Abstract
Notch signaling regulates many cellular processes during development and adult tissue renewal. Upon ligand binding, Notch receptors undergo ectodomain shedding followed by γ-secretase-mediated release of the Notch intracellular domain (NICD), which translocates to the nucleus and associates with the DNA-binding protein CSL (CBF1/RBPjκ/Su(H)/Lag1) to activate gene expression. Mammalian cells contain four Notch receptors that can have both redundant and specific activities. To monitor activation of specific Notch paralogs in live cells and in real time, we developed luciferase complementation imaging (LCI) reporters for NICD/CSL association and validated them as a specific, robust and sensitive assay system that enables structure-function and pharmacodynamic analyses. Detailed kinetic analyses of various mechanistic aspects of Notch signaling, including nuclear translocation, γ-secretase and ADAM inhibition, as well as agonist- and ligand-dependent activation were conducted in live cells. Notch-LCI represents a powerful approach for characterizing modulators that target Notch signaling and for studying pathway dynamics in normal and disease contexts.
INTRODUCTION
The Notch pathway is a short-range signal transducer that is broadly and reiteratively used during embryonic development and adult tissue renewal to regulate cell proliferation and apoptosis, fate specification, differentiation and stem cell maintenance. Mammalian cells contain four Notch receptors and at least five cognate ligands (Dll1,3,4 and Jag1,2). Most Notch-mediated cellular decisions utilize a canonical or core pathway (1) (Fig. 1A): Ligand-binding leads to a conformational change allowing ADAM-mediated ectodomain shedding (S2 cleavage) and subsequent γ-secretase-mediated proteolysis within the transmembrane domain (S3/S4 cleavage) to release the Notch intracellular domain (NICD). NICD translocates to the nucleus and associates with the DNA-binding protein CSL (named after CBF1/RBPjκ in mammals, Su(H) in flies, and Lag1 in worms) to form a composite interface where the coactivator Mastermind (Mastermind-like, or MAML, in mammals) binds, leading to the recruitment of the transcription machinery and target gene expression (Fig. 1A) (1, 2). The biological consequences of Notch signaling are dependent on dose, timing and cellular context and therefore the core pathway is highly regulated. Indeed, abnormal loss or gain in Notch signaling has been associated with a variety of human developmental disorders, late onset diseases and cancers, leading to its emergence as a key therapeutic target (3–6).
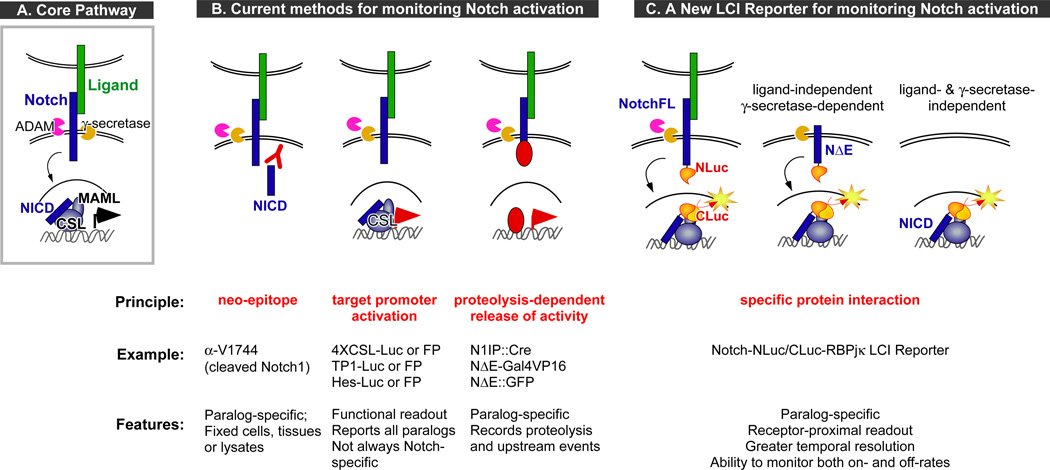
Figure 1. The core Notch pathway and the various approaches for monitoring Notch activation.
A. Schematic of the canonical Notch pathway: Ligand-binding leads to ectodomain shedding (at the S2 site) by ADAM, followed by intramembrane proteolysis (at S3/S4 sites) by γ-secretase. This releases the Notch intracellular domain (NICD), which translocates to the nucleus, interacts with the DNA-binding protein CSL to recruit Mastermind-like proteins (MAML) and other coactivators to activate gene expression. B. Overview of the current methods for monitoring Notch pathway activation: Left, Antibodies can specifically recognize the amino-terminus of NICD that is exposed only after γ-secretase cleavage; Middle, Overall pathway activity can be monitored by reporter proteins (luciferases or fluorescent proteins) under the control of endogenous target promoters (e.g., Hes) or multimerized CSL-binding sites (e.g., 4XCSL, TP1); Right, Regulated proteolysis of Notch-fusion proteins releases various heterologous proteins that can be monitored via transcriptional reporters or nuclear translocation. C. The luciferase complementation imaging (LCI)-based approach for monitoring Notch activation in real time takes advantage of the specific interactions between a particular NICD and CSL (RBPjκ in our studies) to reconstitute activity between fused luciferase fragments (NLuc and CLuc). Variants of the reporter system allow us to interrogate different mechanistic aspects of Notch activation.
Understanding the molecular mechanisms regulating the dose and timing of Notch activity in both normal and disease contexts depends on sensitive methods for monitoring Notch activity in real time and is emerging as a critical need. One strategy utilizes antibodies to detect the γ-secretase-cleaved (i.e., activated) form of Notch (Fig. 1B). Although this has been useful in assessing Notch cleavage in lysates, fixed cells and tissues (7, 8), it is not a dynamic real-time reporting system. Another approach is based on the use of Notch-responsive promoters to drive fluorescent or bioluminescent reporters in cells or animals (Fig. 1B) (9–12). These widely used transcriptional reporters allow sensitive, qualitative/quantitative and dynamic reporting of overall pathway activity. However, these reporters can also record input from other signaling pathways and cannot distinguish which paralog is active in cells where multiple paralogs are expressed. Fusing heterologous proteins (e.g., Gal-VP16, Cre (13–15)) to a particular receptor can help boost sensitivity and/or specificity. Still, due to the time needed for transcription and translation of downstream reporters, a delay is inherent in all promoter-reporter gene approaches. Notch-GFP nuclear translocation assays (16) can address some of these issues, but are not as sensitive due to cellular autofluorescence. Moreover, the need to distinguish the nuclear- vs the membrane-tethered fluorescence presents a greater technical challenge as a high throughput screening (HTS) approach.
We have developed an alternative reporter system for canonical Notch signaling using optimized luciferase complementation imaging (LCI) (17), a protein fragment complementation assay (PCA) system that permitted us to directly monitor interactions between a particular NICD and RBPjκ in real time (Fig. 1C). The key advantages of LCI compared to other PCA or protein interaction-based methods include (1) inherent sensitivity of luminescent enzymatic reporters due to signal amplification and the lack of background cellular luminescence; (2) negligible binding energy between the luciferase fragments, permitting accurate quantification of protein interactions; (3) ability to detect protein interactions in any subcellular compartment; (4) allows non-invasive, repeated imaging in live cells and in vivo; (5) easily deployed with relatively inexpensive, commercially available reagents, and (6) amenability to HTS applications (18, 19). Notably, LCI does not depend on activation of downstream reporters for readout, thereby allowing measurement of reconstituted luciferase activity in near real time and enabling kinetic analyses of pathway activation.
Here we report the extensive validation of the Notch-LCI reporter technology. We confirmed its ability to faithfully reproduce known Notch biochemistry and demonstrated its utility as a non-invasive real-time reporter of Notch activation. We monitored the formation and turnover of the NICD/RBPjκ complex, measured the pharmacodynamics of different γ-secretase and metalloprotease inhibitors, compared the activation kinetics of receptors presented with ligands or agonists in different paradigms and quantified responses to known modifiers. In addition, we have generated several stable cell lines expressing variants of the reporter and validated their robustness for HTS. Thus, the Notch-LCI reporters represent powerful, HTS-ready tools for characterizing cellular activities and pharmacological agents that modulate Notch activation.
RESULTS
Robust and specific complementation between NICD1-NLuc and CLuc-RBPjκ
Complementary luciferase fragments (NLuc and CLuc), which have no activity on their own, can reassemble into an active enzyme when brought into close proximity by the interacting proteins they are fused to (17). To create a real-time assay system for Notch1 activation, we fused NLuc to several Notch1 variants and CLuc to RBPjκ (Fig. 1C and 2A). Notch1-NLuc variants consisted of a full-length (NotchFL or NFL) receptor, truncated inactive Notch molecules lacking the EGF repeats but containing the NRR (NLNG (20)), constitutively active Notch molecules that either lack the entire extracellular domain (NΔE) or contain only the intracellular domain (NICD) and various mutant versions of these receptors (Fig. 2A). Notch activity is retained after addition of heterologous tags or molecules to the Notch C-terminus (21). Still, to minimize steric hindrance, we included a short flexible Gly-Ser linker between the luciferase fragment and the fused protein (17). The integrity of all Notch-NLuc proteins was confirmed by Western blot with a Notch1-specific monoclonal, mAN1, and where appropriate, γ-secretase-dependent proteolysis was confirmed with antibodies against the VLLS epitope (α-V1744), corresponding to the cleaved free amino-terminus of NICD (22) (Fig. 2B). Interestingly, although detectable with mAN1, NICDΔRAM molecules (which were either released from NΔEΔRAM cleavage or produced from a NICDΔRAM expression vector) were not recognized well by some lots of the α-V1744 antibody, highlighting the shortcoming of antibody-based detection of activated Notch proteins. To compare the signaling properties of Notch1-NLuc and CLuc-RBPjκ fusion proteins to their untagged counterparts, each protein was individually evaluated for its ability to activate downstream transcriptional reporters based on multimerized RBPjκ binding sites (i.e., TP1-luciferase) or endogenous Notch target promoters (Hes1- and Hes5-luciferase). The functionality of CLuc-RBPjκ was examined in RBPjκ-null (OT11) cells. The fusion of CLuc to RBPjκ did not change its activity on both the TP1-luciferase and Hes1-luciferase reporters (fig. S1).
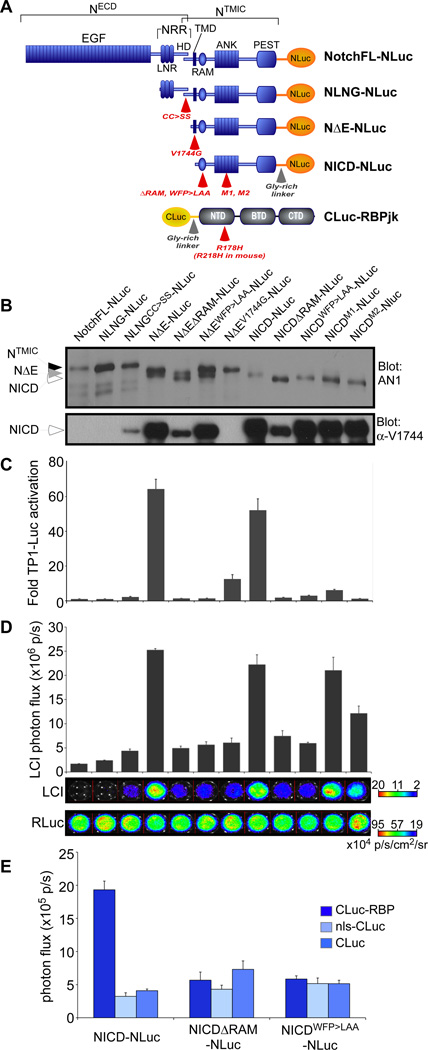
Figure 2. Development and validation of the Notch/RBPjκ LCI reporter.
