana DOD, Muller D. The mycelium-associated Streptomyces reticuli catalase-peroxidase, its gene and regulation by FurS. Microbiology-Uk. 1999;145:549–559. doi: 10.1099/13500872-145-3-549. [DOI] [PubMed] [Google Scholar]
- Schubert J, Wilmer JW. Does hydrogen peroxide exist “free” in biological systems? Free Radic Biol Med. 1991;11:545–55. doi: 10.1016/0891-5849(91)90135-p. [DOI] [PubMed] [Google Scholar]
- Stover NA, Steele RE. Trans-spliced leader addition to mRNAs in a cnidarian. Proc Natl Acad Sci U S A. 2001;98:5693–8. doi: 10.1073/pnas.101049998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Li B, Li X, Wang Y, Xu Y, Jin Y, Piao F, Sun G. Effects of sodium arsenite on catalase activity, gene and protein expression in HaCaT cells. Toxicol In Vitro. 2006;20:1139–44. doi: 10.1016/j.tiv.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Switala J, Loewen PC. Diversity of properties among catalases. Arch Biochem Biophys. 2002;401:145–54. doi: 10.1016/S0003-9861(02)00049-8. [DOI] [PubMed] [Google Scholar]
- Taylor JF, Robinson A, Johnson N, Marroquin-Cardona A, Brattin B, Taylor R, Phillips TD. In vitro evaluation of ferrihydrite as an enterosorbent for arsenic from contaminated drinking water. Environ Sci Technol. 2009;43:5501–6. doi: 10.1021/es803624b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenfield MA, Ng JC, Noller BN, Markich SJ, Dam RA. Dissolved organic carbon reduces uranium bioavailability and toxicity. 2. Uranium[VI] speciation and toxicity to three tropical freshwater organisms. Environ Sci Technol. 2011;45:3082–9. doi: 10.1021/es103349a. [DOI] [PubMed] [Google Scholar]
- Trevisan R, Mello DF, Fisher AS, Schuwerack PM, Dafre AL, Moody AJ. Selenium in water enhances antioxidant defenses and protects against copper-induced DNA damage in the blue mussel Mytilus edulis. Aquat Toxicol. 2011;101:64–71. doi: 10.1016/j.aquatox.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Tripathi BN, Mehta SK, Amar A, Gaur JP. Oxidative stress in Scenedesmus sp. during short- and long-term exposure to Cu2+ and Zn2+. Chemosphere. 2006;62:538–44. doi: 10.1016/j.chemosphere.2005.06.031. [DOI] [PubMed] [Google Scholar]
- van Dam RA, Hogan AC, McCullough CD, Houston MA, Humphrey CL, Harford AJ. Aquatic toxicity of magnesium sulfate, and the influence of calcium, in very low ionic concentration water. Environ Toxicol Chem. 2010;29:410–21. doi: 10.1002/etc.56. [DOI] [PubMed] [Google Scholar]
- Vernouillet G, Eullaffroy P, Lajeunesse A, Blaise C, Gagne F, Juneau P. Toxic effects and bioaccumulation of carbamazepine evaluated by biomarkers measured in organisms of different trophic levels. Chemosphere. 2010;80:1062–8. doi: 10.1016/j.chemosphere.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Wieser R, Adam G, Wagner A, Schuller C, Marchler G, Ruis H, Krawiec Z, Bilinski T. Heat shock factor-independent heat control of transcription of the CTT1 gene encoding the cytosolic catalase T of Saccharomyces cerevisiae. J Biol Chem. 1991;266:12406–11. [PubMed] [Google Scholar]
- Wu C, Zhang W, Mai K, Xu W, Zhong X. Effects of dietary zinc on gene expression of antioxidant enzymes and heat shock proteins in hepatopancreas of abalone Haliotis discus hannai. Comp Biochem Physiol C Toxicol Pharmacol. 2011;154:1–6. doi: 10.1016/j.cbpc.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Zamocky M, Godocikova J, Gasperik J, Koller F, Polek B. Expression, purification, and sequence analysis of catalase-1 from the soil bacterium Comamonas terrigena N3H. Protein Expr Purif. 2004;36:115–23. doi: 10.1016/j.pep.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Zhang J, Shen H, Wang X, Wu J, Xue Y. Effects of chronic exposure of 2,4-dichlorophenol on the antioxidant system in liver of freshwater fish Carassius auratus. Chemosphere. 2004;55:167–74. doi: 10.1016/j.chemosphere.2003.10.048. [DOI] [PubMed] [Google Scholar]
- Zoidis E, Pappas AC, Georgiou CA, Komaitis E, Feggeros K. Selenium affects the expression of GPx4 and catalase in the liver of chicken. Comp Biochem Physiol B Biochem Mol Biol. 2010;155:294–300. doi: 10.1016/j.cbpb.2009.11.017. [DOI] [PubMed] [Google Scholar]