A. Domain organization of the Notch1 receptor and RBPjκ and their respective luciferase fragment fusions used in this study. The different mutations used for assay validation are indicated (red arrows). The full-length receptor (NotchFL) has a large extracellular domain (ECD) composed of EGF repeats and the negative regulatory region (NRR), which can be further subdivided into the Lin-Notch repeats (LNR) and the heterodimerization domain (HD). The NRR keeps the receptor in the ‘off’ state in the absence of ligand. Mature Notch receptors have been furin-processed at the S1 site within HD to generate the NECD and NTMIC (Notch transmembrane and intracellular domain) fragments that are held together by interactions between the N- and C-terminal halves of HD. Following the transmembrane domain (TMD) is a large intracellular region that carries the RAM (RBPjk association module), ANK (7 ankyrin repeats) and PEST (proline/glutamic acid/serine/threonine-rich) domains. NLNG molecules cannot respond to ligand and are completely inactive unless mutations that lead to ligand-independent activation are present (e.g., cc>ss). NΔE and NICD are both constitutively active forms. RBPjκ is composed of the N-terminal domain (NTD), Beta-trefoil domain (BTD) and C-terminal domain (CTD). B. Notch-NLuc proteins were also assessed by Western analyses with the AN1 antibody, which recognizes the ANK domain. Where appropriate, NICD production was also confirmed by the α-V1744 antibody. C. The transactivation profile of the Notch-NLuc receptor variants is consistent with previous studies, with the constitutively active proteins NΔE and NICD exhibiting the highest activity. D. Complementation profile of the different Notch-NLuc fusions with CLuc-RBPjκ. Constitutively active forms exhibit the highest complementation activity. LCI can distinguish the mechanisms of action of the RAM and ANK domain mutants. Cotransfected Renilla Luc (RLuc) activities confirm equivalent transfection efficiency. E. Notch-NLuc/CLuc-RBPjκ complementation is specific: NICD-NLuc produces robust complementation with CLuc-RBPjκ but not with the CLuc alone or nls-CLuc. RAM mutations known to disrupt binding also diminish complementation.
The function of Notch-NLuc fusion proteins was tested in unstimulated 3T3 cells. TP1-luciferase activation was not seen with NotchFL molecules or with NLNG proteins unless they contained amino acid substitutions known to relax the auto-inhibition (NLNGCC>SS)(20). The constitutively active forms, NΔE-NLuc and NICD-NLuc, both activated TP1-luciferase robustly (Fig. 2C). Next, we tested three types of biochemically characterized, inactivating mutations known to greatly diminish or abolish Notch transcriptional activity: (i) a transmembrane domain (TMD) mutant (NΔEV1744G) that produces a destabilized NICD (23, 24); (ii) mutations targeting the RAM domain that abolish interactions with RBPjκ. In this class, we tested a 27-amino acid deletion (ΔRAM) (22) and the WFP to LAA amino acid substitution (NΔEWFP>LAA and NICDWFP>LAA) (25); (iii) Mutations destabilizing the ANK domain (NICDM1, NICDM2) (26, 27) that permit RBPjκ interactions but eliminate transcriptional activity due to failure in recruiting MAML. Based on our analyses, the relative activities of the NLuc-tagged Notch proteins was the same as the untagged molecules (Fig. 2C and fig. S2A). The NLuc fusion reduced the overall transactivation ability of Notch on promoters with multimerized sites (TP1-luciferase; fig. S2, A and B), but had less of an effect on Hes1-luciferase and no effect on Hes5-luciferase activation (fig. S2C). These results suggest that the tag can cause steric hindrance on reporters with tandem repeats of RBPjκ binding sites in head-to-tail orientation. Overall, the data confirmed that the addition of luciferase fragments at the C-terminus of Notch did not greatly alter their signaling properties.
After confirming the proteolytic processing and transcriptional function of Notch-NLuc fusion proteins, we tested their ability to reconstitute luciferase activity with CLuc-RBPjκ. Notch-NLuc variants were coexpressed with CLuc-RBPjκ along with Renilla luciferase (as a transfection control) and then bioluminescence imaging was performed in live cells (Fig. 2D and 2E). A strong signal was readily detected when NICD-NLuc was coexpressed with CLuc-RBPjκ (Fig. 2D and 2E), whereas non-specific interactions between NICD-NLuc and CLuc, nls-CLuc produced only low background luminescence (Fig. 2E). NotchFL-NLuc or NLNG-NLuc (both of which are inactive molecules in this paradigm) did not generate an appreciable bioluminescent signal when coexpressed with CLuc-RBPjκ, indicating that transient ER interactions that could occur during protein synthesis did not contribute to luciferase complementation with this amount of protein. Coexpression of CLuc-RBPjκ with RAM domain mutants (ΔRAM or WFP>LAA) did not result in a signal above background (Fig. 2D and 2E), confirming the dependence of complementation on this high-affinity interaction domain. As anticipated, robust complementation occurred with the dominant-negative form of RBPjκ (CLuc-dnRBPjκ; fig. S3), confirming that unlike transcriptional reporter activation (fig. S1A), NICD/RBPjκ interactions do not depend on the DNA-binding activity of RBPjκ. Importantly, LCI reported the same “signal strength” hierarchy observed with transcriptionally active constructs (Fig. 2D), with the constitutively active molecules equivalently exhibiting the highest luciferase complementation activity (i.e., NΔE=NICD≫NΔEV1744G>NLNGCC>SS).
Notch-LCI can differentiate the mechanisms underlying the effects of RAM vs ANK domain mutations on transcriptional activation. NICD molecules harboring the ANK mutations M1 and M2 failed to activate TP1-luciferase (Fig. 2C), but LCI confirmed their ability to interact with RBPjκ in intact cells (Fig. 2D). This reflects the presence of an intact RAM domain in these molecules and the negligible binding energy contributed by the ANK domain to NICD/RBPjκ interactions relative to its importance in facilitating assembly of the NICD/RBPjκ/MAML complex (2). The ability of LCI to detect the subtle differences in stability of the NICD/RBPjκ complex with some ANK domain mutations confirms the utility of Notch-LCI as a sensitive cell-based reporter of protein-protein interaction. Next we determined whether LCI would allow us to non-invasively monitor changes in NICD/RBPjκ interactions in live cells while modulating the RAM/RBPjκ interface. Soluble RAM peptides have been demonstrated to disrupt NICD/RBPjκ interactions using purified proteins in vitro and in cell lysates (28, 29). Accordingly, the RAM domain polypeptide, but not the mutant RAMWFP>LAA peptide, significantly reduced NICD-NLuc/CLuc-RBPjκ complementation (Fig. 3A).
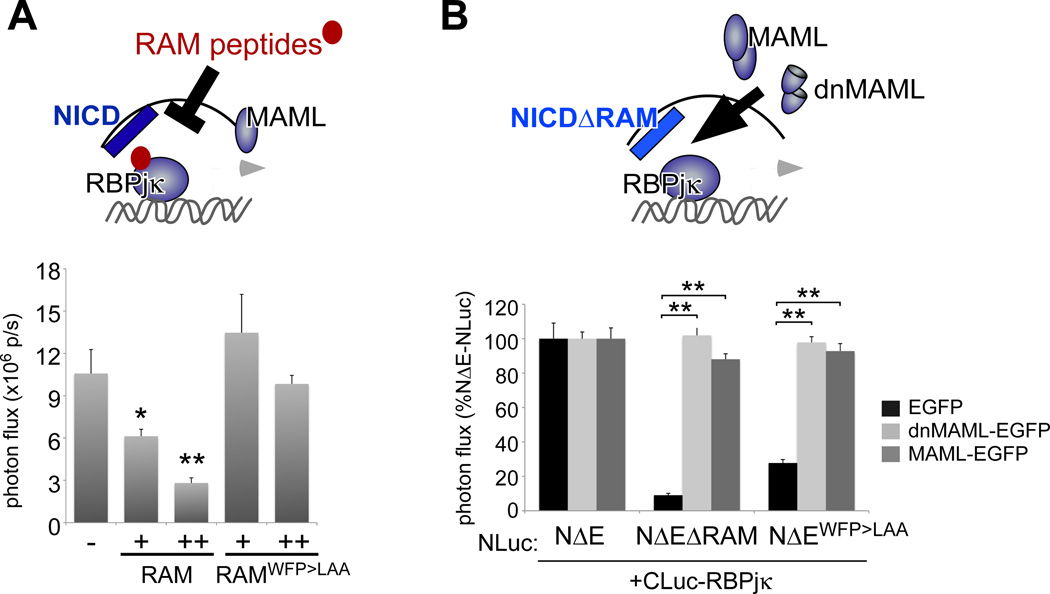
Figure 3. LCI allows direct quantification of the modulation of NICD/RBPjk interactions in live cells.
A. Wild-type (but not WFP>LAA-mutant) RAM polypeptides reduced NΔE-NLuc/CLuc-RBPjκ complementation in a dose-dependent manner. G. MAML-EGFP and dnMAML-EGFP stabilize the interactions between the Notch RAM mutants and RBPjκ. * p<0.01, ** p<0.0001.
The above LCI reporter studies confirm the importance of the RAM domain in mediating NICD/RBPjκ interactions. However, NICDΔRAM molecules can activate reporters or downstream target genes in cells with abundant amounts of Mastermind proteins (30), which act to stabilize the ANK/RBPjκ interface. The aa13–74 fragment of MAML1 (i.e., dominant-negative MAML (dnMAML)) is sufficient to stabilize this interaction in vitro and prevent MAML docking and subsequent target gene expression (31) (fig. S4B). We therefore used our LCI reporters to examine if dnMAML leads to stabilization of ANK/RBPjκ interactions in live cells. As expected, coexpression of MAML-EGFP and dnMAML-EGFP to NICDΔRAM-NLuc rescued its interaction with CLuc-RBPjκ (Fig. 3B). To assess the specificity of these interactions, we tested whether the stabilizing effects of dnMAML-EGFP extended to other RBPjκ partners. The EBNA2 protein interacts with RBPjκ to activate transcription of Epstein-Barr virus genes as well as Notch-responsive reporter genes (32) (fig. S4A). Like NICD, EBNA2 depends on a ΦWΦP motif for its interactions with RBPjκ evidenced by the reduced complementation between CLuc-RBPjκ and EBNA2WW>SR-NLuc (fig. S4C). However, EBNA2 lacks an ANK domain and is not expected to interact with MAML. Indeed, dnMAML-EGFP has no stabilizing effect on EBNA2WW>SR-NLuc/CLuc-RBPjκ complementation and as predicted, no significant inhibitory effect on EBNA2-mediated transcriptional activation (fig. S4, C and D). Taken together, LCI accurately reported in live cells the predicted behavior of NICD/RBPjκ complex formation based on known Notch structure and biochemical analyses of functional domains.
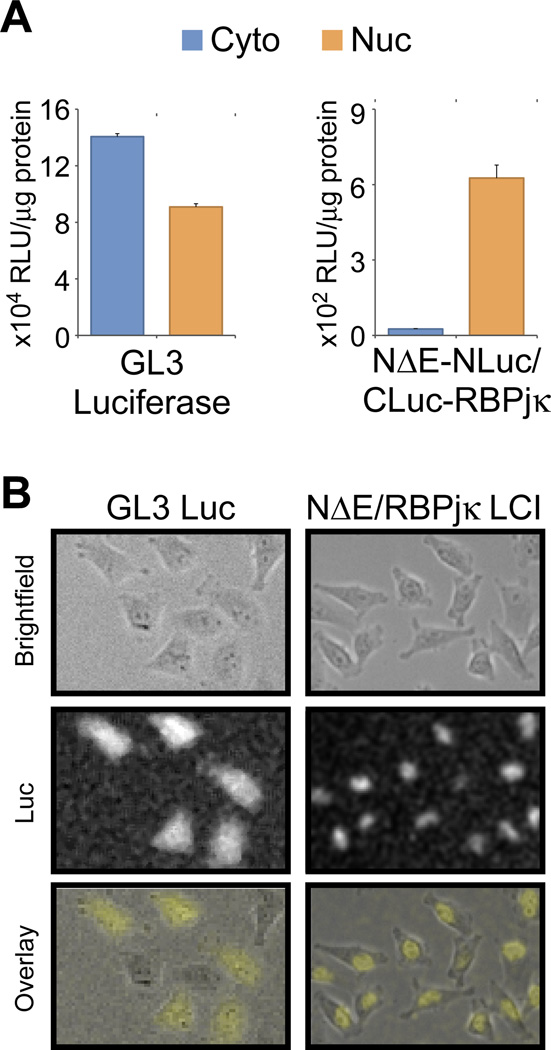
Finally, we wanted to identify the cellular compartment in which luciferase complementation occurred. Once released, NICD translocates to the nucleus, but some have reported that RBPjκ can be found both in the cytoplasm and the nucleus. Cells expressing either GL3 luciferase alone or NΔE-NLuc and CLuc-RBPjκ together were fractionated and assayed for their specific luciferase activities in cytoplasmic and nuclear fractions (Fig. 4A). While the GL3 luciferase-expressing cells showed almost equivalent luciferase activity in both fractions, the NΔE-NLuc/CLuc-RBPjκ-expressing cells displayed substantial luciferase activity only in the nuclear fraction. This subcellular localization was also demonstrated by cellular bioluminescence imaging (Fig. 4B). While full-length luciferase signal can be observed throughout the cell, the Notch/RBP signal is limited to the nucleus, confirming that luciferase complementation occurs after NICD enters the nucleus.
Figure 4. Notch-NLuc/CLuc-RBPjκ complementation occurs in the nucleus.
NΔE-NLuc/CLuc-RBPjκ complementation occurs in the nucleus as demonstrated by subcellular fractionation (A) and cellular bioluminescence imaging (B). This is in contrast to full-length luciferase, which partitions almost equally between the cytoplasmic and nuclear fractions (A) and can be found throughout the cell (B). Cellular bioluminescence images were processed using ImageJ to optimize the brightness and contrast and to reduce the salt-and-pepper noise in the background (i.e., despeckle).
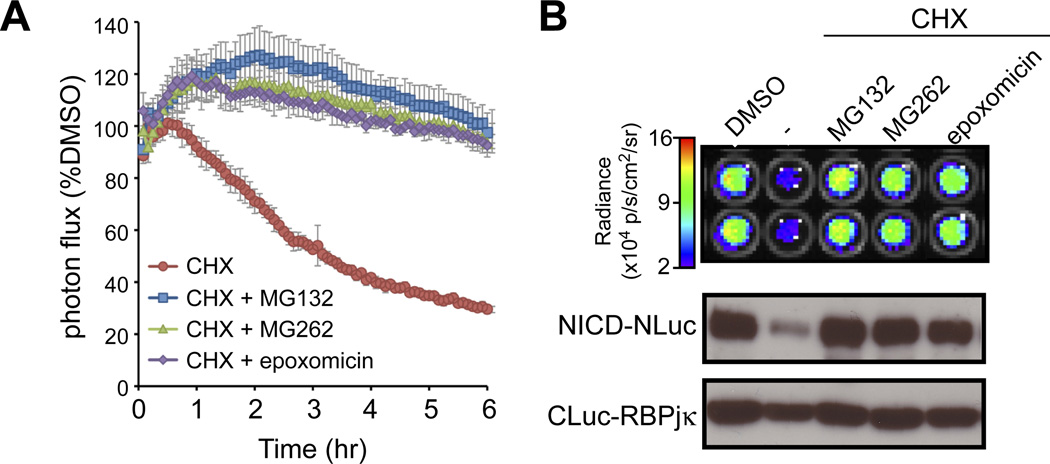
NICD half-life unaffected by the fusion protein
The results above establish LCI as a viable alternative for, or as a complement to biochemical assays for quantifying protein-protein interactions in live cells. However, these studies did not completely rule out the possibility that the addition of luciferase fragments altered the half-life of Notch proteins. To assess half-life, we monitored the stability of the NICD/RBPjκ complex in real time using a HeLaTetOn stable line expressing NΔE-Nluc and CLuc-RBPjκ (line E6; this and additional lines are fully described in fig. S5). Luciferase complementation activity in live NΔE-NLuc/CLuc-RBPjκ-expressing cells was monitored continuously for 6 hrs in the presence or absence of the protein translation inhibitor cycloheximide (CHX; Fig. 5A). The decay in bioluminescence indicated an NICD half-life of ~180 minutes, consistent with the half-life obtained by pulse-chase experiments in HeLa cells (33). As expected, inhibition of the proteasome with either MG132, MG262 or epoxomicin stabilized the NICD/RBPjκ complex (Fig. 5A) and the NICD protein (Fig. 5B). The similarity between NICD half-life determined biochemically and by LCI indicates that the fusion protein does not alter the stability of the NICD/RBPjκ complexes, and confirmed that the stability of the complex is determined by the half-life of NICD. Notch-LCI will therefore enable direct and precise quantitation of both on-rates and off-rates of receptor activation and allow dynamic analyses of receptor-proximal events regulating NICD release and turnover with better temporal resolution over transcription-based reporters.
Figure 5. Monitoring NICD/RBPjk complex stability using LCI.
A. Stable lines expressing NΔE-NLuc and CLuc-RBPjκ were treated with cycloheximide (CHX) to block translation and the degradation of the NICD/RBPjκ complex was monitored by LCI in real time. The half-life of the luciferase complementation activity (t1/2=180 min) was similar with the half-life of NICD determined using pulse-chase. Moreover, complementation activity was stabilized by the indicated proteasome inhibitors, consistent with previous studies demonstrating that NICD is ubiquitinated and degraded by the proteasome. B. Image and Western analyses of cells at 6 hr. Luciferase complementation (degradation and stabilization) correlated closely with NICD-NLuc protein (detected by α-V1744 antibody). These studies confirm that the NICD stability determines the NICD/RBPjk complex half-life. Importantly, because the Luc fusion does not alter the stability of NICD, LCI will us to accurately monitor the off-rates of pathway activation.
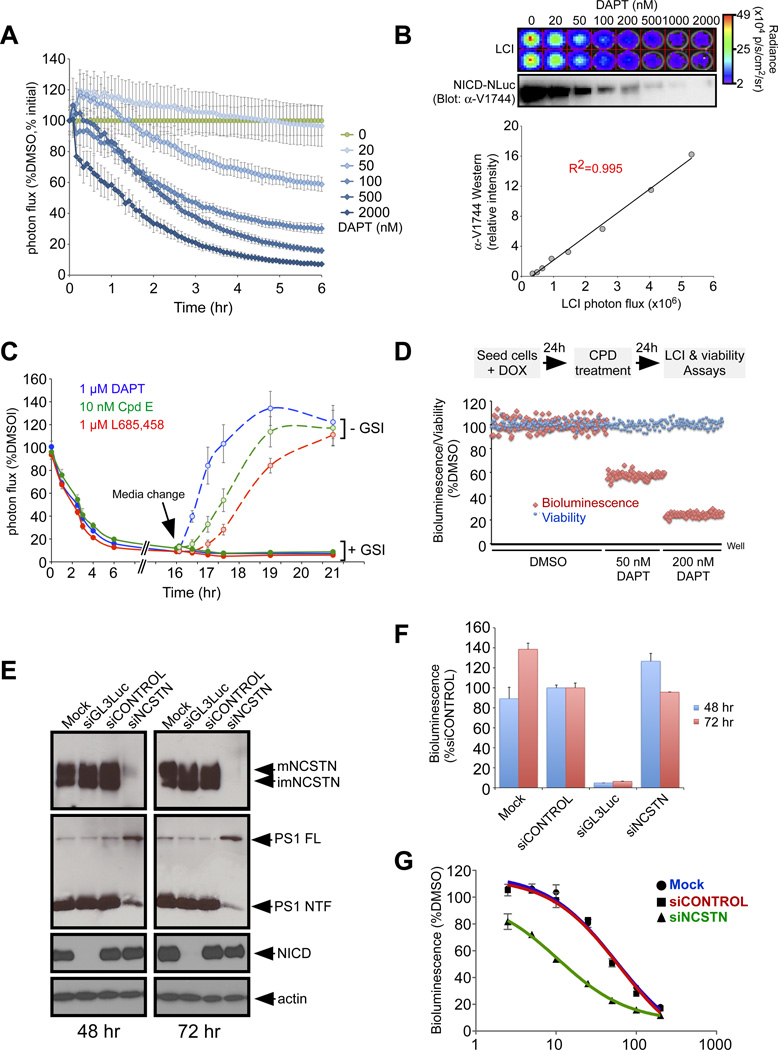
Dynamics of γ-secretase-mediated cleavage of Notch and its response to γ-secretase inhibitors (GSIs)
To test the utility of Notch-LCI to assess modulators of events upstream of the nuclear interactions, we monitored the dynamics of NICD release from NΔE-NLuc, a constitutively active form of Notch that is ligand-independent but proteolysis-dependent (Fig. 1C). Luciferase complementation and NICD accumulation were inhibited in a time- and dose-dependent manner by the γ-secretase inhibitor DAPT (Fig. 6A), whereas complementation between the NICD-NLuc/CLuc-RBPjκ pair was unaffected (fig. S6). Importantly, we observed an excellent correlation (R2=0.995) between the NICD protein amount measured by α-V1744 Western analyses and by luciferase complementation with RBPjκ (Fig. 6B), demonstrating again that Notch-LCI is an outstanding reporter for quantifying NICD amounts. To further demonstrate the utility of Notch-LCI, we conducted dose-response analyses of a blinded set of GSIs (a generous gift from MERCK) on the NΔE-NLuc/CLuc-RBPjκ stable line. LCI-derived IC50 values correctly rank-ordered the GSIs based on efficacy and distinguished active vs inactive enantiomers (fig. S6).
Figure 6. LCI as a real-time reporter of the dynamics of γ-secretase activity and inhibition in live cells.
A. NΔE/RBPjκ LCI reporter cells treated with different concentrations of the γ-secretase inhibitor DAPT exhibited a time- and dose-dependent decrease in complementation activity. B. Complementation activity correlated well with the amount of cleavage product NICD determined by Westerns. C. Kinetics of recovery after removal of inhibitors: All GSIs tested were confirmed to act in a reversible manner with slight differences in recovery half times. D. The NΔE/RBPjκ LCI reporter cells are robust and have been validated for high throughput screening applications. Multiple 96-well plates were seeded and treated with DMSO or DAPT and assayed for bioluminescence and viability. Z’ factors obtained in these experiments were >0.5, indicating that the assay is excellent for HTS. E. Knockdown by siGL3 Luc and siNCSTN was confirmed by Western analyses. siGL3 Luc efficiently targets Notch-NLuc. siNCSTN efficiently reduced NCSTN protein as well as mature γ-secretase complexes, indicated by the reduction in Presenilin1 NTF fragments. F. Luciferase complementation activity was greatly diminished with siGL3 Luc but not with siNCSTN, suggesting that γ-secretase is not limiting in these cells. G. siNCSTN sensitizes cells to sub-inhibitory concentrations of DAPT.
LCI differs from transcriptional reporter assays by enabling the kinetic analysis of GSI effects with precise temporal resolution (Fig. 6A), which permits determination of pharmacodynamic properties of existing and novel GSIs in live cells. To demonstrate this further, we performed a kinetic study of different benchmark GSIs at a single effective dose using the NΔE-NLuc/CLuc-RBPjκ reporter cells. The time-course for γ-secretase inhibition yielded similar half-times (t1/2s) of ~120 min for DAPT, Cpd E and L685,458 (Fig. 6C) and compare favorably with the in vivo data for Aβ inhibition obtained with canulated mice (34). Maximal inhibition was observed at 6 hr and was maintained after an overnight incubation (16 hr). We next tested the kinetics of recovery after release from GSI treatment. Although the inhibition kinetics were similar, LCI differentiated recovery t1/2 values from the different GSIs: 30 min for DAPT, 90 min for Cpd E and 120 min for L685,458. Note that maximal bioluminescence post-recovery exceeded the bioluminescence intensity in cells treated only with DMSO, most likely due to accumulation of the NΔE substrate (20).
Precise, real-time quantitation made possible by this LCI reporter will facilitate the screening of compounds for desired properties (e.g., long-acting or Notch-sparing GSIs). To evaluate the utility and HTS readiness of the Notch-LCI reporter cell lines, we performed large-scale experiments to configure the assay for HTS and to determine the Z’ factor using DAPT as the model compound. The Z’ factor is a statistical measure of assay robustness; a Z’ factor value ≥0.5 is considered to be an indication of an excellent assay for HTS (35). Multiple 96- and 384-well plates were seeded with NΔE-NLuc/CLuc-RBPjκ reporter cells. After 24 hrs, cells were treated with DMSO alone or with different concentrations of DAPT. Cells were then assayed the next day for bioluminescence and viability (using Alamar Blue) (Fig. 6D). The assay was readily automated and highly reproducible. Z’ factors obtained in multiple experiments were 0.60–0.85, indicative of a robust HTS-ready cell line.
Loss-of-function genetic screens using (RNAi) represent a powerful approach to identifying novel pathway modulators. To validate the NΔE-NLuc/CLuc-RBPjκ reporter for RNAi-based applications, we specifically targeted the protein NCSTN, a single-gene component of the γ-secretase complex that displays dosage sensitivity (36). Knockdown with NCSTN siRNA was highly efficient; Western analyses confirmed the reduction in NCSTN protein and mature γ-secretase complexes, as indicated by the presenilin1 N-terminal fragment (PS1 NTF; Fig. 6E). Interestingly, no reduction in NΔE cleavage was detected by LCI (Fig. 6F) or by Western analyses for NICD (Fig. 6E). This may occur if γ-secretase activity was not limiting in these cells: the few surviving γ-secretase molecules would still be sufficient to cleave substrate. To test this hypothesis, we asked if cells treated with siNCSTN would display a greater sensitivity to GSI. Indeed, NCSTN RNAi sensitized the NΔE-NLuc/CLuc-RBPjκ reporter cells to sub-inhibitory amounts of DAPT (note shift in green curve in Fig. 6G), lowering the IC50 for DAPT by 5-fold (based on nonlinear regression analysis). This result is consistent with the notion that γ-secretase amounts are not limiting for Notch cleavage in some tissues and siRNA manipulation may, on its own, be insufficient to generate a phenotype. Therefore, adding a sub-inhibitory amount of a pharmaceutical inhibitor can serve as a chemical genetic screening paradigm for identifying cellular activities that could modulate desired aspects of Notch signaling. To validate the robustness of this assay for high throughput siRNA screening, multiple 96-well plates were seeded with NΔE-NLuc/CLuc-RBPjκ reporter cells and transfected with siCONTROL or siNCSTN followed by treatment with a sub-inhibitory DAPT concentration (10 nM). We obtained Z’ factors of 0.50–0.59 in multiple experiments, indicating that the NΔE-NLuc/CLuc-RBPjκ reporter line is excellent for both small molecule and RNAi HTS applications.
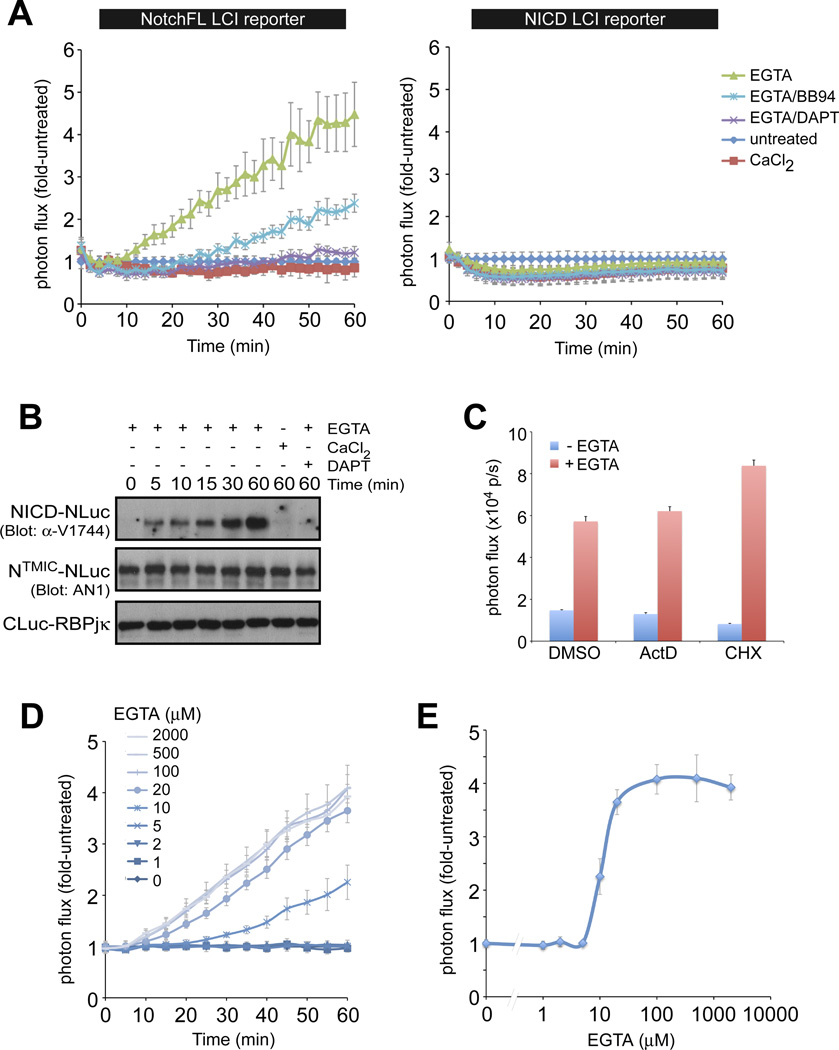
Dynamics of ligand-independent activation of full-length Notch receptors
Next, we established NotchFL-NLuc/CLuc-RBPjκ stable lines and confirmed that the NotchFL fusion proteins undergo normal maturation and S1 processing (line FL2, fig. S5). We used LCI to determine the kinetics of Notch activation in response to the calcium chelators EDTA, BAPTA and EGTA (Fig. 7 and fig. S7). Calcium chelation is thought to disrupt the NRR structure, promoting receptor proteolysis and NICD release independent of ligand (1, 2, 37). Addition of calcium or calcium chelator to the NICD-NLuc/CLuc-RBPjκ reporter cells led to a mild decrease in bioluminescence likely due to the general effects of such treatments on a variety of cellular processes (Fig. 7A and fig. S7A). Importantly, despite these non-specific effects, calcium chelator treatment of NotchFL-NLuc/CLuc-RBPjκ reporter cells still resulted in an increase in photon flux within 10 minutes (Fig. 7A and fig. S7A), which is consistent with the appearance of dissociated NECD in the medium after EDTA treatment of Notch-expressing cells as has been previously reported (37). Bioluminescence increased at a fixed rate for 60 minutes (Fig. 7A and fig. S7A), correlating well with NICD accumulation, as assayed by Westerns (Fig. 7B), indicating that nuclear translocation occurs very rapidly. Indeed, a similar linear increase was observed with a Notch-GFP nuclear translocation assay in EGTA-treated fly S2 cells (16). Chelation may work by dissociation of furin-cleaved Notch molecules (38). To test for dependence on de novo proteolysis, we performed the chelator treatment in the presence of an ADAM inhibitor (BB94, blocks S2 cleavage) or the GSI DAPT. Both abrogated luciferase complementation and NICD production (Fig. 7, A and B and fig. S7, A and B). Taken together, these results confirmed that like ligand, calcium chelation increased bioluminescence via de novo proteolysis of full-length Notch receptors. The chelation-induced increase in bioluminescence was observed in the presence of transcription or translation inhibitors (actinomycin D (ActD) and CHX, respectively; Fig. 7C), demonstrating the real-time nature of the Notch-LCI reporter. In conclusion, non-invasive kinetic analysis with greatly improved temporal resolution distinguishes LCI from conventional transcriptional reporter assays, which usually require hours to assess downstream target transcriptional reporter activity. Using the NotchFL LCI reporter system, we performed detailed quantitative analyses of activation at different EGTA concentrations (Fig. 7D). We show that EGTA-dependent activation behaves as a digital switch with Notch receptors being either inactive or fully activated within a narrow concentration range (5–20 µM; Fig. 7E).
Figure 7. Real-time imaging of the dynamics of ligand-independent activation by the Ca++ chelator EGTA.
A. EGTA treatment resulted in a linear time-dependent increase in bioluminescence from the NotchFL LCI reporter line (but not from the ligand-independent NICD LCI reporter line, right) that was inhibited by both BB94 and DAPT. B. The increase in bioluminescence (fold activation) correlated with the appearance and accumulation of the NICD cleavage product (see fig. S7 for additional Western analyses). C. Monitoring the activation process by LCI does not depend on downstream transcription and translation as it occurs in the presence of 1µg/ml ActinomycinD (ActD) or 10 µg/ml cycloheximide (CHX), demonstrating the real-time nature of the assay. D, E. Using the LCI reporter, we were able to perform detailed temporal and dose response analyses with EGTA. We demonstrate that complete activation occurs within a narrow EGTA concentration range.
The agonist effects of calcium chelation on NotchFL reflect the importance of NRR integrity in keeping Notch in its auto-inhibited state in the absence of ligand. Consistent with previous studies (39, 40), the phorbol ester PMA has no effect on wild-type NotchFL-NLuc/CLuc-RBPjk basal luminescence as the intact NRR prevents access to ADAM proteases (fig. S8A). PMA activates Protein kinase C thereby inducing ADAM protease activity (41), which the intact NRR is insensitive to. Mutations that disrupt the NRR can cause T-cell acute lymphoblastic leukemia (T-ALL) in humans (42) due to “leaky” Notch signaling. Consistent with this, NLNGCC>SS molecules, which carry destabilizing amino acid substitutions in the NRR domain, release a small amount of NICD protein (Fig. 2B) and exhibit ligand-independent basal activity in transcriptional reporter and LCI assays (Fig. 2, C and D). This activity can be further induced by treatment with PMA. To measure the real-time kinetics of this process, we measured NICD release via NLNGCC>SS-NLuc/CLuc-RBPjκ LCI upon addition of PMA and compared the response to that obtained with EGTA. While both NotchFL and NLNGCC>SS can be similarly activated by EGTA treatment (fig. S8C), only NLNGcc>ss can be stimulated by PMA (fig. S8B). The involvement of proteolysis in the PMA-stimulated activation was confirmed by BB94 and DAPT inhibitor treatment. This demonstrates the potential of the LCI assay to perform structure-function analyses on the dynamics of NRR unfolding in live cells.
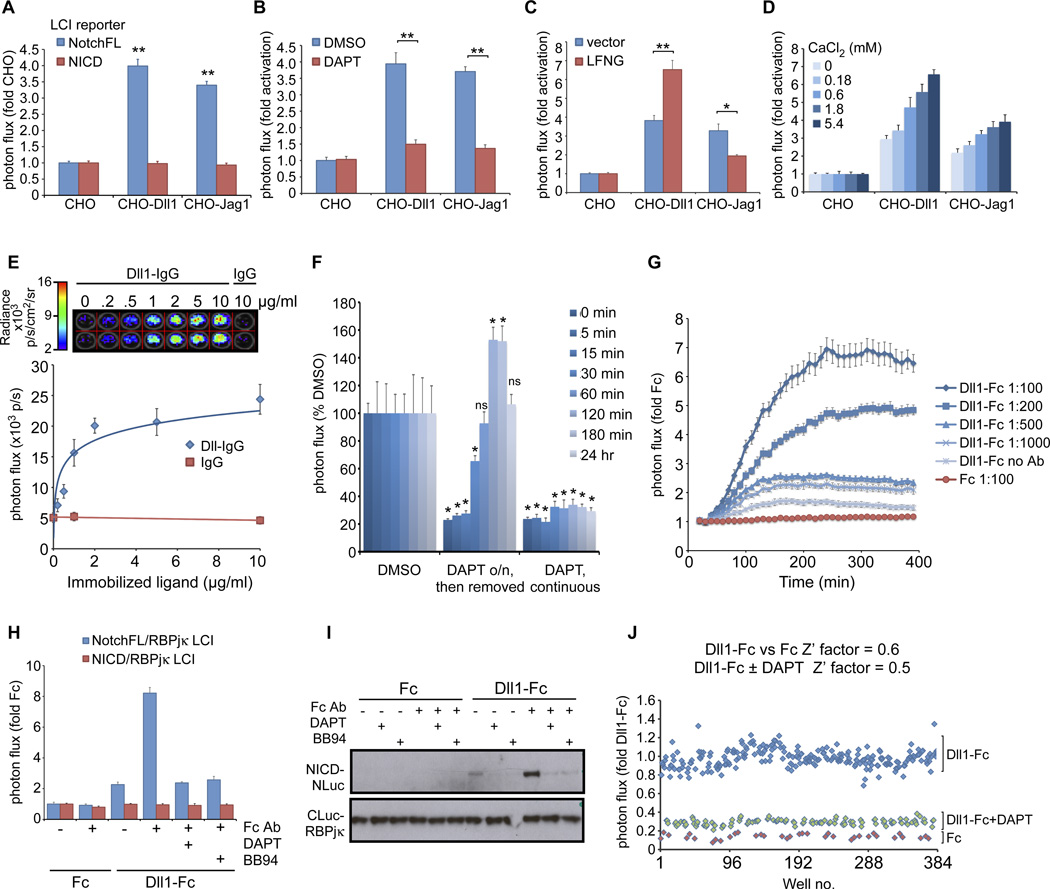
Dynamics of ligand-dependent activation of full-length Notch receptors
We next utilized the NotchFL-LCI reporter to characterize ligand-dependent activation in real time in live cells. We observed a 3.5–4-fold activation of the reporter upon coculturing with CHO-Dll1 or CHO-Jag1 cells (Fig. 8A), which was greatly diminished in the presence of DAPT (Fig. 8B). Coculture with ligand-expressing cells did not affect the activity of the constitutively active NICD-NLuc/CLuc-RBPjκ reporter cells (Fig. 8A), further demonstrating the specificity of the LCI reporter activity. We then assessed the response to known modifiers of ligand-Notch interactions. The fringe family of glycosyltransferases can modify specific EGF repeats of Notch receptors to modulate their responsiveness to ligands in specific contexts (43). Consistent with previous studies (44), overexpression of Lunatic Fringe (LFNG) potentiated the response of the NotchFL LCI reporter to Dll11, but abrogated the response to Jag1 ligand (Fig. 8C). Notch has also been suggested to act as a sensor for extracellular calcium concentrations during the establishment of the left-right axis (45). To examine the effects of extracellular calcium on receptor activation, we cocultured NotchFL LCI reporter cells with CHO-ligand stable lines under varying concentrations of CaCl2 (0–5.4mM; Fig. 8D). We observed a dose-dependent increase in fold activation by either ligand with increasing CaCl2. Thus, extracellular calcium concentration can regulate Notch signal strength.
Figure 8. Real-time imaging of the dynamics of Notch activation by ligands presented in various paradigms.
A. NotchFL (but not NICD) LCI reporter cells are activated when cocultured with either CHO-Dll1 or CHO-Jag1 stable cell lines. B. The activation is diminished in the presence of DAPT, demonstrating the specificity of the response. C. Coexpression of the glycosyltransferase lunatic Fringe (LFNG) enhances activation by Dll1 but reduces responsiveness to Jag1. D. Notch activation can be regulated by the concentration of extracellular calcium. Notch receptors can be activated by ligand ECD-IgG (or -Fc) fusions that have been immobilized (E–F) or preclustered (G–J). E. Dose-dependent activation of NotchFL LCI reporter with immobilized ligands. F. Kinetics of NICD accumulation from NotchFL after DAPT removal was monitored by LCI and found to mimic kinetics of NICD accumulation from NΔE after DAPT removal. Statistical analyses were performed to assess significant differences from the DMSO control at each timepoint. G. Kinetics of Notch activation with Dll1-Fc-containing conditioned medium preclustered with different α-Fc antibody concentrations. H. The response to clustered ligand was specific to activation of full-length Notch receptors: no response is seen with the NICD reporter cells, NotchFL activation can be inhibited by DAPT and BB94 and correlates with NICD protein as assayed by Westerns (I). J. The clustered ligand-dependent activation paradigm was validated for high throughput screening applications. *p<0.001, **p<0.0001.
The mammalian Notch pathway is unique in that it lacks soluble ligands. This presents challenges for biochemical studies on ligand-receptor interactions and for measuring activation rates with ligand in coculture-based systems. To overcome these limitations, other methods for presenting Notch ligands in trans have been developed either by immobilization of ligand ECD-IgG (46) or by preclustering soluble ligand ECD-IgG (47). Both methods have been shown to be biochemically similar to ligands presented by neighboring cells (40) and to lead to biological changes in Notch signal-receiving cells in culture and in vivo (48).
Recent studies measuring the kinetics of Notch activation by immobilized ligands using transcription-based reporters (12XCSL-GFP) determined response times to be greater than 10 hrs, a reflection of transcription, translation and GFP folding and maturation (12). Notably, because these activation rates were measured after release from DAPT inhibition, the apparent on-rate for ligand activation may not be accurate since S2-cleaved (NΔE-like) fragments would accumulate under these conditions and mimic the kinetics of NICD release from NΔE molecules after DAPT removal (Fig. 6C). We therefore used the NotchFL LCI real-time reporter to perform a dose response and kinetic study with immobilized ligands. NotchFL LCI reporter cells exhibited a dose-dependent response to immobilized purified Dll1-IgG ligand, with a linear increase (R2=0.958) from 0 – 2 µg/ml Dll1-IgG that plateaus thereafter (Fig. 8E). We observed a similar dose-dependent response to Dll1-Fc (from conditioned media) immobilized with α-Fc antibodies (fig. S9). After establishing that we can quantify NICD release by immobilized ligands via LCI, we tested the activation kinetics after release from DAPT. In contrast to the slower kinetics of GFP transcriptional reporters, the amount of NICD was equivalent to that of the DMSO-treated control within 1 hour of release from DAPT. After another 1–2 hrs, the amount of NICD in cells released from DAPT exceeded those of control (Fig. 8F), as seen with NΔE after DAPT release. By 24 hrs, luciferase complementation was at steady state. These data imply that therapeutic regimens involving withdrawal from γ-secretase inhibition will potentially include an acute spike in Notch activity following withdrawal from GSI, especially in tissue contexts where ligands are abundant.
An alternative analysis of Notch receptor activation kinetics can be achieved by using preclustered ligand-Fc fusion proteins, which allows the investigator to control the start time (t=0) for ligand addition without using pathway inhibitors. We monitored NotchFL activation in real time via LCI following the addition of Dll1-Fc-containing conditioned media that had been preclustered with different amounts of α-Fc antibody (Fig. 8G). A modest increase in bioluminescence was observed with unclustered Dll1-Fc conditioned media, but higher activation was observed with increasing clustering antibody concentrations, reaching a 6-fold increase in bioluminescence 4 hr after ligand addition (with a 1:100 dilution of α-Fc). This response to clustered ligand is not dependent on downstream transcription and translation (fig. S10). It is not observed with NICD/RBPjκ LCI reporter cells (8H), is blocked by BB94 and DAPT in a dose-dependent manner (Fig. 8H and fig. S11, A and B) and correlates with Western analyses with α-V1744 antibody (Fig. 8I), confirming LCI specificity. Moreover, we can compare the IC50 values of several metalloprotease inhibitors (BB94, GM6001, TAPI-2) by LCI (fig. S11, C and D). These results establish the NotchFL LCI system as an excellent, population-based assay for identifying and characterizing modifiers of Notch activation kinetics. To this end, we have validated the clustered ligand-dependent activation assay for small molecule screening (Z’factors ≥ 0.5) (Fig. 8J).
Discussion
Our studies established LCI to be a specific, robust, flexible and biologically relevant method for probing Notch pathway mechanism and regulation in real time in live cells. We were able to satisfy the key validation criteria for a PCA study (18): (i) robust signal was only observed with relevant interacting fusions (NICD and RBPjκ), but not with non-specific proteins, (ii) mutations or polypeptides expected to inhibit (e.g., RAM domain mutations and polypeptides) or modulate (e.g., ANK mutations, MAML-mediated stabilization) the protein interaction have corresponding effects on complementation, and (iii) specific and robust complementation was also observed with luciferase fragment swapping (fig. S12). Most importantly, we were able to recapitulate known biochemical and regulatory profiles of NICD/RBPjκ complex formation (i.e., subcellular location, half-life, domain contribution, dependence on proteolysis and ligand, sensitivity to known pharmacological agents and modifiers).
The Notch-LCI reporter can be used in several different modes to address mechanistic questions regarding Notch signaling. One mode utilizes LCI for monitoring productive protein-protein interactions relevant to the pathway, with the capacity for performing rapid structure-function analyses in intact cells. While our studies focused on NICD/RBPjκ interactions, the LCI reporter is also amenable to monitoring ternary complex formation (e.g., NICDΔRAM/RBPjκ/MAML; MAML/NICD and MAML/RBPjκ LCI pairs; fig. S13)). Moreover, the reporter can be adapted to probe interactions involving other NICD paralogs (Notch2–4) and, as we demonstrated with EBNA2, to analyze other “non-canonical” interactions and address crosstalk and signal integration with other pathways in real time. The recent development of dual-color luciferase complementation systems (49, 50) would even enable simultaneous monitoring of multiple protein interactions in the same cell.
A second mode utilizes LCI as an enzymatic reporter for nuclear translocation of NICD. As a protein translocation assay, optimized LCI offers advantages over other enzymatic reporters based on β-galactosidase fragment complementation (51, 52) and split intein-mediated Renilla luciferase (53). β-galactosidase is monitored in lysates or in live cells by flow cytometry, making it less applicable as a real-time readout for protein translocation. While Renilla luciferase activity can be imaged non-invasively, the intein-mediated complementation exhibits slower kinetic responses and makes the system irreversible. Our analyses of the kinetics of NICD release (from NΔE and NFL receptors) and complementation with RBPjκ indicate that nuclear translocation occurs very rapidly, with similar kinetics to nuclear translocation assays with Notch-GFP. The sensitivity and real-time quantitative nature of the LCI assay would facilitate analyses of the potential factors that influence the nuclear translocation process, an aspect that has not been explored extensively for the Notch pathway.
A major use for Notch-LCI is based on its ability to quantitatively report on the amount NICD/RBPjκ complexes in live cells with detailed temporal resolution. Since RBPjκ was generally unaffected by the stimuli tested, luciferase complementation highly correlated with NICD amounts and can therefore replace labor-intensive Western analyses, even providing more quantitative information on the net amount of active complexes. Using this LCI mode, we were able to monitor the dynamics of NICD release and accumulation under different conditions (e.g., protein degradation and stabilization, γ-secretase inhibition and recovery, ADAM inhibition), to compare receptor activation kinetics with different agonists, and to examine modifiers of signal strength. The ability to obtain precise temporal information can facilitate efforts in mathematically modeling pathway dynamics, especially when modifiers, target promoters and feedback loops are added to the system.
Finally, the Notch-LCI system is a versatile tool for identifying and characterizing modulators that can target different aspects of the signal transduction pathway. We have validated Notch-LCI reporter lines for HTS applications and established their usefulness in providing pharmacodynamic information and analyzing the mechanism of action of compounds that modulate the pathway. The identification of paralog-specific agents would be useful for manipulating the Notch signaling pathway in cancer, inherited diseases, stem cell differentiation and tissue engineering for both research and potential clinical applications.
Materials and Methods
DNA constructs
To generate the various Notch-NLuc and CLuc-RBPjκ expression plasmids, we replaced the cdc25 and 14-3-3 coding regions within the cdc25-NLuc and CLuc-14-3-3 constructs (a gift from Dr. Helen Piwnica-Worms;(17)), respectively by subcloning from various established mouse Notch1 and human RBPjκ constructs (20, 22, 26). CLuc-dnRBPjκ (R178H, which is equivalent to the previously described R218H substitution in mouse RBPjκ (54)) and NotchWFP>LAA–NLuc mutant constructs were generated by mutagenesis using the QuikChange Kit (Stratagene) following the manufacturer’s protocols. Untagged counterparts for all Notch and RBPjκ variants were prepared by replacing the Luc coding regions with oligo linkers. For stable line generation, we created Tet-inducible pBI dual expression vectors (Clontech) carrying CLuc-RBPjκ and either NotchFL-, NΔE- or NICD-NLuc. EBNA2-NLuc constructs were generated by subcloning from pSG5-EBNA2 and pSG5-EBNA2WW>SR expression plasmids (a gift from Dr. Diane Hayward) into the NICD-NLuc plasmids. The CLuc control plasmid has been previously described(17). nls-CLuc was prepared by inserting an oligonucleotide carrying the SV40 NLS coding sequence downstream of CLuc.
To generate the RAM polypeptide expression plasmids, we amplified the RAM regions (aa1748–1872, similar to RAM polypeptides described previously (28)) from wild-type and WFP>LAA mutant Notch-NLuc plasmids and subcloned the PCR fragments into pCS2+6MT. The dnMAML (aa-12–74)-EGFP was generated by subcloning from MSCV-MAML1(12–74)-EGFP (a gift from Dr. Warren Pear and Dr. Jon Aster). The pcDNA3-MAML-EGFP and CLuc-MAML expression plasmids were prepared by subcloning from the MAML1 cDNA plasmid (a gift from Dr. James Griffin).
To generate the pcDNA4-Dll1-Fc expression construct, we replaced the B7 coding region within the B7-IgG vector (55) (a gift from Dr. Kenneth Murphy) with that of the mouse Dll1 extracellular domain. The resulting construct encodes aa1–535 of mouse Dll1 fused to the CH2–CH3 domain of mouse IgG1 followed by a Myc-6His tag. The corresponding control pcDNA4-Fc expression vector was also prepared by replacing B7 with the signal sequence of mouse Dll1 (aa1–25).
The TP1-Luciferase reporter construct (pGa981-6) was a gift from Dr. Tasuku Honjo. Hes1-Luciferase and Hes5-Luciferase were gifts from Dr. Alain Israel and Dr. Ryoichiro Kageyama, respectively. CS2+Lunatic Fringe (LFNG) was prepared by subcloning the LFNG cDNA from pSPUTK-LFNG (a gift from Dr. Sean Egan) into the CS2+ vector.
Cell lines, Cell culture and plasmid transfections
3T3 cells were cultured in DMEM supplemented with 10% BCS. OT11 (RBPjκ−/−; a gift from Dr. Tasuku Honjo), 293T and CHO-fN2 (a gift from Drs. Shigeru Chiba and Hisamaru Hirai) cells were cultured in DMEM supplemented with 10% FBS. HeLaTetON parental cells (Clontech) and stable line derivatives (E6, IC1, III3 and FL2 lines; see fig. S5 for details) were cultured in DMEM supplemented with 10% Tet-approved FBS. The CHO-GFP, CHO-DLL1-IRES-GFP and CHO-Jag1-IRES-GFP stable lines have been previously described(56) and were cultured in IMDM supplemented with 10%FBS. All cell lines were maintained at 37°C in a humidified atmosphere with 5% CO2.
3T3 cells were transfected using Calcium phosphate with BES-buffered saline pH 6.8. 293T cells were transfected using Fugene6 (Roche), the OT11 cells using Lipofectamine LTX (Invitrogen) and the CHO-fN2, HeLaTetON parental and stable cell lines using Lipofectamine 2000 (Invitrogen), all according to the manufacturer’s recommended protocols.
To generate Fc or Dll1-Fc conditioned media, 293T cells were seeded in P100 dishes, and transfected the next day with pcDNA4-Fc or pcDNA4-Dll1-Fc expression plasmid using Fugene6. After 24 hrs, the cells were fed with fresh 10%FBS-TET/DMEM without phenol red (appropriate for imaging experiments, see below) and incubated for another 48 hrs. Conditioned media were collected, filter-sterilized and stored at 4°C. Production and secretion of the Fc and Dll1-Fc fusion proteins were confirmed by SDS-PAGE/Western analyses with anti-Myc or anti-His antibodies. To precluster the Dll1-Fc ligand, conditioned media were incubated with anti-mouse Fc antibodies (Jackson Immunoresearch) for at least 1 hr and equilibrated to 37°C prior to applying to reporter cells. To immobilize Dll1-Fc from conditioned medium, affinity-purified anti-mouse Fc antibodies (Jackson Immunoresearch; diluted in PBS) were initially adsorbed onto culture plates for 2 hr, aspirated and then incubated with conditioned media for another 2 hr prior to cell seeding.
RNAi reagents and methods
siRNAs were transfected into HeLaTetON stable lines E6, IC1, III3 using Dharmafect3 (Dharmacon) according to the manufacturer’s protocols. All siRNAs used in this study were purchased from Dharmacon: Non-targeting siRNA (siCONTROL) was used as a negative control; siRNAs targeting GL3 luciferase (siGL3 Luc) was used for optimizing transfection conditions and as a positive control for knockdown in our reporter lines; and the siGENOME SMARTpool (siNCSTN) was used to target Nicastrin. Knockdown efficiency was assessed by bioluminescence assays and Western analyses.
Chemicals and Reagents
Doxycycline (DOX), Epoxomicin, Cycloheximide (CHX), Phorbol 12-myristate 13-acetate (PMA), Actinomycin D (ActD), MG-132 and hIgG were all purchased from Sigma. DAPT, BAPTA, MG-262, GM6001, TAPI-2 and L685,458 were purchased from Calbiochem/EMD Chemicals. BB94 was a gift from Dr. Matt Freeman. Compound E was a gift from Dr. Todd Golde. Purified Dll1-IgG was a gift from Dr. Irwin Bernstein.
Western analyses
Cells were washed once with PBS, lysed in Laemmlli buffer and analyzed by SDS-PAGE/Westerns using the following antibodies: anti-Notch1 (AN1 ascites; (23)), anti-activated Notch1 or NICD (α-V1744/cleaved Notch1; Cell Signaling), anti-RBPjκ (CosmoBio), anti-β-actin (AC-15; Sigma), anti-Nicastrin (N-19; Santa Cruz Biotechnology), anti-Presenilin1 (H-70; Santa Cruz Biotechnology), anti-Myc (9E10 ascites), anti-His (Origene).
Bioluminescence Assays
Cells were seeded (and transfected, if needed) in black 96-well or 24-well plates. For the HeLaTetON stable lines, seeding was done in the presence of 0.5 µg/ml DOX to induce reporter expression. For endpoint luciferase complementation imaging assays, cells were imaged in PBS supplemented with 0.1% glucose, 1 mM MgCl2, 0.9 mM CaCl2 and 150 µg/ml D-luciferin (Biosynth) using an IVIS50 or IVIS100 imaging system (Caliper). The following acquisition parameters were used: exposure time, 1–5 min; binning, 4 or 8; no filter; f-stop, 1; field of view, 12 or 15 cm. If Renilla luciferase imaging was to be performed on the same cells, the D-luciferin containing buffer/medium was replaced with buffer containing 400 nM coelenterazine (Biotium) and imaged with the IVIS using the following acquisition parameters: exposure time, 1–2 min; binning, 4 or 8; filter <510; f-stop, 1; field of view, 12 or 15 cm.
For sequence imaging of receptor activation with Ca++ chelators, cells were imaged in HBSS containing 150 µg/ml D-luciferin solution. An initial image was obtained (t=0), after which an equivalent volume of HBSS/D-luciferin containing 2X chelator in HBSS/D-luciferin was added and images were obtained every 5 min for 1 hr. For other sequence imaging experiments (e.g., kinetics of γ-secretase inhibition and recovery), cells were imaged in phenol red-free culture medium containing D-luciferin in an IVIS equilibrated with 5% CO2. An initial image was also obtained before adding the equivalent volume of medium containing 2X inhibitor or PMA in D-luciferin containing medium. For sequence imaging of clustered ligand activation, D-luciferin was directly added to the pre-warmed pre-clustered conditioned media prior to imaging in the IVIS equilibrated with 5% CO2.
Photon flux (photons/second) was quantified on images with regions of interests (ROIs) and analyzed with LivingImage 2.6 (Caliper) and IGOR (Wavemetrics, Lake Oswego, OR, USA) image analysis software.
Most bioluminescence imaging experiments described herein were performed using an IVIS instrument. However, some bioluminescence assays were also performed using the Envision (1 sec/well; enhanced luminescence option; Perkin Elmer) or the Biotek-2 Synergy Microplate reader (1 sec/well), both of which were used for high throughput assay development and importantly, yielded similar results and dynamic range as the IVIS, particularly for the stable lines expressing GL3 luciferase or the LCI reporters of activated forms of Notch (NICD, NΔE).
Imaging results were presented either as mean photon flux (p/s) ± s.d. or normalized to the appropriate controls for a given experiment (as 1 or 100%): e.g., DMSO vehicle (for inhibitor and PMA activation studies), HBSS alone (for Ca++ chelator activation), Fc conditioned medium or purified hIgG (for Dll1-Fc conditioned medium or purified Dll1-IgG-dependent activation, respectively), wild-type Notch construct (vs mutant).
Cellular bioluminescence imaging
HeLaTetON stable lines expressing GL3 Luc alone or coexpressing NΔE-NLuc and CLuc-RBPjκ were seeded in P35 dishes. The next day, the medium was replaced with pre-warmed HEPES-buffered phenol red-free DMEM containing 10%FBS and 150 µg/ml D-luciferin. The cells were then imaged using an Olympus Fluoview1000 inverted microscope equipped with an Andor single-photon level CCD image capturing system and housed within a dark-adapted environmental chamber set at 37°C. Bioluminescence images were obtained with the 10X objective using 30 sec-1 min (GL3) or 5–10 min (Notch-LCI) exposure times.
Transcriptional Reporter Assays
For reporter assays, cells were seeded in 24-well plates and were transfected the next day with TP1-, Hes1- or Hes5-Luc (Notch-responsive reporter plasmids), CS2+ cytoβ gal (used for normalization), test untagged, NLuc- or CLuc-tagged construct and CS2+ vector as carrier DNA (500 ng total DNA/well; run in quadruplicate). Cells were harvested 48 hr after transfection and assayed for luciferase and β-galactosidase activities: Cells were washed once with PBS and lysed in lysis buffer (0.2% Triton X-100, 100 mM potassium phosphate buffer pH 7.8, 1 mM DTT, protease inhibitors) for 15 min. Luciferase activity was measured from 50µl lysate injected with 50 µl 2X assay buffer (30 mM Tricine pH 7.8, 3 mM ATP, 15 mM MgSO4, 10 mM DTT, 0.2 mM Coenzyme A, 1 mM D-luciferin) using a Tropix TR717 luminometer. In parallel, 5µl lysate was used to measure β-galactosidase activity according to the Tropix Galacton Plus chemiluminesence protocols.
Nuclear Fractionation
293T cells were transfected with expression constructs for GL3 luciferase, or a combination of NΔE-NLuc and CLuc-RBPjκ. 48 hr after transfection, cells were lysed and fractionated using the Nuclear Extract Kit (Active Motif) according to the manufacturer’s instructions. Nuclear and cytoplasmic fractions were then assayed for their luciferase activities (as described above for transcriptional reporter assays) and protein concentration (BCA Assay, Pierce).
LCI Assay development for High throughput screening
High throughput, automated procedures for cell seeding, medium replacement, transfections and cell-based assays were optimized at the Washington University Chemical Genetics Screening Core as well as the Molecular Imaging High Throughput Core.
For pilot compound screens, NΔE-NLuc/CLuc-RBPjκ stable lines (E6) were seeded in 96 well plates at 10,000 cells/well (or in 384 well plates at 3,000 cells/well) in medium containing 0.5 µg/ml DOX. After 24 hrs, medium was replaced with fresh DOX-containing medium containing DMSO or DAPT (as the test compound). After another 24 hr incubation, cells were incubated for 15 min at RT with D-luciferin containing buffer and measured for bioluminescence with an Envision or Biotek-2 Synergy (1 sec/well). Afterwards, the cells were fed with fresh medium containing Alamar Blue (1:10 v/v), incubated for 1 hr at 37°C and measured for fluorescence (excitation 544 nm, emission 590 nm; Biotek-2 Synergy or FluoOptima plate reader).
For pilot siRNA screens, NΔE-NLuc/CLuc-RBPjκ-expressing (E6) cells were seeded in 96-well plates and the next day, were transfected with siRNAs (50 nM final conc) using Dharmafect 3. 48 hours later, the cells were fed with fresh medium containing DOX to induce reporter expression. 24 hours later (i.e., 72 hours post-transfection), the cells were assayed for bioluminescence and viability as with the compound screen pilot experiments.
Statistical Analyses
Mean, s.d. and R2 values were calculated and plotted using Microsoft Excel. T-tests, tests for normality and multiple comparison procedures in ANOVA were all performed using JMP software (SAS Institute). Significance was assigned where p<0.05. Non-linear regression analyses and curve-fitting for inhibition curves were performed using GraphPad Prism. Z’ factors were calculated according to Zhang et al (35).
Supplementary Material
Acknowledgements
We would like to thank our colleagues for providing various reagents: Drs. Jon Aster, Irwin Bernstein, Shigeru Chiba, Sean Egan, Matthew Freeman, Todd Golde, James Griffin, Diane Hayward, Hisamaru Hirai, Tasuku Honjo, Alain Israel, Ryoichiro Kageyama, Kenneth Murphy, Warren Pear, Helen Piwnica-Worms as well as Merck. We thank members of the Piwnica-Worms and Kopan laboratories for discussions and technical assistance, especially Chintong Ong for preparing the Dll1-Fc and control constructs. We thank Bill Nolan (Chemical Genetics Screening Core) and Jayne Marasa (Molecular Imaging Center High throughput Screening Core) for their assistance in adapting the Notch-LCI assay for automation and high-throughput screening. We also thank Dennis Oakley (Bakewell NeuroImaging laboratory) for his assistance with the cellular bioluminescence imaging. We acknowledge Shuang Chen and Drs. Matt Hass, Guojun Zhao and Chihiro Sato for their critical reading of the manuscript.
Funding: Support for this work was provided by grants from the National Institutes of Health: a Neuroscience Blueprint Core Grant P30 NS057105 (Washington University), R21-NS06168001 (M.X.G.I.), AG025973P50 (R.K.), CA94056 (D.P.W.) and P50 AG005681 (J. Morris).
Footnotes
Author contributions: M.X.G.I., D.P.W. and R.K. conceptualized the project and designed experiments. M.X.G.I. designed and performed majority of the experiments and data analyses. S.L. performed experiments and analyzed data. M.F. prepared many of the constructs described herein. M.X.G.I., D.P.W. and R.K. contributed new reagents and analytical tools. M.X.G.I. and R.K. wrote the paper, with input from all coauthors.
Competing interests: M.X.G.I., D.P.W., R.K. and Washington University may receive income based on a license of Notch-related technology by the University to Merck. Merck did not support this work.
This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References
- 1.Kopan R, Ilagan MX. The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell. 2009;137:216. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kovall RA, Blacklow SC. Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr Top Dev Biol. 2010;92:31. doi: 10.1016/S0070-2153(10)92002-4. S0070-2153(10)92002-4 [pii]10.1016/S0070-2153(10)92002-4). [DOI] [PubMed] [Google Scholar]
- 3.Gridley T. Notch signaling and inherited disease syndromes. Hum Mol Genet. 2003;12(Spec No 1):R9. doi: 10.1093/hmg/ddg052. published online EpubApr 1. [DOI] [PubMed] [Google Scholar]
- 4.High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat Rev Genet. 2008;9:49. doi: 10.1038/nrg2279. published online EpubJan (nrg2279 [pii]10.1038/nrg2279). [DOI] [PubMed] [Google Scholar]
- 5.Koch U, Radtke F. Notch signaling in solid tumors. Curr Top Dev Biol. 2010;92:411. doi: 10.1016/S0070-2153(10)92013-9. S0070-2153(10)92013-9 [pii]10.1016/S0070-2153(10)92013-9). [DOI] [PubMed] [Google Scholar]
- 6.Pannuti A, Foreman K, Rizzo P, Osipo C, Golde T, Osborne B, Miele L. Targeting Notch to target cancer stem cells. Clin Cancer Res. 2010;16:3141. doi: 10.1158/1078-0432.CCR-09-2823. published online EpubJun 15 (1078-0432.CCR-09-2823 [pii]10.1158/1078-0432.CCR-09-2823). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Del Monte G, Grego-Bessa J, Gonzalez-Rajal A, Bolos V, De La Pompa JL. Monitoring Notch1 activity in development: evidence for a feedback regulatory loop. Dev Dyn. 2007;236:2594. doi: 10.1002/dvdy.21246. published online EpubSep (10.1002/dvdy.21246). [DOI] [PubMed] [Google Scholar]
- 8.Huppert SS, Ilagan MX, De Strooper B, Kopan R. Analysis of Notch function in presomitic mesoderm suggests a gamma-secretase-independent role for presenilins in somite differentiation. Dev Cell. 2005;8:677. doi: 10.1016/j.devcel.2005.02.019. published online EpubMay (S1534-5807(05)00099-7 [pii]10.1016/j.devcel.2005.02.019). [DOI] [PubMed] [Google Scholar]
- 9.Hansson EM, Teixeira AI, Gustafsson MV, Dohda T, Chapman G, Meletis K, Muhr J, Lendahl U. Recording Notch signaling in real time. Dev Neurosci. 2006;28:118. doi: 10.1159/000090758. DNE20060281_2118 [pii]10.1159/000090758). [DOI] [PubMed] [Google Scholar]
- 10.Ong C, Cheng H, Chang LW, Ohtsuka T, Kageyama R, Stormo DG, Kopan R. Target selectivity of vertebrate Notch proteins: collaboration between discrete domains and CSL binding site architecture determine activation probability. J Biol Chem. 2006;281:5106. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- 11.Souilhol C, Cormier S, Monet M, Vandormael-Pournin S, Joutel A, Babinet C, Cohen-Tannoudji M. Nas transgenic mouse line allows visualization of Notch pathway activity in vivo. Genesis. 2006;44:277. doi: 10.1002/dvg.20208. published online EpubMay 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sprinzak D, Lakhanpal A, LeBon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz ME. Cis Interactions beween Notch and Delta generate mutally exclusive signaling states. Nature. 2010;465:86. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon WR, Vardar-Ulu D, L'Heureux S, Ashworth T, Malecki MJ, Sanchez-Irizarry C, McArthur DG, Histen G, Mitchell JL, Aster JC, Blacklow SC. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS One. 2009;4:e6613. doi: 10.1371/journal.pone.0006613. 10.1371/journal.pone.0006613). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649. doi: 10.1016/s0092-8674(00)81193-9. published online EpubMay 15 (S0092-8674(00)81193-9 [pii]). [DOI] [PubMed] [Google Scholar]
- 15.Vooijs M, Ong CT, Hadland B, Huppert S, Liu Z, Korving J, van den Born M, Stappenbeck T, Wu Y, Clevers H, Kopan R. Mapping the consequence of Notch1 proteolysis in vivo with NIP-CRE. Development. 2007;134:535. doi: 10.1242/dev.02733. published online EpubFeb (134/3/535 [pii]10.1242/dev.02733). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawahashi K, Hayashi S. Dynamic intracellular distribution of Notch during activation and asymmetric cell division revealed by functional fluorescent fusion proteins. Genes Cells. 2010;15:749. doi: 10.1111/j.1365-2443.2010.01412.x. published online EpubJun (GTC1412 [pii]10.1111/j.1365-2443.2010.01412.x). [DOI] [PubMed] [Google Scholar]
- 17.Luker KE, Smith MC, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc Natl Acad Sci U S A. 2004;101:12288. doi: 10.1073/pnas.0404041101. published online EpubAug 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Remy I, Michnick SW. Application of protein-fragment complementation assays in cell biology. Biotechniques. 2007;42:137. doi: 10.2144/000112396. published online EpubFeb (000112396 [pii]). [DOI] [PubMed] [Google Scholar]
- 19.Villalobos V, Naik S, Piwnica-Worms D. Current state of imaging protein-protein interactions in vivo with genetically encoded reporters. Annu Rev Biomed Eng. 2007;9:321. doi: 10.1146/annurev.bioeng.9.060906.152044. 10.1146/annurev.bioeng.9.060906.152044). [DOI] [PubMed] [Google Scholar]
- 20.Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates g-secretase-like proteolytic activation of Notch1. Mol. Cell. 2000;5:197. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 21.Vooijs M, Schroeter EH, Pan Y, Blandford M, Kopan R. Ectodomain shedding and intramembrane cleavage of mammalian notch proteins is not regulated through oligomerization. J Biol Chem. 2004;279:50864. doi: 10.1074/jbc.M409430200. published online EpubSep 23. [DOI] [PubMed] [Google Scholar]
- 22.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 23.Huppert SS, Le A, Schroeter EH, Mumm JS, Saxena MT, Milner LA, Kopan R. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature. 2000:405–966. doi: 10.1038/35016111. published online EpubJun 22 (10.1038/35016111). [DOI] [PubMed] [Google Scholar]
- 24.Tagami S, Okochi M, Yanagida K, Ikuta A, Fukumori A, Matsumoto N, Ishizuka-Katsura Y, Nakayama T, Itoh N, Jiang J, Nishitomi K, Kamino K, Morihara T, Hashimoto R, Tanaka T, Kudo T, Chiba S, Takeda M. Regulation of Notch Signaling by Dynamic Changes in the Precision in S3 Cleavage of Notch-1. Mol Cell Biol. 2008;28:165. doi: 10.1128/MCB.00863-07. published online EpubOct 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Curr Biol. 1995;5:1416. doi: 10.1016/s0960-9822(95)00279-x. published online EpubDec 1 (S0960-9822(95)00279-X [pii]). [DOI] [PubMed] [Google Scholar]
- 26.Kopan R, Nye JS, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120:2385. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 27.Kurooka H, Kuroda K, Honjo T. Roles of the ankyrin repeats and C-terminal region of the mouse Notch1 intracellular region. Nucleic Acids Research. 1998;26:5448. doi: 10.1093/nar/26.23.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lubman OY, Ilagan MX, Kopan R, Barrick D. Quantitative Dissection of the Notch:CSL Interaction: Insights into the Notch-mediated Transcriptional Switch. J Mol Biol. 2007;365:577. doi: 10.1016/j.jmb.2006.09.071. published online EpubJan 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson SE, Ilagan MX, Kopan R, Barrick D. Thermodynamic analysis of the CSL:Notch interaction: Distribution of binding energy of the Notch RAM region to the CSL beta-trefoil domain and the mode of competition with the viral transactivator EBNA2. J Biol Chem. 2010;285:6681. doi: 10.1074/jbc.M109.019968. published online EpubDec 23 (M109.019968 [pii]10.1074/jbc.M109.019968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeffries S, Robbins DJ, Capobianco AJ. Characterization of a high-molecular-weight Notch complex in the nucleus of Notch(ic)-transformed RKE cells and in a human T-cell leukemia cell line. Mol Cell Biol. 2002;22:3927. doi: 10.1128/MCB.22.11.3927-3941.2002. published online EpubJun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC, Aster JC. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol. 2003;23:655. doi: 10.1128/MCB.23.2.655-664.2003. published online EpubJan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakai T, Taniguchi Y, Tamura K, Minoguchi S, Fukuhara T, Strobl LJ, Zimberstrobl U, Bornkamm GW, Honjo T. Functional Replacement Of the Intracellular Region Of the Notch1 Receptor By Epstein-Barr Virus Nuclear Antigen 2. Journal of Virology. 1998;72:6034. doi: 10.1128/jvi.72.7.6034-6039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fryer CJ, White JB, Jones KA. Mastermind Recruits CycC:CDK8 to Phosphorylate the Notch ICD and Coordinate Activation with Turnover. Mol Cell. 2004;16:509. doi: 10.1016/j.molcel.2004.10.014. published online EpubNov 19. [DOI] [PubMed] [Google Scholar]
- 34.Cirrito JR, May PC, O'Dell MA, Taylor JW, Parsadanian M, Cramer JW, Audia JE, Nissen JS, Bales KR, Paul SM, DeMattos RB, Holtzman DM. In vivo assessment of brain interstitial fluid with microdialysis reveals plaque-associated changes in amyloid-beta metabolism and half-life. J Neurosci. 2003;23:8844. doi: 10.1523/JNEUROSCI.23-26-08844.2003. published online EpubOct 1 (23/26/8844 [pii]). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 36.Li T, Wen H, Brayton C, Laird FM, Ma G, Peng S, Placanica L, Wu TC, Crain BJ, Price DL, Eberhart CG, Wong PC. Moderate reduction of gamma-secretase attenuates amyloid burden and limits mechanism-based liabilities. J Neurosci. 2007;27:10849. doi: 10.1523/JNEUROSCI.2152-07.2007. published online EpubOct 3 (27/40/10849 [pii]10.1523/JNEUROSCI.2152-07.2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rand MD, Grimm LM, Artavanis-Tsakonas S, Patriub V, Blacklow SC, Sklar J, Aster JC. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol. 2000;20:1825. doi: 10.1128/mcb.20.5.1825-1835.2000. published online EpubMar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nichols JT, Miyamoto A, Olsen SL, D'Souza B, Yao C, Weinmaster G. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J Cell Biol. 2007;176:445. doi: 10.1083/jcb.200609014. published online EpubFeb 12 (jcb.200609014 [pii]10.1083/jcb.200609014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okochi M, Fukumori A, Jiang J, Itoh N, Kimura R, Steiner H, Haass C, Tagami S, Takeda M. Secretion of the Notch-1 Abeta-like peptide during Notch signaling. J Biol Chem. 2006;281:7890. doi: 10.1074/jbc.M513250200. published online EpubMar 24 (M513250200 [pii]10.1074/jbc.M513250200). [DOI] [PubMed] [Google Scholar]
- 40.Yang LT, Nichols JT, Yao C, Manilay JO, Robey EA, Weinmaster G. Fringe glycosyltransferases differentially modulate Notch1 proteolysis induced by Delta1 and Jagged1. Mol Biol Cell. 2005;16:927. doi: 10.1091/mbc.E04-07-0614. published online EpubFeb (E04-07-0614 [pii]10.1091/mbc.E04-07-0614). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, Vooijs M. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J Biol Chem. 2009;284:31018. doi: 10.1074/jbc.M109.006775. published online EpubNov 6 (M109.006775 [pii]10.1074/jbc.M109.006775). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269. doi: 10.1126/science.1102160. published online EpubOct 8 (306/5694/269 [pii]10.1126/science.1102160). [DOI] [PubMed] [Google Scholar]
- 43.Stanley P, Okajima T. Roles of glycosylation in Notch signaling. Curr Top Dev Biol. 2010;92:131. doi: 10.1016/S0070-2153(10)92004-8. S0070-2153(10)92004-8 [pii]10.1016/S0070-2153(10)92004-8). [DOI] [PubMed] [Google Scholar]
- 44.Hicks C, Johnston SH, diSibio G, Collazo A, Vogt TF, Weinmaster G. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nature Cell Biology. 2000;2:515. doi: 10.1038/35019553. [DOI] [PubMed] [Google Scholar]
- 45.Raya A, Kawakami Y, Rodriguez-Esteban C, Ibanes M, Rasskin-Gutman D, Rodriguez-Leon J, Buscher D, Feijo JA, Izpisua Belmonte JC. Notch activity acts as a sensor for extracellular calcium during vertebrate left-right determination. Nature. 2004;427:121. doi: 10.1038/nature02190. published online EpubJan 8. [DOI] [PubMed] [Google Scholar]
- 46.Varnum-Finney B, Wu L, Yu M, Brashem-Stein C, Staats S, Flowers D, Griffin JD, Bernstein ID. Immobilization of Notch ligand, Delta-1, is required for induction of Notch signaling. J Cell Sci. 2000;113:4313. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- 47.Hicks C, Ladi E, Lindsell C, Hsieh JJ, Hayward SD, Collazo A, Weinmaster G. A secreted Delta1-Fc fusion protein functions both as an activator and inhibitor of Notch1 signaling. J Neurosci Res. 2002;68:655. doi: 10.1002/jnr.10263. published online EpubJun 15 (10.1002/jnr.10263). [DOI] [PubMed] [Google Scholar]
- 48.Delaney C, Heimfeld S, Brashem-Stein C, Voorhies H, Manger RL, Bernstein ID. Notch-mediated expansion of human cord blood progenitor cells capable of rapid myeloid reconstitution. Nat Med. 2010;16:232. doi: 10.1038/nm.2080. published online EpubFeb (nm.2080 [pii]10.1038/nm.2080). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hida N, Awais M, Takeuchi M, Ueno N, Tashiro M, Takagi C, Singh T, Hayashi M, Ohmiya Y, Ozawa T. High-sensitivity real-time imaging of dual protein-protein interactions in living subjects using multicolor luciferases. PLoS One. 2009;4:e5868. doi: 10.1371/journal.pone.0005868. 10.1371/journal.pone.0005868). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villalobos V, Naik S, Bruinsma M, Dothager RS, Pan MH, Samrakandi M, Moss B, Elhammali A, Piwnica-Worms D. Dual-color click beetle luciferase heteroprotein fragment complementation assays. Chem Biol. 2010;17:1018. doi: 10.1016/j.chembiol.2010.06.018. published online EpubSep 24 (S1074-5521(10)00311-X [pii]10.1016/j.chembiol.2010.06.018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fung P, Peng K, Kobel P, Dotimas H, Kauffman L, Olson K, Eglen RM. A homogeneous cell-based assay to measure nuclear translocation using beta-galactosidase enzyme fragment complementation. Assay Drug Dev Technol. 2006;4:263. doi: 10.1089/adt.2006.4.263. published online EpubJun (10.1089/adt.2006.4.263). [DOI] [PubMed] [Google Scholar]
- 52.Wehrman TS, Casipit CL, Gewertz NM, Blau HM. Enzymatic detection of protein translocation. Nat Methods. 2005;2:521. doi: 10.1038/nmeth771. published online EpubJul (nmeth771 [pii]10.1038/nmeth771). [DOI] [PubMed] [Google Scholar]
- 53.Kim SB, Ozawa T, Watanabe S, Umezawa Y. High-throughput sensing and noninvasive imaging of protein nuclear transport by using reconstitution of split Renilla luciferase. Proc Natl Acad Sci U S A. 2004;101:11542. doi: 10.1073/pnas.0401722101. published online EpubAug 10 (10.1073/pnas.04017221010401722101 [pii]). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chung CN, Hamaguchi Y, Honjo T, Kawaichi M. Site-directed mutagenesis study on DNA binding regions of the mouse homologue of Suppressor of Hairless, RBP-J kappa. Nucleic Acids Res. 1994;22:2938. doi: 10.1093/nar/22.15.2938. published online EpubAug 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe N, Gavrieli M, Sedy JR, Yang J, Fallarino F, Loftin SK, Hurchla MA, Zimmerman N, Sim J, Zang X, Murphy TL, Russell JH, Allison JP, Murphy KM. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat Immunol. 2003;4:670. doi: 10.1038/ni944. published online EpubJul (10.1038/ni944ni944 [pii]). [DOI] [PubMed] [Google Scholar]
- 56.Ong CT, Sedy JR, Murphy KM, Kopan R. Notch and presenilin regulate cellular expansion and cytokine secretion but cannot instruct Th1/Th2 fate acquisition. PLoS One. 2008;3:e2823. doi: 10.1371/journal.pone.0002823. 10.1371/journal.pone.0002823). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.